Abstract
Sex hormones play important roles in the onset and progression of several cancers, such as breast, ovarian, and prostate cancer. Although drugs targeting sex hormone function are useful in treating cancer, tumors often develop resistance. Thus, we need to define the downstream effectors of sex hormones in order to develop new treatment strategies for these cancers. Recent studies unearthed one potential mediator of steroid hormone action in tumors: growth regulation by estrogen in breast cancer 1 (GREB1). GREB1 is an early estrogen-responsive gene, and its expression is correlated with estrogen levels in breast cancer patients. Additionally, GREB1 responds to androgen in prostate cancer cells, and can stimulate the proliferation of breast, ovarian, and prostate cancer cells. Recent studies have shown that GREB1 also responds to progesterone in human endometrial cells, suggesting that GREB1 is a pan steroid-responsive gene. This mini-review examines evidence that GREB1 participates in several hormone-dependent cancers and could be targeted to treat these cancers.
Keywords: GREB1, steroid hormone, estrogen, progesterone, androgen, breast cancer, ovarian cancer, prostate cancer
1. Hormone-Dependent Cancers
Steroid hormones are a selective branch of signaling molecules, which are biosynthesized from cholesterol derivatives in the mitochondria [1]. Steroid hormones are broadly classified into two subcategories based on their biosynthesis: corticosteroids and sex steroids [2]. Produced in the adrenal cortex, corticosteroids regulate a wide array of physiological processes, including immune, inflammatory, and stress responses. Notable corticosteroids include the glucocorticoid, hydrocortisone, mineralocorticoid, and aldosterone hormones. Produced in the gonads, hormones in the sex steroid class (i.e., estrogen, progesterone, and androgen) are vital for the development and maintenance of sexual characteristics as well as for general reproductive function [2]. Despite their role in normal reproductive development, sex steroids have also co-opted to play a role in the development and progression of several human cancers.
Sex-steroid hormones exert their physiologic and pathological function through their cognate receptors (steroid receptors), including estrogen receptors (ERs), androgen receptors (ARs), and progesterone receptors (PRs). All of these receptors belong to the nuclear receptor superfamily and consist of tissue-specific subtypes [3,4]. For example, produced differentially in various tissues, two distinct receptors, ERα and ERβ, exert distinct physiological and pathophysiological functions. ERα is the predominant receptor in the uterus, breasts, and ovaries, whereas ERβ plays significant roles in the bone, kidney, and cardiovascular systems [5]. Importantly, numerous findings revealed a key role for these hormones and their cognate receptors in human cancer onset and progression [4,6,7]. For example, the majority of breast cancer cell lines express the estrogen receptor [8,9,10], and the first-line therapy Tamoxifen selectively antagonizes ER [11,12]. Likewise, the risk of epithelial ovarian cancer is increased by long-term exposure to estrogen, as occurs in early menarche, late menopause, and in the context of hormone replacement therapy [13,14]. Indeed, one study of 394 serous epithelial ovarian cancer patients reported that 86% of tumors were ER-positive [15]. Moreover, when ER-positive epithelial ovarian cancer cells were implanted in the peritoneal cavity of mice, estrogen significantly increased tumorigenesis, promoted lymph node metastasis, and decreased survival [16,17]. Additionally, prostate cancer initiation and progression is regulated by androgen [18]. Thus, common treatment options for prostate cancer include orchiectomy and hormonal therapy, both of which function by restricting the androgen/androgen receptor interaction [4].
Although anti-hormonal therapeutics have been used to treat these hormone-dependent cancers, such drugs are often ineffective in the long term as the cancers develop drug resistance. For example, Tamoxifen, a selective estrogen receptor modulator (SERM), is widely used in breast cancer therapy [11]. However, about one-third of patients will acquire drug resistance and suffer from tumor recurrence [19]. Overcoming this challenge will require defining the mechanisms by which hormones drive these cancers and identifying new therapeutic targets.
2. Nuclear Receptor Coactivators in Hormone-Dependent Cancers
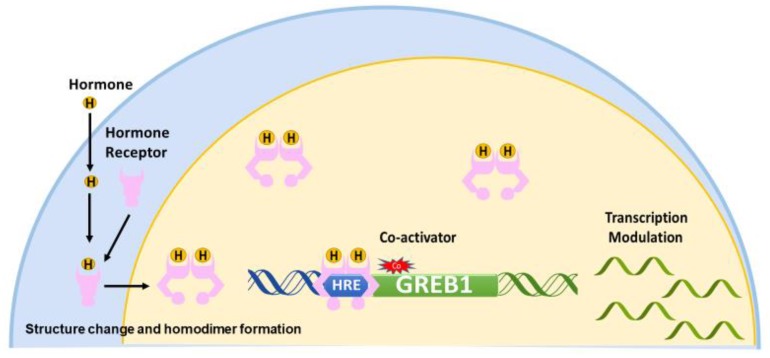
Nuclear receptors, including ER, AR, and PR, share a common mechanism of regulation. Upon the binding of a hormone to its cognate receptors, the receptor undergoes a conformational change to expose the hormone response element binding site and form a homodimer [5]. Upon interaction of the receptor with a coactivator, the homodimer binds to hormone response elements and modulates the transcription of hormone-dependent genes [3,20] (Figure 1).
Figure 1.
Model of the classic hormone-signaling pathway. The hormone enters the cell andbinds to a hormone receptor (HR). The arrows represent steps involved in activation of HR by its hormone. The HR forms a homodimer, undergoes a structural change to expose the hormone response element (HRE) binding domain, and enters the nucleus. Then, in conjunction with coactivators, HR binds an HRE to modulate the transcription of downstream genes such as GREB1. The arrows represent steps involved in activation of HR by its hormone.
The steroid receptor coactivator (SRC) family was the first identified coactivator class for nuclear receptors [21]. Three members, SRC-1, SRC-2, and SRC-3, are part of this key coactivator family. All three isoforms share conserved domains, including the N-terminal basic helix-loop-helix-Per/ARNT/Sim (b-HLH-PAS), the central nuclear receptor interacting domain, and two C-terminal activation domains, AD1 and AD2 [22]. The N-terminal b-HLH-PAS domain is highly conserved in the SRC family. The central nuclear receptor interacting domain contains three Leu-X-X-Leu-Leu (LXXLL) motifs, which are crucial for the coactivator’s function toward nuclear receptors. The X-ray crystal structure shows that LXXLL motifs have a two-turn α-helical conformation. The three conserved leucines align on the face of the helix and fit into the hydrophobic channels of the ligand-binding domain (LBD) on nuclear receptors [23]. The C-terminal activation domain can recruit different coactivators to modulate the transcription [22,24].
Recent advancements revealed an indispensable role for SRCs in steroid hormone-dependent functions in hormone-dependent carcinogenesis. In hormone-dependent cancers, all three members of the SRC family are expressed and are associated with prognosis [25,26,27]. In addition, SRC-1 and SRC-3 can stimulate cancer cell proliferation and promote cancer metastasis [28,29]. The expression of SRC-1 in breast cancer patients is associated with acquired drug resistance and relapse after endocrine therapy [30]. Likewise, in ovarian cancer, SRC-3 is elevated in high-grade ovarian epithelial cancer tissue [31]. SRC-3 considered as a poor prognostic factor and confers platinum resistance in ovarian epithelial cancer [32]. In endometrial cancers, the high expression of SRC-3 mRNA correlates with the advanced stage of cancer and poor prognosis [33]. Recent advancements highlighted a central role for SRC members in prostate cancer cells [34]. For example, all three family members were found to be essential for cancer cell survival and promote metastasis [35,36,37]. In contrast, the inhibition of the expression of SRC-3 can improve the sensitivity of human breast cancer cell line MDA-MB-231 to histone deacetylase inhibitors [38]. Further, the downregulation of SRC-3 reduces ovarian cancer cell migration and metastasis [31]. These aforementioned findings highlight an indispensable role for SRCs in hormone-dependent cancer onset and progression.
Interestingly, SRC-3 is reported to interact with growth regulation by estrogen in breast cancer 1 (GREB1) and increase estrogen-induced transcription in breast cancer cell line MCF-7 cells [39]. We found that GREB1 is transcriptionally activated by SRC-2 via the progesterone–PR axis in endometrium [40], and a novel fusion gene, GREB1-SRC-2, was detected in uterine sarcoma [41]. These evidences point out that GREB1 may play a vital role in hormone-dependent cancers. In this review, we will argue that GREB1 is a good candidate for development of therapeutics for breast, ovarian, prostate, and perhaps even uterine and testicular cancers.
3. GREB1 Structure and Function
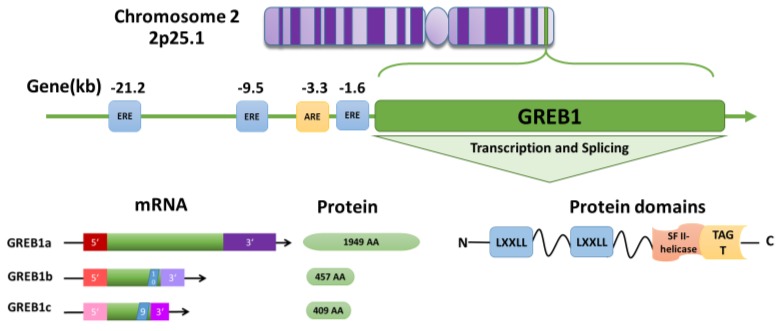
GREB1 was first discovered in brain tissue [42] and later identified as an early estrogen-responsive gene in breast cancer [43]. The GREB1 gene is highly conserved across species and is expressed in many tissues, including the brain [42], mammary gland [43], ovary [16], prostate [44], and endometrium [40]. In humans, the GREB1 gene is located at 2p25.1, spans 108.68 kb, and contains 38 exons and 40 introns [43,45] (Figure 2). GREB1 is expressed as three distinct isoforms: GREB1a, GREB1b, and GREB1c [43]. GREB1a is the full-length isoform, whereas variant splicing and the introduction of early termination codons result in GREB1b and GREB1c, which terminate after exons 10 and 9, respectively [46] (Figure 2).
Figure 2.
Model of growth regulation by estrogen in breast cancer 1 (GREB1) structure and transcription. The human GREB1 gene is located on 2p25.1. Three upstream estrogen response elements (EREs) located at −21.2, −9.5, and −1.6 kb and one androgen response element (ARE) located at −3.3 kb can modulate GREB1 transcription. The GREB1 gene produces at least three isoforms: GREB1a (NM_014668.3 → NP_055483.2), GREB1b (NM_033090.2 → NP_149081.1), and GREB1c (NM_148903.2 → NP_683701.2), which have alternate 5′ and 3′ UTRs. Splicing after exons 10 and 9 results in short GREB1b and GREB1c isoforms, respectively. The GREB1a protein contains two LXXLL motifs and a superfamily II (SFII) helicase module fused with a glycosyltransferase (TAGT) motif in the C-terminus.
Very little is known on the functional differences between these three isoforms. Much of our understanding of the GREB1 function comes from the analysis of its protein domains. At the C-terminus, GREB1 contains a circularly permuted superfamily II (SFII) helicase fused to a ten-eleven translocation/J binding protein (TET/JBP) and glycosyltransferase (TAGT) domain [47] (Figure 2). The SFII helicase is presumed to bind nucleic acids, but has no catalytic activity [47], and TAGTs can glycosylate pyrimidines in DNA [47]. The glycosylation is a key modification observed in different types of cancers, including breast, ovarian, and prostate cancers [48]. Glycosylation facilitates the cell adhesion, proliferation, and metastasis of breast cancer [49]. Six glycosylated genes—FUT1, FUCA1, POFUT1, MAN1A1, RPN1 and DPM1—are reported to be associated with the prognosis of breast cancer [50]. Further, glycosyltransferase gene expression is correlated with the prognosis of ER-positive breast cancer [51]. However, the importance of GREB1-mediated DNA modification is unknown. At the N-terminus, GREB1 contains two LXXLL motifs, which are commonly found in coactivators and mediate interactions with nuclear receptors [52,53] (Figure 2). However, the LXXLL motifs are not necessary for GREB1 to interact with ER [46], and although all three GREB1 isoforms bind to ER [53], they may not function as coactivators. Instead, exogenously expressed GREB1a can independently enhance the expression of ER target genes PS2 and SDF1, while exogenously expressed GREB1b had no effect [46]. These findings suggest that the GREB1 isoforms might have differential effects on ER driven gene expression.
4. Hormone Regulation of GREB1
The classic hormone-signaling pathway begins with a hormone binding to its cognate hormone receptor, causing the receptor to homodimerize [5] and undergo a structural change to expose the hormone response element binding site [54]. While the hormone-binding domain is located at the C-terminus, the hormone response element binding domain is in the central region of the receptor. The hormone-bound homodimer enters the nucleus where, in conjunction with a coactivator, it binds to hormone response elements in the upstream of GREB1 and then modulates the transcription of target genes [3,20] (Figure 1).
Several lines of evidence indicate that GREB1 is regulated by the hormones estrogen, progesterone, and androgen [54,55,56,57,58]. First, GREB1 expression was markedly elevated in the mammary tissue of female cynomolgus macaques during the pubertal stages, when estrogen levels were high, and decreased after menopause, when estrogen levels were low [56]. Second, in breast cancer patients, GREB1 expression correlated with estrogen levels [57]. Third, throughout women’s reproductive years, GREB1 expression in the endometrium fluctuates in accordance with estrogen levels [59]. Fourth, GREB1 expression increased during progesterone-mediated endometrial stromal cell decidualization, and progesterone receptor knockdown led to a decrease in GREB1 transcript levels [40]. Further, three estrogen response elements (EREs) [54,58] and at least one androgen response element (ARE) [44,53] are located upstream of the GREB1 transcription start site (Figure 2). Consistent with this, the expression of GREB1 correlated with the expression of hormone receptors in reproductive tissues in human (Table 1). The known functions of GREB1 in reproductive cancers will be detailed in the following sections of this review.
Table 1.
The Protein Expression of GREB1 and Hormone Receptor in Different Tissue Types. ER: Estrogen Receptor, AR: Androgen Receptor, PR: Progesterone Receptor.
| Tissue Type and Expression | GREB1 | ER | AR | PR | ||||
|---|---|---|---|---|---|---|---|---|
| Normal | Cancer | Normal | Cancer | Normal | Cancer | Normal | Cancer | |
| Breast | + | + | ER-α + | ER-α + | + | + | + | + |
| ER-β − | ER-β + | |||||||
| Ovary | + | + | ER-α − | ER-α + | + | + | − | + |
| ER-β − | ER-β + | |||||||
| Endometrium | + | + | ER-α + | ER-α + | + | + | + | + |
| ER-β − | ER-β + | |||||||
| Myometrium of Uterus | + | ? | ER-α + | ER-α ? | + | ? | + | ? |
| ER-β − | ER-β ? | |||||||
| Prostate | + | + | ER-α − | ER-α + | − | + | − | + |
| ER-β − | ER-β + | |||||||
| Testis | + | + | ER-α − | ER-α + | + | + | − | + |
| ER-β + | ER-β + | |||||||
+: expression; −: no expression; ?: not mentioned. (The expression of tissue is according to the data shown on the website of human protein atlas: https://www.proteinatlas.org/).
5. Role of GREB1 in Breast Cancer
Estrogen and progesterone play a vital role in mammary gland growth and cancer development [60]. Breast cancer is the most prevalent form of cancer in women [61]. In the mammary gland, ERα is the predominant subclass of ER, and is expressed in the majority of breast cancer cells. Estrogen is a key mediator in the development and progression of breast cancer [7], and several studies indicate that ER regulates GREB1 expression. Cancer cells expressing the ER are classified as ER-positive breast cancers [8]. In ER-positive breast cancers, the plasma level of estradiol and the expression of ERs are positively correlated with GREB1 expression [62]. The younger (age ≤ 45 years) group of breast cancer patients who had higher estrogen levels showed significantly higher GREB1 expression than the older (age ≥ 70 years) group [63]. Additionally, the majority of breast cancer cell lines, including MCF-7, T-47D, and BT-474 [9,10], are ER-positive and express GREB1 at high levels. Conversely, ER-negative breast cancer cell lines MDA-MB 231 and SUM 225 express GREB1 at lower levels [64]. GREB1 appears to help regulate the transcription of ER target genes, as the chromatin immunoprecipitation (ChIP) sequencing analysis of MCF-7 cells revealed that approximately 95% of the GREB1-bound regions were also bound by ER [53]. Upon GREB1 knockdown in MCF-7 cells, nearly half of the estrogen-responsive genes were no longer differentially expressed, and the cells were less able to form colonies [53]. To assess the in vivo effect of estrogen on GREB1 expression, Rae et al. ovariectomized athymic nude mice to prevent endogenous estrogen production and implanted MCF-7 cells in the peritoneal cavity [10]. As expected, the levels of GREB1 and estrogen were reduced upon ovariectomization. Conversely, GREB1 expression increased when exogenous estrogen was administered. Although estrogen treatment did not affect tumor size during the 48-hour treatment period [10], longer treatment could result in enhanced tumor growth, as suggested by in vitro work showing that estrogen induced the proliferation of MCF-7, BT-474, and T47D cell lines [10].
Further evidence for the role of GREB1 in breast cancer comes from studies of the cells’ responses to Tamoxifen. Breast cancer cells initially respond to Tamoxifen, which antagonizes the ER and leads to decreased GREB1 expression [65]. However, in Tamoxifen-resistant breast cancer tissues, the GREB1 promoter becomes more methylated via the action of the histone lysine N-methyltransferase EZH2, leading to the epigenetic downregulation of GREB1 [65].
6. Role of GREB1 in Ovarian Cancer
GREB1 also plays a significant role in ovarian cancer, which is a gynecologic malignancy that causes an estimated 12,700 deaths in the United States (US) alone [66]. Unlike breast tumors, where the tissue is easily accessible for diagnostic purposes, the ovary is located deep in the pelvic cavity. Therefore, the early detection of ovarian cancer proves difficult due to the subtle symptomology of cancer and poor screening methods [67]. As a result, nearly 75% of ovarian cancers are diagnosed only in later (advanced) stages of cancer [66,67]. Ovarian cancers are traditionally classified into three main categories with additional subtypes based on their histogenesis and subsequent differentiation [68]. Sex cord stromal tumors develop in the connective tissue of the ovaries and account for approximately 5% of ovarian cancers in women. Similarly, germ cell tumors account for about 5% of ovarian cancers, and are typically diagnosed in young women aged aroundearly 20 years. Accounting for almost 90% of cases, epithelial ovarian cancers are classified into several subtypes, of which serous tumors are the most notable [66]. A key study reported that 86% of serous epithelial ovarian tumors are ER-positive [15]. In vitro, estrogen can stimulate the proliferation of ER-positive ovarian cancer cell lines [69]. Several lines of evidence point to a role for GREB1 in the progress of epithelial ovarian cancer. In a tissue microarray analysis of ovarian cancer cases, GREB1 was expressed in 75–85% of epithelial ovarian cancer cells [55]. Tumor xenograft studies using mouse ovarian cancer ascites (MAS) cells with elevated GREB1 expression indicated an increase in tumor growth and reduced median survival time [16,55]. In vitro, GREB1 knocked down in MAS cells results in decreased proliferation and favorable morphologic changes [16,55]. These findings support a key role for GREB1 in estrogen-dependent actions in epithelial ovarian cancers.
7. Role of GREB1 in Prostate Cancer
Accounting for nearly 366,000 deaths globally per year, prostate cancer is the second most prevalent cancer in males in the US, and the majority of cases are adenocarcinomas [70,71]. Similar to the hormonal regulation of reproductive tract development and sexual function by estrogen in women, androgen controls the central reproductive function in males, including the prostate [72]. The prostate is an excretory gland that secretes prostate fluid, and the prostatic glands are androgen-dependent in nature. The initiation and progression of prostate cancer are regulated by androgen [73]. The common treatment options for prostate cancer include orchiectomy and hormonal-deprived therapy, both of which function by restricting androgen/androgen receptor (AR) functions [4]. Androgen can regulate GREB1 expression by binding to ARs, which then bind the ARE upstream of the GREB1 transcriptional start site (Figure 2) [44]. Rae et al. showed that GREB1 was highly expressed in benign prostatic hypertrophy, prostate cancer, and AR-positive prostatic carcinoma cell lines, and that GREB1 responded to androgen in a dose-dependent manner. Additionally, they reported that GREB1 knockdown reduced the proliferation of lymph node carcinoma of the prostate (LNCaP) cells [44]. Similarly, Ferreira, et al. found that treatment with the androgen dihydrotestosterone upregulated GREB1 expression in androgen-responsive LNCaP cells [74].
8. The Emerging Role for GREB1 in Other Hormone-Dependent Cancers
Although not as well studied as in breast, ovary, and prostate cancers, GREB1 may also play a role in uterine and testicular cancer. The uterus is composed of the endometrium and myometrium. Among women of reproductive age, the cyclical fluctuation of estrogen and progesterone in the endometrium promotes the monthly proliferation, differentiation, and shedding of tissue, which is commonly known as menstruation [72]. In the uterus, GREB1 is highly expressed in the endometrium [59]. Throughout women’s reproductive age, GREB1 expression fluctuates in accordance with estrogen levels [59]. Endometrial tissues under the long-term influence of estrogen are disposed to endometrial hyperplasia and endometrial adenocarcinoma. In addition to being regulated by estrogen, recent evidence indicates that GREB1 is regulated by progesterone in human endometrial cells [40]. GREB1 expression increased during progesterone-mediated endometrial stromal cell decidualization, and progesterone receptor knockdown led to a decrease in GREB1 transcript levels [40]. Given that endometrium exposed to estrogen in the absence of progesterone is prone to hyperplasia and carcinogenesis [72] and that GREB1 is regulated by both hormones, it seems likely that GREB1 contributes to endometrial cancer. One study reported a positive correlation between GREB1 expression and ERα activation in endometrial cancer [75]. Additionally, endometrial cancer appears to overlap genetically with endometriosis [76,77], and GREB1 mRNA levels are significantly higher in ectopic endometrium (endometriosis lesions) than in eutopic endometrium [78]. Furthermore, single nucleotide polymorphisms around the GREB1 locus have been identified in endometriosis patients [59,79,80,81]. Further, GREB1 polymorphic variants are associated with bone mineral density in a cohort of Caucasians [82]. Low bone mineral density is a critical manifestation of osteoporosis, and estrogen is known to control bone remodeling. Thus, future work should address the possibility that mutations in GREB1 predispose women to hormone-related diseases including endometriosis and postmenopausal osteoporosis.
GREB1 may also participate in uterine sarcomas, which constitute 8% of uterine corpus cancers [83] and are derived from myometrial, mesenchymal, or undifferentiated cells [84]. Brunetti et al. reported a translocation (2;8)(p25;q13) in a patient with undifferentiated uterine sarcoma and lung metastases. Transcriptome sequencing analysis revealed that this translocation created a novel GREB1–nuclear receptor coactivator 2 (NCOA2) fusion gene in which the first three exons of GREB1 (NM_014668.3) were fused to exon 15 of NCOA2 (NM_001321703.1) [41]. Given that NCOA2, which is also called SRC-2, regulates the progesterone-dependent transcriptional regulation of GREB1 in human endometrial cells [40], additional studies are required to determine the role of this fusion gene in endometrial cancer onset and progression.
The testes, which both produces gametes and secretes androgen [72] to maintain sexual characteristics [85], relies on androgen signaling for many of its functions. No work has been done to assess the role of GREB1 in testicular cancer, but some findings suggest that GREB1 participates in testicular functions [86,87]. The Sertoli cells, which establish the blood–testis barrier in the seminiferous tubules, express both androgen and estrogen receptors [88]. Two papers suggested that GREB1 is expressed in mouse Sertoli cells as a result of estrogen binding to the ER [86,87]. Given these findings, future work should be directed at assessing whether or not GREB1 participates in testicular functions and testicular cancer.
9. Translational Relevance
Understanding the underlying mechanism of hormone actions will offer novel diagnostic and/or treatment strategies in the clinical management of hormone-dependent cancers. Considering the role of GREB1 in hormone-dependent cancers, GREB1 should be considered as a diagnostic and therapeutic target. In breast cancer, Mohammed et al. found that the level of GREB1 was a favorable prognostic factor independent of the Nottingham prognostic index, which predicts breast cancer prognosis after surgery on the basis of lesion size, the number of involved lymph nodes, and tumor grade [53,89]. Similarly, in ovarian cancer, the hypomethylation of GREB1 was positively correlated with progression-free survival in ovarian cancer patients [90]. Additionally, a microarray analysis of prostate cancer samples from 33 patients showed that high GREB1 expression was associated with organ-confined disease, which is lower stage and a more favorable prognosis in prostate cancer [91]. It important to note that elevated expressions of GREB1 have been observed in hormone-dependent cancers, which are more sensitive to hormone therapy than receptor-negative tumors. Therefore, GREB1 expression in lower stage receptor-positive cancers might be more favorable toward prognosis. The majority of higher stage tumors from the aforementioned report [91] might be receptor-negative, which are resistant to therapy, and thus are not favorable for prognosis. Nonetheless, future work should conduct similar studies in larger cohorts of patients from multiple hormone-dependent cancers.
In addition to informing prognosis, the improved understanding of GREB1 may reveal novel strategies to combat drug resistance. In Tamoxifen-resistant breast cancer tissues, the GREB1 promoter becomes more methylated via the action of the histone lysine N-methyltransferase EZH2, leading to the epigenetic downregulation of GREB1 [65]. EZH2-mediated histone methylation drives the epigenetic programming in tumor cells, including breast, ovarian, and prostate cancers [92,93,94,95]. These epigenetic changes activate multiple oncogenes and promote tumor progression and metastasis [96]. Thus, it is plausible that GREB1 methylation confers drug resistance for hormone-dependent cancers. Based on the aforementioned studies, strategies to target GREB1 could lead to improved hormone-dependent cancer prognosis and treatment.
Interestingly, the expression analysis of GREB1 demonstrated in Oncomine, the Cancer Cell Line Encyclopedia (CCLE), and protein atlas indicate that GREB1 has much wider expression than in hormone-related organs or tissues. Generally, the expression level of GREB1 is much higher in pathologic status than in healthy status. Thus, a small molecular inhibitor or monoclonal antibody against GREB1 that abolishes the overexpressed level of GREB1 with minimal effect on physiological actions would be a viable option to target GREB1. Thus, an in-depth functional analysis of GREB1 in hormone-dependent cancers is warranted prior to targeting GREB1 in hormone-dependent cancers.
10. Conclusions and Open Questions
The pan hormone-responsive gene GREB1 plays important roles in the initiation and progression of some sex hormone-driven cancers. However, many open questions remain, such as: (1) Do GREB1 isoforms have differential actions in hormone-dependent cancers? If so, what are the underlying mechanisms? (2) Is GREB1 epigenetically regulated in cancer? (3) Does GREB1 function via common or hormone-specific functions? (4) Are mutations in GREB1 associated with the occurrence of hormone-dependent cancers? By addressing such questions, the field will be able to fully elucidate the normal functions of GREB1, define its role in cancers, and assess its potential as a prognostic and therapeutic target.
Acknowledgments
Meng Cheng is supported by a fellowship from the West China Second University Hospital, Sichuan University. We would like to thank Sangappa B. Chadchan, Deborah Frank, and InPrint editor Elizabeth Mueller (all at Washington University in St. Louis) for critical and editorial review of the text and figures.
Abbreviations
| AD | Activation Domain |
| AR | Androgen Receptors |
| ARE | Androgen Response Element |
| b-HLH-PAS | Basic Helix-Loop-Helix-Per/ARNT/Sim |
| CCLE | Cancer Cell Line Encyclopedia |
| ChIP | Chromatin Immunoprecipitation |
| ER | Estrogen Receptors |
| ERE | Estrogen Response Element |
| GREB1 | Growth Regulation by Estrogen in Breast Cancer 1 |
| HR | Hormone Receptors |
| HRE | Hormone Response Element |
| LBD | Ligand Binding Domain |
| LNCaP cell | Lymph Node Carcinoma of the Prostate cell |
| LXXLL | Leu-X-X-Leu-Leu |
| MAS cell | Mouse Ovarian Cancer Ascites Cell |
| NCOA2 | Nuclear Receptor Coactivator 2 |
| PR | Progesterone Receptors |
| SERM | Selective Estrogen Receptor Modulator |
| SF II | Superfamily II |
| SRC | Steroid Receptor Coactivator |
| TAGT | TET/JBP-Associated Glycosyltransferase |
| TET/JBP | Ten-Eleven Translocation/J Binding Protein |
Author Contributions
M.C. wrote the original draft. S.M. and R.K. reviewed and edited the manuscript. M.C. and R.K. created the figures. R.K. supervised the work.
Funding
This work is supported by funding from National Institutes of Health/National Institute of Child Health and Human Development grant R00 HD080742 and Washington University School of Medicine start-up funds to R.K.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Miller W.L., Bose H.S. Early steps in steroidogenesis: Intracellular cholesterol trafficking. J. Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marceau K., Ruttle P.L., Shirtcliff E.A., Essex M.J., Susman E.J. Developmental and contextual considerations for adrenal and gonadal hormone functioning during adolescence: Implications for adolescent mental health. Dev. Psychobiol. 2015;57:742–768. doi: 10.1002/dev.21214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nwachukwu J.C., Srinivasan S., Zheng Y., Wang S., Min J., Dong C., Liao Z., Nowak J., Wright N.J., Houtman R., et al. Predictive features of ligand-specific signaling through the estrogen receptor. Mol. Syst. Biol. 2016;12:864. doi: 10.15252/msb.20156701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan M.H., Li J., Xu H.E., Melcher K., Yong E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015;36:3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyster K.M. Estrogen Receptor. Springer; Hertfordshire, UK: 2016. [Google Scholar]
- 6.Chuffa L.G., Lupi-Junior L.A., Costa A.B., Amorim J.P., Seiva F.R. The role of sex hormones and steroid receptors on female reproductive cancers. Steroids. 2017;118:93–108. doi: 10.1016/j.steroids.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Ray S., Johnston R., Campbell D.C., Nugent S., McDade S.S., Waugh D., Panov K.I. Androgens and estrogens stimulate ribosome biogenesis in prostate and breast cancer cells in receptor dependent manner. Gene. 2013;526:46–53. doi: 10.1016/j.gene.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Dall G.V., Hawthorne S., Seyed-Razavi Y., Vieusseux J., Wu W., Gustafsson J.A., Byrne D., Murphy L., Risbridger G.P., Britt K.L. Estrogen receptor subtypes dictate the proliferative nature of the mammary gland. J. Endocrinol. 2018;237:323–336. doi: 10.1530/JOE-17-0582. [DOI] [PubMed] [Google Scholar]
- 9.Lin C.Y., Strom A., Vega V.B., Kong S.L., Yeo A.L., Thomsen J.S., Chan W.C., Doray B., Bangarusamy D.K., Ramasamy A., et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rae J.M., Johnson M.D., Scheys J.O., Cordero K.E., Larios J.M., Lippman M.E. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 11.Bogush T.A., Polezhaev B.B., Mamichev I.A., Bogush E.A., Polotsky B.E., Tjulandin S.A., Ryabov A.B. Tamoxifen Never Ceases to Amaze: New Findings on Non-Estrogen Receptor Molecular Targets and Mediated Effects. Cancer Investig. 2018;36:211–220. doi: 10.1080/07357907.2018.1453933. [DOI] [PubMed] [Google Scholar]
- 12.Singletary S.E., Allred C., Ashley P., Bassett L.W., Berry D., Bland K.I., Borgen P.I., Clark G., Edge S.B., Hayes D.F., et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J. Clin. Oncol. 2002;20:3628–3636. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen L.H., Schnack T.H., Buchardi K., Hummelshoj L., Missmer S.A., Forman A., Blaakaer J. Risk factors of epithelial ovarian carcinomas among women with endometriosis: A systematic review. Acta Obstet. Gynecol. Scand. 2017;96:761–778. doi: 10.1111/aogs.13010. [DOI] [PubMed] [Google Scholar]
- 14.Webb P.M., Jordan S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Shen F., Zhang X., Zhang Y., Ding J., Chen Q. Hormone receptors expression in ovarian cancer taking into account menopausal status: A retrospective study in Chinese population. Oncotarget. 2017;8:84019–84027. doi: 10.18632/oncotarget.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laviolette L.A., Hodgkinson K.M., Minhas N., Perez-Iratxeta C., Vanderhyden B.C. 17β-Estradiol upregulates GREB1 and accelerates ovarian tumor progression in vivo. Int. J. Cancer Suppl. 2014;135:1072–1084. doi: 10.1002/ijc.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spillman M.A., Manning N.G., Dye W.W., Sartorius C.A., Post M.D., Harrell J.C., Jacobsen B.M., Horwitz K.B. Tissue-specific pathways for estrogen regulation of ovarian cancer growth and metastasis. Cancer Res. 2010;70:8927–8936. doi: 10.1158/0008-5472.CAN-10-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attard G., Parker C., Eeles R.A., Schroder F., Tomlins S.A., Tannock I., Drake C.G., de Bono J.S. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 19.Ojo D., Wei F., Liu Y., Wang E., Zhang H., Lin X., Wong N., Bane A., Tang D. Factors Promoting Tamoxifen Resistance in Breast Cancer via Stimulating Breast Cancer Stem Cell Expansion. Curr. Med. Chem. 2015;22:2360–2374. doi: 10.2174/0929867322666150416095744. [DOI] [PubMed] [Google Scholar]
- 20.Deschenes J., Bourdeau V., White J.H., Mader S. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 2007;282:17335–17339. doi: 10.1074/jbc.C700030200. [DOI] [PubMed] [Google Scholar]
- 21.Onate S.A., Tsai S.Y., Tsai M.J., O’Malley B.W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 22.Dasgupta S., Lonard D.M., O’Malley B.W. Nuclear receptor coactivators: Master regulators of human health and disease. Annu. Rev. Med. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savkur R.S., Burris T.P. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 24.Johnson A.B., O’Malley B.W. Steroid receptor coactivators 1, 2, and 3: Critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol. Cell. Endocrinol. 2012;348:430–439. doi: 10.1016/j.mce.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Duan C., Bian C., Xiong Y., Zhang J. Steroid receptor coactivator-1: A versatile regulator and promising therapeutic target for breast cancer. J. Steroid Biochem. Mol. Biol. 2013;138:17–23. doi: 10.1016/j.jsbmb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Song X., Zhang C., Zhao M., Chen H., Liu X., Chen J., Lonard D.M., Qin L., Xu J., Wang X., et al. Steroid Receptor Coactivator-3 (SRC-3/AIB1) as a Novel Therapeutic Target in Triple Negative Breast Cancer and Its Inhibition with a Phospho-Bufalin Prodrug. PLoS ONE. 2015;10:e0140011. doi: 10.1371/journal.pone.0140011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenne I.S., Helland T., Flageng M.H., Dankel S.N., Mellgren G., Sagen J.V. Downregulation of steroid receptor coactivator-2 modulates estrogen-responsive genes and stimulates proliferation of MCF-7 breast cancer cells. PLoS ONE. 2013;8:e70096. doi: 10.1371/journal.pone.0070096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin L., Wu Y.L., Toneff M.J., Li D., Liao L., Gao X., Bane F.T., Tien J.C., Xu Y., Feng Z., et al. NCOA1 Directly Targets M-CSF1 Expression to Promote Breast Cancer Metastasis. Cancer Res. 2014;74:3477–3488. doi: 10.1158/0008-5472.CAN-13-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lydon J.P., O’Malley B.W. Minireview: Steroid receptor coactivator-3: A multifarious coregulator in mammary gland metastasis. Endocrinology. 2011;152:19–25. doi: 10.1210/en.2010-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleming F.J., Myers E., Kelly G., Crotty T.B., McDermott E.W., O’Higgins N.J., Hill A.D., Young L.S. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J. Clin. Pathol. 2004;57:1069–1074. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida H., Liu J., Samuel S., Cheng W., Rosen D., Naora H. Steroid receptor coactivator-3, a homolog of Taiman that controls cell migration in the Drosophila ovary, regulates migration of human ovarian cancer cells. Mol. Cell. Endocrinol. 2005;245:77–85. doi: 10.1016/j.mce.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri C., Gojis O., Rudraraju B., Stamp-Vincent C., Wilson D., Langdon S., Gourley C., Faratian D. Expression of steroid receptor coactivator 3 in ovarian epithelial cancer is a poor prognostic factor and a marker for platinum resistance. Br. J. Cancer. 2013;108:2039–2044. doi: 10.1038/bjc.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaguchi H., Fujimoto J., Sun W.S., Tamaya T. Clinical implications of steroid receptor coactivator (SRC)-3 in uterine endometrial cancers. J. Steroid Biochem. Mol. Biol. 2007;104:237–240. doi: 10.1016/j.jsbmb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Culig Z., Santer F.R. Studies on Steroid Receptor Coactivators in Prostate Cancer. Methods Mol. Biol. 2018;1786:259–262. doi: 10.1007/978-1-4939-7845-8_15. [DOI] [PubMed] [Google Scholar]
- 35.Dasgupta S., Putluri N., Long W., Zhang B., Wang J., Kaushik A.K., Arnold J.M., Bhowmik S.K., Stashi E., Brennan C.A., et al. Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. J. Clin. Investg. 2015;125:1174–1188. doi: 10.1172/JCI76029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agoulnik I.U., Vaid A., Bingman W.E., 3rd, Erdeme H., Frolov A., Smith C.L., Ayala G., Ittmann M.M., Weigel N.L. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 37.Tien J.C., Liu Z., Liao L., Wang F., Xu Y., Wu Y.L., Zhou N., Ittmann M., Xu J. The steroid receptor coactivator-3 is required for the development of castration-resistant prostate cancer. Cancer Res. 2013;73:3997–4008. doi: 10.1158/0008-5472.CAN-12-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Z., Luo X., Nie P., Wu B., Zhang T., Wei Y., Wang W., Geng G., Jiang J., Mi Y. Inhibition of SRC-3 enhances sensitivity of human cancer cells to histone deacetylase inhibitors. Biochem. Biophys. Res. Commun. 2016;478:227–233. doi: 10.1016/j.bbrc.2016.07.063. [DOI] [PubMed] [Google Scholar]
- 39.Panigrahi A.K., Foulds C.E., Lanz R.B., Hamilton R.A., Yi P., Lonard D.M., Tsai M.J., Tsai S.Y., O’Malley B.W. SRC-3 Coactivator Governs Dynamic Estrogen-Induced Chromatin Looping Interactions during Transcription. Mol. Cell. 2018;70:679–694. doi: 10.1016/j.molcel.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camden A.J., Szwarc M.M., Chadchan S.B., DeMayo F.J., O’Malley B.W., Lydon J.P., Kommagani R. Growth regulation by estrogen in breast cancer 1 (GREB1) is a novel progesterone-responsive gene required for human endometrial stromal decidualization. Mol. Hum. Reprod. 2017;23:646–653. doi: 10.1093/molehr/gax045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunetti M., Panagopoulos I., Gorunova L., Davidson B., Heim S., Micci F. RNA-sequencing identifies novel GREB1-NCOA2 fusion gene in a uterine sarcoma with the chromosomal translocation t(2;8)(p25;q13) Gene. Chromosome Cancer. 2018;57:176–181. doi: 10.1002/gcc.22518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagase T., Ishikawa K., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh M.G., Thompson D.A., Weigel R.J. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 44.Rae J.M., Johnson M.D., Cordero K.E., Scheys J.O., Larios J.M., Gottardis M.M., Pienta K.J., Lippman M.E. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate. 2006;66:886–894. doi: 10.1002/pros.20403. [DOI] [PubMed] [Google Scholar]
- 45.Gene-NCBI. [(accessed on 20 May 2018)]; Available online: https://www.ncbi.nlm.nih.gov/gene/
- 46.Haines C.N., Braunreiter K.M., Mo X.M., Burd C.J. GREB1 isoforms regulate proliferation independent of ERalpha co-regulator activities in breast cancer. Endocr. Relat. Cancer. 2018;25:735–746. doi: 10.1530/ERC-17-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer L.M., Zhang D., Burroughs A.M., Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drake R.R. Glycosylation and cancer: Moving glycomics to the forefront. Adv. Cancer Res. 2015;126:1–10. doi: 10.1016/bs.acr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Singh C., Shyanti R.K., Singh V., Kale R.K., Mishra J.P.N., Singh R.P. Integrin expression and glycosylation patterns regulate cell-matrix adhesion and alter with breast cancer progression. Biochem. Biophys. Res. Commun. 2018;499:374–380. doi: 10.1016/j.bbrc.2018.03.169. [DOI] [PubMed] [Google Scholar]
- 50.Milde-Langosch K., Karn T., Schmidt M., Zu Eulenburg C., Oliveira-Ferrer L., Wirtz R.M., Schumacher U., Witzel I., Schutze D., Muller V. Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res. Treat. 2014;145:295–305. doi: 10.1007/s10549-014-2949-z. [DOI] [PubMed] [Google Scholar]
- 51.Ashkani J., Naidoo K.J. Glycosyltransferase Gene Expression Profiles Classify Cancer Types and Propose Prognostic Subtypes. Sci. Rep. 2016;6:26451. doi: 10.1038/srep26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plevin M.J., Mills M.M., Ikura M. The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 2005;30:66–69. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 53.Mohammed H., D’Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L., et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun J., Nawaz Z., Slingerland J.M. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 2007;21:2651–2662. doi: 10.1210/me.2007-0082. [DOI] [PubMed] [Google Scholar]
- 55.Hodgkinson K., Forrest L.A., Vuong N., Garson K., Djordjevic B., Vanderhyden B.C. GREB1 is an estrogen receptor-regulated tumour promoter that is frequently expressed in ovarian cancer. Oncogene. 2018 doi: 10.1038/s41388-018-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stute P., Sielker S., Wood C.E., Register T.C., Lees C.J., Dewi F.N., Williams J.K., Wagner J.D., Stefenelli U., Cline J.M. Life stage differences in mammary gland gene expression profile in non-human primates. Breast Cancer Res. Treat. 2012;133:617–634. doi: 10.1007/s10549-011-1811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haakensen V.D., Bjoro T., Luders T., Riis M., Bukholm I.K., Kristensen V.N., Troester M.A., Homen M.M., Ursin G., Borresen-Dale A.L., et al. Serum estradiol levels associated with specific gene expression patterns in normal breast tissue and in breast carcinomas. BMC Cancer. 2011;11:332. doi: 10.1186/1471-2407-11-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourdeau V., Deschenes J., Metivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J.H., Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 59.Fung J.N., Holdsworth-Carson S.J., Sapkota Y., Zhao Z.Z., Jones L., Girling J.E., Paiva P., Healey M., Nyholt D.R., Rogers P.A., et al. Functional evaluation of genetic variants associated with endometriosis near GREB1. Hum. Rep. 2015;30:1263–1275. doi: 10.1093/humrep/dev051. [DOI] [PubMed] [Google Scholar]
- 60.Kariagina A., Xie J., Leipprandt J.R., Haslam S.Z. Amphiregulin mediates estrogen, progesterone, and EGFR signaling in the normal rat mammary gland and in hormone-dependent rat mammary cancers. Horm. Cancer. 2010;1:229–244. doi: 10.1007/s12672-010-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Razzaghi H., Quesnel-Crooks S., Sherman R., Joseph R., Kohler B., Andall-Brereton G., Ivey M.A., Edwards B.K., Mery L., Gawryszewski V., et al. Leading Causes of Cancer Mortality—Caribbean Region, 2003–2013. Morb. Mortal. Wkly. Rep. 2016;65:1395–1400. doi: 10.15585/mmwr.mm6549a3. [DOI] [PubMed] [Google Scholar]
- 62.Dunbier A.K., Anderson H., Ghazoui Z., Folkerd E.J., A’Hern R., Crowder R.J., Hoog J., Smith I.E., Osin P., Nerurkar A., et al. Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor-positive breast cancer in postmenopausal women. J. Clin. Oncol. 2010;28:1161–1167. doi: 10.1200/JCO.2009.23.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yau C., Fedele V., Roydasgupta R., Fridlyand J., Hubbard A., Gray J.W., Chew K., Dairkee S.H., Moore D.H., Schittulli F., et al. Aging impacts transcriptomes but not genomes of hormone-dependent breast cancers. Breast Cancer Res. 2007;9:R59. doi: 10.1186/bcr1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hnatyszyn H.J., Liu M., Hilger A., Herbert L., Gomez-Fernandez C.R., Jorda M., Thomas D., Rae J.M., El-Ashry D., Lippman M.E. Correlation of GREB1 mRNA with protein expression in breast cancer: Validation of a novel GREB1 monoclonal antibody. Breast Cancer Res. Treat. 2010;122:371–380. doi: 10.1007/s10549-009-0584-x. [DOI] [PubMed] [Google Scholar]
- 65.Wu Y., Zhang Z., Cenciarini M.E., Proietti C.J., Amasino M.F., Hong T., Yang M., Liao Y., Chiang H.C., Kaklamani V., et al. Tamoxifen resistance in breast cancer is regulated by the EZH2-ERalpha-GREB1 transcriptional axis. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peres L.C., Cushing-Haugen K.L., Kobel M., Harris H.R., Berchuck A., Rossing M.A., Schildkraut J.M., Doherty J.A. Invasive Epithelial Ovarian Cancer Survival by Histotype and Disease Stage. J. Natl. Cancer Inst. 2018 doi: 10.1093/jnci/djy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.National Collaborating Centre for Cancer (UK) Ovarian Cancer: The Recognition and Initial Management of Ovarian Cancer. National Collaborating Centre for Cancer; Cardiff, UK: 2011. [PubMed] [Google Scholar]
- 68.Committee on the State of the Science in Ovarian Cancer Research. Board on Health Care Services. Institute of Medicine. National Academies of Sciences and Medicine . Ovarian Cancers: Evolving Paradigms in Research and Care. National Academies Press (US); Washington, DC, USA: 2016. [PubMed] [Google Scholar]
- 69.Langdon S.P., Hirst G.L., Miller E.P., Hawkins R.A., Tesdale A.L., Smyth J.F., Miller W.R. The regulation of growth and protein expression by estrogen in vitro: A study of 8 human ovarian carcinoma cell lines. J. Steroid Biochem. Mol. Biol. 1994;50:131–135. doi: 10.1016/0960-0760(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 70.Brawley O.W. Prostate cancer epidemiology in the United States. World J. Urol. 2012;30:195–200. doi: 10.1007/s00345-012-0824-2. [DOI] [PubMed] [Google Scholar]
- 71.Pernar C.H., Ebot E.M., Wilson K.M., Mucci L.A. The Epidemiology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2003;30:209–217. doi: 10.1101/cshperspect.a030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephen Nussey S.W. Endocrinology: An Integrated Approach. BIOS Scientific Publishers; Oxford, UK: 2001. [PubMed] [Google Scholar]
- 73.Banerjee P.P., Banerjee S., Brown T.R., Zirkin B.R. Androgen action in prostate function and disease. Am. J. Clin. Exp. Urol. 2018;6:62–77. [PMC free article] [PubMed] [Google Scholar]
- 74.Ferreira L.B., Palumbo A., de Mello K.D., Sternberg C., Caetano M.S., de Oliveira F.L., Neves A.F., Nasciutti L.E., Goulart L.R., Gimba E.R. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer. 2012;12:507. doi: 10.1186/1471-2407-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang W., Chen J.H., Aguilera-Barrantes I., Shiau C.W., Sheng X., Wang L.S., Stoner G.D., Huang Y.W. Urolithin A suppresses the proliferation of endometrial cancer cells by mediating estrogen receptor-alpha-dependent gene expression. Mol. Nutr. Food Res. 2016;60:2387–2395. doi: 10.1002/mnfr.201600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Painter J.N., O’Mara T.A., Morris A.P., Cheng T.H.T., Gorman M., Martin L., Hodson S., Jones A., Martin N.G., Gordon S., et al. Genetic overlap between endometriosis and endometrial cancer: Evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med. 2018;7:1978–1987. doi: 10.1002/cam4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anglesio M.S., Papadopoulos N., Ayhan A., Nazeran T.M., Noe M., Horlings H.M., Lum A., Jones S., Senz J., Seckin T., et al. Cancer-Associated Mutations in Endometriosis without Cancer. N. Engl. J. Med. 2017;376:1835–1848. doi: 10.1056/NEJMoa1614814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pellegrini C., Gori I., Achtari C., Hornung D., Chardonnens E., Wunder D., Fiche M., Canny G.O. The expression of estrogen receptors as well as GREB1, c-MYC, and cyclin D1, estrogen-regulated genes implicated in proliferation, is increased in peritoneal endometriosis. Fertil. Steril. 2012;98:1200–1208. doi: 10.1016/j.fertnstert.2012.06.056. [DOI] [PubMed] [Google Scholar]
- 79.Sapkota Y., Fassbender A., Bowdler L., Fung J.N., Peterse D., O D., Montgomery G.W., Nyholt D.R., D’Hooghe T.M. Independent Replication and Meta-Analysis for Endometriosis Risk Loci. Twin Res. Hum. Genet. 2015;18:518–525. doi: 10.1017/thg.2015.61. [DOI] [PubMed] [Google Scholar]
- 80.Sapkota Y., Vivo I., Steinthorsdottir V., Fassbender A., Bowdler L., Buring J.E., Edwards T.L., Jones S., O D., Peterse D., et al. Analysis of potential protein-modifying variants in 9000 endometriosis patients and 150000 controls of European ancestry. Sci. Rep. 2017;7:11380. doi: 10.1038/s41598-017-10440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rahmioglu N., Nyholt D.R., Morris A.P., Missmer S.A., Montgomery G.W., Zondervan K.T. Genetic variants underlying risk of endometriosis: Insights from meta-analysis of eight genome-wide association and replication datasets. Hum. Reprod. Update. 2014;20:702–716. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hegarty K.G., Drummond F.J., Daly M., Shanahan F., Molloy M.G. GREB1 genetic variants are associated with bone mineral density in Caucasians. J. Bone Miner. Metab. 2018;36:189–199. doi: 10.1007/s00774-017-0823-x. [DOI] [PubMed] [Google Scholar]
- 83.Cronin K.A., Lake A.J., Scott S., Sherman R.L., Noone A.M., Howlader N., Henley S.J., Anderson R.N., Firth A.U., Ma J., et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer. 2018 doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chern J.Y., Boyd L.R., Blank S.V. Uterine Sarcomas: The Latest Approaches for These Rare but Potentially Deadly Tumors. Oncology. 2017;31:229–236. [PubMed] [Google Scholar]
- 85.Shores M.M., Matsumoto A.M. Testosterone, aging and survival: Biomarker or deficiency. Curr. Opin. Endocrinol. Diabetes Obes. 2014;21:209–216. doi: 10.1097/MED.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin J., Zhu J., Li X., Li S., Lan Z., Ko J., Lei Z. Expression of genomic functional estrogen receptor 1 in mouse sertoli cells. Reprod. Sci. 2014;21:1411–1422. doi: 10.1177/1933719114527355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin J., Lei Z. Chromatin Immunoprecipitation with Estrogen Receptor 1 and the Promoter of Greb1 in TM4 Sertoli Cells. Methods Mol. Biol. 2016;1366:67–77. doi: 10.1007/978-1-4939-3127-9_7. [DOI] [PubMed] [Google Scholar]
- 88.Sneddon S.F., Walther N., Saunders P.T. Expression of androgen and estrogen receptors in sertoli cells: Studies using the mouse SK11 cell line. Endocrinology. 2005;146:5304–5312. doi: 10.1210/en.2005-0914. [DOI] [PubMed] [Google Scholar]
- 89.Fong Y., Evans J., Brook D., Kenkre J., Jarvis P., Gower-Thomas K. The Nottingham Prognostic Index: Five- and ten-year data for all-cause survival within a screened population. Ann. R. Coll. Surg. Engl. 2015;97:137–139. doi: 10.1308/003588414X14055925060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bauerschlag D.O., Ammerpohl O., Brautigam K., Schem C., Lin Q., Weigel M.T., Hilpert F., Arnold N., Maass N., Meinhold-Heerlein I., et al. Progression-free survival in ovarian cancer is reflected in epigenetic DNA methylation profiles. Oncology. 2011;80:12–20. doi: 10.1159/000327746. [DOI] [PubMed] [Google Scholar]
- 91.Antunes A.A., Leite K.R., Reis S.T., Sousa-Canavez J.M., Camara-Lopes L.H., Dall’oglio M.F., Srougi M. GREB1 tissue expression is associated with organ-confined prostate cancer. Urol. Oncol. 2012;30:16–20. doi: 10.1016/j.urolonc.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 92.Simon J.A., Lange C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 93.Wu J., Crowe D.L. The histone methyltransferase EZH2 promotes mammary stem and luminal progenitor cell expansion, metastasis and inhibits estrogen receptor-positive cellular differentiation in a model of basal breast cancer. Oncol. Rep. 2015;34:455–460. doi: 10.3892/or.2015.4003. [DOI] [PubMed] [Google Scholar]
- 94.Yang Y.A., Yu J. EZH2, an epigenetic driver of prostate cancer. Protein Cell. 2013;4:331–341. doi: 10.1007/s13238-013-2093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones B.A., Varambally S., Arend R.C. Histone Methyltransferase EZH2: A Therapeutic Target for Ovarian Cancer. Mol. Cancer Ther. 2018;17:591–602. doi: 10.1158/1535-7163.MCT-17-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsang D.P., Cheng A.S. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J. Gastroenterol. Hepatol. 2011;26:19–27. doi: 10.1111/j.1440-1746.2010.06447.x. [DOI] [PubMed] [Google Scholar]