Abstract
α-thalassemia X-linked intellectual disability (ATR-X) syndrome is caused by mutations in ATRX. An ATR-X model mouse lacking Atrx exon 2 displays phenotypes that resemble symptoms in the human intellectual disability: cognitive defects and abnormal dendritic spine formation. We herein target activation of sigma-1 receptor (Sig-1R) that can induce potent neuroprotective and neuroregenerative effects by promoting the activity of neurotrophic factors, such as brain-derived neurotrophic factor (BDNF). We demonstrated that treatment with SA4503, a potent activator of Sig-1R, reverses axonal development and dendritic spine abnormalities in cultured cortical neurons from ATR-X model mice. Moreover, the SA4503 treatment rescued cognitive deficits exhibited by the ATR-X model mice. We further found that significant decreases in the BDNF-protein level in the medial prefrontal cortex of ATR-X model mice were recovered with treatment of SA4503. These results indicate that the rescue of dendritic spine abnormalities through the activation of Sig-1R has a potential for post-diagnostic therapy in ATR-X syndrome.
Keywords: ATR-X syndrome, cognitive function, dendritic spine, sigma-1 receptor, brain-derived neurotrophic factor
1. Introduction
α-thalassemia X-linked mental retardation (ATR-X, OMIM Entry #301040) syndrome is caused by ATRX mutations [1,2]. ATR-X syndrome is characterized by various clinical manifestations, including severe intellectual disability, facial dysmorphism, genital abnormalities, and epileptic seizures [3]. ATR-X syndrome is very rare, probably with an incidence of less than 1/100,000 live-born males [4]. ATRX encodes a protein containing two signature domains. The helicase domain has chromatin remodeling activity and DNA translocaze ability. The ATRX-DNMT3-DNMT3L (ADD) domain binds histone H3 tails at H3K4me0K9me2/3 [5,6,7,8,9]. However, the functions of ATRX protein remain unclear.
Several ATRX mutations or low ATRX gene expression have been identified in patients with ATR-X syndrome [8,9,10,11]. ATR-X patients with an Arg37Stop (R37X) mutation in exon 2 exhibit mild intellectual disability, which is accompanied by reduced expression of ATRX protein in lymphoblastic cells [12,13]. We previously reported that Atrx mutant mice lacking exon 2 (AtrxΔE2 mice) express a mutant protein that corresponds to a variant of an R37X mutation seen in human ATR-X syndrome [14]. AtrxΔE2 mice exhibited an 80% reduction in ATRX protein levels [14,15,16], similar to outcomes observed in 27 individuals with ATR-X syndrome [9]. The mice also show cognitive defects and abnormal dendritic spine formation in the medial prefrontal cortex (mPFC) [15] and, consistent with features of intellectual disability [17,18], show longer and thinner dendritic spines compared with wild-type (WT) mice [15].
The morphology of dendritic spines relates closely to the function and plasticity of the synapses. For example, the volume of the spine head is directly proportional to the area of postsynaptic density and the number of postsynaptic receptors, and, hence, the size of synaptic currents and synaptic strength [19,20,21,22]. Such spine anomalies have been reported in several neurological disorders associated with cognitive dysfunction, including Alzheimer’s diseases, schizophrenia, and intellectual disability [17,18,23,24].
Activation of the sigma-1 receptor (Sig-1R) can induce potent neuroprotective and neuroregenerative effects by promoting the activity of neurotrophic factors, including brain-derived neurotrophic factor (BDNF) [25,26]. BDNF is an important neurotrophic factor linked with neuronal development, synaptic maturation and neural plasticity [27,28,29]. The activation of BDNF pathways improved learning and memory in intellectual disability model mice, such as Rett syndrome and Fragile-X syndrome [30,31]. Here, we show that Sig-1R activation with SA4503, a potent activator of Sig-1R [32], features potential as a therapeutic intervention for diminished cognitive function in ATR-X syndrome.
2. Results
2.1. Treatment with SA4503 Reverses Abnormality of Axonal Development and Dendritic Filopodia in Cultured Cortical Neurons from AtrxΔE2 (Atrx Mutant Mice Lacking Exon 2) Mice
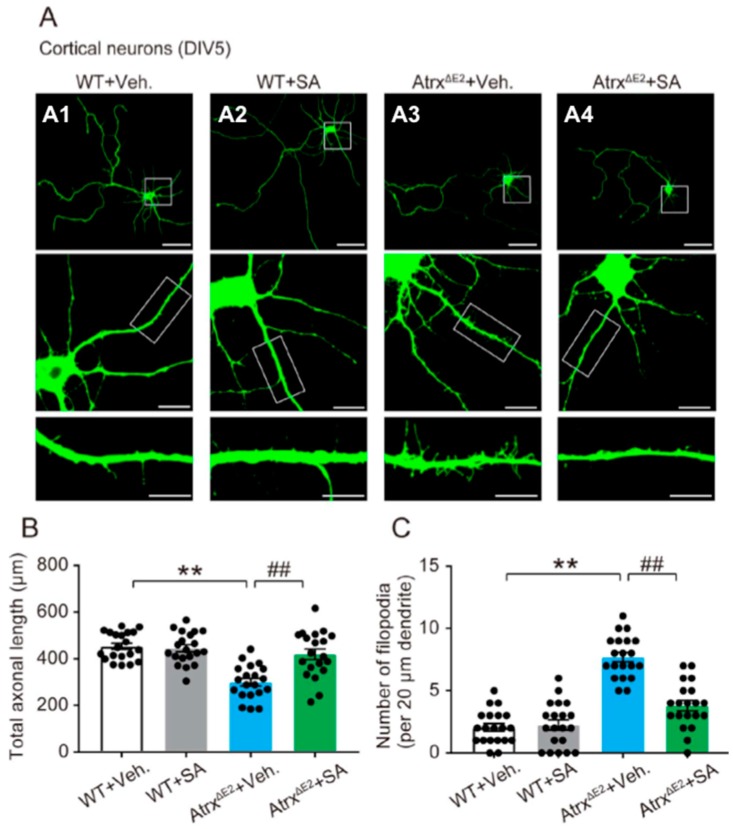
We investigated changes in synaptic morphology using cultured cortical neurons from AtrxΔE2 mice. To analyze axon development and dendritic filopodia, we labeled cultured neurons by transfection with EGFP at DIV0 and investigated these morphologies at DIV5. We first examined the effect of the Atrx gene mutation on the outgrowth of axons (Figure 1A3). Quantification revealed that the lengths of the axons were significantly reduced in AtrxΔE2 compared to those of WT neurons. The decreases of axon length in AtrxΔE2 neurons were significantly restored at 48 h after SA4503 treatment (Figure 1B) (The axon length; WT + vehicle, 453.5 ± 13.3 µm; WT + SA4503, 445.5 ± 15.1 µm; AtrxΔE2 + vehicle, 300.1 ± 14.5 µm; AtrxΔE2 + SA4503, 419.6 ± 22.3 µm). As dendritic spines originate from dendritic filopodia, we hypothesized that AtrxΔE2 neurons change the development of these structures. The number of filopodia was significantly increased in cultured neurons of AtrxΔE2 mice relative to the number measured in WT mice (Figure 1C), and cultured neurons from AtrxΔE2 mice treated with SA4503 showed a significant decrease in the number of filopodia (Figure 1A4,C) (The number of filopodia per 20 µm dendritic length; WT + vehicle, 2.1 ± 0.3; WT + SA4503, 2.1 ± 0.4; AtrxΔE2 + vehicle, 7.7 ± 0.4; AtrxΔE2 + SA4503, 3.8 ± 0.4).
Figure 1.
Treatment with SA4503 reverses abnormality of axonal development and dendritic filopodia in cultured AtrxΔE2 (Atrx mutant mice lacking exon 2) neurons. (A) Representative images of EGFP-transfected cortical neurons at DIV5. Neurons are stained for anti-GFP. Images in the middle and bottom panels are enlarged from the corresponding boxed areas. Scale bars: top panels, 50 µm; middle panels, 10 µm; bottom panels, 5 µm. (B) Total axonal length. ** p < 0.01, versus vehicle-treated WT (wild type) neurons; ## p < 0.01, versus vehicle-treated AtrxΔE2 neurons by one-way ANOVA with post-hoc Tukey’s test; F (3, 76) = 17.4; n = 20 neurons for each group. The experiments were repeated three times with similar results. (C) Data show the number of filopodia per 20 µm dendritic length. ** p < 0.01, versus vehicle-treated WT neurons; ## p < 0.01, versus vehicle-treated AtrxΔE2 neurons by one-way ANOVA with post hoc Tukey’s test; F (3, 76) = 48.26; n = 20 neurons for each group. The experiments were repeated three times with similar results. Each bar represents the mean ± SEM. Abbreviations: Veh., vehicle; SA, SA4503.
2.2. Treatment with SA4503 Ameliorates Dendritic Spine Abnormality in Cultured Cortical Neurons from AtrxΔE2 Mice
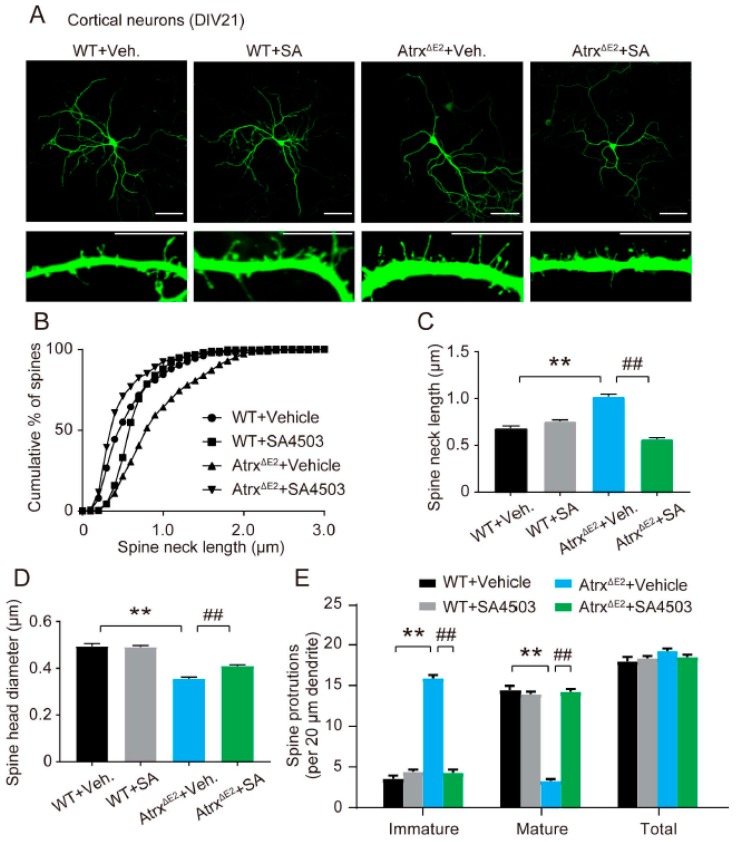
To investigate synaptogenesis, EGFP-transfected cortical neurons were harvested at DIV21. These neurons were fixed and stained with anti-GFP. We then performed spine morphological analysis (Figure 2A). In cultured neurons of AtrxΔE2 mice, spines resembling filopodia were abnormally thin and long relative to those observed in WT neurons. The spine length distribution was shifted to the right in AtrxΔE2 compared to WT neurons, indicating a higher proportion of long and filopodia-like spines in the former. Moreover, SA4503-treated AtrxΔE2 neurons restored the spine abnormality (Figure 2B,C) (spine length; WT + vehicle, 0.68 ± 0.01 µm; WT + SA4503, 0.76 ± 0.01 µm; AtrxΔE2 + vehicle, 1.02 ± 0.01 µm; AtrxΔE2 + SA4503, 0.56 ± 0.01 µm). The spine-head diameter was significantly decreased in AtrxΔE2 compared to WT neurons, while SA4503 treatment ameliorated the decrease observed in the former (Figure 2D) (The spine-head diameter; WT + vehicle, 0.49 ± 0.01 µm; WT + SA4503, 0.49 ± 0.01 µm; AtrxΔE2 + vehicle, 0.36 ± 0.01 µm; AtrxΔE2 + SA4503, 0.41 ± 0.01 µm). As expected, AtrxΔE2 neurons showed increased numbers of immature-appearing and fewer mature-appearing spines than did their WT counterparts. SA4503 treatment in AtrxΔE2 neurons significantly reduced the number of immature spines and increased the number of mature spines, without changing total spine numbers (Figure 2E). (The number of mature spines per 20 µm dendritic length: WT + vehicle, 14.4 ± 0.5; WT + SA4503, 13.9 ± 0.3; AtrxΔE2 + vehicle, 3.3 ± 0.2; AtrxΔE2 + SA4503, 14.2 ± 0.4. The number of immature spines per 20 µm dendritic length: WT + vehicle, 3.6 ± 0.4; WT + SA4503, 4.4 ± 0.3; AtrxΔE2 + vehicle, 15.9 ± 0.4; AtrxΔE2 + SA4503, 4.3 ± 0.4.)
Figure 2.
Treatment with SA4503 ameliorates dendritic spine abnormality in cultured AtrxΔE2 neuron. (A) Representative images of EGFP-transfected cortical neurons at DIV21. Neurons are stained for anti-GFP. Images in the bottom panels are enlarged from each dendrite. Scale bars: top panels, 100 µm; bottom panels, 10 µm. (B) Relationship between cumulative percentage of spines and spine length. p < 0.01 in WT versus AtrxΔE2 neurons and in AtrxΔE2 versus SA4503-treated AtrxΔE2 neurons by the Kolmogorov–Smirnov test. (C) Data show the number of spines. ** p < 0.01 versus vehicle-treated WT neurons; ## p < 0.01, versus vehicle-treated AtrxΔE2 neurons by one-way ANOVA with post hoc Tukey’s test; F (3, 1494) = 89.38. (D) Data show the spine-head diameter. ** p < 0.01 versus vehicle-treated WT neurons; ## p < 0.01, versus vehicle-treated AtrxΔE2 neurons by one-way ANOVA with post hoc Tukey’s test; F (3, 76) = 76.38. (E) Spine protrusions per 20 µm dendritic length. ** p < 0.01 versus vehicle-treated WT neurons; ## p < 0.01, versus vehicle-treated AtrxΔE2 neurons by two-way ANOVA with post hoc Bonferroni’s test; F (3, 228) = 2.549, p = 0.0566 (group); F (2, 228) = 1008, p < 0.01 (spine protrusions), F (6, 228) = 243.6, p < 0.01 (interaction between group and spine protrusions); n = 20 neurons for each group. The experiments were repeated three times with similar results. WT, n = 360 spines; SA4503-treated WT, n = 367 spines; AtrxΔE2, n = 384 spines; SA4503-treated AtrxΔE2, n = 370 spines. Each bar represents the mean ± SEM. Abbreviations: Veh., vehicle; SA, SA4503.
2.3. Treatment with SA4503 Rescues Memory and Cognitive Deficits Seen in AtrxΔE2 Mice
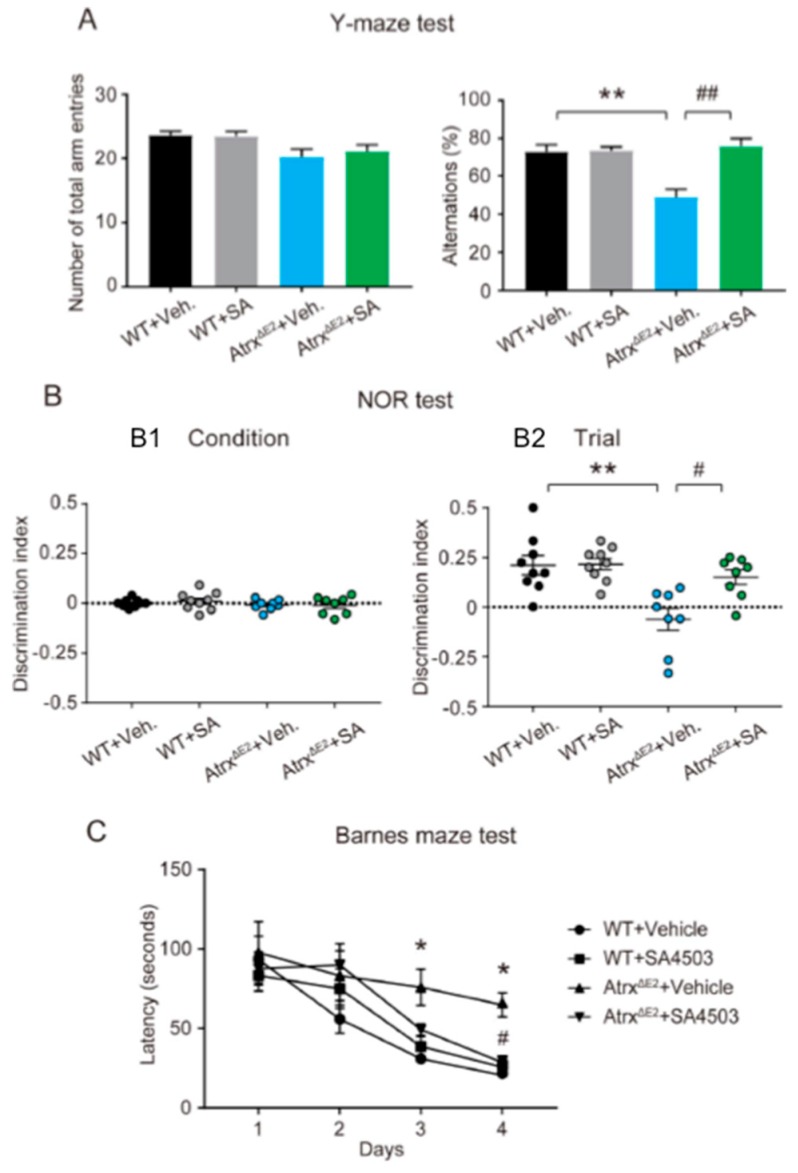
In the Y-maze test, measurement of impairment was based on the percentage of alternation behaviors relative to WT mice without a change in the total number of arm entries. SA4503 treatment increased the percentage of spontaneous alternation behaviors in AtrxΔE2 mice (Figure 3A) (the percentage of alternation behaviors: WT + vehicle, 73.2 ± 3.5; WT + SA4503, 73.7 ± 1.9; AtrxΔE2 + vehicle, 49.1 ± 4.0; AtrxΔE2 + SA4503, 75.9 ± 4.0). In the novel object recognition task, we observed no differences in the discrimination index using the same object across training trials in all groups (Figure 3B1). After a 24 h retention interval between the trial and test sessions, AtrxΔE2 mice revealed a significantly lower discrimination index for novel objects than did WT mice. The discrimination index of AtrxΔE2 mice treated with SA4503 was significantly higher than that of vehicle-treated AtrxΔE2 groups (Figure 3B2) (the discrimination index in test session: WT + vehicle, 0.21 ± 0.05; WT + SA4503, 0.22 ± 0.03; AtrxΔE2 + vehicle, −0.06 ± 0.06; AtrxΔE2 + SA4503, 0.15 ± 0.04). In the Barnes maze test, WT mice featured a significant decline in latency time to enter the target hole across the four days of training, indicating a normal improvement of cognitive performance. By contrast, the cognitive performance of AtrxΔE2 mice did not improve, because longer latency times was required compared to WT mice. SA4503 administration significantly rescued the latency time (Figure 3C) (the latency time in Day 4: WT + vehicle, 20.6 ± 1.0 s; WT + SA4503, 25.7 ± 3.1 s; AtrxΔE2 + vehicle, 64.8 ± 7.7 s; AtrxΔE2 + SA4503, 24.8 ± 2.8 s).
Figure 3.
SA4503 treatment decreased cognitive deficits in AtrxΔE2 mice based on memory-related tests. (A) Number of total arm entries (left) and alternations (right) in a Y-maze test. ** p < 0.01, versus vehicle-treated WT mice; ## p < 0.01, versus vehicle-treated AtrxΔE2 mice by one-way ANOVA with post hoc Tukey’s test; F (3, 18) = 12.98. (B) Novel object recognition task. Discrimination index of object exploration during the sample phase (left), and while exploring familiar and new objects in the test phase after 24 h (right). ** p < 0.01, versus vehicle-treated WT mice; # p < 0.05, versus vehicle-treated AtrxΔE2 mice by one-way ANOVA with post hoc Tukey’s test; F (3, 30) = 8.898. (C) Barnes maze test measurement of total latency (s) to reach the target hole. * p < 0.05 by two-way ANOVA with post hoc Bonferroni’s test; F (3, 120) = 6.535, p < 0.01 (group); F (3, 120) = 25.16, p < 0.01 (day), F (9, 120) = 1.184, p = 0.3112 (interaction between group and day). Each bar represents the mean ± SEM. Abbreviations: Veh., vehicle; SA, SA4503. WT + Vehicle: n = 9 mice, WT + SA4503: n = 9 mice, AtrxΔE2 + Vehicle: n = 8 mice, AtrxΔE2 + SA4503: n = 8 mice.
2.4. Treatment with SA4503 Increases the BDNF (Brain-Derived Neurotrophic Factor) Protein Level in mPFC of AtrxΔE2 Mice
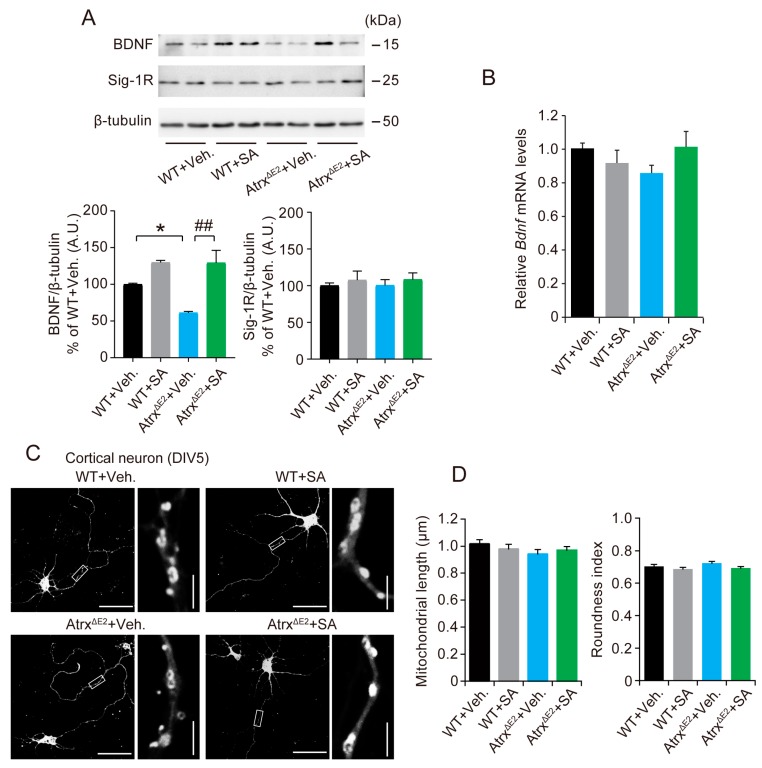
Next, we explored whether the expression of the BDNF protein was altered in AtrxΔE2 mice. We observed significant decreases in the levels of the BDNF protein in the mPFC of AtrxΔE2 relative to WT mice without changes in the mRNA level; treatment with SA4503 reversed the decrease (Figure 4A,B). However, there was no significant change in NR1, a down stream target of BDNF, or in the Sig-1R protein expression between groups. Sig-1Rs act on restoring Ca2+ transferring into mitochondria and ATP productions in Chinese hamster ovary (CHO) cells [33] and neuroblastoma Neuro-2a cells [34], as well as changes in mitochondrial morphology in SA4503-treated cardiomyocytes [35]. We investigated mitochondrial morphology in axons of cultured cortical neurons at DIV5 with the mitochondrial-specific marker, MitoTracker Red. There were no significant changes in mitochondrial length or the roundness index between the groups (Figure 4D), suggesting that Sig-1Rs have no effect on either mitochondrial degradation or enhanced mitochondrial fusion in Atrx depletion or cultured cortical neurons.
Figure 4.
Treatment with SA4503 increases BDNF protein level in AtrxΔE2 mouse brain. (A) Representative immunoblot of mouse medial prefrontal cortex lysates probed with the indicated antibodies (top panel). Densitometry analysis of indicated proteins to β-tubulin (arbitrary units; (A.U.)) (bottom panel). * p < 0.05, versus vehicle-treated WT mice; ## p < 0.01, versus vehicle-treated AtrxΔE2 mice by one-way ANOVA with post hoc Tukey’s test; F (3, 16) = 13.7 in BDNF; n = 5 mice each. (B) Real-time RT-qPCR showing total BDNF mRNA (using exon IX primers) expression in mouse medial prefrontal cortex lysates. BDNF mRNA were determined relative to Gapdh mRNA. n = 5 mice for each group. The experiments were repeated two times with similar results. (C) Representative images of cultured cortical neurons at DIV5 with the mitochondrial-specific marker, MitoTracker Red. Images in the right panels are enlarged from the corresponding boxed areas. Scale bars: left panels, 50 µm; right panels, 3 µm. (D) Data show the mitochondrial length (left) and the roundness index (right); n = 20 cells each. Abbreviations: Veh., vehicle; SA, SA4503.
3. Discussion
In this study, we demonstrated that postnatal activation of Sig-1Rs in AtrxΔE2 mice ameliorates some of the intellectual disability-related abnormalities present at the levels of synaptic morphology and behavior. We have also provided evidence that activation of Sig-1Rs enhanced BDNF protein expression in mPFC of AtrxΔE2 mice. These results indicate that the pharmacological rescue of abnormal synaptic morphology and behavior in the adult ATR-X mouse model and identified postnatal Sig-1R activation as a potential therapeutic strategy for countering the various debilitating symptoms of intellectual disability, including those of ATR-X syndrome.
Dendritic spines can assume various shapes by which they are classified; these include filopodia, thin, stubby, or mushroom-like [36,37]. Small spines (filopodia and thin) change their form rapidly, either disappearing or growing into large spines during intense neuronal activity in the mouse brain [38,39,40,41]. Small spines are often short-lived, usually representing weak or silent synapses [42]. Such observations suggest that structural alterations of small spines underlie adaptive and learning processes [43]. Conversely, large spines (stubby and mushroom) are relatively stable and survive for long periods of time: more than a month [40] or even for a year [41] in the mouse cortex in vivo. This observation suggests that memory is maintained in a structural form for extended periods in the brain. The link between spine abnormalities, increases of small spines and intellectual disability has been reported in Down, Fragile X, and Rett syndromes [17,18]. The present report found diminished axonal development and increases of dendritic filopodia in cultured AtrxΔE2 neurons at early stage DIV5 (Figure 1). Moreover, we have demonstrated that neurons in AtrxΔE2 mice exhibit longer, thinner dendritic spines relative to WT neurons at DIV24. This finding is consistent with an in vivo study of adult AtrxΔE2 mice [15], suggesting that the immature patterning of dendritic spines occurs during postnatal brain development of AtrxΔE2 mice. Notably, short-term treatment with SA4503 (48 h) reversed the axonal degeneration and dendritic spine immaturity in cultured AtrxΔE2 neurons in both DIV5 and DIV24 (Figure 1 and Figure 2). The short-term in vivo treatment with SA4503 (i.p., daily from Postnatal Day 70 to Postnatal Day 84) to AtrxΔE2 mice also rescued decreased cognitive deficits (Figure 3). These results indicate that the maturation of dendritic spines provides a potential avenue for post-diagnostic therapy in ATR-X syndrome, as even in mature neurons the size and shape of spines can change. The plastic nature of dendritic spines might be a key component of cognitive function therapy that address intellectual disabilities. In addition, we reported that aberrant expression of Xlr3b is involved in cognitive dysfunction in AtrxΔE2 mice [16]. Interestingly, overexpression of Xlr3b shows abnormalities in dendrite branching, spine development, and synapse formation in Layer II–III neurons of the mouse cerebral cortex [44]. In the future, we will examine that involvement of Xlr3b in abnormality of spine formation in ATR-X syndrome.
Sig-1R plays an important role in the development and stability of dendritic spines. Knockdown of Sig-1Rs by small interfering RNA (siRNA) causes a deficit in the formation of dendritic spines in rat hippocampal primary neurons [45]. In addition, SA4503 induces the upregulation of BDNF protein in the rat hippocampus [46]. SA4503 does not merely increase the total amount of BDNF but potentiates the posttranslational processing of BDNF proteins related to protein secretion in rat neuroblastoma B104 cells [47]. In addition, knockdown of Sig-1Rs decreases the release of mature BDNF without significantly affecting protein synthesis of BDNF [47]. BDNF promotes dendritic and axonal growth, increases dendritic spine density, and enhances synaptic plasticity [27,28,29]. The human BDNF gene product having a methionine substitution for valine at codon 66 (Val66Met) impairs BDNF trafficking and its activity-dependent release, thereby impairing cognition in the general population [48]. Dendritic complexity is reduced in dentate granule cells of Val66Met knock-in mice [49], indicating that proper secretory trafficking of BDNF is essential to dendritic spine development and plasticity. We found significant decreases of BDNF protein levels in the mPFC of AtrxΔE2 mice, and treatment with SA4503 recovered the BDNF protein levels without changing the levels of mRNA level (Figure 4). Although we could not reveal the exact molecular mechanism underlying decreases of BDNF protein level in AtrxΔE2 brain, our results indicate that the activation of Sig-1Rs leads to an increased release of BDNF, resulting in the recovery of synaptic morphology and behavior abnormalities of AtrxΔE2 mouse. Similarly, we could not find an association between Ca2+/calmodulin-dependent protein kinase II (CaMKII) signaling pathway and Sif-1R/BDNF changes, suggesting that multiple signaling pathways may contribute to altered spine morphology in AtrxΔE2 mice.
Sig-1R is a stress- and ligand-regulated, endoplasmic reticulum (ER) chaperone protein [50]. Its ability to modulate the actions of neurotransmitter receptors and ion channels explains its involvement in neuroprotection, neuroplasticity, and the release of neurotransmitters [26], especially the activation of the Sig-1R enhanced BDNF secretion in primary rat astrocytes [51], in the mouse striatum [52] and in rat neuroblastoma B104 cells [47]. In addition, Sig-1R dissociates from the mitochondrion-associated ER membrane by Sig-1R agonist and redistributes widely within the cell [26]. The agonist-induced redistribution of Sig-1R is linked to an increased intracellular trafficking of BDNF required for brain repair in rat models of stroke [53] and in 6-hydroxydopamine lesion mouse model of Parkinson’s disease [52]. Interestingly, a Sig-1R-active antidepressant, imipramine, potentiates BDNF-induced activation of the phospholipase C-gamma (PLC-gamma)/inositol 1,4,5-trisphosphate (IP3)/Ca2+ pathway that triggers glutamate release from cortical neurons [54]. The other selective Sig-1R agonist, PRE084 (2-(4-morpholinethyl)1 phenylcyclohexanecarboxylate), elevates the expression of glial cell-derived neurotrophic factor (GDNF) protein in astrocytes after root avulsion injury in rats [55]. The prototypic Sig-1R agonist, (+)-pentazocine, as well as the Sig-1R-active antidepressants, imipramine and fluvoxamine, potentiate nerve growth factor (NGF)-induced neurite sprouting of PC12 cells [56]. Sig-1Rs also potentiate epidermal growth factor (EGF) signaling toward neuritogenesis in PC12 cells [57]. Thus, the synergistic effects of some growth factors through Sig-1Rs may relate to the observed amelioration of dendritic spine abnormalities and cognitive dysfunction in AtrxΔE2 mice.
In conclusion, although future studies will be necessary to further characterize the precise molecular mechanisms of the role of BDNF in ATR-X syndrome, our findings clearly demonstrate that Sig-1R activation can exert actions that reverse several key neuronal and behavioral symptoms of ATR-X syndrome. Taken together, these results indicate a primary, and potentially causal, role for defects in spine morphogenesis in intellectual disability. By examining common clinical phenotypes correlated to spine and synaptic abnormalities in intellectual disability, we can elucidate causalities of dysgenesis and identify potential targets for therapeutic intervention.
4. Materials and Methods
4.1. Animals
Mice (C57BL/6J) were housed under climate-controlled conditions with a 12-h light/dark cycle and were provided with standard food and water ad libitum. Animal studies were conducted in accordance with the Tohoku University institutional guidelines. Ethical approval (9 November 2015) was obtained from the Institutional Animal Care and Use Committee of the Tohoku University Environmental and Safety Committee. The generation of homozygous AtrxΔE2 mice is described in a previous study [14]. In brief, to construct an Atrx targeting vector, a 2-kb EcoRV-SpeI genomic fragment of the Atrx gene and a 5.5-kb XhoI-BamHI fragment were used as short and long homologous regions, respectively. The IRES-beta-geo cassette with a splicing acceptor site was used to replace the SpeI-XhoI fragment, which contains exon 2. The targeting vector was transfected into J1 ES cells, and G418-resistant clones were selected. Homologous recombinants were identified by Southern blot analysis. AtrxΔE2 mice were backcrossed six generations onto C57BL/6J. For RT-qPCR, immunoblotting and behavioral analyses, male mice at 10–12 weeks of age were used.
4.2. Cell Culture and Transfection
Primary cultures of neurons were obtained using previously described methods [16]. Briefly, cortical tissue was dissected from Embryonic-Day-18 mice and dissociated by trypsin treatment and trituration through a Pasteur pipette. Neurons were plated on coverslips coated with poly-l-lysine in MEM (Invitrogen, Carlsbad, CA, USA). After cell attachment, coverslips were transferred to dishes containing a glial cell monolayer and maintained in Neurobasal Medium (Invitrogen) containing 2% B27 supplement (Invitrogen) and 1% GlutaMax (Invitrogen). Cytosine β-d-arabinofuranoside (5 µM; Sigma-Aldrich, St. Louis, MO, USA) was added to cultures at 3 days in vitro (DIV3) after plating to inhibit glial proliferation. Primary neurons were transfected with GFP (green fluorescent protein) expression vector using electroporation at DIV0. For electroporation, mixture of the single cell suspension with plasmid DNA made its final concentration reach 3 × 105 cells and 5 µg plasmid DNA (1 µg/µL) in 100 µL Opti-MEM® medium. Two pulses of 275 V, 0.5 ms each at 50 ms intervals, were delivered through electroporation cuvettes (2 mm gap) with an electroporator (NEPA21; NEPAGENE, Chiba, Japan). Cortical neurons were harvested at DIV5 and DIV24. These time points correspond to neuronal developmental stages of axon formation and dendrite outgrowth (stage 4) and synaptogenesis and maturation (stage 5), respectively [58,59]. Differential distributions of axons and dendrites could be detectable from DIV3, because axons elongate 5–10 times faster than dendrites; longer neurites could thus be identified as axons [60].
4.3. Drugs
SA4503 was synthesized in the Laboratory of Medicinal Chemistry, Zhejiang University according to previously described methods [61]. SA4503 (1 µM, dissolved in distilled water) was treated for 48 h in cultured cortical neurons in both DIV5 and DIV24. Mice were randomized into four groups corresponding to differential drug administration as follows: We administered SA4503 over the course of 2 weeks (1 mg/kg, intraperitoneally (i.p.) daily from postnatal day (P) 70 to P84) to AtrxΔE2 mice and subsequently assessed memory-related behaviors. In studies on learning and memory, effective doses of SA4503 are commonly 1.0 mg/kg or less for rodents [62,63]. A diagram of the experimental schedule is given as Figure 5.
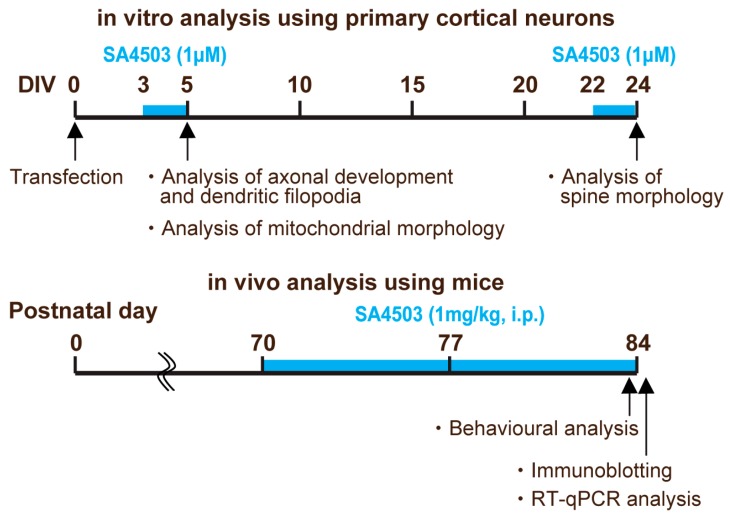
Figure 5.
Diagrams of the experimental schedule. In in vitro analysis using primary cortical neurons, SA4503 (1 µM, dissolved in distilled water) was treated for 48 h in cultured cortical neurons in both DIV5 and DIV24. In in vivo analysis using mice, we administered SA4503 over the course of two weeks (1 mg/kg, intraperitoneally (i.p.) daily from postnatal day (P) 70 to P84) to AtrxΔE2 mice and subsequently assessed memory-related behaviors and biological studies.
4.4. Antibodies
For Western blot analysis, we used the following: anti-BDNF antibody (1:1000, AB1779SP, Millipore, Burlington, MA, USA); anti-Sig-1R antibody (1:1000, ab53852, Abcam, Cambridge, UK); anti-β-Tubulin antibody (1:1000, T0198, Sigma-Aldrich); goat anti-mouse IgG (H + L) and human ads-HRP (1:500, 1031-05, SouthernBiotech, Birmingham, AL, USA); goat anti-rabbit IgG (H + L); and mouse/human ads-HRP (1:500, 4050-05, SouthernBiotech). For immunohistochemistry, we used the following: living colors® Full-Length GFP polyclonal antibody (anti-GFP) (1:300, 632592, Clontech, Mountain View, CA, USA); donkey anti-Rabbit IgG (H + L) highly cross-adsorbed secondary antibody; and Alexa Fluor 488 (1:500, A21206, Invitrogen).
4.5. RT-qPCR Analysis
Total RNA was purified from mice brains using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA was reverse transcribed into single-stranded cDNA using an oligo(dT) primer (Promega, Madison, WI, USA) and Moloney murine leukemia virus reverse transcriptase (Invitrogen) and then subjected to RT-PCR with gene-specific primers. RT-qPCR analysis was performed as described previously [16] in 48-well plates (Mini Opticon Real-Time PCR system, Bio-Rad, Hercules, CA, USA) using iQ SYBR Green Supermix 2× (Bio-Rad). Gene expression was assessed using the differences in normalized CT (cycle threshold) (∆∆Ct) method after normalization to GAPDH. Fold change was calculated by 2−∆∆Ct. The following primers were used for RT-qPCR:
Mouse Bdnf in exon IX (FW) (5′-AAGGACGCGGACTTGTACAC-3′)
Mouse Bdnf in exon IX (RV) (5′-CGCTAATACTGTCACACACGC-3′)
Mouse Gapdh (FW) (5′-TGTGTCCGTCGTGGATCTGA-3′)
Mouse Gapdh (RV) (5′-CACCACCTTCTTGATGTCATCATAC-3′).
4.6. Immunoblotting
Immunoblotting analysis was performed as previously described [16]. Briefly, tissues were homogenized in a buffer containing 50 mM Tris-HCl (pH 7.5), 0.5% Triton X-100, 0.15 M NaCl, 4 mM EDTA, 4 mM EGTA, 1 mM Na3VO4, 50 mM NaF, 1 mM dithiothreitol and protease inhibitors (2 µg/mL pepstatin A, 1 µg/mL leupeptin, and 2 µg/mL trypsin inhibitor), and then treated with Laemmli’s sample solution and boiled for 3 min. Bradford protein assay was used to measure the concentration of total protein in a sample. Equivalent amounts of protein (5 µg) were electrophoresed on SDS-PAGE and proteins were then transferred to an Immobilon polyvinylidene difluoride membrane. The procedure of immunoblotting analyses were carried out as previously described [16]. For housekeeping protein, β-tubulin was used as loading control.
4.7. Immunohistochemistry
Immunohistochemistry was performed as described previously [16]. Briefly, fixed cells with 4% paraformaldehyde in Phosphate buffered saline (PBS) were treated with PBS containing 0.1% Triton X-100 for 30 min. Samples were incubated overnight at 4 °C with enhanced green fluorescence protein (EGFP) antibody and then washed in PBS and incubated with the secondary antibody, Alexa 488-conjugated donkey anti-rabbit (1:500; A-21206, Invitrogen). Fluorescence images were analyzed by confocal laser scanning microscopy (LSM700, Carl Zeiss).
4.8. Spine Morphological Analysis
Spine-head diameters were measured along an axis perpendicular to the spine neck, approximately in the middle of the spine head. Spine length was calculated as the radial distance from the tip of the spine head to the dendritic shaft. The spine diameter corresponded to the maximum dimension of the spine head. In cases where the spine head showed irregular morphology, or if the spine-head maximum diameter was not perpendicular to the neck, we measured the maximum head diameter. Spines were categorized according to a previously described method [64] along an immature- to mature-appearing spine continuum. Measurements were obtained using ImageJ (National Institutes of Health freeware).
4.9. Analysis of Mitochondrial Morphology
Analysis of mitochondrial morphology was performed as described previously [65]. Briefly, cells were treated with MitoTracker™ Red CMXRos (final concentration 350 nM, M7512, Invitrogen) for 15 min and then treated with 4% paraformaldehyde in PBS (phosphate buffer saline). After images were acquired by confocal laser scanning microscopy (LSM700, Carl Zeiss, Oberkochen, Germany), they were analyzed using ImageJ software. ImageJ counts each signal and simultaneously determines the pixel area, perimeter, and maximum length for each signal. From these measurements, ImageJ calculates a score for roundness index: roundness index = 4π × (area/perimeter2).
4.10. Behavioral Analysis
Mice were subjected to behavioral tests including the Y-maze, novel object recognition and Barnes maze tasks. The videotapes for all behavioral analyses were scored by a trained observer blind to the genotype and treatment. In the Y-maze task using apparatus consisted of three identical arms (50 × 16 × 32 cm3) of black Plexiglas, spontaneous alternation behavior in a Y-maze was assessed as a task of spatial reference memory. An alternation was defined as consecutive entries into all three arms. The maximum number of alternations was defined as the total number of arms entered minus two, and the percentage of alternations was calculated as actual alternations/maximum alternations × 100. The total number of arms entered during the session was also determined. The novel object recognition task used an open-field box (35 × 25 × 35 cm3). During the acquisition phase, two objects of the same material were placed symmetrically in the center of the chamber for 10 min. After 24 h, one object was replaced by a novel object, and exploratory behavior was analyzed again for 5 min. Exploration of an object was defined as rearing on the object or sniffing it at a distance of <1 cm, touching it with the nose or both. Discrimination of spatial novelty was assessed by comparing the difference between exploratory contacts of novel and familiar objects and the total number of contacts with both, making it possible to adjust for differences in total exploration contacts. The Barnes maze consists of a circular platform (92 cm in diameter) with 20 holes (hole diameter: 5 cm) along its perimeter. At the beginning of each trial, the mouse was placed in the middle of the maze in a cylindrical start chamber (7.5 cm). After 10 s, the chamber was lifted and the mouse was free to explore the maze. The test ended when the mouse entered the goal tunnel or after 3 min elapsed. Immediately after the mouse entered the tunnel, the mouse was allowed to stay in the tunnel for 1 min. Mice were trained for two trials per day for 4 days. Errors and search time required to escape into the tunnel were recorded as the test parameters; errors were defined as nose pokes and head deflections over any hole that did not lead to a tunnel.
4.11. Statistical Analysis
All values are expressed as means ± SEM. Statistical significance for differences among groups was tested by one-way analysis of variance (ANOVA) with post-hoc Tukey’s multiple comparison test or two-way ANOVA with post-hoc Bonferroni’s multiple comparison test. Cumulative percentage of spines was tested by the Kolmogorov–Smirnov test. p < 0.05 was considered significant. All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
We thank Isao Kitajima for kindly providing AtrxΔE2 mice, deposited in the RIKEN BioResource Center (RBRC04937). This research was partially supported by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED) (N.S. and K.F.). This work was also supported by MEXT/JSPS KAKENHI (25110705) to N.S and KAKENHI (24659024) to K.F.
Author Contributions
Investigation and Formal Analysis, K.Y. and Y.Y.; Drug Synthesis, C.Z. and F.H.; Writing—Original Draft Preparation, N.S.; Writing—Review and Editing, Supervision, K.F.; and Funding Acquisition, N.S. and K.F.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gibbons R.J., Suthers G.K., Wilkie A.O., Buckle V.J., Higgs D.R. X-linked α-thalassemia/mental retardation (ATR-X) syndrome: Localization to Xq12-q21.31 by X inactivation and linkage analysis. Am. J. Hum. Genet. 1992;51:1136–1149. [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 3.Gibbons R.J., Wada T., Fisher C.A., Malik N., Mitson M.J., Steensma D.P., Fryer A., Goudie D.R., Krantz I.D., Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum. Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons R.J. A thalassaemia-mental retardation, X linked. Orphanet J. Rare Dis. 2006;1:15. doi: 10.1186/1750-1172-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argentaro A., Yang J.-C., Chapman L., Kowalczyk M.S., Gibbons R.J., Higgs D.R., Neuhaus D., Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhayalan A., Tamas R., Bock I., Tattermusch A., Dimitrova E., Kudithipudi S., Jeltsch A. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 2011;20:2195–2203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwase S., Xiang B., Ghosh S., Ren T., Lewis P.W., Cochrane J.C., Allis C.D., Picketts D.J., Patel D.J., Li H., et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picketts D.J., Higgs D.R., Bachoo S., Blake D.J., Quarrell O.W., Gibbons R.J. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 9.Mitson M., Kelley L.A., Sternberg M.J.E., Higgs D.R., Gibbons R.J. Functional significance of mutations in the Snf2 domain of ATRX. Hum. Mol. Genet. 2011;20:2603–2610. doi: 10.1093/hmg/ddr163. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons R.J., Bachoo S., Picketts D.J., Aftimos S., Asenbauer B., Bergoffen J., Berry S.A., Dahl N., Fryer A., Keppler K., et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 11.Villard L., Bonino M.C., Abidi F., Ragusa A., Belougne J., Lossi A.M., Seaverb L., Bonnefontd J.P., Romanoc C., Ficherac M., et al. Evaluation of a mutation screening strategy for sporadic cases of syndrome. J. Med. Genet. 1999;36:183–186. [PMC free article] [PubMed] [Google Scholar]
- 12.Howard M.T., Malik N., Anderson C.B., Voskuil J.L.A., Atkins J.F., Gibbons R.J. Attenuation of an amino-terminal premature stop codon mutation in the ATRX gene by an alternative mode of translational initiation. J. Med. Genet. 2004;41:951–956. doi: 10.1136/jmg.2004.020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abidi F.E., Cardoso C., Lossi A.M., Lowry R.B., Depetris D., Mattéi M.G., Lubs H.A., Stevenson R.E., Fontes M., Chudley A.E., et al. Mutation in the 5′ alternatively spliced region of the XNP/ATR-X gene causes Chudley-Lowry syndrome. Eur. J. Hum. Genet. 2005;13:176–183. doi: 10.1038/sj.ejhg.5201303. [DOI] [PubMed] [Google Scholar]
- 14.Nogami T., Beppu H., Tokoro T., Moriguchi S., Shioda N., Fukunaga K., Ohtsuka T., Ishii Y., Sasahara M., Shimada Y., et al. Reduced expression of the ATRX gene, a chromatin-remodeling factor, causes hippocampal dysfunction in mice. Hippocampus. 2011;21:678–687. doi: 10.1002/hipo.20782. [DOI] [PubMed] [Google Scholar]
- 15.Shioda N., Beppu H., Fukuda T., Li E., Kitajima I., Fukunaga K. Aberrant calcium/calmodulin-dependent protein kinase II (CaMKII) activity is associated with abnormal dendritic spine morphology in the atrx mutant mouse brain. J. Neurosci. 2011;31:346–358. doi: 10.1523/JNEUROSCI.4816-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shioda N., Yabuki Y., Yamaguchi K., Onozato M., Li Y., Kurosawa K., Tanabe H., Okamoto N., Era T., Sugiyama H., et al. Targeting G-quadruplex DNA as cognitive function therapy for ATR-X syndrome. Nat. Med. 2018;24:802–813. doi: 10.1038/s41591-018-0018-6. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann W.E., Moser H.W. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 18.Levenga J., Willemsen R. Perturbation of dendritic protrusions in intellectual disability. Prog. Brain Res. 2012;197:153–168. doi: 10.1016/B978-0-444-54299-1.00008-X. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki M., Ellis-Davies G.C.R., Nemoto T., Miyashita Y., Iino M., Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtmaat A.J.G.D., Trachtenberg J.T., Wilbrecht L., Shepherd G.M., Zhang X., Knott G.W., Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi J., Matsuzaki M., Ellis-Davies G.C.R., Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46:609–622. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasai H., Fukuda M., Watanabe S., Hayashi-Takagi A., Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Fiala J.C., Spacek J., Harris K.M. Dendritic spine pathology: Cause or consequence of neurological disorders? Brain Res. Rev. 2002;39:29–54. doi: 10.1016/S0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- 24.Forrest M.P., Parnell E., Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 2018;19:215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kourrich S., Su T.P., Fujimoto M., Bonci A. The sigma-1 receptor: Roles in neuronal plasticity and disease. Trends Neurosci. 2012;35:762–771. doi: 10.1016/j.tins.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su T.P., Su T.C., Nakamura Y., Tsai S.Y. The Sigma-1 Receptor as a Pluripotent Modulator in Living Systems. Trends Pharmacol. Sci. 2016;37:262–278. doi: 10.1016/j.tips.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- 28.Lewin G.R., Barde Y.A. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 29.Horch H.W. Local effects of BDNF on dendritic growth. Rev. Neurosci. 2004;15:117–129. doi: 10.1515/REVNEURO.2004.15.2.117. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee A., Castro J., Sur M. Rett syndrome: Genes, synapses, circuits, and therapeutics. Front. Psychiatry. 2012;3:34. doi: 10.3389/fpsyt.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauterborn J.C., Rex C.S., Kramár E., Chen L.Y., Pandyarajan V., Lynch G., Gall C.M. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuno K., Nakazawa M., Okamoto K., Kawashima Y., Mita S. Binding properties of SA4503, a novel and selective σ1 receptor agonist. Eur. J. Pharmacol. 1996;306:271–279. doi: 10.1016/0014-2999(96)00201-4. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi T., Su T.P. Sigma-1 Receptor Chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Shioda N., Ishikawa K., Tagashira H., Ishizuka T., Yawo H., Fukunaga K. Expression of a truncated form of the endoplasmic reticulum chaperone protein, σ1 receptor, promotes mitochondrial energy depletion and apoptosis. J. Biol. Chem. 2012;287:23318–23331. doi: 10.1074/jbc.M112.349142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagashira H., Zhang C., Lu Y.M., Hasegawa H., Kanai H., Han F., Fukunaga K. Stimulation of σ1-receptor restores abnormal mitochondrial Ca2+ mobilization and ATP production following cardiac hypertrophy. Biochim. Biophys. Acta. 2013;1830:3082–3094. doi: 10.1016/j.bbagen.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 36.Hering H., Sheng M. Dentritic spines: Structure, dynamics and regulation. Nat. Rev. Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 37.Holtmaat A., Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 38.Maletic-Savatic M., Malinow R., Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 39.Engert F., Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 40.Trachtenberg J.T., Chen B.E., Knott G.W., Feng G., Sanes J.R., Welker E., Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 41.Grutzendler J., Kasthuri N., Gan W. Long-term dendritic spine stabilityin the adult cortex. Nature. 2002;420:1–5. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- 42.Rochefort N.L., Konnerth A. Dendritic spines: From structure to in vivo function. EMBO Rep. 2012;13:699–708. doi: 10.1038/embor.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasai H., Matsuzaki M., Noguchi J., Yasumatsu N., Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- 44.Cubelos B., Sebastián-Serrano A., Beccari L., Calcagnotto M.E., Cisneros E., Kim S., Dopazo A., Alvarez-Dolado M., Redondo J.M., Bovolenta P., et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai S.-Y., Hayashi T., Harvey B.K., Wang Y., Wu W.W., Shen R.-F., Zhang Y., Becker K.G., Hoffer B.J., Su T.P. Sigma-1 receptors regulate hippocampal dendritic spine formation via a free radical-sensitive mechanism involving Rac1xGTP pathway. Proc. Natl. Acad. Sci. USA. 2009;106:22468–22473. doi: 10.1073/pnas.0909089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikuchi-Utsumi K., Nakaki T. Chronic treatment with a selective ligand for the sigma-1 receptor chaperone, SA4503, up-regulates BDNF protein levels in the rat hippocampus. Neurosci. Lett. 2008;440:19–22. doi: 10.1016/j.neulet.2008.05.055. [DOI] [PubMed] [Google Scholar]
- 47.Fujimoto M., Hayashi T., Urfer R., Mita S., Su T.P. Sigma-1 receptor chaperones regulate the secretion of brain-derived neurotrophic factor. Synapse. 2012;66:630–639. doi: 10.1002/syn.21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egan M.F., Kojima M., Callicott J.H., Goldberg T.E., Kolachana B.S., Bertolino A., Zaitsev E., Gold B., Goldman D., Dean M., et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/S0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z.Y., Jing D., Bath K.G., Ieraci A., Khan T., Siao C.-J., Herrera D.G., Toth M., Yang C., McEwen B.S., et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su T.P., Hayashi T., Maurice T., Buch S., Ruoho A.E. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 2010;31:557–566. doi: 10.1016/j.tips.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malik M., Rangel-Barajas C., Sumien N., Su C., Singh M., Chen Z., Huang R.Q., Meunier J., Maurice T., Mach R.H., et al. The effects of sigma (σ1) receptor-selective ligands on muscarinic receptor antagonist-induced cognitive deficits in mice. Br. J. Pharmacol. 2015;172:2519–2531. doi: 10.1111/bph.13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francardo V., Bez F., Wieloch T., Nissbrandt H., Ruscher K., Cenci M.A. Pharmacological stimulation of sigma-1 receptors has neurorestorative effects in experimental parkinsonism. Brain. 2014;137:1998–2014. doi: 10.1093/brain/awu107. [DOI] [PubMed] [Google Scholar]
- 53.Ruscher K., Shamloo M., Rickhag M., Ladunga I., Soriano L., Gisselsson L., Toresson H., Ruslim-Litrus L., Oksenberg D., Urfer R., et al. The sigma-1 receptor enhances brain plasticity and functional recovery after experimental stroke. Brain. 2011;134:732–746. doi: 10.1093/brain/awq367. [DOI] [PubMed] [Google Scholar]
- 54.Yagasaki Y., Numakawa T., Kumamaru E., Hayashi T., Su T.P., Kunugi H. Chronic antidepressants potentiate via sigma-1 receptors the brain-derived neurotrophic factor-induced signaling for glutamate release. J. Biol. Chem. 2006;281:12941–12949. doi: 10.1074/jbc.M508157200. [DOI] [PubMed] [Google Scholar]
- 55.Penas C., Pascual-Font A., Mancuso R., Forés J., Casas C., Navarro X. Sigma receptor agonist 2-(4-morpholinethyl)1 phenylcyclohexanecarboxylate (Pre084) increases GDNF and BiP expression and promotes neuroprotection after root avulsion injury. J. Neurotrauma. 2011;28:831–840. doi: 10.1089/neu.2010.1674. [DOI] [PubMed] [Google Scholar]
- 56.Takebayashi M., Hayashi T., Su T., Unit C.P., Neurobiology C. Nerve growth factor-induced neurite sprouting in PC12 cells involves σ-1 receptors: Implications for antidepressants. Syst. Res. 2002;303:1227–1237. doi: 10.1124/jpet.102.041970. [DOI] [PubMed] [Google Scholar]
- 57.Takebayashi M., Hayashi T., Su T.P. σ-1 receptors potentiate epidermal growth factor signaling towards neuritogenesis in PC12 cells: Potential relation to lipid raft reconstitution. Synapse. 2004;53:90–103. doi: 10.1002/syn.20041. [DOI] [PubMed] [Google Scholar]
- 58.Dotti C.G., Sullivan C., Banker G. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Spronsen M., Van Battum E.Y., Kuijpers M., Vangoor V.R., Rietman M.L., Pothof J., Gumy L.F., Van Ijcken W.F., Akhmanova A., Pasterkamp R.J., et al. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS ONE. 2013;8:e74907. doi: 10.1371/journal.pone.0074907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahnert-Hilger G., Höltje M., Große G., Pickert G., Mucke C., Nixdorf-Bergweiler B., Boquet P., Hofmann F., Just I. Differential effects of Rho GTPases on axonal and dendritic development in hippocampal neurones. J. Neurochem. 2004;90:9–18. doi: 10.1111/j.1471-4159.2004.02475.x. [DOI] [PubMed] [Google Scholar]
- 61.Fujimura K., Matsumoto J., Niwa M., Kobayashi T., Kawashima Y., In Y., Ishida T. Synthesis, structure and quantitative structure-activity relationships of σ receptor ligands, 1-[2-(3,4-dimethoxyphenyl)ethyl]-4-(3-phenylpropyl)piperazines. Bioorg. Med. Chem. 1997;5:1675–1683. doi: 10.1016/S0968-0896(97)00093-X. [DOI] [PubMed] [Google Scholar]
- 62.Maurice T., Privat A. SA4503, a novel cognitive enhancer with σ1 receptor agonist properties, facilitates NMDA receptor-dependent learning in mice. Eur. J. Pharmacol. 1997;328:9–18. doi: 10.1016/S0014-2999(97)83020-8. [DOI] [PubMed] [Google Scholar]
- 63.Zou L.B., Yamada K., Sasa M., Nakata Y., Nabeshima T. Effects of σ1 receptor agonist SA4503 and neuroactive steroids on performance in a radial arm maze task in rats. Neuropharmacology. 2000;39:1617–1627. doi: 10.1016/S0028-3908(99)00228-2. [DOI] [PubMed] [Google Scholar]
- 64.Irwin S.A., Idupulapati M., Gilbert M.E., Harris J.B., Chakravarti A.B., Rogers E.J., Crisostomo R.A., Larsen B.P., Mehta A., Alcantara C.J., et al. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am. J. Med. Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- 65.Wiemerslage L., Lee D. Quantification of mitochondrial morphology in neurites of dopaminergic neurons using multiple parameters. J. Neurosci. Methods. 2016;262:56–65. doi: 10.1016/j.jneumeth.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]