Abstract
Smoking is a major risk factor for cardiovascular diseases and has been implicated in the regulation of the G protein-coupled receptor 15 (GPR15) by affecting CpG methylation. The G protein-coupled receptor 15 is involved in angiogenesis and inflammation. An effect on GPR15 gene regulation has been shown for the CpG site CpG3.98251294. We aimed to analyze the effect of smoking on GPR15 expression and methylation sites spanning the GPR15 locus. DNA methylation of nine GPR15 CpG sites was measured in leukocytes from 1291 population-based individuals using the EpiTYPER. Monocytic GPR15 expression was measured by qPCR at baseline and five-years follow up. GPR15 gene expression was upregulated in smokers (beta (ß) = −2.699, p-value (p) = 1.02 × 10−77) and strongly correlated with smoking exposure (ß = −0.063, p = 2.95 × 10−34). Smoking cessation within five years reduced GPR15 expression about 19% (p = 9.65 × 10−5) with decreasing GPR15 expression over time (ß = 0.031, p = 3.81 × 10−6). Additionally, three novel CpG sites within GPR15 affected by smoking were identified. For CpG3.98251047, DNA methylation increased steadily after smoking cessation (ß = 0.123, p = 1.67 × 10−3) and strongly correlated with changes in GPR15 expression (ß = 0.036, p = 4.86 × 10−5). Three novel GPR15 CpG sites were identified in relation to smoking and GPR15 expression. Our results provide novel insights in the regulation of GPR15, which possibly linked smoking to inflammation and disease progression.
Keywords: GPR15, smoking, biomarker, DNA methylation
1. Introduction
Cigarette smoking severely increases the risk for life-threatening diseases such as cardiovascular diseases, cancer, and chronic respiratory diseases with over 7 million deaths attributed to tobacco worldwide [1]. Cigarette smoke contains more than 5000 compounds with many of them, upon entering the blood stream, negatively affecting organs and cells [2]. However, the exact molecular mechanisms showing how tobacco substances influence disease onset and progression remain elusive.
Within the last several years, the field of epigenetics has emerged and has provided new approaches and insights into the regulation of gene expression. It was shown that the modification of a cytosine followed by a guanine nucleotide (CpG) to 5-methylcytosine in the gene promoter region may result in a decrease of transcriptional activity of the corresponding gene [3]. Several studies have been published by focusing on the effect of cigarette smoke on methylation and changes in methylation status in response to smoking have been shown for several genes such as AHRR and F2RL3 [4,5]. These genes have been implicated in cell proliferation, immune response, and detoxification, which can contribute to the pathogenesis of smoking-associated diseases [6].
The CpG site CpG3.98251294 was identified in relation to smoking within the GPR15 gene, which encodes the G protein-coupled receptor 15 [7,8,9]. Smoking was associated with decreased CpG3.98251294 methylation and increased G protein-coupled receptor 15 gene (GPR15) expression [8,9,10,11]. This effect was slowly reversible after smoking cessation [7,11,12]. Additionally, CpG3.98251294 methylation correlated with increased cumulative smoking exposure [13]. The transmembrane receptor GPR15 is mainly expressed on T cells and acts as a coreceptor for human immunodeficiency virus and simian immunodeficiency virus [14,15,16]. The G protein-coupled receptor 15 has also been shown to be involved in angiogenesis and inflammation [17,18,19,20,21,22], which indicated a role of GPR15 in smoking-associated diseases such as cardiovascular diseases and cancer. Hence, GPR15 poses an interesting new candidate gene of smoking-related diseases and smoking-induced molecular pathways. However, to date, the influence of smoking on only one CpG site (CpG3.98251294) within the GPR15 gene region has been described [7,8,9,10,11,13].
The aim of the present study was to analyze the association between smoking, GPR15 methylation and expression and to gain further knowledge of additional DNA methylation sites spanning the GPR15 locus. This study identified three novel DNA methylation sites within the GPR15 locus related to smoking as well as GPR15 expression.
2. Materials and Methods
2.1. Study Participants
In this study, 1291 individuals in the population-based Gutenberg Health Study were analyzed [23]. RNA and DNA were collected, as described previously [24]. Self-reported smoking statuses were classified as follows: current smokers (including occasional smokers), ex smokers (smoking cessation at least six weeks before study participation), and never smokers. Cumulative smoking exposure was evaluated by pack years (one pack year = smoking of 20 cigarettes per day for one year) for current smokers and ex smokers. For longitudinal gene expression (follow up), participants visited the same study center five years after baseline recruitment. Written informed consent was obtained from all study participants. The study protocols and sampling design were approved by the local ethics committee of the Medical Chamber of Rhineland-Palatinate, Germany (ethical approval code 837.020.07 (5555)).
2.2. RNA and DNA Isolation
Monocytic RNA was isolated as described previously [24]. Blood was collected using the Vacutainer CPT Cell Preparation Tube System (BD Biosciences, San Jose, CA, USA) and monocytes were enriched by negative selection with the Rosette Sep Monocyte Enrichment Cocktail (StemCell Technologies, Vancouver, BC, Canada), which leads to 72% to 85% of enriched monocytes. Total RNA was isolated using Trizol extraction and purification by silica-based columns. Genomic DNA from leukocytes was extracted as described by Miller et al. from buffy-coated ethylenediamine tetraacetic acid blood samples [24,25].
2.3. Analysis of GPR15 Gene Expression
Monocytic GPR15 mRNA expression was measured by real-time quantitative polymerase chain reaction (qPCR) using the 7900 TaqMan system (Applied Biosystems, Vancouver, BC, Canada). RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to manufacturers’ protocols. For GPR15 gene expression analyses, the GPR15 Hs00922903_s1 gene expression assay was used (Applied Biosystems). Quantification of the housekeeping gene GAPDH (Hs99999905_m1, Applied Biosystems) as an internal control was performed for each sample. Expression of GPR15 mRNA was normalized to GAPDH mRNA expression by calculating ΔCt values.
2.4. Analysis of DNA Methylation
Analysis of GPR15 DNA methylation was carried out with the EpiTYPER MassARRAY technology (Agena Bioscience, San Diego, CA, USA), according to the manufacturer’s protocols, which allowed region-specific quantification of CpG sites [26]. Genomic DNA was bisulfite modified to convert un-methylated cytosines into uracils. One μg of DNA was treated with sodium bisulfite using the EZ methylation kit (Zymo Research, Irvine, CA, USA), according to the manufacturers’ protocols. Ten ng of bisulfite DNA was amplified by polymerase chain reaction (PCR) using the following primers: 5′-aggaagagagGTTTTTTGGTGATGGATTTAGAAGA-3′ and 5′-cagtaatacgactcactatagggag aaggctTAAACAAAAAAATAAACAACCCCAA-3′, which results in a 523 bp-fragment. Subsequently, DNA was reverse transcribed, fragmented, and analyzed by mass spectrometry. The GPR15 gene includes 15 CpG sites. Nine of these CpG sites (CpG3.98250924 to CpG3.98251294) were located within the 523 bp-fragment and were labeled, according to Saffery et al. [27]. Six of these CpG sites could be analyzed. Due to the low mass of the cleavage product, which leads to unreliable methylation values, CpG3.98250924 and CpG3.98251081 could not be covered by the assay. CpG3.98251268 was not included in the analyses since it was fully methylated in 90% of the samples. CpG3.98251294 corresponds to the previously described cg19859270 site. The GPR15 locus including the CpG sites CpG3.98250924 to CpG3.98251294 is depicted in Figure S1. More detailed information on the mass fragments detected by the EpiTYPER are given in Table S1.
2.5. Statistical Analysis
Associations between cigarette smoking status, GPR15 mRNA expression, and GPR15 DNA methylation were calculated by linear mixed regression models and adjusted for age and sex. Changes in GPR15 gene expression between the baseline and a five-years follow up visit were analyzed by a paired t-test. The threshold for statistical significance was set at p-value (p ≤ 0.05). Bonferroni correction was performed for linear mixed regression models to adjust for multiple testing. Statistical analyses were performed and figures were prepared using the R version 3.4.3 [28] and GraphPad prism version 6.05 for Windows (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Characteristics of the Study Population
Baseline characteristics of the study population are provided in Table 1. Out of the 1291 individuals, 46% were never smokers, 37% were ex smokers, and 17% were current smokers. Smoking cessation was 18.6 years for ex smokers when calculating the mean value. Current smokers differed from never smokers in terms of sex (8% women compared to 27%) and hypertension (6% compared to 23%).
Table 1.
Characteristics of study individuals.
| Characteristic | All | Never Smokers | Ex Smokers | Current Smokers |
|---|---|---|---|---|
| n (%) | 1291 | 593 (46) | 477 (37) | 221 (17) |
| Females, n (%) | 643 (50) | 354 (27) | 187 (15) | 102 (8) |
| Age, years | 55 (46–64) | 57 (45–66) | 57 (48–65) | 50 (45–57) |
| Time since quitting, years | 18 (7–28) | NA | 18 (7–28) | NA |
| Pack years | 0.1 (0.0–3.6) | NA | 1.6 (0.6–3.7) | 23.1 (11.7–36.0) |
Continuous variables are described by median values (25th percentile to 75th percentile). Dichotomous variables are presented as total numbers (%). NA = data not available.
3.2. Smoking Increases GPR15 mRNA Expression
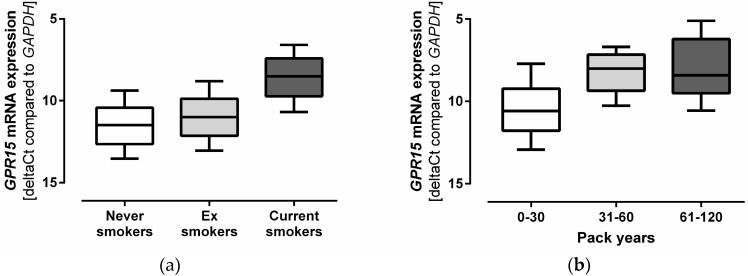
Gene expression was measured to determine the effect of smoking status on GPR15 expression. GPR15 mRNA expression strongly correlated with current smoking (beta (ß) = −2.699, p = 1.02 × 10−77) and was 7.6-fold higher in current smokers when compared to never smokers (Figure 1a). Furthermore, the effect of cumulative smoking exposure as indicated by the number of pack years significantly correlated with GPR15 mRNA expression levels (ß = −0.0631, p = 5.95 × 10−34) (Figure 1b).
Figure 1.
Smoking increases G protein-coupled receptor 15 (GPR15) mRNA expression. (a) GPR15 mRNA expression is depicted for never smokers, ex smokers, and current smokers. It was significantly associated with current smoking (beta (ß) = −2.699, p-value (p) = 1.02 × 10−77). n = 191–542; (b) GPR15 mRNA expression levels depending on cumulative smoking exposure are plotted for current smokers and ex smokers combined. They correlated significantly with the number of pack years smoked (ß = –0.0631, p = 5.95 × 10−34). One pack equals smoking 20 cigarettes per day for one year. n = 6-509. The linear mixed regression model was adjusted for age and sex. Box plot whiskers represent 10–90th percentile. GPR15 mRNA expression is depicted as ΔCt values normalized to GAPDH mRNA expression. Lower ΔCt values indicate higher GPR15 mRNA expression.
3.3. Smoking Cessation Alters GPR15 Expression
To determine the longitudinal effect of smoking behavior on GPR15 expression, gene expression levels at baseline and after a follow up time of five years were compared. Smokers who quit smoking within five years (n = 39) showed a significant decrease in GPR15 expression (ß = 1.182, p = 9.65 × 10−5) in Figure 2a, which indicated that the change in smoking behavior affected GPR15 mRNA expression. Furthermore, the time since smoking cessation significantly correlated with GPR15 mRNA expression (ß = 0.031, p = 3.81 × 10−6) (Figure 2b), which shows that, within the first years after smoking cessation, GPR15 mRNA expression decreased more rapidly (Figure S2).
Figure 2.
Effect of smoking cessation on G protein-coupled receptor 15 (GPR15) mRNA expression. (a) GPR15 mRNA expression was compared between baseline (BL) and the five-years follow up (FU) visit. Quitting smoking significantly decreased GPR15 mRNA expression about 19% (p) = 9.65 × 10−5). n = 39, paired t-test. (b) Time course of GPR15 mRNA expression after smoking cessation. Time since smoking cessation significantly correlated with GPR15 mRNA expression (ß = 0.031, p = 3.81 × 10−6). n = 20–186, linear mixed regression model adjusted for age and sex. Box plot whiskers represent 10–90th percentile. GPR15 mRNA expression is depicted as ΔCt values normalized to GAPDH mRNA expression. Lower ΔCt values indicate higher GPR15 mRNA expression.
3.4. Novel GPR15 DNA Methylation Sites Associated with Smoking
In previous studies, only the GPR15 CpG3.98251294 site had been described in relation to smoking and GPR15 expression [7,8,9,10,11,13]. To enhance the understanding of the molecular mechanisms regulating GPR15 expression and to identify new GPR15 CpG sites in relation to smoking, DNA methylation of nine additional CpG sites within the GPR15 gene was measured.
Out of nine sites, DNA methylation of CpG3.98251047, CpG3.98251179, and CpG3.98251219 showed strong associations with smoking (ß = −3.376, p = 1.37 × 10−6; ß = −4.655, p = 1.78 × 10−4; and ß = −3.609, p = 2.24 × 10−3, respectively) (Figure 3). For sites CpG3.98251063, CpG3.98251070, and CpG3.98251294, no association with the smoking status was identified in our analyses.
Figure 3.
Decreased G protein-coupled receptor 15 (GPR15) DNA methylation in smokers. Methylation of GPR15 DNA negatively correlated with smoking in the three newly identified sites CpG3.98251047, CpG3.98251179, and CpG3.98251219 (n = 114–582). Linear mixed regression model adjusted for age and sex with a significance threshold of 0.0083 after Bonferroni correction, the box plot whiskers represent 10–90th percentile.
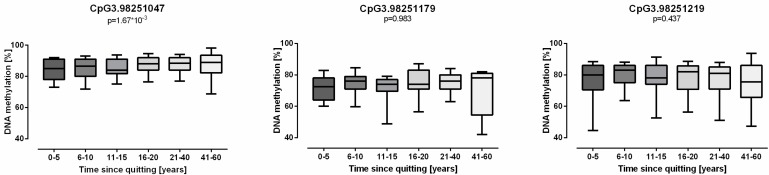
As expected, following the decrease in GPR15, mRNA expression after smoking cessation (Figure 2) and GPR15 DNA methylation at site CpG3.98251047 correlated with time since quitting in ex smokers (ß = 0.123, p = 1.67 × 10−3) (Figure 4), which showed increasing levels of GPR15 methylation. For CpG sites CpG3.98251179 and CpG3.98251219, no significant change over time could be observed.
Figure 4.
Methylation of G protein-coupled receptor 15 (GPR15) DNA slowly increased after smoking cessation. In ex smokers, GPR15 DNA methylation of CpG3.98251047 gradually rose within years after quitting whereas CpG3.98251179 and CpG3.98251219 methylation was not associated with time since quitting (n = 9–153). Linear mixed regression model adjusted for age and sex with a significance threshold of 0.0083 after Bonferroni correction, Box plot whiskers represent 10–90th percentile.
3.5. Decreased Levels of GPR15 DNA Methylation Correlate with Increased GPR15 mRNA Expression
Our data showed decreased levels of novel GPR15 DNA methylation as well as increased levels of GPR15 mRNA expression in smokers. Consequently, GPR15 mRNA expression significantly correlated with GPR15 DNA methylation at sites CpG3.98251047 (ß = 0.036, p = 4.86 × 10−5) and CpG3.98251179 (ß = 0.024, p = 4.72 × 10−4) (Figure 5). Site CpG3.98251219 showed a similar pattern. However, it did not reach statistical significance. In contrast to never smokers, CpG3.98251179 methylation was significantly correlated to GPR15 mRNA expression in current smokers (ß = 0.032, p = 6.30 × 10−3).
Figure 5.
Methylation of G protein-coupled receptor 15 (GPR15) DNA was associated with GPR15 mRNA expression. DNA methylation of CpG3.98251047 and CpG3.98251179 significantly correlated with GPR15 mRNA expression. DNA methylation were divided in tertiles (T) to distinguish low, medium, and high DNA methylation levels. Subjects with lower DNA methylation had higher GPR15 mRNA expression levels. n = 32–164, linear mixed regression model adjusted for age and sex with a significance threshold of 0.0083 after Bonferroni correction, Box plot whiskers represent 10–90th percentile. GPR15 mRNA expression is depicted as ΔCt values normalized to GAPDH mRNA expression. Lower ΔCt values indicate higher GPR15 mRNA expression.
4. Discussion
The association between smoking and multiple life-threatening diseases such as cardiovascular diseases has been well-known [1]. This is partly caused by the promotion of inflammatory processes [29]. In the present study, we investigated the effects of smoking on DNA methylation and mRNA expression of the GPR15 locus in the population-based Gutenberg Health Study. Our results are manifold: (i) we identified associations between DNA methylation and smoking status in three previously unstudied CpG sites and (ii) showed that smokers had increased levels of GPR15 expression, which slowly decreased after smoking cessation. In addition, (iii) smokers had decreased methylation levels of the three novel CpG sites within the GPR15 gene and (iv) DNA methylation of the novel methylation sites negatively correlated with GPR15 expression.
The GPR15 gene comprises 15 CpG sites in total. However, up to now, only CpG3.98251294 (cg19859270) has been investigated [7,8,9,13]. Previous studies showed decreased CpG3.98251294 methylation in smokers compared to never smokers, which was associated with cumulative smoking exposure [7,8,9,30,31]. In this study, we discovered three novel methylation sites in relation to smoking: CpG3.98251047, CpG3.98251179, and CpG3.98251219, with CpG3.98251047 being the most strongly influenced by smoking. The three novel CpG sites presented an even stronger difference in DNA methylation of 4% to 6% between smokers and never smokers. Contrary to CpG3.98251294, methylation of none of the three new CpG sites was significantly associated with cumulative smoking exposure (Table S2).
Beside DNA methylation, mRNA expression of GPR15 had been described to be influenced by smoking status [10,11,32,33]. Consistently, we showed that current smokers had 7.6-fold higher GPR15 mRNA expression levels compared to never smokers. GPR15 mRNA levels slowly decreased after smoking cessation. Additionally, our data showed that GPR15 mRNA expression correlated with the number of pack years. Our results implicate that GPR15 expression depends on the amount of cigarettes smoked per day and the duration of smoking.
Analyzing five years longitudinal gene expression data, we examined whether smoking changes GPR15 mRNA expression. GPR15 mRNA expression decreased after smoking cessation. Consistently, GPR15 mRNA expression decreased with increasing time since quitting, which is in line with current knowledge [7]. Furthermore, our data showed that decreasing methylation levels of CpG3.98251047 and CpG3.98251179 were linked to increasing levels of GPR15 mRNA, which indicated a direct influence of methylation status on GPR15 gene expression. Even though the methylation sites analyzed in our study are not directly located in the GPR15 promoter region but are within the single exon of GPR15 and since GPR15 consists of only 1252 bp, DNA methylation in close proximity to the promoter can influence transcription by inhibiting transcription factor binding [34]. Hence, as a potential mechanism, smoking might decrease GPR15 DNA methylation in the progenitor cells, which could lead to an increase in GPR15 positive cells in the blood, which results in increased GPR15 mRNA expression.
Taken together, our data indicate that smoking decreases GPR15 DNA methylation, which, in turn, leads to increased GPR15 mRNA expression. Thereby, GPR15 DNA methylation or GPR15 mRNA expression might have a potential to act as new biomarkers for smoking behavior since factors such as second-hand smoke exposure, irregular smoking behavior, electrical cigarettes, and smokeless tobacco are almost impossible to estimate accurately by questionnaire.
The strengths of the presented work are the measurement of previously undescribed CpG methylation sites within the GPR15 gene and its integration with mRNA expression. Our results originate from a large sample size with individuals from a population-based cohort and include longitudinal data. In this study, RNA was only available from enriched monocytes and DNA was available from leukocytes. GPR15 RNA expression levels were low and the detected increase in GPR15 expression in smokers could result from an increase in the number of GPR15 positive cells in the negatively selected monocyte fraction, as suggested by Bauer et al., rather than an upregulation of the gene expression [33]. The measurement of limited numbers of CpG sites, however, is a limitation of this study. Out of 15 CpG sites within GPR15, six sites could be analyzed due to technical limitations in the EpiTYPER amplicon design. Three sites were associated with smoking. Therefore, we cannot exclude the possibility that more methylation sites within the GPR15 gene are present that are also associated with smoking. Contrary to previous results from microarray data [7,8,9,30,31], the CpG3.98251294 DNA methylation site was neither significantly associated with smoking nor GPR15 expression in our analyses possibly because the EpiTYPER assay is less sensitive [26]. However, microarrays only include a limited selection of CpG sites. Using the EpiTYPER assay, we not only identified three novel CpG sites in relation to smoking, but the alterations in DNA methylation for these sites were even higher compared to Cpg3.98251294 [7,8,9,30,31]. Kõks et al. also determined DNA methylation of GPR15 with the EpiTYPER assay. Comparably to our results, a different methylation between smokers and non-smokers was shown for CpG3.98251179 but not for CpG3.98251294 [10].
In summary, we identified three novel methylation sites within the GPR15 gene whose methylation was affected by smoking, which led to an altered GPR15 gene expression. These smoking-related changes in GPR15 methylation and expression could perturb immune function and increase the risk for complex diseases with inflammatory pathogenesis, which could contribute to cardiovascular disease.
Acknowledgments
The authors thank Simone Schnella for editorial assistance.
Supplementary Materials
The following are available online at http://www.mdpi.com/2218-273X/8/3/74/s1. Table S1: Mass fragments detected by the EpiTYPER assay, Table S2: Results from linear mixed regression analyses, Figure S1: The GPR15 locus in the UCSC Genome Browser human GRCh37/hg19 assembly, Figure S2: GPR15 mRNA expression after smoking cessation. Availability of primary data is restricted due to limitations in ethical approvals.
Author Contributions
Conceptualization, T.H. and T.Z. Methodology, T.H., C.M., J.K., C.R., J.S., M.W., and S.K. Formal Analysis, T.H. and C.M. Resources, P.S.W., M.K., and K.J.L. Data Curation, P.S.W., KJ.L., C.M., and T.H. Writing-Original Draft Preparation, T.H. and T.Z. Writing-Review & Editing, T.H., C.M., J.K., C.R., J.S., S.K., M.W., T.M., N.P., P.S.W., M.M., F.M., M.K., K.J.L., S.B., and T.Z. Visualization, T.H. and C.M. Supervision, T.Z. Project Administration, T.H. and T.Z. Funding Acquisition, T.H. and T.Z.
Funding
This research was funded by the German Centre for Cardiovascular Research (DZHK e.V.) grant number [81X2710163_81604/151 (TH), B17-036 SE (MW) and 81Z1710101 (TZ)] and the Research Promotion Fund of the Faculty of Medicine (FFM) of the University Medical Center Hamburg Eppendorf (UKE) to TH. The Gutenberg Health Study is funded through the government of Rhineland-Palatinate (“Stiftung Rheinland-Pfalz für Innovation,” contract AZ 961-386261/733), the research programs “Wissen schafft Zukunft” and “Center for Translational Vascular Biology (CTVB)” of the Johannes Gutenberg-University of Mainz and its contract with Boehringer Ingelheim and PHILIPS Medical Systems including an unrestricted grant for the Gutenberg Health Study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- 1.GBD 2016 Risk Factors Collaborators Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet. 2017;390:1345–1422. doi: 10.1016/S0140-6736(17)32366-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talhout R., Schulz T., Florek E., van Benthem J., Wester P., Opperhuizen A. Hazardous compounds in tobacco smoke. Int. J. Environ. Res. Public Health. 2011;8:613–628. doi: 10.3390/ijerph8020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirodkar A.V., Marsden P.A. Epigenetics in cardiovascular disease. Curr. Opin. Cardiol. 2011;26:209–215. doi: 10.1097/HCO.0b013e328345986e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27k discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. Coordinated changes in ahrr methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X., Jia M., Zhang Y., Breitling L.P., Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: A systematic review of DNA methylation studies. Clin. Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan E.S., Qiu W., Baccarelli A., Carey V.J., Bacherman H., Rennard S.I., Agusti A., Anderson W., Lomas D.A., Demeo D.L. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y.V., Smith A.K., Conneely K.N., Chang Q., Li W., Lazarus A., Smith J.A., Almli L.M., Binder E.B., Klengel T., et al. Epigenomic association analysis identifies smoking-related DNA methylation sites in african americans. Hum. Genet. 2013;132:1027–1037. doi: 10.1007/s00439-013-1311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dogan M.V., Shields B., Cutrona C., Gao L., Gibbons F.X., Simons R., Monick M., Brody G.H., Tan K., Beach S.R., et al. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from african american women. BMC Genomics. 2014;15:151. doi: 10.1186/1471-2164-15-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koks G., Uudelepp M.L., Limbach M., Peterson P., Reimann E., Koks S. Smoking-induced expression of the GPR15 gene indicates its potential role in chronic inflammatory pathologies. Am. J. Pathol. 2015;185:2898–2906. doi: 10.1016/j.ajpath.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Vink J.M., Jansen R., Brooks A., Willemsen G., van Grootheest G., de Geus E., Smit J.H., Penninx B.W., Boomsma D.I. Differential gene expression patterns between smokers and non-smokers: Cause or consequence? Addict. Biol. 2017;22:550–560. doi: 10.1111/adb.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson R., Wahl S., Pfeiffer L., Ward-Caviness C.K., Kunze S., Kretschmer A., Reischl E., Peters A., Gieger C., Waldenberger M. The dynamics of smoking-related disturbed methylation: A two time-point study of methylation change in smokers, non-smokers and former smokers. BMC Genomics. 2017;18:805. doi: 10.1186/s12864-017-4198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su D., Wang X., Campbell M.R., Porter D.K., Pittman G.S., Bennett B.D., Wan M., Englert N.A., Crowl C.L., Gimple R.N., et al. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS ONE. 2016;11:e0166486. doi: 10.1371/journal.pone.0166486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer M., Fink B., Thurmann L., Eszlinger M., Herberth G., Lehmann I. Tobacco smoking differently influences cell types of the innate and adaptive immune system-indications from cpg site methylation. Clin. Epigenetics. 2015;7:83. doi: 10.1186/s13148-016-0249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H.K., Unutmaz D., KewalRamani V.N., Littman D.R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 16.Farzan M., Choe H., Martin K., Marcon L., Hofmann W., Karlsson G., Sun Y., Barrett P., Marchand N., Sullivan N., et al. Two orphan seven-transmembrane segment receptors which are expressed in cd4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer A., Zundler S., Atreya R., Rath T., Voskens C., Hirschmann S., Lopez-Posadas R., Watson A., Becker C., Schuler G., et al. Differential effects of α4β7 and GPR15 on homing of effector and regulatory T cells from patients with uc to the inflamed gut in vivo. Gut. 2016;65:1642–1664. doi: 10.1136/gutjnl-2015-310022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamczyk A., Gageik D., Frede A., Pastille E., Hansen W., Rueffer A., Buer J., Buning J., Langhorst J., Westendorf A.M. Differential expression of GPR15 on T cells during ulcerative colitis. JCI Insight. 2017;2:90585. doi: 10.1172/jci.insight.90585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmutz C., Hulme A., Burman A., Salmon M., Ashton B., Buckley C., Middleton J. Chemokine receptors in the rheumatoid synovium: Upregulation of CXCR5. Arthritis Res. Ther. 2005;7:217–229. doi: 10.1186/ar1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartwright A., Schmutz C., Askari A., Kuiper J.H., Middleton J. Orphan receptor GPR15/BOB is up-regulated in rheumatoid arthritis. Cytokine. 2014;67:53–59. doi: 10.1016/j.cyto.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suply T., Hannedouche S., Carte N., Li J., Grosshans B., Schaefer M., Raad L., Beck V., Vidal S., Hiou-Feige A., et al. A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia. Sci. Signal. 2017;10:eaal0180. doi: 10.1126/scisignal.aal0180. [DOI] [PubMed] [Google Scholar]
- 22.Pan B., Wang X., Nishioka C., Honda G., Yokoyama A., Zeng L., Xu K., Ikezoe T. G-protein coupled receptor 15 mediates angiogenesis and cytoprotective function of thrombomodulin. Sci. Rep. 2017;7:692. doi: 10.1038/s41598-017-00781-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild P.S., Zeller T., Beutel M., Blettner M., Dugi K.A., Lackner K.J., Pfeiffer N., Munzel T., Blankenberg S. The gutenberg health study. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55:824–829. doi: 10.1007/s00103-012-1502-7. [DOI] [PubMed] [Google Scholar]
- 24.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond—The transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunze S. Quantitative region-specific DNA methylation analysis by the epityper technology. Methods Mol. Biol. 2018;1708:515–535. doi: 10.1007/978-1-4939-7481-8_26. [DOI] [PubMed] [Google Scholar]
- 27.Saffery R., Gordon L. Time for a standardized system of reporting sites of genomic methylation. Genome Biol. 2015;16:85. doi: 10.1186/s13059-015-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team . R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: 2017. R Foundation for Statistical Computing; Vienna, Austria: 2017. [(accessed on 23 May 2018)]. Available online: http://www.R-project.org. [Google Scholar]
- 29.Colak Y., Afzal S., Lange P., Nordestgaard B.G. Smoking, systemic inflammation, and airflow limitation: A mendelian randomization analysis of 98,085 individuals from the general population. Nicotine Tob. Res. 2018:nty077. doi: 10.1093/ntr/nty077. [DOI] [PubMed] [Google Scholar]
- 30.Zeilinger S., Kuhnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A., et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dogan M.V., Xiang J., Beach S.R., Cutrona C., Gibbons F.X., Simons R.L., Brody G.H., Stapleton J.T., Philibert R.A. Ethnicity and smoking-associated DNA methylation changes at hiv coreceptor GPR15. Front. Psychiatry. 2015;6:132. doi: 10.3389/fpsyt.2015.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsaprouni L.G., Yang T.P., Bell J., Dick K.J., Kanoni S., Nisbet J., Vinuela A., Grundberg E., Nelson C.P., Meduri E., et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9:1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer M., Linsel G., Fink B., Offenberg K., Hahn A.M., Sack U., Knaack H., Eszlinger M., Herberth G. A varying t cell subtype explains apparent tobacco smoking induced single cpg hypomethylation in whole blood. Clin. Epigenetics. 2015;7:81. doi: 10.1186/s13148-015-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E., et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.