Abstract
CLBQ14 is an 8-hydroxyquinoline analogue that inhibits methionine aminopeptidase (MetAP), an enzyme responsible for the post-translational modification of several proteins and polypeptides. MetAP has been validated as druggable target for some infectious diseases, and its inhibitors have been investigated as potential therapeutic agents. In this study, we developed and validated a liquid chromatography tandem-mass spectrometry (LC-MS/MS) method for the quantification of CLBQ14 in solution, and in rat plasma and urine. This method was applied to the pharmacokinetic evaluation of CLBQ14 in adult male Sprague Dawley (SD) rats. Chromatographic separation was achieved using an ultra-high-performance liquid chromatography (UHPLC) system equipped with Waters XTerra MS C18 column (3.5 μm, 125Å, 2.1 × 50 mm) using 0.1% formic acid in acetonitrile/water gradient system as mobile phase. Chromatographic analysis was performed with a 4000 QTRAP® mass spectrometer using MRM in positive mode for CLBQ14 transition [M+H]+ m/z 257.919 → m/z 151.005, and IS (clioquinol) transition [M+H]+ m/z 305.783 → m/z 178.917. CLBQ14 was extracted from plasma and urine samples by protein precipitation. The retention times for CLBQ14 and IS were 1.31 and 1.40 minutes respectively. The standard curves were linear for CLBQ14 concentration ranging from 1 – 1000 ng/mL. The intra-day and inter-day accuracy and precision were found to be within 15 % of the nominal concentration. Extraction recoveries were greater than 96.3 % and 96.6 % from rat plasma and urine respectively, and there was no significant matrix effect from the biological matrices. CLBQ14 is stable in samples subjected to expected storage, preparation, and handling conditions. Pharmacokinetic studies revealed that CLBQ14 has a bi-exponential disposition in SD rats, is extensively distributed with a long plasma half-life and is eliminated primarily by liver metabolism.
Keywords: 8-hydroxyquinoline, LC-MS/MS, bio-analytical method development, pharmacokinetics
1. INTRODUCTION
Methionine amino peptidase (MetAP) has been investigated as a potential target for the development of novel chemotherapeutic agents [1–7]. MetAP is a metalloprotease that catalyzes the cleavage of N-terminal methionine, the initiating amino acid for protein and polypeptide synthesis [8–10]. This process, referred to as N-terminal methionine excision (NME), is universal and essential for the localization, stability and post-translational modification of several important nascent proteins and polypeptides[9, 10]. The essentiality of the NME process in prokaryotes makes MetAP a very appealing target for infectious diseases. Two classes of MetAP have been identified in eukaryotes and prokaryotes[11]. While eukaryotes possess both classes (MetAP1 and MetAP2), prokaryotes have homologs of either classes [11–13].
MetAPs have been explored as a potential druggable target for many different diseases. Various classes of MetAPs have been validated as targets for infectious diseases caused by Mycobacterium tuberculosis (Mtb)[1], Cryptosporidium parvum[6], Escherichia coli and Staphylococcus aureus[4], Acinetobacter baumannii[3], and Plasmodium falciparium[5]. In addition to infectious diseases, MetAP inhibitors have been explored as potential therapeutic agents for treating other diseases such as cancer and rheumatoid arthritis[14–16].
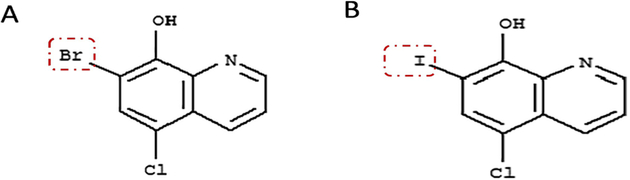
Analogues of the 8-hydroxyquinoline have been also been reported as highly effective inhibitors of MetAP, showing in vitro activity against various pathogens [2] Using high-throughput screening, 7-bromo-5-chloroquinolin-8-ol (CLBQ14) (Figure 1), a congener of clioquinol (CQ) was previously identified and reported as a potent and selective inhibitor of two MetAP in Mycobacterium tuberculosis (MtMetAP1a and MtMetAP1c) [2, 17]. Further in vitro studies have demonstrated some of the potential therapeutic advantages of CLBQ14 [2]. It has great potency against replicating Mtb; an increased potency against aged non-growing Mtb and exhibited great selectivity for both MtMetAPs over human MetAPs.
Figure 1.
Chemical structure of (A) CLBQ14 and (B) Clioquinol. CLBQ14 and Clioquinol are congeners of 8-hydroxyquinoline varying only by the halogen attached at position C7. CLBQ14 has bromine attached at position C7, while Clioquinol has iodine. CLBQ14 is acidic at 25 °C with a calculated pKa of 2.23± 0.30. It has a clog P value of 3.92 ± 0.39 at room temperature.
Once a promising lead molecule like CLBQ14 has been identified, further pre-clinical studies will be needed to develop these molecules for clinical applications. Bioanalytical assays are extremely important during the process of drug development. These assay methods are imperative for the quantification of the lead compound at various stages of the development. A full validation of the bioanalytical method is required by the FDA when a new method is developed for the quantification of a new drug entity, and for revisions to an existing method. With respect to CLBQ14, a simple, specific, and sensitive assay for its determination in biological matrices such as plasma and urine is important for further pre-clinical pharmacokinetic evaluation. Although reverse phase high-performance liquid chromatography (HPLC) methods have been reported for halogenated 8-hydroxyquinolines[18], sulfonated hydroxyquinolines[19], and 8-hydroxyquinoline sulfate[20], no bioanalytical method for the determination of CLBQ14 has been reported.
In this study, we developed and validated a simple, sensitive, and reliable LCMS/MS method for the quantification of CLBQ14 in solution, and in rat plasma and urine. The linearity, selectivity, sensitivity, and reproducibility were validated according to the FDA guidance document on bioanalytical method validation. The stability of CLBQ14 in rat plasma and urine samples subjected to expected sample storage, handling, preparation, and analysis conditions was also assessed. The validated method was subsequently applied to the pharmacokinetic study of CLBQ14 in adult male Sprague Dawley (SD) rats.
2. MATERIALS AND METHODS
2.1. Materials
CLBQ14 (purity ≥ 98%) was purchased from TCI Chemicals (Tokyo, Japan). Clioquinol, LC-MS grade acetonitrile, water, and formic acid were purchased from Sigma Aldrich (St. Louis, MO). Heparin sodium injection (1000 units/mL) was purchased from Hospira (Lake Forest, IL). Adult male SD rats were purchased from Envigo RMS, Inc. Freshly obtained rat plasma and urine were collected from male SD rats from Envigo and stored at −80 °C until use. All chemicals and reagents were used as received.
2.2. Instruments and Conditions
HPLC analysis was performed with a Shimadzu Nexera X2 UHPLC System (Columbia, MD) equipped with a Waters XTerra® MS C18 column (3.5 μm, 125Å, 2.1 × 50 mm, Milford, MA) at room temperature. A flow rate of 0.5 mL/min and sample injection volume of 10 μL was employed. Chromatographic separation was achieved with a binary solvent system: Solvent A was 0.2% formic acid in LC-MS grade water and Solvent B – 0.2% formic acid in LC-MS grade acetonitrile. All samples were analyzed using gradient elution: initial 20% B, 70% B at 0.80 min, 95% from 2.80 – 3.80 min, and 40% B from 4.00 – 5.50 min (Table 1).
Table 1.
Summary of gradient elution profile applied to the chromatographic separation of CLBQ14 and IS from matrix.
| Time (min) | Flow rate (mL/min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|---|
| 0 | 0.5 | 80 | 20 |
| 0.8 | 0.5 | 30 | 70 |
| 2.8 | 0.5 | 5 | 95 |
| 3.8 | 0.5 | 5 | 95 |
| 4.0 | 0.5 | 60 | 40 |
| 5.5 | 0.5 | 60 | 40 |
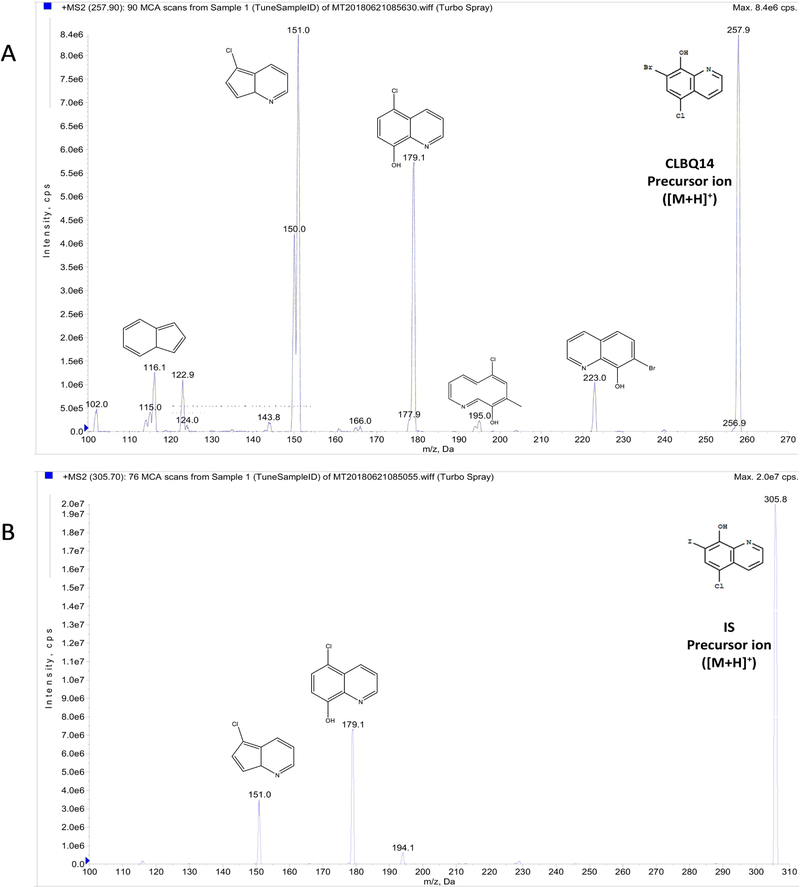
Chromatographic analysis was performed using a 4000 QTRAP® LC-MS/MS system (AB Sciex, Redwood City, CA). The hybrid triple quadrupole LIT (linear ion trap) mass spectrometer is equipped with a Turbo V™ ion source. Pure nitrogen used as curtain gas, and source and exhaust gases were generated by a Peak Scientific GENIUS ABN2ZA Tri Gas Generator. The quantification of the analyte and IS was performed by multiple reaction monitoring (MRM) operated in positive mode, with electrospray ionization (ESI). The transitions from specific precursor ion to product ion for CLBQ14 ([M+H]+ m/z 257.919 →m/z 151.005) and IS ([M+H]+ m/z 305.783 →m/z 178.900) were selected based on being the most intense peaks for the final MRM method. Clioquinol (CQ) was used as internal standard (IS). The ion spray heater was maintained at 550 °C and nebulizer gas and heater gas pressure of 55.0 and 50.0 psi respectively was employed. The ion spray voltage of 5500 V, curtain gas pressure of 25.0 psi and high collision “CAD” gas pressure were applied. The collision energy for CLBQ14 and IS were 53.00 eV and 39.00 eV respectively. The compound dependent parameters were optimized with entrance potential (EP) of 10 V, and dwell time of 150 milliseconds for the analyte and IS applied. Analyst® Software 1.6.2 (AB Sciex, Redwood City, CA) was used to control the LC-MS/MS system and analyze the data. The compound dependent electronic parameters for the MS/MS acquisition of CLBQ14 and IS are summarized in Table 2. The chemical structures and fragmentation patterns of CLBQ14 and IS are presented in Figure 1 and 2 respectively.
Table 2.
Electronic parameters for MS/MS acquisition of CLBQ14 and IS.
| Parent | Transition (m/z) | Dwell Time (msec) | DP (Volts) | EP (Volts) | CE (Volts) | CXP (Volts) |
|---|---|---|---|---|---|---|
| CLBQ14 | 257.9→151.0 | 150 | 101.00 | 10.00 | 53.00 | 8.00 |
| Clioquinol (IS) | 305.7→178.9 | 150 | 91.00 | 10.00 | 39.00 | 8.00 |
Figure 2.
Product ion spectra with predicted structures of fragments: (A) CLBQ14 precursor (m/z 257.9) and product ions; (B) Clioquinol (IS) precursor (m/z 305.7) and product ions. Their exact masses are 256.9243 and 304.9104 respectively. [M+H]+ was selected as precursor ion, while fragment with highest intensity was selected as product ion for the MRM acquisition method.
2.3. Standard and Quality Control Samples
Stock solutions of the analyte, CLBQ14, and IS (1 mg/mL) were prepared separately by dissolving each compound in acetonitrile and stored at −20 °C until use. Standard samples were prepared in rat plasma and urine at concentrations between 1 – 1000 ng/mL. Quality control (QC) samples were prepared in rat plasma and urine at lower limit of quantitation (LLOQ; 1 ng/mL), low (2.5 ng/mL), medium (400 ng/mL) and high (800 ng/mL) concentrations of CLBQ14. Plasma and samples were processed by protein precipitation. A 45 μL aliquot of plasma or urine spiked with 5 μL of standard solutions of CLBQ14 (in pure acetonitrile) was extracted with 200 μL of acetonitrile containing 150 ng/mL of IS. The mixture was vortex mixed for 30 seconds and then centrifuged at 14,000 rpm (18,879 g) and 4°C for 10 minutes. The resulting supernatant was then transferred to an auto-sampler vial for LCMS/MS analysis.
2.4. Plasma and Urine Sample Preparation
The rat plasma and urine samples were collected and stored at –80°C until analysis. The plasma and urine samples were processed by protein precipitation as described above. Briefly, a 50 μL aliquot of the plasma or urine sample was deproteinized/extracted with 200 μL of acetonitrile containing 150 ng/mL of IS followed by vortex mixing for 30 seconds. The mixture was centrifuged at 14,000 rpm (18,879 g) and 4 °C for 10 minutes. The resulting supernatant was then transferred to an auto-sampler vial for LC-MS/MS analysis.
2.5. Method Validation
The LC-MS/MS assay described herein was validated according to the “United States Food and Drug Administration, Center for Drug Evaluation and Research: Guidance for Industry – Bioanalytical Method Validation” document [21].
2.5.1. Linearity and Sensitivity
Linear calibration curves in rat plasma and urine were generated by plotting the peak area ratio of CLBQ14 to IS against known standard concentrations of CLBQ14. The slope, intercept, and coefficient of determination were estimated using least squares linear regression method with a weighting of 1/x2. The lower limit of quantification (LLOQ) was evaluated based on the signal-to-noise ratio of at least 5:1.
2.5.2. Selectivity and Specificity
The selectivity and specificity of the assay was evaluated by analyzing blank plasma and urine samples from six individual sources for interference with the analyte. The IS response in the blank was compared to the average IS responses of standard curve and QCs.
2.5.3. Accuracy and Precision
The intra-day and inter-day accuracy and precision of the assay for the determination of CLBQ14 rat plasma and urine were determined by measuring the concentration of CLBQ14 in six replicates of LLOQ, low, medium, and high QC samples. The intra-day accuracy and precision were computed by analyzing the QC samples using calibration curve constructed on the same day, while the inter-day accuracy and precision were obtained using calibration curved constructed on 3 different days. The accuracy of the assay was established by the relative error from the theoretical CLBQ14 concentrations, while the assay precision was reflected by the coefficient of variation.
2.5.4. Dilution Integrity
The dilution integrity of the assay was evaluated by analyzing six replicates per dilution factor (5, 10, 20, and 50) of blank rat plasma spiked with CLBQ14 at 5000 ng/mL. The accuracy and precision of the measurements of the diluted samples was estimated.
2.5.5. Extraction Recovery and Matrix Effect
The extraction recovery of CLBQ14 from rat plasma and urine was determined by analyzing two sets of QC samples (n = 3) spiked with CLBQ14 either pre- or post- protein precipitation. The extraction recovery of CLBQ14 was calculated as follows:
| (1) |
where Responsepre-extraction sample is the average area count of CLBQ14 in a biological matrix spiked with the analyte prior to protein precipitation, and Responsepost-extraction spiked sample is the average area count of CLBQ14 in a sample spiked with the analyte after the protein precipitation.
The effect of the biological matrix on the determination of CLBQ14 was evaluated by analyzing two sets of QC samples (n = 3) containing either a biological matrix or neat solution (pure acetonitrile). The matrix factor was calculated as follows:
| (2) |
where Responsepost-extraction spike sample is the average area count of CLBQ14 in a biological matrix spiked with the analyte after the protein precipitation, and Response neat sample is the average peak area count for the same concentration of CLBQ14 prepared in a neat solution.
Furthermore, signal interference by PEG 400 (used as a dosing vehicle excipient in pharmacokinetic studies) was evaluated by estimating the recovery, accuracy, and precision of six replicates of QC samples prepared by spiking CLBQ14 into rat plasma containing 0.1% PEG 400 and 1% PEG 400 respectively. These samples were compared to samples to which PEG 400 was not incorporated[22].
2.5.6. Stability
The stability of CLBQ14 in rat plasma and urine samples during sample storage, handling, preparation, and analysis was evaluated by determining the recovery of the analyte in such samples subjected to short-term bench-top storage, freeze and thaw cycles, and storage on the auto-sampler prior to analysis. These experiments were conducted in triplicate.
The short-term (bench-top) stability of CLBQ14 in rat plasma and urine samples was evaluated by analyzing three sets each of freshly prepared plasma and urine samples placed on the bench-top for 2, 4, and 6 h, respectively. All the samples were compared with freshly prepared samples of the same concentration.
The long-term stability of CLBQ14 in freezer stored rat plasma and urine samples was evaluated by comparing samples stored at – 80°C for 14 days to freshly prepared samples of the same concentration.
The stability of CLBQ14 in frozen and thawed rat plasma and urine samples was evaluated by analyzing three sets of QC plasma and urine samples (low, medium and high CLBQ14 concentration) exposed to three cycles of freeze (– 80°C) and thaw (room temperature). The freeze-thawed samples were compared to freshly prepared samples.
The stability of CLBQ14 in samples placed on the instrument (auto-sampler maintained at 15 °C) was evaluated by comparing freshly prepared samples to protein extracted plasma and urine QC samples placed on the auto-sampler for 2, 4, and 6 h respectively. One set of the QC samples was extracted with acetonitrile containing IS, and the other set with pure acetonitrile without IS.
2.6. Application to Pharmacokinetic Study
The validated LC-MS/MS method was applied to the quantification of CLBQ14 in rat plasma and urine in a pharmacokinetic study. The animal experiment and protocol were reviewed and approved by the Institutional Animal Care and Use Committee at Texas Southern University. All experimental procedures were performed in accordance with the “National Institute of Health: Guide for the Care and Use of Laboratory Animals, 8th Edition”[23].
Adult male SD rats weighing 300 – 350 g were cannulated through the jugular vein under anesthesia one day before the study. The rats divided into three groups were administered either a 2 mg/kg, 5 mg/kg or 10 mg/kg intravenous (IV) bolus dose of CLBQ14 in a co-solvent formulation comprising of 5% N, N – dimethyl acetamide, 35% PEG 400, 60% Tween 80 and 7.5 mg/mL of CLBQ14. The formulation was diluted 5 times with normal saline prior to dosing. Serial heparinized blood samples (approximately 250 μL) were withdrawn from each rat (through the cannulated jugular vein) at 0.08, 0.25, 0.5, 1, 2. 4, 6, 8, 12, 24 and 48 hours after injection. The blood samples were centrifuged, and the supernatant plasma obtained and stored at – 80 °C until analysis. Urine samples were collected up to 24 hours after the injection and stored under same conditions until analysis. The plasma and urine samples were analyzed using the validated LC-MS/MS method to determine the concentration of CLBQ14. The pharmacokinetic parameters for each rat were estimated using Phoenix WinNonlin v7.0 software (Pharsight Corporation, Mountain View, CA, USA).
2.7. Pharmacokinetic and Statistical Analyses
The pharmacokinetic parameters for CLBQ14 were computed with Phoenix WinNonlin v7.0 using compartmental analysis. Compartmental pharmacokinetic analysis was performed to estimate the area under the plasma concentration – time curve (AUC), mean residence time (MRT), and the plasma concentration at time zero (Co), beta phase elimination half-life (T1/2β), volume of distribution of the central compartment (VD), and total plasma clearance (Cl). The Akaike inclusion criteria (AIC), Schwartz criteria (SC), sums of square residuals (SSR), weighted sum of square residuals (WSSR), and the observed versus predicted plasma concentration – time profile were considered when selecting the most suitable compartmental model for the estimation of the pharmacokinetic parameters. The model yielding the lowest AIC and SC values was selected as the most appropriate model [24].
The C0 and AUC0-t(last) data were analyzed non-parametrically to study the dose proportionality. The parameters were dose-normalized and compared using a Friedman test for each dose comparison. A 0.05 level of significance was used.
3. RESULTS AND DISCUSSION
3.1. LC-MS/MS Method Development
A reliable, sensitive, and robust bioanalytical method is essential for the quantification of lead molecules in solutions and biological matrices during pre-clinical and clinical studies. Such a quantitative method must be considerably sensitive, selective, rapid, and reproducible in accordance with the FDA’s regulatory standards. No chromatographic method has been previously reported for quantification of CLBQ14, hence it was imperative to develop and validate an LCMS/MS method for the quantification of CLBQ14 in future studies.
3.1.1. Chromatographic Conditions
The mobile phase and column were optimized until an optimal intensity and peak shape was attained. Optimal response and symmetrical peak shape was achieved with a Waters ® Xterra MS C18 column (50 × 2.1 mm, 3.5 μm, 125Å) using 0.2% formic acid in water and acetonitrile as mobile phase compared to 0 – 0.1% formic acid. Positive mode and electronic spray ionization (ESI) were found to most suitable for an optimal peak intensity compared to negative mode and atmospheric chemical ionization (APCI). The source parameters were optimized to obtain the most suitable conditions for the determination of the analyte. A column chamber temperature of 25 °C were found to be most suitable.
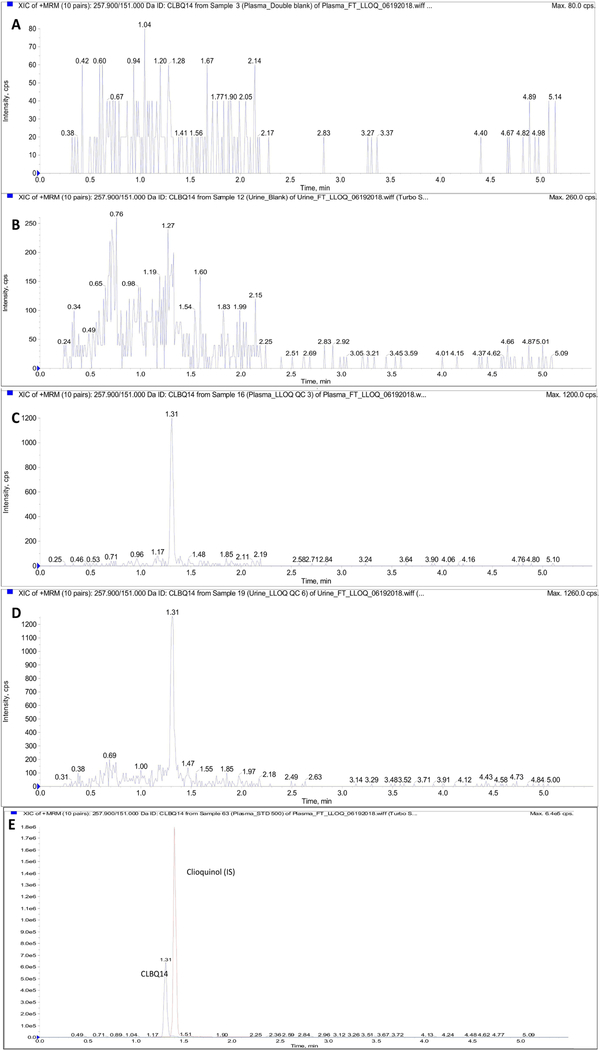
The chromatographic run time for the assay was 5.5 minutes with retention times of 1.31 and 1.40 minutes observed for analyte and IS respectively. Figure 3 illustrates the chromatograms obtained from double blank rat plasma sample, CLBQ14 spiked rat plasma sample, and rat plasma sample obtained 30 minutes after the administration of CLBQ14 in a pharmacokinetic study. These chromatograms demonstrate the absence of matrix interference and carryover effects. Carryover effects or needle contamination was avoided by using a strong organic solvent mixture comprising of 40% isopropyl alcohol, 50% acetonitrile, 10% acetone and 0.05% formic acid as needle wash between each injection. No peak with retention comparable to CLBQ14 were detected in double blank samples injected immediately after injection of sample with high CLBQ14 concentration.
Figure 3.
Representative LC-MS/MS chromatograms (XIC of MRM) for (A) Blank rat plasma sample; (B) Blank rat urine; (C) Rat plasma sample spiked with LLOQ concentration of CLBQ14; (D) Rat urine sample spiked with LLOQ concentration of CLBQ14; (E) Rat plasma sample showing spiked analyte and IS. CLBQ14 transition depicted in blue and IS transition depicted in red.
3.2. Method Validation
3.2.1. Linearity and Sensitivity
The calibration curves for the quantification of CLBQ14 in solution, and in rat plasma and urine were linear for CLBQ14 concentration ranging from 1 – 1000 ng/mL. Linear correlation coefficient greater than 0.99 and 0.999 were considered acceptable for the determination of the analyte in biological matrices. The lower limit of quantification (LLOQ) was determined based on a signal-to-noise ratio of at least 5:1. Based on this, the LLOQ of the assay was 1 ng/mL in rat plasma and urine respectively.
3.2.2. Selectivity and Specificity
The specificity and selectivity of the method was assessed from cross-reacting molecules, and component of biological matrices by analyzing replicates of blank samples from multiple sources. Blank and zero calibrators were free of interference at the retention times of the analyte and IS. The IS response in the blank samples were 2.1% of the average IS response of the calibrators and QCs.
3.2.3. Accuracy and Precision
The intra-day and inter-day accuracy and precision, presented as percentage relative error and percentage co-efficient of variation respectively, were within the acceptable limit of ≤ 20% for LLOQ QC and ≤ 15 % for other QCs. The data (Table 3) indicates that the LC-MS/MS method is accurate and precise for the quantification of the CLBQ14 in rat plasma and urine at concentrations ranging from 1 to 1000 ng/mL.
Table 3.
Intra- and Inter-day accuracy and precision of the LC-MS/MS method for the quantification of CLBQ14 in various matrices. Values ≤ 20% for LLOQ and ≤ 15 % for other QCs are considered acceptable.
| Biological Matrix | QC | Nominal Concentration (ng/mL) | Intra-day (n=6) | Inter-day (n=6) | ||
|---|---|---|---|---|---|---|
| Accuracy (RE, %) | Precision (CV, %) | Accuracy (RE, %) | Precision (CV, %) | |||
| Plasma | LLOQ | 1 | 94.5 | 5.8 | 96.7 | 5.6 |
| LQC | 2.5 | 88.7 | 6.4 | 94.2 | 7.3 | |
| MQC | 400 | 95.2 | 1.6 | 100.4 | 3.3 | |
| HQC | 800 | 92.0 | 2.5 | 95.9 | 3.2 | |
| Urine | LLOQ | 1 | 96.1 | 7.6 | 100.9 | 7.2 |
| LQC | 2.5 | 99.0 | 9.2 | 96.4 | 4.7 | |
| MQC | 400 | 99.6 | 2.3 | 100.6 | 2.6 | |
| HQC | 800 | 97.9 | 2.6 | 99.8 | 2.5 | |
3.2.4. Dilution Integrity
The accuracy and precision of diluted plasma samples was evaluated to establish the dilution integrity of the assay. The accuracy and precision of plasma samples diluted up to 50 times with blank plasma were within 15% of the nominal concentration. The data summarized in Table 5 suggests that the LC-MS/MS method can be used to determine the concentration of CLBQ14 in rat plasma sample diluted up to 50 times with blank rat plasma.
Table 5:
Effect of dilution on the quantification of CLBQ14 using this LC-MS/MS method (n = 6). Values ≤ 15 % are considered acceptable.
| Biological Matrix | Dilution Factor | Accuracy (RE, %) | Precision (CV, %) |
|---|---|---|---|
| Plasma | 5 | 99.7 | 3.5 |
| 10 | 94.6 | 8.6 | |
| 20 | 95.6 | 10.7 | |
| 50 | 106.7 | 6.4 |
3.2.5. Extraction Recovery and Matrix Effect
The extraction recovery of CLBQ14, expressed as a percentage recovered in Table 5, was calculated as a ratio of the response obtained from a sample of biological matrix spiked with CLBQ14 before protein precipitation to the response from sample spiked after protein precipitation. The percentage recoveries of CLBQ14 spiked to rat plasma and urine were consistent and found to be greater than 96.3 ± 3.85 and 96.6 ± 1.59 % respectively.
The matrix factor indicates the likelihood of ion enhancement or suppression by co-eluting biological matrix components during LC-MS/MS analysis. A positive value indicates the enhancement of the analyte signal, while a negative value indicates suppression of the analyte signal [25]. Matrix effect is considered significant if the matrix factor is greater than ± 15%. Table 5 shows the average matrix factors obtained for low, medium, and high QC concentrations of CLBQ14 in rat plasma and urine respectively. The data suggest that there was no measurable matrix effect interfering with the determination of CLBQ14 in rat plasma and urine using this LC-MS/MS method.
The accuracy for rat plasma QC samples spiked with 0.1% and 1% PEG 400 were 99.7% and 101.1% respectively. The precision for the samples were 4.3% and 6.4% accordingly. Accuracy and precision within 15% of the nominal concentration suggests that there is no significant signal interference by PEG 400 which is used as dosing formulation excipient in the pharmacokinetic study of the analyte.
3.2.6. Stability
The stability of the analyte under expected sample handling, storage and preparation conditions prior to LC-MS/MS analysis was evaluated by determining the recovery of CLBQ14 from plasma and urine samples placed on the bench-top, and auto-sampler, frozen at – 80 °C and subjected to freeze and thaw cycles. The results, expressed as mean percentage of nominal concentration remaining, are summarized in Table 6.
Table 6:
Stability of CLBQ14 in Sprague Dawley Rat Plasma Samples for LC-MS/MS Analysis [n = 3; mean (± SD)]
| Biological Matrix | Time | Mean Recovery ± SD | |
|---|---|---|---|
| Short-term/Long-term Stability | Plasma | 2h | 91.6 ± 4.06 |
| 4h | 95.1± 3.24 | ||
| 6h | 93.7 ± 1.17 | ||
| 14 days | 91.3 ± 5.45 | ||
| Urine | 2h | 94.4 ± 1.83 | |
| 4h | 93.2 ± 4.10 | ||
| 6h | 95.6 ± 1.44 | ||
| 14 days | 101.6 ± 4.29 |
| Time (hr) | Mean Recovery ± SD | |||
|---|---|---|---|---|
| No IS | With IS | |||
| Processed sample or Auto-sampler stability | Plasma | 2 | 97.8 ± 1.11 | 95.9 ± 1.87 |
| 4 | 97.1 ± 5.42 | 94.7 ± 4.20 | ||
| 6 | 95.5 ± 6.69 | 97.3 ± 5.55 | ||
| Urine | 2 | 92.3 ± 1.32 | 102.1 ± 1.35 | |
| 4 | 92.8 ± 2.31 | 99.5 ± 1.89 | ||
| 6 | 94.2 ± 1.59 | 103.2 ± 2.33 | ||
| Nominal Concentration (ng/mL) | Mean Recovery ± SD | ||
|---|---|---|---|
| Freeze-thaw Cycle Stability | Plasma | 2.5 | 104.0 ± 4.72 |
| 400 | 95.9 ± 1.89 | ||
| 800 | 96.8 ± 1.25 | ||
| Urine | 2.5 | 95.5 ± 2.51 | |
| 400 | 87.4 ± 2.57 | ||
| 800 | 87.6 ± 2.26 |
CLBQ14 is stable for up to 6 hours in rat plasma and urine samples at room temperature, thus ruling out concerns about the degradation of the analyte during sample preparation. The long-term stability of CLBQ14 in rat plasma and urine samples stored at – 80 °C was assessed over 14 days. Up to 91.3 ± 5.45 % and 101.6 ± 4.29 % of CLBQ14 was recovered from rat plasma and urine respectively, demonstrating that CLBQ14 is stable in biological samples stored under this condition.
The mean recovery of CLBQ14 from rat plasma and urine samples after three freeze-thaw cycles was greater than 95.9 % and 87.4 % respectively, suggesting that the analyte is not affected by re-freezing after thawing at room temperature.
The auto-sampler stability is expressed as average recovery of CLBQ14 from extracted plasma and urine samples placed on the auto-sampler (temperature maintained at 15 °C) for up to 6 h before analysis. The samples were extracted with either pure acetonitrile or acetonitrile containing IS to assess the effect of IS on the analyte recovery and stability. The data (Table 6) indicates that CLBQ14 is stable in the processed samples placed on the instrument for up to 6 hours, and its stability is independent of the presence of internal standard.
3.3. Pharmacokinetic Study
A pharmacokinetic study of CLBQ14 was performed to demonstrate the applicability of the validated LC-MS/MS assay method. In this study, a single 2, 5 or 10 mg per kg IV bolus dose of CLBQ14 was administered to adult male SD rats, cannulated under anesthesia prior to dosing. Blood and urine samples collected were analyzed using the validated L-MS/MS assay for CLBQ14 levels.
Based on our pharmacokinetic model selection criteria, a two compartment was selected for the compartmental analysis of the data. The two-compartment model is described in equation 3:
| (3) |
where A and B are the coefficients; α and β are alpha and beta phase rate constants respectively, and Ct is the plasma concentration of CLBQ14 at time, t. The pharmacokinetic analyzes were performed using four different weighting schemes (uniform, 1/Y, 1/Y2, and 1/Yhat*Yhat) which were evaluated based on either correlation coefficients, or observed and predicted fits of the plasma concentration-time plot, AIC and SC. The weighting factors 1/Yhat*Yhat was applied to the compartmental analysis of the PK data.
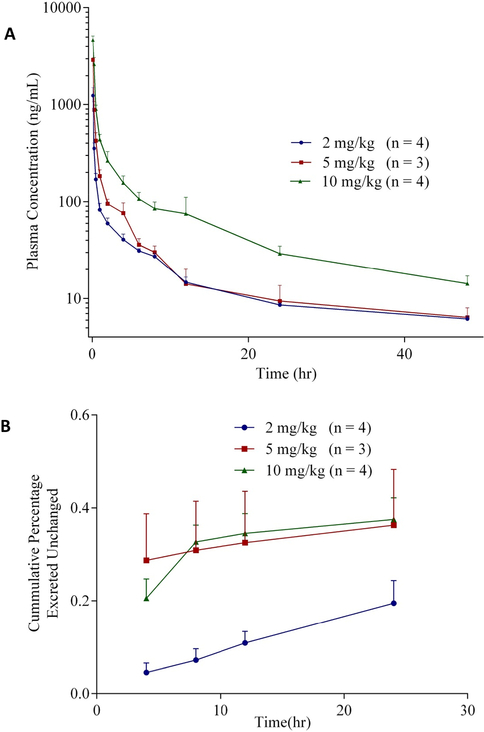
All three dose levels showed bi-exponential disposition with an initial rapid distribution, followed by a slow elimination process. The observed vs predicted (not shown) fits indicate that the data fits a 2-compartment model adequately. The mean pharmacokinetic parameters are summarized in Table 7. Following the single IV bolus injections of 2, 5 and 10 mg/kg of CLBQ14, estimated initial plasma concentrations were 1.44 ± 0.27 mg/L, 3.24 ± 0.44 mg/L, and 5.27 ± 1.43 mg/L for the respective dose levels. Beta phase elimination half-lives of 14.8 ± 0.53 hr, 10.0 ± 4.60 hr and 13.4 ± 3.59 hr were observed from the different groups. Estimated total plasma clearance was ≤ 2.62 ± 0.50 L/hr/kg and steady state volume of distribution ≤ 25.3 ± 11.2 L/kg across the 3 groups. The mean AUC0−∞ of CLBQ14 increased proportionally from 1.36 ± 0.06 to 4.83 ± 0.74 hr*mg/L as the dose was increased from 2 mg/kg to 10 mg/kg. There was no statistically significant difference between dose-normalized C0 and AUC0-t(last) estimated for each dose level. The data suggest that the pharmacokinetics of CLBQ14 is linear for IV doses under 10 mg/kg.
Table 7:
Pharmacokinetic parameters (mean ± SD) of CLBQ14 following the administration of 2, 5 and 10 mg/kg single IV bolus doses.
| Pharmacokinetic Parameter (Unit) | 2 mg/kg (n = 4) | 5 mg/kg (n = 3) | 10 mg/kg (n = 4) |
|---|---|---|---|
| C0(mg/L) | 1.44 ± 0.27 | 3.24 ± 0.44 | 5.27 ± 1.43 |
| AUC0−∞(hr*mg/L) | 1.36 ± 0.06 | 1.96 ± 0.40 | 4.83 ± 0.74 |
| A (mg/L) | 1.39 ± 0.27 | 3.14 ± 0.42 | 5.09 ± 1.36 |
| T1/2α(hr) | 0.16 ± 0.04 | 0.17 ± 0.03 | 0.25 ± 0.09 |
| B (mg/L) | 0.05 ± 0.00 | 0.09 ± 0.04 | 0.17 ± 0.07 |
| T1/2β(hr) | 14.8 ± 0.53 | 10.0 ± 4.60 | 13.4 ± 3.59 |
| Vss(L/kg) | 24.3 ± 1.28 | 23.2 ± 11.5 | 27.3 ± 11.6 |
| Cl (L/hr/kg) | 1.48 ± 0.07 | 2.62 ± 0.50 | 2.11 ± 0.32 |
C0 = concentration at time zero; AUC0−∞ = area under curve from time zero to infinity;
A = coefficient of distributional phase; T1/2α = half-life of distributional phase;
B = coefficient of terminal phase; T1/2β = elimination half-life of the beta elimination phase;
Vss = volume of distribution at steady state; Cl = total clearance
From the plasma concentration – time profiles (Fig. 4), it appears that CLBQ14 is rapidly distributed to the body tissues after administration but is slowly eliminated after the distribution phase. Also, CLBQ14 seems to be extensively metabolized as evident by 0.19 ± 0.04 % of the administered dose being excreted unchanged in urine after 24 hr. The sample size in this pharmacokinetic study is small and hence these estimations should be interpreted with caution.
Figure 4.
(A) Plasma concentration – time profile following the administration of 2, 5 and 10 mg/kg single IV bolus doses of CLBQ14 to male SD rats, respectively, (B) Cumulative percentage of the administered dose of CLBQ14 excreted unchanged in urine following the IV bolus administration.
4. CONCLUSION
A simple, specific, reliable, and reproducible LC-MS/MS method for the quantification of 7-bromo-5-chloro-8-hydroxyquinoline in rat plasma and urine was developed and validated. The method was validated to be linear, accurate and precise over the concentration range 1 – 1000 ng/mL for the quantification of CLBQ14. The stability of the analyte was not impacted by the expected sample handling, storage, preparation and analysis conditions. Pharmacokinetic evaluation of CLBQ14 in adult male SD rats revealed that it has a bi-exponential disposition in SD rats, is extensively distributed with a long plasma half-life and is eliminated primarily by liver metabolism. This method can be applied to the future studies of CLBQ14.
Table 4:
Extraction recovery and matrix effect on the quantification of CLBQ14 using this LC-MS/MS method. [n = 3; mean (± SD)]
| Biological Matrix | QC | Nominal Concentration (ng/mL) | Extraction Recovery | *Matrix Effect |
|---|---|---|---|---|
| Plasma | LQC | 2.5 | 96.8 ± 7.88 | 3.9 ± 2.23 |
| MQC | 400 | 99.0 ± 1.54 | 7.5 ± 2.29 | |
| HQC | 800 | 96.3 ± 3.85 | 3.6 ± 1.88 | |
| Urine | LQC | 2.5 | 97.0 ± 5.39 | 6.4 ± 3.79 |
| MQC | 400 | 96.6 ± 2.48 | 8.2 ± 2.76 | |
| HQC | 800 | 96.6 ± 1.59 | 2.5 ± 1.28 |
Matrix effect is considered significant if matrix factor is > °15%
Acknowledgments
5. Funding:
This work was supported in part by the National Institute of Health SC3 (grant number 1SC3GM102018) and by the National Institute of Health’s Research Centers in Minority Institutes Program (grant number 2G12MD007605–22A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olaleye O, et al. , Methionine aminopeptidases from Mycobacterium tuberculosis as novel antimycobacterial targets. Chem Biol, 2010. 17(1): p. 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olaleye O, et al. , Characterization of clioquinol and analogues as novel inhibitors of methionine aminopeptidases from Mycobacterium tuberculosis. Tuberculosis (Edinb), 2011. 91 Suppl 1: p. S61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan H, et al. , Two methionine aminopeptidases from Acinetobacter baumannii are functional enzymes. Bioorg Med Chem Lett, 2011. 21(11): p. 3395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai SC, et al. , Growth inhibition of Escherichia coli and methicillin-resistant Staphylococcus aureus by targeting cellular methionine aminopeptidase. Eur J Med Chem, 2011. 46(8): p. 3537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, et al. , Inhibitors of Plasmodium falciparum methionine aminopeptidase 1b possess antimalarial activity. Proc Natl Acad Sci U S A, 2006. 103(39): p. 14548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang JM, et al. , Characterization of the biochemical properties of two methionine aminopeptidases of Cryptosporidium parvum. Parasitol Int, 2012. 61(4): p. 707–10. [DOI] [PubMed] [Google Scholar]
- 7.John SF, et al. , Characterization of 2-hydroxy-1-naphthaldehyde isonicotinoyl hydrazone as a novel inhibitor of methionine aminopeptidases from Mycobacterium tuberculosis. Tuberculosis. 101: p. S73–S77. [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw RA, Brickey WW, and Walker KW, N-Terminal processing: the methionine aminopeptidase and Nα-acetyl transferase families. Trends in Biochemical Sciences, 1998. 23(7): p. 263–267. [DOI] [PubMed] [Google Scholar]
- 9.Giglione C, Vallon O, and Meinnel T, Control of protein life-span by N-terminal methionine excision. The EMBO Journal, 2003. 22(1): p. 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giglione C, Boularot A, and Meinnel T, Protein N-terminal methionine excision. Cell Mol Life Sci, 2004. 61(12): p. 1455–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowther WT and Matthews BW, Structure and function of the methionine aminopeptidases.Biochim Biophys Acta, 2000. 1477(1–2): p. 157–67. [DOI] [PubMed] [Google Scholar]
- 12.Keeling PJ and Doolittle WF, Methionine aminopeptidase-1: the MAP of the mitochondrion? Trends Biochem Sci, 1996. 21(8): p. 285–6. [PubMed] [Google Scholar]
- 13.Arfin SM, et al. , Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proceedings of the National Academy of Sciences of the United States of America, 1995. 92(17): p. 7714–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith EC, et al. , Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem Biol, 1997. 4(6): p. 461–71. [DOI] [PubMed] [Google Scholar]
- 15.Chun E, et al. , Novel inhibitors targeted to methionine aminopeptidase 2 (MetAP2) strongly inhibit the growth of cancers in xenografted nude model. Int J Cancer, 2005. 114(1): p. 124–30. [DOI] [PubMed] [Google Scholar]
- 16.Bernier SG, et al. , A methionine aminopeptidase-2 inhibitor, PPI-2458, for the treatment of rheumatoid arthritis. Proc Natl Acad Sci U S A, 2004. 101(29): p. 10768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, et al. , Expression and characterization of two functional methionine aminopeptidases from Mycobacterium tuberculosis H37Rv. Curr Microbiol, 2009. 59(5): p. 520–5. [DOI] [PubMed] [Google Scholar]
- 18.Wojtowicz EJ, Reverse-phase high-performance liquid chromatographic determination of halogenated 8-hydroxyquinoline compounds in pharmaceuticals and bulk drugs. J Pharm Sci, 1984. 73(10): p. 1430–3. [DOI] [PubMed] [Google Scholar]
- 19.Soroka K, et al. , Fluorescence properties of metal complexes of 8-hydroxyquinoline-5-sulfonic acid and chromatographic applications. Analytical Chemistry, 1987. 59(4): p. 629–636. [Google Scholar]
- 20.Mihajlovic A, et al. , High-performance liquid chromatographic assay of 8-hydroxyquinoline sulfate and its stability in immunobiological preparations. J Chromatogr A, 1998. 798(1–2): p. 173–7. [DOI] [PubMed] [Google Scholar]
- 21. United States Food and Drug Administration, Center for Drug Evaluation and Research and Center for Veterinay Medicine, Editors. 2013. Guidance for Industy - Bioanalytical Method Validation, in Available from: http://www.fda.gov/cder/guidance C.f.D.E.a. Research and C.f.V. Medicine, Editors. 2013.
- 22.Tong XS, et al. , Effect of Signal Interference from Dosing Excipients on Pharmacokinetic Screening of Drug Candidates by Liquid Chromatography/Mass Spectrometry. Analytical Chemistry, 2002. 74(24): p. 6305–6313. [DOI] [PubMed] [Google Scholar]
- 23.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals Guide for the Care and Use of Laboratory Animals. 8th edition ed. 2011, Washington (DC): National Academies Press. [PubMed] [Google Scholar]
- 24.Yamaoka Kiyoshi, Nakagawa Terumichi, and Uno T, Application of Akaike’s information criterion (AIC) in the evaluation of linear pharmacokinetic equations. Journal of Pharmacokinetics and Biopharmaceutics 1978. 6(2): p. 165–175. [DOI] [PubMed] [Google Scholar]
- 25.Matuszewski BK, Constanzer ML, and Chavez-Eng CM, Strategies for the Assessment of Matrix Effect in Quantitative Bioanalytical Methods Based on HPLC-MS/MS. Analytical Chemistry, 2003. 75(13): p. 3019–3030. [DOI] [PubMed] [Google Scholar]