Abstract
AIM
To investigate the effect of PNPLA3 polymorphisms on serum lipidomics and pathological characteristics in nonalcoholic fatty liver disease (NAFLD).
METHODS
Thirty-four biopsy-proven NAFLD patients from Northern, Central, and Southern China were subjected to stratification by genotyping their single nucleotide polymorphisms (SNPs) in PNPLA3. Ultra performance liquid chromatographytandem mass spectrometry was then employed to characterize the effects of PNPLA3 SNPs on serum lipidomics. In succession, correlation analysis revealed the association of PNPLA3-related lipid profile and hepatic pathological characteristics on a basis of steatosis, activity, and fibrosis assessment. The variant-based scoring of hepatocyte steatosis, ballooning, lobular inflammation, and liver fibrosis was finally performed so as to uncover the actions of lipidomics-affecting PNPLA3 SNPs in NAFLD-specific pathological alterations.
RESULTS
PNPLA3 SNPs (rs139051, rs738408, rs738409, rs 2072906, rs2294918, rs2294919, and rs4823173) demonstrated extensive association with the serum lipidomics, especially phospholipid metabolites [lysophosphatidylcholine (LPC), lysophosphatidylcholine plasmalogen (LPCO), lysophosphatdylethanolamine (LPE), phosphatidylcholine (PC), choline plasmalogen (PCO), phosphatidylethanolamine (PE), ethanolamine plasmalogen (PEO)], of NAFLD patients. PNPLA3 rs139051 (A/A genotype) and rs2294918 (G/G genotype) dominated the up-regulatory effect on phospholipids of LPCs (LPC 17:0, LPC 18:0, LPC 20:0, LPC 20:1, LPC 20:2) and LPCOs (LPC O-16:1, LPC O-18:1). Moreover, subjects with high-level LPCs/LPCOs were predisposed to low-grade lobular inflammation of NAFLD (rho: -0.407 to -0.585, P < 0.05-0.001). The significant correlation of PNPLA3 rs139051 and inflammation grading [A/A vs A/G + G/G: 0.50 (0.00, 1.75) vs 1.50 (1.00, 2.00), P < 0.05] further demonstrated its pathological role based on the modulation of phospholipid metabolite profile.
CONCLUSION
The A/A genotype at PNPLA3 rs139051 exerts an up-regulatory effect on serum phospholipids of LPCs and LPCOs, which are associated with low-grade lobular inflammation of NAFLD.
Keywords: Nonalcoholic fatty liver disease, Patatin-like phospholipase domain containing 3, Single nucleotide polymorphism, Phospholipid, Inflammation
Core tip: PNPLA3 single nucleotide polymorphisms reflect an important genetic basis of serum lipidomics, especially phospholipid metabolites, in nonalcoholic fatty liver disease (NAFLD) patients. PNPLA3 rs139051 (A/A genotype) exerts an up-regulatory effect on phospholipids of lysophosphatidylcholines (LPCs) and lysophosphatidylcholine plasmalogens (LPCOs). Moreover, both the A/A genotype at PNPLA3 rs139051 and high-level LPCs/LPCOs share an association with the low-grade lobular inflammation of NAFLD. Therefore, PNPLA3 rs139051 may underlie the inflammatory progress of NAFLD with its modulation of phospholipid metabolite profiles.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is characterized by excess triglyceride accumulation and lobular inflammatory infiltration and is a leading cause of most chronic liver diseases in European, Asian-Pacific, and American patients[1]. According to the diagnosis and management guidelines, hyperlipidemia (e.g., hypertriglyceridemia or hypercholesterolemia) has been identified as one of the most important risk factors for NAFLD[2]. Approximately 50% of patients with hyperlipidemia have ultrasonographic evidence of fatty infiltration in the liver[3]. Patients with a spectrum of NAFLD, ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), show prevalence of dyslipidemia, including increased serum triglycerides and low-density lipoprotein (LDL), and low levels of high-density lipoprotein cholesterol (HDL-C)[4]. Thus, the lipidemic properties are closely associated with the initiation and progression of NAFLD.
Recently, genome-wide association analysis and clinical investigations have found that single nucleotide polymorphisms (SNPs) of patatin-like phospholipase domain containing 3 (PNPLA3) (e.g., rs738409, rs 1010023, rs2281135, rs139051 and rs2294918) underlie the genetic susceptibility of NAFLD, independent of gender, age and ethnic background[5-11]. Functional studies have highlighted that adiponutrin, the encoded product of PNPLA3, is a crucial regulator of lipid metabolism in the liver via its activities on triacylglycerol lipase and acyl glycerol O-acyltransferase[12]. Given the central role of the liver in systemic lipid homeostasis, these loss-of-function SNPs in PNPLA3 are suggested to predispose individuals to NAFLD, probably by dysregulation of hepatic and serum lipid profiles[4]. This lipid-metabolism-regulating role of PNPLA3 SNPs has been confirmed in the liver of NAFLD patients[13]. However, understanding of the SNP-specific impact on serum lipids and their correlation with pathological characteristics is still limited and controversial[14-17].
We therefore stratified Chinese Han patients with biopsy-proven NAFLD by genotyping their PNPLA3 SNPs. Ultra-high performance liquid chromatographytandem mass spectrometry (UPLC-MS/MS) was used to characterize the effects of PNPLA3 SNPs on serum lipidomics of NAFLD patients. Correlation analysis uncovered the association of PNPLA3-related lipid profile and hepatic pathological characteristics by assessment of steatosis, activity and fibrosis (SAF). Finally, variant-based scoring of steatosis, ballooning, inflammation, and liver fibrosis was performed to reveal the actions of lipidomics-affecting PNPLA3 SNPs on NAFLD-specific pathological alterations.
MATERIALS AND METHODS
Study population
Thirty-four Chinese Han patients with biopsy-proven NAFLD were enrolled from Shanghai Xinhua Hospital (n = 17), Zhengxing Hospital (n = 8) and Tianjin Hospital of Infectious Diseases (n = 9) between January 2012 and June 2013. The exclusion criteria were as follows: historic or current high alcohol consumption equivalent to > 20 g/d for men and > 10 g/d for women[18,19], viral hepatitis, drug-induced liver disease, Wilson’s disease, autoimmune liver diseases and other diseases that lead to steatosis. The study was approved by the Xinhua Hospital Research Ethics Committee, and informed consent was obtained from all patients.
Anthropometric and biochemical assessments
The baseline data of age (41.03 ± 14.81 years), gender, height (167.44 ± 7.82 cm), weight (75.34 ± 9.49 kg), and body mass index (BMI) (26.90 ± 3.13) were characterized for the study population. Fasting blood samples were collected from the NAFLD patients, and a multichannel automatic analyzer (Advia 1650; Bayer, Moss, Norway) was used to test biochemical indexes of alanine aminotransferase (58.33 ± 33.89 U/L), aspartate aminotransferase (24.05 ± 28.63 U/L), alkaline phosphatase (101.90 ± 110.49 U/L), and γ-glutamyltransferase (115.73 ± 278.59 U/L). Fasting blood glucose (42.70 ± 28.07 μIU/mL), total cholesterol (4.70 ± 0.61 mg/dL), and triglyceride (1.70 ± 0.68 mg/dL) were subjected to assessment using Wako Bioproducts (Wako Pure Chemical Industries, Richmond, VA, United States).
Hepatic histopathological assessment
Liver samples from each NAFLD patient were obtained by needle biopsy after obtaining informed consent. Liver tissues were fixed in formalin, paraffin embedded, and 5-μm sections were cut. Both hematoxylin and eosin and Masson’s trichrome staining were used for pathological characterization, including hepatocyte steatosis, lobular inflammation, ballooning, and liver fibrosis according to the SAF scoring method[20-22], by three pathologists who were not aware of the study.
Genotyping of PNPLA3 SNPs
Blood samples from NAFLD patients were treated as follows: (1) centrifugation at 1500 rpm for 10 min; (2) separation of buffy-coat layer; and (3) extraction of genomic DNA from the buffy coat lymphocytes by QiAamp DNA Mini Kit (Qiagen, Venlo, Netherlands). A custom Ion AmpliSeq panel (Life Technologies Thermo Fisher Scientific, Waltham, MA, United States) of human PNPLA3 was designed for the emulation polymerase chain reaction of template DNA using a Ion OneTouch 2 System (Life Technologies). PNPLA3 SNPs were genotyped by the Ion 318 Chip (Life Technologies) according to the Ion PGM 200 Sequencing kit protocol[23].
UPLC-MS/MS
After 12-h fasting, serum lipidomics of the enrolled NAFLD patients were analyzed with a combination of UPLC (Waters, Milford, MA, United States) and Triple TOF 5600 mass spectrometer (AB SCIEX, Framingham, MA, United States)[24]. The column temperature was set to 55°C. The ratios of acetonitrile/water (mobile phase A) and propanol/acetonitrile (mobile phase B) were 3/2 and 9/1, respectively, with 10 mmol/L ammonium acetate added to all mobile phases. The elution gradient program was carried out as follows: 0-1.5 min: 32% B; 1.5-14 min: 85% B; 15.5-15.6 min: 97% B; 15.6- 18 min: 97% B; 18-20 min: 32% B (flow rate: 0.26 mL/min). MS was applied by electrospray ionization operating in the positive and negative ion modes. The temperature of the interface heater was 600°C (–) and 500°C (+), with 4500 V (–) and 5500 V (+) ion spray voltage. The declustering potential was 100 V (–) and 100 V (+), and collision energy was 10 V (–) and 10 V (+). Thirteen quality control (QC) samples, being randomly inserted into the sequence, were analyzed for data precision. The raw data obtained from UPLC-MS/MS by Analyst TF 1.6 software (AB SCIEX) were subjected to identification of lipid composition using LipidView/PeakView and quantification of lipid concentrations using MultiQuant 2.0. The relative standard deviation (RSD) of 239 serum lipids in QC samples was evaluated against the internal standards.
Statistical analysis
The clinical data were expressed as mean ± SD (continuous, normally distributed variables) or medians (interquartile range) (discontinuous, non-normally distributed variables). Differences in serum lipidomics among the groups of PNPLA3 SNPs were investigated by unpaired Student’s independent t tests. Correlation analysis of phospholipid metabolite profile and hepatic pathological characteristics was assessed by Spearman’s correlation. The Mann-Whitney U test was performed to evaluate the differences in pathological grading among groups of PNPLA3 SNPs. SPSS version 19.0 (SPSS Inc., Chicago, IL, United States) was used for statistical analysis with a two-side significant criterion at P < 0.05.
RESULTS
PNPLA3 SNPs associated with serum lipidomics in NAFLD patients
With the stable distribution and limited RSD of QC samples (Figure 1 and Table 1), an obvious association of PNPLA3 SNPs (rs139051, rs738408, rs738409, rs2072906, rs2294918, rs2294919 and rs4823173) and serum lipidomic characteristics was observed in Chinese Han NAFLD patients by combination of PNPLA3 genotyping and UPLC-MS/MS (Table 2). In detail, these PNPLA3 SNPs significantly correlated to the serum level of various members of cholesteryl ester (CE), free fatty acid (FFA), lysophosphatidylcholine (LPC), lysophosphatidylcholine plasmalogen (LPCO), lysophosphatidylethanolamine (LPE), phosphatidylcholine (PC), choline plasmalogen (PCO), phosphatidylethanolamine (PE), ethanolamine plasmalogen (PEO), and triacylglycerol (TAG) (Table 2).
Figure 1.
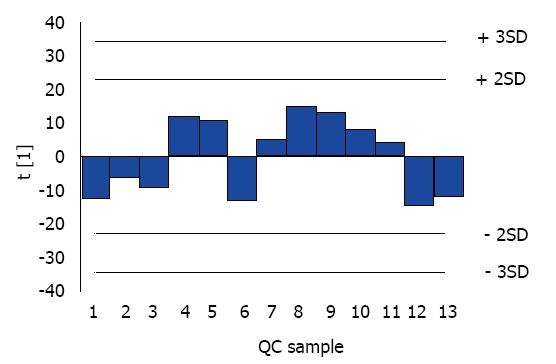
Dynamic distribution of quality control samples throughout the Ultra-high performance liquid chromatography-tandem mass spectrometry analysis. Colored bars indicate the quality control samples that are projected onto the first principal component with a range from -2 SD to +2 SD. QC: Quality control; SD: Standard deviation.
Table 1.
Relative standard deviation distribution in quality control samples
| Percentage |
Relative SD |
||||
| <10% | 10%-15% | 15%-20% | 20%-30% | 30%-50% | |
| Peak number in total | 10 | 47 | 32 | 10 | 1 |
| Sum of peak number in total | 10 | 57 | 89 | 99 | 100 |
Table 2.
PNPLA3 single nucleotide polymorphisms associated with serum lipidomics of nonalcoholic fatty liver disease patients
| SNPs |
Number of SNP-associated serum lipids |
|||||||||
| CE | FFA | LPC | LPCO | LPE | PC | PCO | PE | PEO | TAG | |
| rs139051 (A/A: A/G + G/G ) | 1 | 1 | 5 | 2 | 1 | 0 | 0 | 1 | 1 | 0 |
| rs738408 (T/T: C/T + C/C) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| rs738409 (G/G: C/G + C/C) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| rs2072906 (G/G: A/G + A/A) | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| rs2294918 (G/G: A/A + A/G) | 0 | 0 | 3 | 3 | 1 | 1 | 0 | 9 | 0 | 5 |
| rs2294919 (C/C: C/T + T/T) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| rs4823173 (A/A: A/G + G/G) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
CE: Cholesteryl ester; FFA: Free fatty acid; LPC: Lysophosphatidylcholine; LPCO: Lysophosphatidylcholine plasmalogen; LPE: Lysophosphatdylethanolamine; NAFLD: Nonalcoholic fatty liver disease; PC: Phosphatidylcholine; PCO: Choline plasmalogen; PE: Phosphatidylethanolamine; PEO: Ethanolamine plasmalogen; PNPLA3: Patatin-like phospholipase domain containing 3; SNP: Single nucleotide polymorphisms; TAG: Triacylglycerol.
PNPLA3 rs139051 and rs2294918 exerted upregulatory effects on LPCs and LPCOs
PNPLA3 rs139051 and rs2294918, with their effects on 12 and 22 different serum lipids, respectively, dominated the PNPLA3 SNP-lipidomics association (Table 2). Compared to those with the A/G or G/G genotype, NAFLD patients carrying the A/A genotype at PNPLA3 rs139051 demonstrated significantly higher serum levels of LPC 17:0, LPC 18:0, LPC 20:0, LPC 20:1, LPC 20:2, LPC O-16:1, LPC O-18:1, and significantly lower levels of LPE 20:4, PE 34:0 and PE O-36:5 (Table 3). Quantitative analysis showed significantly increasing levels of LPC 17:0, LPC 20:0, LPC 20:1, LPC O-16:0, LPC O-16:1, and LPC O-18:1 in the NAFLD patients with the G/G phenotype compared to the A/A or A/G phenotype at PNPLA3 rs2294918 (Table 3). Nevertheless, there was lower serum levels of LPE 22:6, PC 32:1, PE 34:0, PE 34:2, PE 36:2, PE 36:4, PE 38:4, PE 38:5, PE 38:6, PE 40:5 and PE 40:6 in these patients (Table 3).
Table 3.
Effects of PNPLA3 rs139051 and rs2294918 on serum profile of phospholipid metabolites in nonalcoholic fatty liver disease patients
| Phospholipid metabolites |
SNPs |
|||||
|
rs139051 |
rs2294918 |
|||||
| A/A | A/G + G/G | P value | G/G | A/A + A/G | P value | |
| LPC 17:0 | 0.148 ± 0.119 | 0.089 ± 0.032 | 0.045a | 0.143 ± 0.113 | 0.084 ± 0.023 | 0.023a |
| LPC 18:0 | 5.501 ± 4.064 | 3.482 ± 1.032 | 0.045a | 5.199 ± 3.888 | 3.564 ± 0.941 | 0.068 |
| LPC 20:0 | 0.019 ± 0.012 | 0.011 ± 0.005 | 0.013a | 0.018 ± 0.011 | 0.010 ± 0.005 | 0.008a |
| LPC 20:1 | 0.035 ± 0.028 | 0.020 ± 0.072 | 0.031a | 0.033 ± 0.026 | 0.018 ± 0.007 | 0.017a |
| LPC 20:2 | 0.046 ± 0.040 | 0.025 ± 0.090 | 0.037a | 0.042 ± 0.038 | 0.027 ± 0.010 | 0.091 |
| LPC O-16:0 | 0.128 ± 0.109 | 0.078 ± 0.016 | 0.058 | 0.123 ± 0.102 | 0.075 ± 0.016 | 0.038a |
| LPC O-16:1 | 0.111 ± 0.088 | 0.068 ± 0.016 | 0.046a | 0.106 ± 0.083 | 0.067 ± 0.014 | 0.041a |
| LPC O-18:1 | 0.076 ± 0.071 | 0.041 ± 0.011 | 0.039a | 0.072 ± 0.067 | 0.040 ± 0.011 | 0.036a |
| LPE 20:4 | 0.087 ± 0.037 | 0.116 ± 0.035 | 0.029a | 0.092 ± 0.040 | 0.113 ± 0.034 | 0.143 |
| LPE 22:6 | 0.091 ± 0.044 | 0.113 ± 0.037 | 0.138 | 0.090 ± 0.036 | 0.122 ± 0.046 | 0.035a |
| PC 32:1 | 1.574 ± 0.941 | 1.912 ± 1.167 | 0.358 | 1.450 ± 0.949 | 2.263 ± 1.036 | 0.030a |
| PE 34:0 | 0.312 ± 0.018 | 0.325 ± 0.016 | 0.041a | 0.310 ± 0.015 | 0.332 ± 0.015 | 0.001a |
| PE 34:2 | 0.335 ± 0.187 | 0.309 ± 0.159 | 0.673 | 0.280 ± 0.131 | 0.418 ± 0.219 | 0.028a |
| PE 36:2 | 0.758 ± 0.349 | 0.702 ± 0.324 | 0.634 | 0.650 ± 0.271 | 0.913 ± 0.398 | 0.030a |
| PE 36:4 | 0.282 ± 0.142 | 0.324 ± 0.169 | 0.435 | 0.246 ± 0.105 | 0.410 ± 0.181 | 0.015a |
| PE 38:4 | 0.746 ± 0.349 | 0.830 ± 0.348 | 0.493 | 0.661 ± 0.228 | 1.031 ± 0.423 | 0.018a |
| PE 38:5 | 0.079 ± 0.036 | 0.079 ± 0.034 | 0.960 | 0.070 ± 0.026 | 0.098 ± 0.043 | 0.025a |
| PE 38:6 | 0.621 ± 0.431 | 0.613 ± 0.332 | 0.953 | 0.493 ± 0.232 | 0.878 ± 0.517 | 0.036a |
| PE 40:5 | 0.053 ± 0.026 | 0.064 ± 0.033 | 0.305 | 0.047 ± 0.022 | 0.080 ± 0.030 | 0.001a |
| PE 40:6 | 0.445 ± 0.316 | 0.451 ± 0.220 | 0.952 | 0.354 ± 0.174 | 0.644 ± 0.351 | 0.003a |
| PE O-36:5 | 0.529 ± 0.192 | 0.699 ± 0.276 | 0.041a | 0.547 ± 0.171 | 0.707 ± 0.331 | 0.071 |
LPC: Lysophosphatidylcholine; LPCO: Lysophosphatidylcholine plasmalogen; LPE: Lysophosphatdylethanolamine; NAFLD: Nonalcoholic fatty liver disease; PC: Phosphatidylcholine; PCO: Choline plasmalogen; PE: Phosphatidylethanolamine; PEO: Ethanolamine plasmalogen; PNPLA3: Patatin-like phospholipase domain containing 3. Values are expressed as mean ± SD. aP < 0.05.
High levels of LPCs and LPCOs demonstrated a correlation with low-grade hepatic inflammation
To shed light on the influence of PNPLA3 SNP-related lipidomic characteristics, correlations of phospholipid metabolites and NAFLD-specific pathological disorders (hepatocyte steatosis, lobular inflammation, ballooning, and liver fibrosis) were analyzed. Various LPCs (LPC 17:0, LPC 18:0, LPC 20:0, LPC 20:1 and LPC 20:2) and LPCOs (LPC O-16:1 and LPC O-18:1) showed negative correlations with the grade of lobular inflammation. The serum levels of these phospholipid metabolites were high in NAFLD patients with the A/A genotype at PNPLA3 rs139051 and/or the G/G genotype at rs2294918. Most of them shared similar Spearman’s rank correlation coefficients (Table 4). In contrast, pathological indexes other than lobular inflammation correlated to neither high-level phospholipid metabolites (LPCs and LPCOs) nor low-level ones (LPEs, PC 32:1, PEs and PEO 36:5) in these NAFLD patients (Table 4). Given their impact on the serum profile of phospholipid metabolites, both PNPLA3 rs139051 and rs2294918 are proposed to act in hepatic inflammation of NAFLD by targeting, at least to a large extent, LPCs and LPCOs.
Table 4.
Phospholipid metabolites correlated with pathological characteristics of nonalcoholic fatty liver disease
| Phospholipid metabolites |
Steatosis |
Lobular inflammation |
Ballooning |
Fibrosis |
||||
| rho | P value | rho | P value | rho | P value | rho | P value | |
| LPC 17:0 | 0.149 | 0.399 | -0.525 | 0.001a | 0.093 | 0.599 | 0.095 | 0.592 |
| LPC 18:0 | 0.024 | 0.892 | -0.478 | 0.004a | 0.136 | 0.442 | 0.089 | 0.618 |
| LPC 20:0 | -0.071 | 0.692 | -0.585 | 0.000a | 0.010 | 0.956 | 0.082 | 0.645 |
| LPC 20:1 | -0.061 | 0.734 | -0.489 | 0.003a | -0.165 | 0.352 | -0.072 | 0.686 |
| LPC 20:2 | 0.092 | 0.604 | -0.453 | 0.007a | 0.145 | 0.415 | 0.042 | 0.812 |
| LPC O-16:0 | 0.297 | 0.088 | -0.296 | 0.089 | 0.025 | 0.886 | 0.146 | 0.410 |
| LPC O-16:1 | 0.070 | 0.695 | -0.425 | 0.012a | 0.028 | 0.874 | 0.027 | 0.881 |
| LPC O-18:1 | -0.114 | 0.521 | -0.407 | 0.017a | -0.018 | 0.921 | 0.107 | 0.546 |
| LPE 20:4 | -0.079 | 0.658 | 0.107 | 0.547 | 0.071 | 0.690 | -0.071 | 0.692 |
| LPE 22:6 | -0.274 | 0.117 | 0.054 | 0.764 | 0.125 | 0.481 | 0.173 | 0.327 |
| PC 32:1 | 0.020 | 0.913 | 0.086 | 0.630 | 0.107 | 0.547 | -0.069 | 0.699 |
| PE 34:0 | -0.012 | 0.946 | 0.193 | 0.275 | -0.173 | 0.327 | -0.146 | 0.410 |
| PE 34:2 | -0.054 | 0.762 | -0.025 | 0.888 | -0.043 | 0.809 | -0.153 | 0.388 |
| PE 36:2 | -0.050 | 0.777 | -0.079 | 0.659 | -0.118 | 0.506 | -0.257 | 0.143 |
| PE 36:4 | -0.091 | 0.608 | 0.150 | 0.398 | 0.121 | 0.495 | -0.052 | 0.770 |
| PE 38:4 | -0.112 | 0.529 | 0.214 | 0.224 | 0.110 | 0.537 | -0.065 | 0.716 |
| PE 38:5 | -0.234 | 0.182 | 0.178 | 0.313 | -0.026 | 0.885 | -0.096 | 0.587 |
| PE 38:6 | -0.151 | 0.393 | 0.096 | 0.588 | 0.176 | 0.320 | 0.105 | 0.554 |
| PE 40:5 | -0.013 | 0.944 | 0.132 | 0.457 | 0.049 | 0.783 | -0.194 | 0.273 |
| PE 40:6 | -0.192 | 0.277 | 0.132 | 0.457 | 0.150 | 0.397 | 0.067 | 0.705 |
| PE O-36:5 | 0.021 | 0.905 | 0.178 | 0.313 | -0.205 | 0.245 | -0.131 | 0.461 |
LPC: Lysophosphatidylcholine; LPCO: Lysophosphatidylcholine plasmalogen; LPE: Lysophosphatdylethanolamine; NAFLD: Nonalcoholic fatty liver disease; PC: Phosphatidylcholine; PCO: Choline plasmalogen; PE: Phosphatidylethanolamine; PEO: Ethanolamine plasmalogen; rho: Spearman's rank correlation coefficient. aP < 0.05.
Low-grade hepatic inflammation occurred in NAFLD patients with the A/A genotype at PNPLA3 rs139051
After the stratification of NAFLD patients by PNPLA3 genotypes, comparison of steatosis, lobular inflammation, ballooning, and liver fibrosis was carried out to assess the pathological role of PNPLA3 rs139051 and rs2294918. By the SAF-based scoring, the A/A genotype at PNPLA3 rs139051 conferred significantly lower lobular inflammation than the A/G + G/G genotypes (Figure 2). However, PNPLA3 rs139051 did not seem to exert any significant impact on NAFLD-specific steatosis, ballooning, and fibrosis (Figure 2). NAFLD patients with PNPLA3 rs2294918, another SNP related to the inflammation-associated phospholipid metabolites, showed a comparable grade in each of these pathological indexes regardless of genotype (Figure 3).
Figure 2.
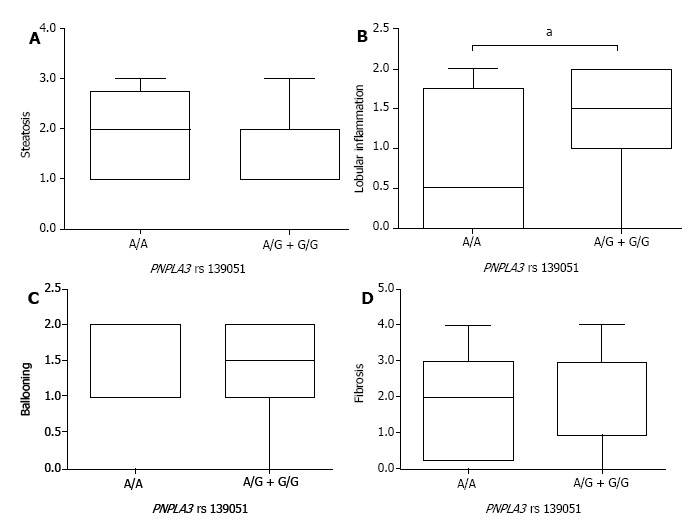
Nonalcoholic fatty liver disease patients carrying the A/A genotype at PNPLA3 rs139051 demonstrated low-grade lobular inflammation. Box plots indicate the difference in pathologic characteristics of steatosis (A); lobular inflammation (B); ballooning (C); and fibrosis (D) between nonalcoholic fatty liver disease patients with the A/A or A/G + G/G genotype at PNPLA3 rs139051. Results are presented as medians and Interquartile Range. aP < 0.05.
Figure 3.
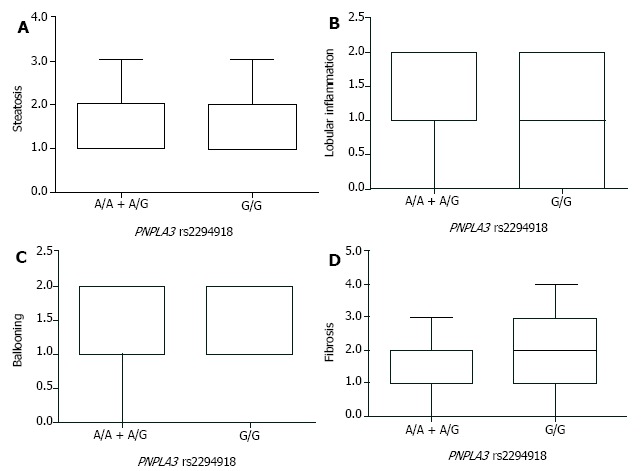
PNPLA3 rs2294918 showed no association with pathologic characteristics of nonalcoholic fatty liver disease patients. Box plots indicate the grade of steatosis (A); lobular inflammation (B); ballooning (C); and fibrosis (D) in nonalcoholic fatty liver disease patients with the G/G or A/A + A/G genotype at PNPLA3 rs2294918. Results are presented as medians and Interquartile Range.
DISCUSSION
PNPLA3 is the liver-enriched member of the PNPLA family, which is located on the membrane of lipid droplets and endoplasmic reticulum in hepatocytes[25,26]. Its conserved patatin-like domain demonstrates hydrolase activity against TAG, diacylglycerol and monoacylglycerol[15]. Activities of thioesterase and acylglycerol transacylase reflect the other aspects of the lipometabolic action of PNPLA3[15]. PNPLA3 was recently identified to promote the transfer of very-long-chain polyunsaturated fatty acids from TAG to phospholipids[27]. These effects shed light on an important regulatory role of PNPLA3 in lipid homeostasis[13], which is supposed to be affected by PNPLA3-related genetic variants.
In the present study, genotype-based, widespread effects of PNPLA3 SNPs in lipid profiles were found in the sera of NAFLD patients from Northern (Tianjin), Central (Shanghai) and Southern China (Zhangzhou, Fujian). Being similar to previous reports[13,28,29], members of TAG, CE and FFA were identified in the differential serum lipids associated with PNPLA3 SNPs (e.g., rs738408, rs738409, rs2072906, rs2294919 and rs4823173). In contrast, PNPLA3 rs139051 and rs2294918 primarily exerted their lipidomic impact on phospholipid metabolites. LPCs, LPCOs and PEs were confirmed to dominate the differential serum lipids because of their high abundance. Therefore, dysregulation of phospholipid metabolite profile reflects the major role of PNPLA3 SNPs in serum lipidomics.
To gain further insight into the PNPLA3-related phospholipid characteristics, differences in serum levels of LPC, LPCO and PE were analyzed between various genotypes of PNPLA3 rs139051 and rs2294918. NAFLD patients with the A/A instead of A/G + G/G genotype at PNPLA3 rs139051 exhibited significantly higher levels of LPCs and LPCOs. The G/G, but not the A/A + A/G, genotype of PNPLA3 rs2294918 also predisposed NAFLD patients to statistical elevation of serum LPCs and LPCOs. Various types of LPCs (e.g., LPC 17:0 and LPC 18:0) have been shown to play a critical role in the spectrum of NAFLD[30,31]. Their reduction in blood samples was detected in patients with 1H-MRS- or biopsy-proven hepatic steatosis[30]. Similarly, significant decreases in serum palmitoyl-, stearoyl- and oleoyl-LPC were obtained in an experimental rodent model of NASH[32]. In contrast, uptake of LPC metabolite (PC) ameliorates the hepatic injury of NAFLD with a reduction of serum aspartate aminotransferase and alanine aminotransferase[33]. Given the similarity in LPC- and LPCO-based lipid metabolism[32,34-37], PNPLA3 rs139051 and rs2294918 are suggested to protect the liver from NAFLD-related impairment by their upregulatory effect on both LPCs and LPCOs.
In contrast to its correlation with LPCs and LPCOs upregulation, the G/G genotype at PNPLA3 rs2294918 conferred significantly lower levels of PEs in the NAFLD patients compared to those with the A/A + A/G genotype. A similar decrease in serum PE 34:0 was found in these NAFLD patients in association with the A/A genotype at PNPLA3 rs139051. Recent studies have highlighted a growing increase in PE concentration during progression of NAFLD (healthy control < simple steatosis < NASH)[38]. Nevertheless, individuals carrying the Val175Met variant allele of PE N-methyltransferase (PEMT), which inhibits PE to PC conversion, show high susceptibility to NASH[39]. PEMT-/- mice also suffer from NASH after being fed a choline-deficient or high-fat diet[40,41]. Thus, PNPLA3 rs139051 and rs2294918 represent a novel layer of genetic regulation that downregulates the harmful components of lipidomics.
Metabolically, LPCs are catabolized into PCs by the lysophosphatidylcholine acyltransferase 1/2/4[32]. Glycerophosophocholine, the lysoplasmalogenase-dependent catalysate of LPCOs, serves as another precursor of PCs[36,37]. As a result, the upregulation of LPCs and LPCOs, together with the downregulation of PEs, may lead to an increased ratio of PC to PE in NAFLD patients with the A/A genotype at PNPLA3 rs139051 and/or G/G at PNPLA3 rs2294918. Acting as the major source of serum phospholipids, hepatic PCs and PEs play an essential part in the integrity of cellular and organelle membranes[41,42]. An abnormally low molar ratio of PC/PE has been linked to steatohepatitis due to its induction of membrane leakage[43]. On the contrary, administration of polyene PC prevents patients from hepatic injury, with an improvement in PC/PE ratio[44]. Because of the antioxidant propensity of plasmalogen, LPCO deficiency is responsible for additional effects of NASH that sensitize patients to reactive-oxygen-species-dependent peroxidation, and successive lobular inflammation[45,46]. An inflammation-attenuating effect of PNPLA3 rs139051 and rs2294918 is then proposed on the basis of LPCs and LPCOs augmentation.
Indeed, our experimental observations relating to NAFLD patients revealed that an increase in LPCs and LPCOs significantly correlated with an attenuation of hepatic inflammation. Both high-level LPCs/LPCOs and low-grade lobular inflammation characterized patients with the A/A genotype at PNPLA3 rs139051 and/or the G/G genotype at rs2294918. However, pathological characteristics other than hepatic inflammation, including hepatocyte steatosis, ballooning, and liver fibrosis, were not associated with either of these phospholipid metabolites. PNPLA3 genotyping and SAF-based pathological grading confirmed the phospholipid-mediated effect of PNPLA3 SNPs on NAFLD. There was a lower grade of lobular inflammation in patients with the A/A vs A/G + G/G genotype at PNPLA3 rs139051. Similar, yet mild, amelioration of lobular inflammation occurred in NAFLD patients carrying the G/G rather than other genotypes at PNPLA3 rs2294918.
In conclusion, PNPLA3 SNPs reflect an important genetic basis of lipidomic characteristics in NAFLD patients, with the focus on phospholipid metabolite profile. The A/A genotype at PNPLA3 rs139051 exerts an upregulatory effect on serum LPCs and LPCOs. Both the genotype of PNPLA3 rs139051 and the increased levels of LPCs/LPCOs share an association with the low-grade lobular inflammation of NAFLD. PNPLA3 rs139051, therefore, may underlie the inflammatory progress of NAFLD by its modulation of the phospholipid metabolite profile.
ARTICLE HIGHLIGHTS
Research background
Genome-wide association analysis and clinical investigations have found that single nucleotide polymorphisms (SNPs) of patatin-like phospholipase domain containing 3 (PNPLA3) underlie the genetic susceptibility of nonalcoholic fatty liver disease (NAFLD), independent of gender, age and ethnic background.
Research motivation
The understanding of the SNP-specific impact on serum lipids and their correlation with pathological characteristics is still limited and controversial. In this study, the authors stratified Chinese Han patients with biopsy-proven NAFLD by genotyping their PNPLA3 SNPs.
Research objectives
In this study, the authors investigated the effect of PNPLA3 polymorphisms on serum lipidomics and pathological characteristics of NAFLD.
Research methods
Thirty-four biopsy-proven NAFLD patients from China were subjected to stratification by genotyping their SNPs in PNPLA3. Ultra-performance liquid chromatographytandem mass spectrometry was employed to characterize the effects of PNPLA3 SNPs on serum lipidomics. The variant-based scoring of hepatocyte steatosis, ballooning, lobular inflammation, and liver fibrosis was performed to uncover the actions of lipidomics-affecting PNPLA3 SNPs in NAFLD-specific pathological alternations.
Research results
PNPLA3 SNPs demonstrated extensive association with the serum lipidomics, especially phospholipid metabolites of NAFLD patients. The significant correlation of PNPLA3 rs139051 and inflammation grading further convinced its pathological role that was based on the modulation of phospholipid metabolite profile.
Research conclusions
The authors found that the A/A genotype at PNPLA3 rs139051 exerts an up-regulatory effect on serum phospholipids of lysophosphatidylcholine (LPC) and lysophosphatidylcholine plasmalogen (LPCO), which are associated with low-grade lobular inflammation of NAFLD.
Research perspectives
These experimental observations relating to NAFLD patients revealed that an increase in LPCs and LPCOs significantly correlated with an attenuation of hepatic inflammation. However, the pathological characteristics other than hepatic inflammation displayed no association with either of these phospholipid metabolites.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Xinhua Hospital Ethics Committee Affiliated to Shanghai Jiaotong University School of Medicine.
Conflict-of-interest statement: No potential conflicts of interest.
Manuscript source: Unsolicited manuscript
Peer-review started: June 19, 2018
First decision: July 11, 2018
Article in press: August 4, 2018
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Schwarz SM S- Editor: Dou Y L- Editor: Filipodia E- Editor: Wu YXJ
Contributor Information
Ji-Jun Luo, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Hai-Xia Cao, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Rui-Xu Yang, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Rui-Nan Zhang, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Qin Pan, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China. panqin@xinhuamed.com.cn.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ; American Gastroenterological Association; American Association for the Study of Liver Diseases; American College of Gastroenterologyh. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–1609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Assy N, Kaita K, Mymin D, Levy C, Rosser B, Minuk G. Fatty infiltration of liver in hyperlipidemic patients. Dig Dis Sci. 2000;45:1929–1934. doi: 10.1023/a:1005661516165. [DOI] [PubMed] [Google Scholar]
- 4.Cohen DE, Fisher EA. Lipoprotein metabolism, dyslipidemia, and nonalcoholic fatty liver disease. Semin Liver Dis. 2013;33:380–388. doi: 10.1055/s-0033-1358519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J, Kiviluoto T, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 7.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Q, Chen MM, Zhang RN, Wang YQ, Zheng RD, Mi YQ, Liu WB, Shen F, Su Q, Fan JG. PNPLA3 rs1010023 Predisposes Chronic Hepatitis B to Hepatic Steatosis but Improves Insulin Resistance and Glucose Metabolism. J Diabetes Res. 2017;2017:4740124. doi: 10.1155/2017/4740124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Qu HQ, Rentfro AR, Grove ML, Mirza S, Lu Y, Hanis CL, Fallon MB, Boerwinkle E, Fisher-Hoch SP, et al. PNPLA3 polymorphisms and liver aminotransferase levels in a Mexican American population. Clin Invest Med. 2012;35:E237–E245. doi: 10.25011/cim.v35i4.17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng XE, Wu YL, Lin SW, Lu QQ, Hu ZJ, Lin X. Genetic variants in PNPLA3 and risk of non-alcoholic fatty liver disease in a Han Chinese population. PLoS One. 2012;7:e50256. doi: 10.1371/journal.pone.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donati B, Motta BM, Pingitore P, Meroni M, Pietrelli A, Alisi A, Petta S, Xing C, Dongiovanni P, del Menico B, et al. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology. 2016;63:787–798. doi: 10.1002/hep.28370. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 13.Peter A, Kovarova M, Nadalin S, Cermak T, Königsrainer A, Machicao F, Stefan N, Häring HU, Schleicher E. PNPLA3 variant I148M is associated with altered hepatic lipid composition in humans. Diabetologia. 2014;57:2103–2107. doi: 10.1007/s00125-014-3310-0. [DOI] [PubMed] [Google Scholar]
- 14.Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, Yang K, Kumari M, Gross RW, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyysalo J, Gopalacharyulu P, Bian H, Hyötyläinen T, Leivonen M, Jaser N, Juuti A, Honka MJ, Nuutila P, Olkkonen VM, et al. Circulating triacylglycerol signatures in nonalcoholic fatty liver disease associated with the I148M variant in PNPLA3 and with obesity. Diabetes. 2014;63:312–322. doi: 10.2337/db13-0774. [DOI] [PubMed] [Google Scholar]
- 17.Sharma M, Mitnala S, Vishnubhotla RK, Mukherjee R, Reddy DN, Rao PN. The Riddle of Nonalcoholic Fatty Liver Disease: Progression From Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis. J Clin Exp Hepatol. 2015;5:147–158. doi: 10.1016/j.jceh.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinert DF, Allen JP. The Alcohol Use Disorders Identification Test (AUDIT): a review of recent research. Alcohol Clin Exp Res. 2002;26:272–279. [PubMed] [Google Scholar]
- 19.Pérula de Torres LA, Fernández-García JA, Arias-Vega R, Muriel-Palomino M, Márquez-Rebollo E, Ruiz-Moral R. [Validation of the AUDIT test for identifying risk consumption and alcohol use disorders in women] Aten Primaria. 2005;36:499–506. doi: 10.1016/S0212-6567(05)70552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedossa P; FLIP Pathology Consortium. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic fatty liver disease. Hepatology. 2014;60:565–575. doi: 10.1002/hep.27173. [DOI] [PubMed] [Google Scholar]
- 21.Munteanu M, Tiniakos D, Anstee Q, Charlotte F, Marchesini G, Bugianesi E, Trauner M, Romero Gomez M, Oliveira C, Day C, et al. Diagnostic performance of FibroTest, SteatoTest and ActiTest in patients with NAFLD using the SAF score as histological reference. Aliment Pharmacol Ther. 2016;44:877–889. doi: 10.1111/apt.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman ZD. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol. 2007;47:598–607. doi: 10.1016/j.jhep.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Pan Q, Zhang RN, Wang YQ, Zheng RD, Mi YQ, Liu WB, Shen F, Chen GY, Lu JF, Zhu CY, et al. Linked PNPLA3 polymorphisms confer susceptibility to nonalcoholic steatohepatitis and decreased viral load in chronic hepatitis B. World J Gastroenterol. 2015;21:8605–8614. doi: 10.3748/wjg.v21.i28.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang RX, Hu CX, Sun WL, Pan Q, Shen F, Yang Z, Su Q, Xu GW, Fan JG. Serum Monounsaturated Triacylglycerol Predicts Steatohepatitis in Patients with Non-alcoholic Fatty Liver Disease and Chronic Hepatitis B. Sci Rep. 2017;7:10517. doi: 10.1038/s41598-017-11278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, Hobbs HH. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsche MA, Hobbs HH, Cohen JC. Patatin-like phospholipase domain-containing protein 3 promotes transfer of essential fatty acids from triglycerides to phospholipids in hepatic lipid droplets. J Biol Chem. 2018;293:6958–6968. doi: 10.1074/jbc.RA118.002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamoun Z, Vacca F, Parton RG, Gruenberg J. PNPLA3/adiponutrin functions in lipid droplet formation. Biol Cell. 2013;105:219–233. doi: 10.1111/boc.201200036. [DOI] [PubMed] [Google Scholar]
- 29.Perttilä J, Huaman-Samanez C, Caron S, Tanhuanpää K, Staels B, Yki-Järvinen H, Olkkonen VM. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am J Physiol Endocrinol Metab. 2012;302:E1063–E1069. doi: 10.1152/ajpendo.00125.2011. [DOI] [PubMed] [Google Scholar]
- 30.Orešič M, Hyötyläinen T, Kotronen A, Gopalacharyulu P, Nygren H, Arola J, Castillo S, Mattila I, Hakkarainen A, Borra RJ, et al. Prediction of non-alcoholic fatty-liver disease and liver fat content by serum molecular lipids. Diabetologia. 2013;56:2266–2274. doi: 10.1007/s00125-013-2981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman A, Eder SK, Felder TK, Kedenko L, Paulweber B, Stadlmayr A, Huber-Schönauer U, Niederseer D, Stickel F, Auer S, et al. Clinical and Metabolic Characterization of Lean Caucasian Subjects With Non-alcoholic Fatty Liver. Am J Gastroenterol. 2017;112:102–110. doi: 10.1038/ajg.2016.318. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka N, Matsubara T, Krausz KW, Patterson AD, Gonzalez FJ. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. 2012;56:118–129. doi: 10.1002/hep.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HS, Nam Y, Chung YH, Kim HR, Park ES, Chung SJ, Kim JH, Sohn UD, Kim HC, Oh KW, et al. Beneficial effects of phosphatidylcholine on high-fat diet-induced obesity, hyperlipidemia and fatty liver in mice. Life Sci. 2014;118:7–14. doi: 10.1016/j.lfs.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 34.Taniyama Y, Shibata S, Kita S, Horikoshi K, Fuse H, Shirafuji H, Sumino Y, Fujino M. Cloning and expression of a novel lysophospholipase which structurally resembles lecithin cholesterol acyltransferase. Biochem Biophys Res Commun. 1999;257:50–56. doi: 10.1006/bbrc.1999.0411. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian VS, Goyal J, Miwa M, Sugatami J, Akiyama M, Liu M, Subbaiah PV. Role of lecithin-cholesterol acyltransferase in the metabolism of oxidized phospholipids in plasma: studies with platelet-activating factor-acetyl hydrolase-deficient plasma. Biochim Biophys Acta. 1999;1439:95–109. doi: 10.1016/s1388-1981(99)00072-4. [DOI] [PubMed] [Google Scholar]
- 36.Wu LC, Pfeiffer DR, Calhoon EA, Madiai F, Marcucci G, Liu S, Jurkowitz MS. Purification, identification, and cloning of lysoplasmalogenase, the enzyme that catalyzes hydrolysis of the vinyl ether bond of lysoplasmalogen. J Biol Chem. 2011;286:24916–24930. doi: 10.1074/jbc.M111.247163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veldhuizen RA, Mok A, McMurray WC, Possmayer F. Examination of the potential role of the glycerophosphorylcholine (GPC) pathway in the biosynthesis of phosphatidylcholine by liver and lung. Biochim Biophys Acta. 1989;1005:157–161. doi: 10.1016/0005-2760(89)90181-1. [DOI] [PubMed] [Google Scholar]
- 38.Ma DW, Arendt BM, Hillyer LM, Fung SK, McGilvray I, Guindi M, Allard JP. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutr Diabetes. 2016;6:e220. doi: 10.1038/nutd.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H, Wang J, Li C, Hirose A, Nozaki Y, Takahashi M, Ono M, Akisawa N, Iwasaki S, Saibara T, et al. The phosphatidylethanolamine N-methyltransferase gene V175M single nucleotide polymorphism confers the susceptibility to NASH in Japanese population. J Hepatol. 2007;46:915–920. doi: 10.1016/j.jhep.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Vance DE. Physiological roles of phosphatidylethanolamine N-methyltransferase. Biochim Biophys Acta. 2013;1831:626–632. doi: 10.1016/j.bbalip.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 41.van der Veen JN, Kennelly JP, Wan S, Vance JE, Vance DE, Jacobs RL. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim Biophys Acta. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Kent C, Carman GM. Interactions among pathways for phosphatidylcholine metabolism, CTP synthesis and secretion through the Golgi apparatus. Trends Biochem Sci. 1999;24:146–150. doi: 10.1016/s0968-0004(99)01365-1. [DOI] [PubMed] [Google Scholar]
- 43.Arendt BM, Ma DW, Simons B, Noureldin SA, Therapondos G, Guindi M, Sherman M, Allard JP. Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl Physiol Nutr Metab. 2013;38:334–340. doi: 10.1139/apnm-2012-0261. [DOI] [PubMed] [Google Scholar]
- 44.Gundermann KJ, Kuenker A, Kuntz E, Droździk M. Activity of essential phospholipids (EPL) from soybean in liver diseases. Pharmacol Rep. 2011;63:643–659. doi: 10.1016/s1734-1140(11)70576-x. [DOI] [PubMed] [Google Scholar]
- 45.Zoeller RA, Lake AC, Nagan N, Gaposchkin DP, Legner MA, Lieberthal W. Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem J. 1999;338(Pt 3):769–776. [PMC free article] [PubMed] [Google Scholar]
- 46.Nagan N, Zoeller RA. Plasmalogens: biosynthesis and functions. Prog Lipid Res. 2001;40:199–229. doi: 10.1016/s0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]


