Abstract
FGF signaling is known to play a critical role in the specification of primitive endoderm (PrE) and epiblast (Epi) from the inner cell mass (ICM) during mouse preimplantation development, but how FGFs synergize with other growth factor signaling pathways is unknown. Because PDGFRα signaling has also been implicated in the PrE, we investigated the coordinate functions of PDGFRα together with FGFR1 or FGFR2 in PrE development. PrE development was abrogated in Pdgfra; Fgfr1 compound mutants, or significantly reduced in Pdgfra; Fgfr2 or PdgfraPI3K; Fgfr2 compound mutants. We provide evidence that both Fgfr2 and Pdgfra play roles in PrE cell survival while Fgfr1 controls PrE cell specification. Our results suggest a model where FGFR1-engaged ERK1/2 signaling governs PrE specification while PDGFRα-and by analogy possibly FGFR2- engaged PI3K signaling regulates PrE survival and positioning in the embryo. Together, these studies indicate how multiple growth factors and signaling pathways can cooperate in preimplantation development.
Keywords: Preimplantation, cell specification, survival, ERK1/2, PI3K
Introduction
During mouse preimplantation development, the embryo undergoes two sequential cell fate decisions. At the morula stage (embryonic day (E) 2.5), outside cells differentiate to trophectoderm (TE) while inner cells constitute the inner mass cells (ICM). At the blastocyst stage, starting at E3.5, ICM cells undergo the second fate decision and become either the pluripotent epiblast (Epi), which forms the embryo proper, or the primitive endoderm (PrE) destined to produce components of the yolk sac. FGF4 signaling is critically required for the differentiation of ICM cells into PrE (Chazaud et al., 2006; Feldman et al., 1995; Goldin and Papaioannou, 2003; Kang et al., 2013; Nichols et al., 2009; Yamanaka et al., 2010). The FGF family includes 18 ligands and four receptors, FGFR1–4 (Brewer et al., 2016; Ornitz and Itoh, 2015). While all four FGFRs are expressed in the late mouse blastocyst (Guo et al., 2010; Ohnishi et al., 2014), only FGFR1 and FGFR2 play a critical role in PrE development (Brewer et al., 2015; Kang et al., 2017; Molotkov et al., 2017). Fgfr1 is expressed in all cell lineages of the mouse embryo during preimplantation development (Kang et al., 2017; Molotkov et al., 2017; Ohnishi et al., 2014). Genetic ablation of Fgfr1 on a 129S4 co-isogenic background results in decreased numbers of PrE cells and peri-implantation lethality (Brewer et al., 2015). Fgfr2 is expressed in both the TE and PrE but is not found in Epi cells (Kang et al., 2017; Molotkov et al., 2017; Ohnishi et al., 2014). Fgfr2−/− embryos on the same 129S4 genetic background do not show any defects in PrE development, but die at E10.5 from defects in placenta development (Molotkov et al., 2017; Xu et al., 1998; Yu et al., 2003). Despite the lack of PrE defects in Fgfr2−/− embryos, targeted inactivation of both Fgfr1 and Fgfr2 prevents development of the PrE and phenocopies the Fgf4−/− mutant phenotype (Kang et al., 2017; Molotkov et al., 2017). Upregulation of Fgfr2 expression in ICM cells directed them toward a PrE fate (Morris et al., 2013). This indicates that Fgfr2 plays a role in PrE development and can compensate at some level for the lack of Fgfr1.
FGF4, the only FGF ligand expressed during preimplantation (Feldman et al., 1995; Ohnishi et al., 2014) is produced by Epi precursor cells in the ICM, binds to FGFRs and induces the PrE fate in some ICM cells. Culture of wild-type E2.5 embryos in the presence of FGF4 induced the PrE fate in all ICM cells and rescued PrE development in Fgf4−/− embryos (Grabarek et al., 2012; Kang et al., 2013; Krawchuk et al., 2013). Surprisingly, FGF4 treatment of Fgfr1−/− and Fgfr2−/− compound mutant embryos revealed an absolute requirement for FGF4 signaling through FGFR1 to induce PrE fate in ICM cells, since even at extremely high doses FGF4 was unable to induce PrE specification through FGFR2 (Molotkov et al., 2017). These results indicate that both FGFR1 and FGFR2 are required for PrE development, but that signaling through FGFR1 has a key role in FGF4-induced PrE fate specification.
In addition to the FGFRs, PDGFRα is expressed in the PrE (Artus et al., 2010; Plusa et al., 2008). Analysis of Pdgfra−/− blastocysts revealed an increase in PrE cell apoptosis, resulting in a decrease of the number of PrE cells (Artus et al., 2013), although loss of PDGFRα results in a range of abnormalities only at mid-gestation (Soriano, 1997). The decrease in the number of PrE cells in Pdgfra−/− embryos was not accompanied by a compensatory increase in the number of Epi cells, which is observed in Fgf4−/−, Fgfr1−/− and Fgfr1−/−; Fgfr2−/− embryos (Brewer et al., 2015; Kang et al., 2017; Kang et al., 2013; Molotkov et al., 2017). This suggests that PDGFRαsignaling by PDGFA, the cognate ligand expressed at this stage (Ohnishi et al., 2014), plays a role in the survival of developing PrE cells rather than in PrE specification, which is controlled mainly by FGF4 signaling through FGFR1.
FGF ligands bind to FGFRs to engage multiple downstream signaling pathways, including ERK1/2 and PI3K / AKT, which mediate cellular responses (for a review, see (Brewer et al., 2016). Culturing wild-type E2.5 embryos in the presence of a MEK inhibitor (PD0325901, 1 μM) leads to a dramatic reduction in the number of PrE cells and a compensatory increase in the number of Epi cells (Nichols et al., 2009). This closely resembles the phenotype observed in Fgf4−/− and Fgfr1−/−; Fgfr2−/− embryos. This suggests that FGF activation of MEK / ERK signaling is critical for control of PrE cell fate specification. In contrast, studies using an allelic series of mouse knock-ins designed to disable binding of Src, SHP2, PLCγ and PI3K to PDGFRs indicate that PI3K is a major signaling pathway downstream of PDGFRα (Klinghoffer et al., 2002). It is thus possible that signaling through FGFR2 and/or PDGFRα engaged PI3K signaling plays a role in PrE cell survival.
In this manuscript, we demonstrate that both FGFR2 and PDGFRα play a role in PrE cell survival while only FGFR1 is involved in ICM cell fate specification. We also hypothesize that ERK and PI3K play sequential roles in PrE development: activation of ERK1/2 by FGF4 / FGFR1 is required for ICM cell fate decision towards the PrE, while subsequent activation of PI3K by PDGFA / PDGFRα and possibly FGF4 / FGFR2 is needed for PrE cells to survive and adopt their proper position in the embryo.
Materials and methods
Mice
Fgfr1+/− mice (Hoch and Soriano, 2006), Fgfr2+/− mice (Molotkov et al., 2017), Fgfr2T2A-H2B-mCherry/T2A-H2B-mCherry mice referred to as Fgfr2mCherry (Molotkov et al., 2017), Pdgfra+/H2B-GFP mice referred to as Pdgfra+/− (Hamilton et al., 2003) and Pdgfra+/tm5Sor mice referred to as Pdgfra+/PI3K (Klinghoffer et al., 2002) were maintained on a 129S4/SvJaeSor (MGI:3044540) co-isogenic background, further referred to as 129S4. All animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai.
Embryo dissection and embryonic culture
Female mice were super-ovulated by i.p. injection of 5 IU of PMSG (Pregnant Mare Serum Gonadotropin) followed 48 h later by i.p. injection of 5 IU of hCG (human Chorionic Gonadotropin) prior to mating with stud males. Vaginal plugs were checked the following day (E0.5). E2.5 embryos (8-cell stage) were flushed from oviducts and cultured for 72h in DMEM (11965–118, ThermoFisher Scientific, USA) with 1/100 of 100 mM sodium pyruvate solution (11360–070, ThermoFisher Scientific, USA), 1/100 of MEM non-essential amino acid solution (11140–050, ThermoFisher Scientific, USA) and 1/200 of penicillin / streptomycin mix (15140–122, ThermoFisher Scientific, USA) at 37°C and 5% CO2; E3.5 embryos were flushed from uteri and cultured for 48h. All embryos were fixed for 30 min. in 4% PFA and stored in 30% Ethanol in PBST at 4°C until analysis.
Embryos immunostaining
Blastocyst immunostaining was performed as described previously (Nichols et al., 2009). Briefly, embryos were washed once in PBST and once in PBS/PVP (P0930, Sigma), permeabilized for 45 min. in 0.5% Triton X100 (T9284, Sigma) in PBS/PVP, blocked in 0.1% BSA with 2% donkey serum in PBST for 2h and incubated overnight with primary antibodies used at 1/100 dilution. Primary antibodies used were: NANOG (RCAB0002P-F, Cosmo Bio Co., Ltd.), CDX2 (MU392A-UC, BioGenex), GATA4 (sc-1237, Santa Cruz), mCHERRY (ab167453, Abcam) and GFP (A6455, Invitrogen). On the following day, embryos were washed in PBST and incubated for 2hours with secondary Alexa Fluor (Invitrogen) conjugated antibodies diluted 1/400 in 1% donkey serum in PBST. Blastocysts were counterstained with DAPI and mounted individually in 20 μl PBS droplets on glass-bottom Petri dishes for imaging.
Image analysis
Optical section images were taken on a Zeiss Axio-Observer Z1 with an Apotome 2 attachment, using a Hamamatsu Orca Flash 4.0 LT camera and ZenPro 2015 software. For cell counting, we imported Zeiss z-stack images into Metamorph software (version 7.8.13.0) and manually counted individual cells by scrolling through the stack.
Quantification and statistical analysis
Statistical analysis was performed using Prism 6.0. All data are represented as mean ± standard error. Statistical p-values were calculated using two-tailed Student’s t-test for unpaired samples.
RESULTS
PDGFRα and FGFR2 are expressed in PrE
We analyzed expression of Fgfr2 and Pdgfra during preimplantation development in Fgfr2+/mCherry; Pdgfra+/H2B-GFP double heterozygous embryos between E2.5 and E3.5. We also cultured a subset of E3.5 blastocysts for 48h to obtain expanded blastocysts (E3.5+48h). In Fgfr2mCherry embryos, a T2A self-cleavage peptide links H2B-mCherry to Fgfr2, which puts it under the control of Fgfr2 regulatory elements to allow reporting of Fgfr2 expression without affecting FGFR2 levels (Molotkov et al., 2017). In Pdgfra+/H2B-GFP knock-in embryos, H2B-GFP is expressed under control of the Pdgfra promoter. Pdgfra+/H2B-GFP blastocysts do not display any PrE phenotype (Artus et al., 2010).
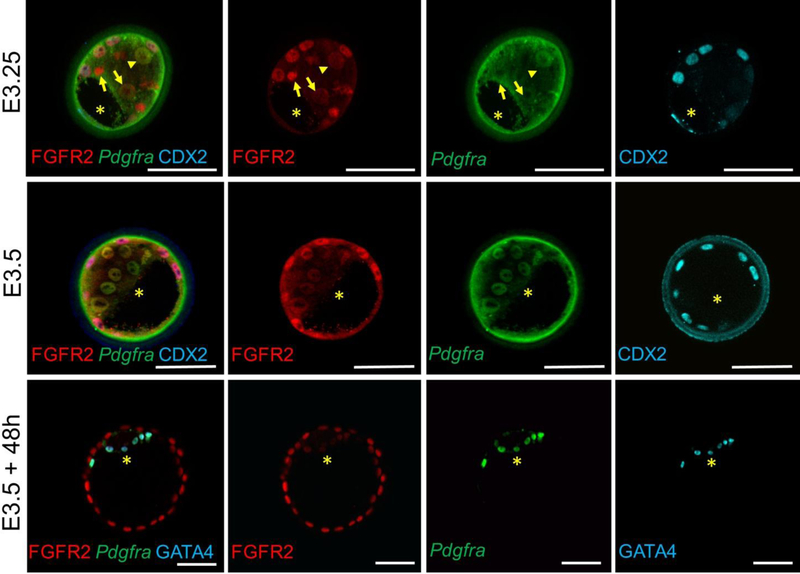
We found that at E3.25, Fgfr2mCherry is expressed at various levels in all ICM cells (red in Fig. 1), whereas PdgfraH2B-GFP expression (green in Fig. 1) is observed in some but not all ICM cells (compare cells labeled with arrow and arrowhead in Fig.1). We also noted a lack of correlation between the levels of Fgfr2mCherry and PdgfraH2B-GFP expression at this stage, suggesting that these genes may be independently regulated. While Pdgfra is expressed exclusively in the PrE, Fgfr2 expression is also observed in TE cells in addition to ICM cells (TE cells are specifically labeled with Cdx2 antibody, cyan, in Fig. 1). By E3.5, Fgfr2mCherry and PdgfraH2B-GFP co-localize and are uniformly expressed in all developing PrE cells. In expanded blastocysts, as expected, Fgfr2 and Pdgfra are both co-expressed in the same PrE cells (E3.5+48h, Fig.1), and can be co-labeled with antibodies to GATA4, a cell-type specific marker of mature PrE cells.
Figure 1.
Expression of Fgfr2 and Pdgfra in E3.0 and E3.5 preimplantation embryos. Note, while Fgfr2mCherry and PdgfraH2B-GFP are differentially expressed in ICM cells at E3.0, both receptors are expressed in the same cells in E3.5 expanded embryos (E3.5 + 48h). Green background fluorescence in E3.0 and E3.5 embryos is due to the presence of the Zona Pellucida that surrounds the embryos. *, blastocoel cavity. Scale bars, 50 μm.
Both PDGFRα and FGFR2 play roles in survival of PrE cells
The number of PrE cells has been shown previously to not change in Pdgfra+/− embryos, but it is reduced by ~40% in Pdgfra−/− embryos by E4.5 due to apoptosis (Artus et al.,2010). To determine if Fgfr2 contributes to the survival of these remaining PrE cells and can partially rescue Pdgfra−/− embryos, we bred Fgfr2+/− and Pdgfra+/− mice to generate double heterozygous mice. We next intercrossed these mice to obtain E3.5 blastocysts deficient for both Fgfr2 and Pdgfra (Fig. 2A).
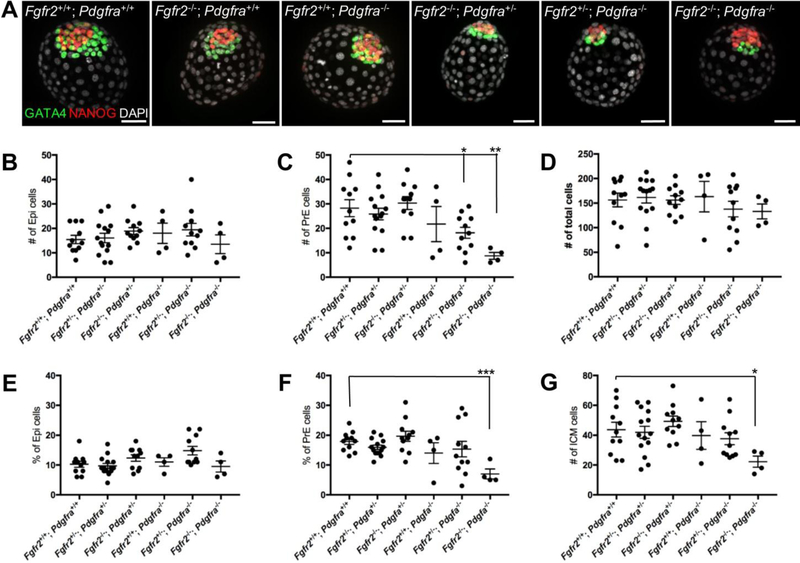
Figure 2.
PrE development is deficient in Fgfr2−/−; Pdgfra−/− embryos. (A) E3.5 embryos were cultured for 48h in DMEM and stained with antibodies to NANOG (red) and GATA4 (green). DAPI (white) was used to counter-stain nuclei. Number of Epi (B), PrE (C) and total cells (D) were counted in E3.5 embryos cultured for 48h in DMEM. Percentages of Epi (E) and PrE (F) cells relative to the total number of cells are shown. (G) Number of ICM cells is shown. Data represented as mean + SEM. *, p=0.02; **, p=0.006. Scale bars, 50 μm.
We and others have previously shown that PrE development is normal in Fgfr2−/− embryos (Kang et al., 2017; Molotkov et al., 2017). We noted a reduction of the number of PrE cells (red GATA4+ cells in Fig. 2A) in Fgfr2+/+; Pdgfra−/− embryos, consistent with a previous report (Artus et al., 2010). This deficiency in the number of PrE cells was additionally enhanced in Fgfr2+/−; Pdgfra−/− and was particularly severe in Fgfr2−/−; Pdgfra−/− double homozygous embryos, which had only a few remaining PrE cells (Fig. 2A). To further analyze the defect in PrE development in these mutant embryos, we counted the total number of Epi and PrE cells (Fig. 2B–G and Table 1). We found that the average number of Epi, PrE and total cells in the embryos were similar between Fgfr2+/+; Pdgfra+/+ (wild-type control), Fgfr2+/−; Pdgfra+/− and Fgfr2−/−; Pdgfra+/− embryos (Fig. 2B, C, D and Table 1). The number of PrE cells was slightly decreased in the Fgfr2+/+; Pdgfra−/− embryos, it was significantly lower in Fgfr2+/−; Pdgfra−/− (p=0.02), and most reduced in Fgfr2−/−; Pdgfra−/− (p=0.006) embryos. Interestingly, the number of Epi cells trended higher but did not change significantly in any of the mutant embryos we analyzed. This is different from the phenotype observed earlier in Fgfr1 and Fgfr2 compound mutants (Kang et al., 2017; Molotkov et al., 2017) or in Fgf4−/− embryos (Kang et al., 2013; Krawchuk et al., 2013). We observed in Fgfr2−/−; Pdgfra+/−, Fgfr2+/−; Pdgfra−/− and Fgfr2−/−; Pdgfra−/− embryos an increased incidence of pyknotic nuclei, consistent with apoptosis (Table 2 and supplementary Fig. S1). To exclude an effect of embryo size on the relative size of the Epi, we calculated the percentage of Epi and PrE cells in the embryos and the size of the ICM (Fig. 2 E–G). We found that the relative size of the Epi was similar in all mutant classes except for Fgfr2−/−; Pdgfra−/− embryos, where it was significantly reduced (Fig. 2F). Similar to the relative size of the PrE, the combined number of Nanog+ Epi cells and Gata4+ PrE cells was considerably decreased in Fgfr2−/−; Pdgfra−/− embryos, while it remained unchanged in the rest of the mutants we analyzed (Fig. 2G).
Table 1.
Cell lineage composition in Fgfr2; Pdgfra compound mutants
| Number of cells | ||||
|---|---|---|---|---|
| # | Epi | PrE | Total | |
| Fgfr2+/+; Pdgfra+/+ | 11 | 15 ± 6 | 28 ± 3 | 156 ± 14 |
| Fgfr2+/−; Pdgfra+/− | 14 | 16 ± 2 | 26 ± 2 | 161 ± 11 |
| Fgfr2−/−; Pdgfra+/− | 11 | 19 ± 1 | 30 ± 3 | 156 ± 9 |
| Fgfr2+/+; Pdgfra−/− | 4 | 18 ± 4 | 22 ± 7 | 163 ± 31 |
| Fgfr2+/−; Pdgfra−/− | 11 | 19 ± 3 | 18 ± 2* | 138 ± 16 |
| FgfrZ−/−; Pdgfra−/− | 4 | 13 ± 4 | 9 ± 1** | 133 ±15 |
Notes:
, p=0.02;
, p=0.006 when compared to wild-type control
Table 2.
Number of pyknotic nuclei in Fgfr2; Pdgfra compound mutants
| # | Epi | |
|---|---|---|
| Fgfr2+/+; Pdgfra+/+ | 11 | 1.3 ± 0.4 |
| Fgfr2+/+; Pdgfra−/− | 4 | 1.8 ± 0.6 |
| Fgfr2−/−; Pdgfra+/− | 11 | 2.1 ± 0.7 |
| Fgfr2+/−; Pdgfra−/− | 11 | 2.4 ± 0.6 |
| Fgfr2−/−; Pdgfra−/− | 4 | 2.8 ± 0.3 |
PrE development is abrogated in Fgfr1−/−; Pdgfra−/− embryos
Previously we (Molotkov et al., 2017) and others (Kang et al., 2017) demonstrated that both FGFR1 and FGFR2 play roles in PrE development, and that genetic inactivation of both receptors leads to the total absence of PrE development. Our results also suggested that FGFR1 plays a major role in PrE fate specification, while FGFR2 is involved in PrE cell survival / proliferation. Our data on Fgfr2 and Pdgfra compound mutants suggest that while both FGFR2 and PDGFRα play roles in PrE cell survival, only Pdgfra−/− embryos exhibit deficient PrE development, whereas the PrE is normal in Fgfr2−/− embryos (Fig. 2). This raises the intriguing possibility that Fgfr1−/−; Pdgfra−/− mutants might exhibit a phenotype similar to the one we described earlier for Fgfr1−/−; Fgfr2−/− embryos (Molotkov et al., 2017). To test this hypothesis, we crossed Fgfr1+/− and Pdgfra+/− mice to obtain double heterozygous Fgfr1+/−; Pdgfra+/− mice. We were unable to recover any Fgfr1+/−; Pdgfra+/− double heterozygous mice at weaning on the 129S4 genetic background (0 out of 55 pups), similar to the background dependency problem we encountered previously with Fgfr1+/−; Fgfr2+/− double heterozygous mice (Molotkov et al., 2017). To overcome this difficulty, we intercrossed Pdgfra+/− mice on a 100% 129S4 background with Fgfr1+/− mice on a 75% B6 / 25% 129S4 background, allowing us to obtain Fgfr1+/−; Pdgfra+/− mice on a mixed 129S4 / B6 genetic background.
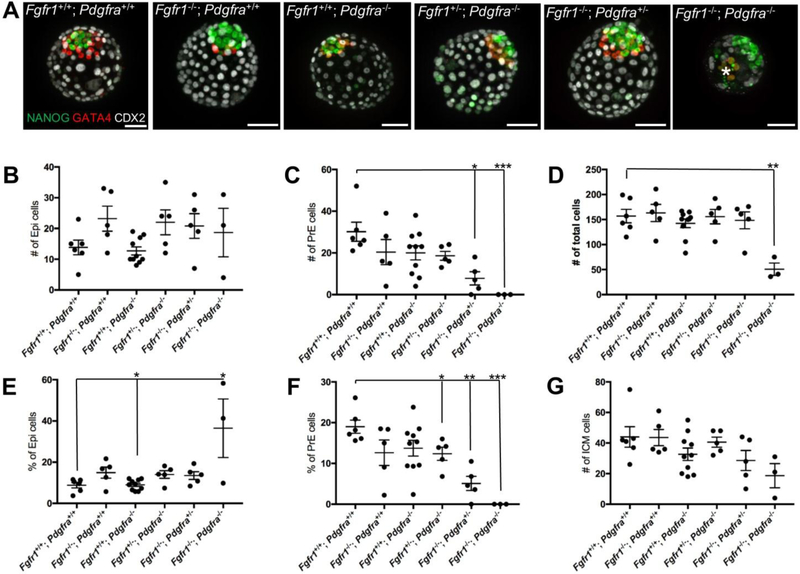
We intercrossed Fgfr1+/−; Pdgfra+/− compound mutants, and isolated E3.5 blastocysts that we cultured for 48h prior to analysis. We found that simultaneous deletion of both copies of Fgfr1 and Pdgfra disrupts PrE development in double homozygous embryos (n=3; Fig. 3 and Table 3). Interestingly, in 2 out of 3 Fgfr1−/−; Pdgfra−/− embryos we observed a group of 2 (embryo 1) or 6 (embryo 2, shown with asterisks on Fig. 3A) cells that expressed GATA4 faintly, and that were also located at a considerable distance from the Epi. We did not score these cells as PrE, due to their weak GATA4 expression level and ectopic location. To assess the individual contributions of Fgfr1 and Pdgfra to this phenotype, we counted the number of Epi (NANOG+ cells), PrE (GATA4+ cells) and the total number of cells in embryos of different genotypes (Fig. 3B–D). We found that in all embryos deficient for one or two Fgfr1 alleles, the number of Epi cells was increased compared to wild-type control, while Pdgfra−/− embryos showed no change in Epi number (Fig. 3B and Table 3). This further reinforces the concept that FGFR1 plays a major role in ICM cell fate decision towards the PrE lineage. Fgfr1 deficiency causes fewer ICM cells to become PrE, with a compensatory increase in the number of Epi cells. Whereas PDGFRα plays a more important role than FGFR1 in the survival of formed PrE cells, Pdgfra deficiency does not directly affect the number of Epi cells. The number of PrE cells (Fig. 3C) trended lower in Fgfr1−/−; Pdgfra+/+, Fgfr1+/+; Pdgfra−/−, Fgfr1+/−; Pdgfra−/− embryos and was significantly lower in Fgfr1−/−; Pdgfra+/− (n=5, p=0.004) embryos. Fgfr1−/−; Pdgfra−/− embryos were also considerably smaller (p=0.002) than the other embryos (Fig. 3D). Preliminary results suggest that TE development is compromised in these mutants (data not shown).
Figure 3.
PrE development is disrupted in Fgfr1−/−; Pdgfra−/− embryos. (A) Embryos were dissected at E3.5, cultured for 48h in DMEM and stained with antibodies to NANOG (green), GATA4 (red) and CDX2 (white). Number of Epi (B), PrE (C) and total number of cells (D) were counted and are shown for the individual embryos. The proportion of Epi (E) and PrE (F) cells in the embryos are shown. (G) Number of ICM cells. Data represented as mean + SEM. *, p=0.002; **, p=0.003. Scale bars, 50 μm.
Table 3.
Cell lineage composition in Fgfr1; Pdgfra compound mutants
| Number of cells | ||||
|---|---|---|---|---|
| # | Epi | PrE | Total | |
| Fgfr1+/+; Pdgfra+/+ | 6 | 14 ± 2 | 30 ± 5 | 156 ± 14 |
| Fgfrr−/−; Pdgfra+/+ | 5 | 23 ± 4 | 20 ± 6 | 161 ± 11 |
| Fgfr1+/+; Pdgfra−/− | 10 | 13 ± 1 | 20 ± 3 | 156 ± 9 |
| Fgfr1+/−; Pdgfra−/− | 5 | 22 ± 4 | 19 ± 2 | 163 ± 31 |
| Fgfr1−/−; Pdgfra+/− | 5 | 21 ± 4 | 8 ± 3* | 148 ± 17 |
| Fgfr1−/−; Pdgfra−/− | 3 | 19 ± 8 | 0 ± 0** | 51 ± 12 |
Notes:
, p=0.002;
, p=0.003 when compared to wild-type control
To exclude an effect of embryo size on our results, we calculated the percentage of Epi and PrE relative to the total number of cells (Fig. 3E–G). The proportion of Epi cells significantly increased in Fgfr1+/−; Pdgfra−/− (p=0.04), Fgfr1−/−; Pdgfra+/− (p=0.05) and Fgfr1−/−; Fgfr2−/− (p=0.02) embryos (Fig. 3E), with a corresponding decrease in the number of PrE cells (Fig. 3G). The combined number of Epi and PrE cells trended lower in Fgfr1+/+; Pdgfra−/− and Fgfr1+/−; Pdgfra−/− embryos and was significantly lower in Fgfr1−/−; Pdgfra−/− embryos.
PI3K is downstream of PDGFR signaling in PrE development
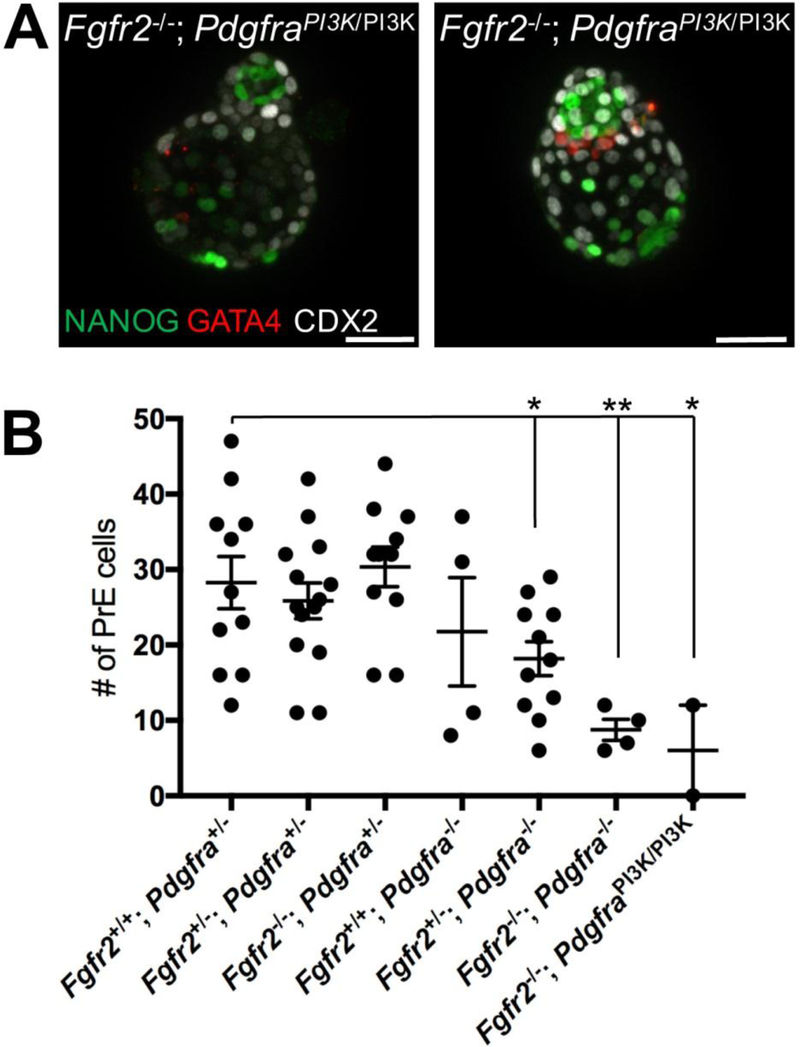
Previous genetic studies in our lab have demonstrated that PI3K is the main signaling pathway downstream of PDGFRα (Fantauzzo and Soriano, 2014, 2016; Klinghoffer et al., 2001; Vasudevan et al., 2015). The PI3K / AKT pathway is active in preimplantation embryos (Riley et al., 2005). To test if PDGFRα-mediated PI3K signaling plays a critical role in PrE development, we intercrossed Pdgfra+/PI3K mice (Klinghoffer et al., 2001), in which PI3K cannot bind to the PDGFRα and become activated upon PDGFRα engagement, with Fgfr2+/− mice to obtain Fgfr2+/−; Pdgfra+/PI3K double heterozygous animals. We crossed Fgfr2+/−; Pdgfra+/PI3K mice to obtain E3.5 embryos, which we further cultured for 48h (Fig. 4A). We found that double homozygous Fgfr2−/−; PdgfraPI3K/PI3K embryos (n=2) have severely reduced number of PrE cells, similar to Fgfr2−/−; Pdgfra−/− embryos (Fig. 4B). This result strongly supports the concept that PDGF-engaged PI3K signaling plays an important role in PrE development.
Figure 4.
PI3K is the main signaling pathway downstream PDGFRα in PrE development. (A) Two E3.5 Fgfr1−/−; PdgfraPI3K/PI3K embryos are shown. NANOG (green) labels Epi cells, GATA4 (red) labels PrE. DAPI (blue) used to counter label cell nuclei. (B) Number of PrE cells in Fgfr1−/−; PdgfraPI3K/PI3K embryos (red box) are shown compared to Fgfr1 & Pdgfra compound mutants. Scale bars, 50 μm.
DISCUSSION
Both Pdgfra and Fgfr2 are expressed in the PrE of the implanting blastocyst. Pdgfra is expressed exclusively in the PrE at this stage (Plusa et al., 2008), while Fgfr2 starts to be expressed in the future TE cells at the 16-cell stage and is later expressed in both the PrE and TE (Kang et al., 2017; Molotkov et al., 2017; Ohnishi et al., 2014). Interestingly, we observed that while both Pdgfra and Fgfr2 are expressed in the same PrE cells in the late blastocyst, Fgfr2 and Pdgfra expression does not entirely overlap at the morula and midblastocyst stages. This suggests that induction of Fgfr2 and Pdgfra expression in ICM cells is controlled by independent mechanisms.
Pdgfra−/− blastocysts have reduced numbers of PrE cells due to increased apoptosis of the developing PrE cells (Artus et al., 2013). PrE development in Fgfr2−/− blastocysts is normal (Kang et al., 2017; Molotkov et al., 2017). Culturing blastocysts in the presence of high doses of external FGF4 failed to convert ICM cells into PrE in embryos deficient for both copies of Fgfr1, while external FGF4 induces the PrE fate in all wild-type and Fgfr1+/− ICM cells (Molotkov et al., 2017). This demonstrated a primary role for FGFR1 but not for FGFR2 in the determination of the PrE fate in ICM cells during development (Kang et al., 2017; Molotkov et al., 2017). However, while around 30% of Fgfr1−/− embryos did not contain any PrE cells, PrE development was still maintained in 70% of Fgfr1−/− blastocysts, with some mutant embryos having normal number of the PrE cells (Brewer et al., 2015). We and others recently demonstrated that simultaneous deletion of both Fgfr1 and Fgfr2 is required to completely block PrE development in all embryos (Kang et al., 2017; Molotkov et al., 2017). This strongly suggests that while FGFR2 may not play a critical role in ICM cell fate decision toward PrE, it may play an auxiliary role. At that time we speculated that, similar to PDGFRα (Artus et al., 2013), FGFR2 might be involved in the control of PrE cell survival and proliferation (Molotkov et al., 2017). The results presented here demonstrate that deletion of both FGFR2 and PDGFRα receptors further reduced the number of PrE cells and was associated with increased apoptosis. Interestingly, this reduction in the number of PrE cells was not accompanied by a compensatory increase in the number of Epi cells, observed in Fgfr1−/−; Fgfr2−/− or Fgf4−/− embryos (Kang et al., 2017; Kang et al., 2013; Krawchuk et al., 2013; Molotkov et al., 2017). This suggests that the reduction in PrE cell number in Fgfr2−/−; Pdgfra−/− embryos is due to decreased survival of these cells rather than a switch in the fate of the developing ICM toward Epi, which is controlled by FGF4 signaling to FGFR1.
The critical role of FGFR1 in the control of PrE fate was further substantiated by our analysis of Fgfr1−/−; Pdgfra−/− embryos. We found that, similar to Fgfr1−/−; Fgfr2−/− embryos, deletion of Pdgfra together with Fgfr1 prevented PrE development. In line with this observation, we observed a compensatory increase in the number of Epi cells in Fgfr1−/−; Pdgfra−/− embryos, also seen previously in some Fgfr1−/− and in all Fgfr1−/−; Fgfr2−/− embryos. This further supports the hypothesis that FGF4 signaling through FGFR1 is required for PrE fate specification. Interestingly, two out of three Fgfr1−/−; Pdgfra−/− embryos contained a few cells that expressed GATA4 faintly, but these cells were located far from NANOG+ Epi cells, suggesting that they do not acquire a “classical” PrE cell fate and location adjacent to Epi. Such cells were never observed in Fgfr1−/−; Fgfr2−/− embryos. We hypothesize that these few faintly expressing Fgfr1−/−; Pdgfra−/− GATA4+ cells retain sufficient levels of Fgfr2 that allowed them to survive, and that in the absence of both FGFR1 and PDGFRα they are unable to migrate to their proper position adjacent to Epi, leaving them in ectopic locations in the embryo.
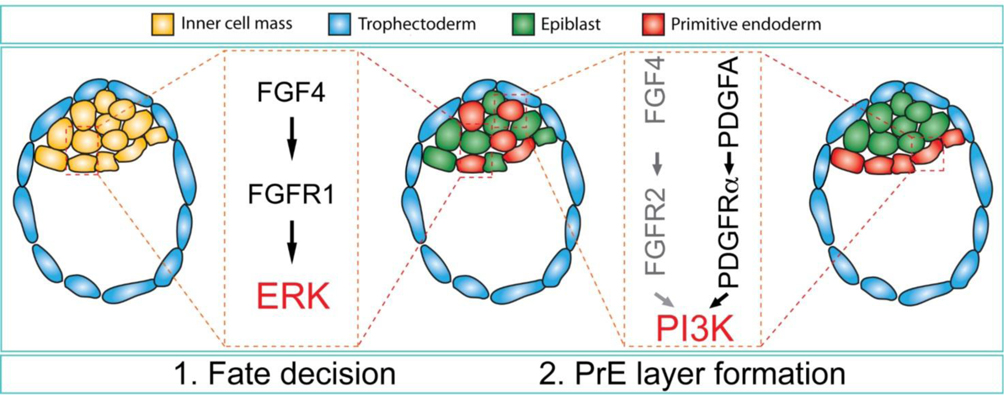
PI3K is the main signaling pathway downstream of the PDGFRα (Fantauzzo and Soriano, 2014, 2016; Klinghoffer et al., 2001). In contrast, FGFRs have long been thought to be dependent on downstream activation of ERK for their biological activity, but they are also known to engage PI3K signaling (Brewer et al., 2016; Brewer et al., 2015; Vasudevan et al., 2015). Our data now suggests that both ERK and PI3K signaling might be involved in PrE development (Fig. 5). First, in the mid-blastocyst, FGF4 produced by a subpopulation of ICM cells binds to FGFR1 on neighboring ICM cells and activates ERK1/2, which induces the PrE fate. The involvement of ERK1/2 in PrE fate determination was demonstrated earlier by studies using selective inhibitors (Nichols et al., 2009). Second, selective inhibition of PI3K significantly reduced number of embryos developing to the blastocyst stage and increased the proportion of cells with a fragmented nuclei (Lu et al., 2004). In the expanded blastocyst, PDGFA (Ohnishi et al., 2014) signaling to PDGFRα, and potentially FGF4 signaling to FGFR2, activates PI3K in newly forming PrE cells, which may control their survival and possibly position (Brewer et al., 2016; Manning and Toker, 2017). Our results thus suggest a sequential role for multiple signaling pathways in establishing cell fates in the preimplantation embryo.
Figure 5.
Model for FGF and PDGF signaling in PrE development. At the mid-blastocyst stage, FGF4 is produced by a subpopulation of the ICM cells and signals in a paracrine fashion through ERK1/2 to induce PrE cells. In expanded blastocysts, PDGFA signaling to PDGFRα, and potentially FGF4 signaling to FGFR2, activates PI3K in newly forming PrE cells, which controls their survival.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal to CDX2 | BioGenex | MU392A-UC |
| Goat polyclonal to GATA4 | Santa Cruz | sc-1237 |
| Goat polyclonal to GATA6 | R&D Systems | AF1700 |
| Rabbit polyclonal to GFP | Invitrogen | A6455 |
| Rabbit polyclonal to mCHERRY | Abeam | ab167453 |
| Rabbit polyclonal to NANOG | Cosmo Bio Co., Ltd. | RCAB0002P-F |
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| N/A | ||
| Critical Commercial Assays | ||
| N/A | ||
| Deposited Data | ||
| N/A | ||
| Experimental Models: Cell Lines | ||
| N/A | ||
| Experimental Models: Organisms/Strains | ||
| Fgfr1+/− mouse strain | This lab | N/A |
| Fgfr2+/− mouse strain | This lab | N/A |
| Pdgfra+/H2B-GFP mouse strain | This lab | N/A |
| Pdgfra+/tm5Sor mouse strain | This lab | N/A |
| Fgfr2 T2A-H2B-mCherry/T2A-H2B-mCherry mouse strain | This lab | N/A |
| Oligonucleotides | ||
| N/A | ||
| Recombinant DNA | ||
| N/A | ||
| Software and Algorithms | ||
| Excel | Microsoft | 2016 |
| FIJI (ImageJ) | NIH | ImageJ ver 2 |
| MetaMorph Image Analysis | Molecular Devices | Ver 7.8.13.0 |
| Prism | GraphPad | Ver 6 |
| Other | ||
| N/A | Ibidi | 80826 |
Supplementary Material
Figure S1. Fragmented cell nuclei observed in Fgfr2−/−; Pdgfra−/− embryo (bottom row). Top row, wild-type blastocyst (control). Combined image (left) and individual channel images for Pdgfra (green), NANOG (red) and DAPI (grey) are shown. The yellow arrow points to an example of a fragmented nucleus. Scale bars, 50 μm.
Highlights.
PDGF and FGF signaling are both critical in primitive endoderm (PrE) development in the mouse.
FGF4/FGFR1 engaged ERK1/2 signaling regulates cell allocation towards the PrE.
PDGFA/PDGFRα engaged PI3K signaling regulates PrE survival and positioning.
FGF4/FGFR2 also regulates cell survival, thus suggesting a role for PI3K signaling in this process.
Acknowledgements
We thank our laboratory colleagues for helpful discussions and critical comments on the manuscript. This work was supported in part by the Tisch Cancer Institute at Mount Sinai (P30 CA196521 Cancer Center Support Grant) and by grants from NYSTEM (IIRP N11G-131) and NIH/NIDCR (RO1 DE022778) to P.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artus J, Kang M, Cohen-Tannoudji M, Hadjantonakis AK, 2013. PDGF Signaling Is Required for Primitive Endoderm Cell Survival in the Inner Cell Mass of the Mouse Blastocyst. Stem cells (Dayton, Ohio) 31, 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Panthier JJ, Hadjantonakis AK, 2010. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development (Cambridge, England) 137, 3361–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Mazot P, Soriano P, 2016. Genetic insights into the mechanisms of Fgf signaling. Genes & development 30, 751–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Molotkov A, Mazot P, Hoch RV, Soriano P, 2015. Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes & development 29, 1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J, 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental cell 10, 615–624. [DOI] [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P, 2014. PI3K-mediated PDGFR alpha signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes & development 28, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P, 2016. PDGFR beta regulates craniofacial development through homodimers and functional heterodimers with PDGFRalpha. Genes & development 30, 2443–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M, 1995. Requirement of FGF-4 for postimplantation mouse development. Science (New York, N.Y.) 267, 246–249. [DOI] [PubMed] [Google Scholar]
- Goldin SN, Papaioannou VE, 2003. Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis (New York, N.Y.: 2000) 36, 40–47. [DOI] [PubMed] [Google Scholar]
- Grabarek JB, Zyzynska K, Saiz N, Piliszek A, Frankenberg S, Nichols J,Hadjantonakis AK, Plusa B, 2012. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development (Cambridge, England) 139, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P, 2010Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental cell 18, 675–685. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P, 2003. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and cellular biology 23, 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch RV, Soriano P, 2006. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development (Cambridge, England) 133, 663–673. [DOI] [PubMed] [Google Scholar]
- Kang M, Garg V, Hadjantonakis AK, 2017. Lineage Establishment and Progression within the Inner Cell Mass of the Mouse Blastocyst Requires FGFR1 and FGFR2. Developmental cell 41, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK, 2013. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development (Cambridge, England) 140, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P, 2002. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Developmental cell 2, 103–113. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Mueting-Nelsen PF, Faerman A, Shani M, Soriano P, 2001. The two PDGF receptors maintain conserved signaling in vivo despite divergent embryological functions. Molecular cell 7, 343–354. [DOI] [PubMed] [Google Scholar]
- Krawchuk D, Honma-Yamanaka N, Anani S, Yamanaka Y, 2013. FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Developmental biology 384, 65–71. [DOI] [PubMed] [Google Scholar]
- Lu DP, Chandrakanthan V, Cahana A, Ishii S, O’Neill C, 2004. Trophic signals acting via phosphatidylinositol-3 kinase are required for normal pre-implantation mouse embryo development. Journal of cell science 117, 1567–1576. [DOI] [PubMed] [Google Scholar]
- Manning BD, Toker A, 2017. AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Mazot P, Brewer JR, Cinalli RM, Soriano P, 2017. Distinct Requirements for FGFR1 and FGFR2 in Primitive Endoderm Development and Exit from Pluripotency. Developmental cell 41, 511–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Graham SJ, Jedrusik A, Zernicka-Goetz M, 2013. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open biology 3, 130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A, 2009. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development (Cambridge, England) 136, 3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oles AK, Arauzo-Bravo MJ, Saitou M, Hadjantonakis AK, Hiiragi T, 2014. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nature cell biology 16, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N, 2015. The Fibroblast Growth Factor signaling pathway. Wiley interdisciplinary reviews. Developmental biology 4, 215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK, 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst.Development (Cambridge, England) 135, 3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JK, Carayannopoulos MO, Wyman AH, Chi M, Ratajczak CK, Moley KH,2005. The PI3K/Akt pathway is present and functional in the preimplantation mouse embryo. Developmental biology 284, 377–386. [DOI] [PubMed] [Google Scholar]
- Soriano P, 1997. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development (Cambridge, England) 124, 2691–2700. [DOI] [PubMed] [Google Scholar]
- Vasudevan HN, Mazot P, He F, Soriano P, 2015. Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways.eLife 4, e07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C, 1998. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development (Cambridge,England) 125, 753–765. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J, 2010. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development (Cambridge, England) 137, 715–724. [DOI] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM, 2003Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development (Cambridge, England) 130, 3063–3074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Fragmented cell nuclei observed in Fgfr2−/−; Pdgfra−/− embryo (bottom row). Top row, wild-type blastocyst (control). Combined image (left) and individual channel images for Pdgfra (green), NANOG (red) and DAPI (grey) are shown. The yellow arrow points to an example of a fragmented nucleus. Scale bars, 50 μm.