Abstract
Monocytes/macrophages are important in orchestrating inflammatory responses. However, knowledge of the long noncoding RNA (lncRNA) regulation of monocytic cell differentiation and diseases remains limited. We aimed to elucidate the role of the 17 kb lncRNA noncoding transcript in T cells (NTT) in monocyte functions. Knockdown and chromatin immunoprecipitation (ChIP) assays in THP-1 cells (human monocytic leukemia cell line) revealed that NTT is regulated by the monocyte key transcription factor C/EBPβ and that it binds to the promoter of nearby gene PBOV1 via hnRNP-U. Overexpression of PBOV1 in THP-1 cells resulted in cell cycle G1 arrest, differentiation into macrophages, a marked increase in IL-10 and CXCL10 mRNA levels, and upregulation of the costimulatory molecules. In contrast to the downregulated NTT observed in lipopolysaccharide (LPS)-treated THP-1 cells, the C/EBPβ/NTT/PBOV1 axis was found to be hyperactivated in peripheral blood mononuclear cells (PBMCs) of first-time diagnosed untreated early rheumatoid arthritis (RA) patients, and their gene expression levels decreased markedly after treatment. Higher initial C/EBPβ/NTT/PBOV1 expression levels were associated with a trend of higher disease activity DAS28 scores. In conclusion, our study suggests that the lncRNA NTT is a regulator of inflammation in monocytes, and its activation participates in monocyte/macrophage differentiation and the pathogenesis of RA.
Keywords: long noncoding RNA, NTT, C/EBPβ, PBOV1, monocyte differentiation, rheumatoid arthritis
1. Introduction
Monocytes are central players in orchestrating complex immune responses. Upon sensing stimuli from infectious or aseptic sources, monocytes can differentiate into dendritic cells (DCs) or macrophages [1]. Being trafficked to the sites of inflammation, macrophages exert their functions by phagocytosis, cytokine secretions, and interactions with cells of adaptive immunity, and their functions may change along the progression of inflammation [2,3]. Macrophage differentiation and polarization to pro-inflammatory or immunosuppressive phenotypes are tightly regulated by environmental factors (i.e., cytokines, metabolic stresses), transcription factors, and noncoding RNAs [2,4]. Among noncoding RNAs, micro-RNAs (miRNAs) have been extensively studied as mediators that intricately regulate multiple functions of monocytes/macrophages [5,6]. Dysregulations of miRNAs has been implicated in a variety of diseases, including infection, atherosclerosis, obesity, cancer, and autoimmune/autoinflammtory diseases [5,7]. Long noncoding RNAs (lncRNAs) are noncoding RNAs that are more than 200 nucleotides long. Accumulated evidence has shown that lncRNA can affect different stages of gene regulation via RNA–DNA, RNA–RNA, or RNA–protein interactions [8]. Similarly, lncRNAs have been reported to fine-tune both innate and adaptive immune responses [9,10,11,12,13]. However, knowledge of the mechanisms of lncRNA regulation for monocyte differentiation/activation and their roles in inflammatory diseases remains limited.
It was presented by Satpathy A.T. and Chang H.Y. in Immunity 2015 that several lncRNAs are involved in the differentiation of myeloid cells and their activations after stimulation with different Toll-like receptor (TLR) ligands [14]. Lnc-DC was reported to be upregulated by the transcription factor PU.1 during monocyte differentiation to classical dendritic cells (cDCs), while HOTAIRM1 was found to participate in terminal granulocyte differentiation [15,16]. Upon TLR2 stimulation, lincRNA-Cox2 has been demonstrated to regulate immune gene transcriptions via interactions with the heterogeneous nuclear ribonucleoproteins (hnRNP)-A/B and hnRNP-A2/B1, while THRIL was shown to regulate TNF-α production via interaction with hnRNP-L in THP-1 macrophages [17,18]. Noncoding transcript in T cells (NTT) is a 17 kb lncRNA which is expressed in the nucleus. It was discovered in 1997 in activated CD4+ T cells and in peripheral blood mononuclear cells (PBMCs) stimulated with HIV peptides, suggesting its role in adaptive immunity [19,20]. Yet, the mechanisms underlying the NTT-mediated potential regulation of T-cell functions are still unclear. Furthermore, whether and how NTT is involved in innate immunity are currently unknown.
NTT is located at chromosome 6q23–q24, which is close to several genes related to immune function, hematopoiesis, and cell proliferation, including IFNGR1, TNFAIP3, MYB, and PBOV1 [20]. IFNGR1 is associated with macrophage TNF-α production and susceptibility to Listeria monocytogenes infection [21]. TNFAIP3 is an essential negative regulator of inflammation, and dysregulation of TNFAIP3 is associated with autoimmune diseases [22]. MYB is a transcription factor reported to regulate myeloid hematopoiesis and usually is aberrantly expressed in leukemia [23]. Similarly, PBOV1 has been reported to be overexpressed in breast, prostate, and ovarian cancer tissues, regulating tumor proliferation; however, the function of PBOV1 on immune cells has not been discovered [24,25,26,27]. Due to its large size (17 kb nt) and proximity to these potentially immune-related genes, it has been suggested that NTT might exert its function via regulating the nearby genes [20].
In this study, we aimed to investigate the role of the lncRNA NTT in monocyte functions, how NTT is regulated in inflammation, and its potential dysregulation in a chronic inflammatory autoimmune disease, rheumatoid arthritis (RA).
2. Results
2.1. NTT (Noncoding Transcript in T Cells) Is Expressed in Human Monocytic Cells and Is Regulated by C/EBPβ
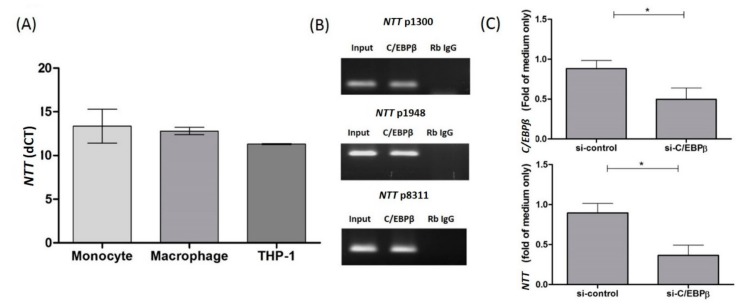
NTT was found to be expressed in resting human primary monocytes, monocyte-derived macrophages, and the THP-1 cell line (Figure 1A). By examining the promoter sequence of NTT, we identified the potential binding motif of C/EBPβ, a key transcription factor in monocytes. To further check if C/EBPβ binds to the NTT promoter and regulates NTT expression, we performed a chromatin immunoprecipitation assay (ChIP) and siRNA knockdown for C/EBPβ in the THP-1 cell line. C/EBPβ binding was detected on three positions of the NTT promoter (Figure 1B), and C/EBPβ knockdown in THP-1 resulted in decreased NTT expression (Figure 1C).
Figure 1.
Expression of noncoding transcript in T cells (NTT) in immune cells and regulation of NTT during lipopolysaccharide (LPS) stimulation. (A) NTT is expressed in human primary monocytes, monocyte-derived macrophages, and THP-1 cells; n = 3, bars represent mean ± SEM. (B) THP-1 cell nuclear lysates were fixed and immunoprecipitated by anti-C/EBPβ or isotype control IgG antibody (ChIP), and the antibody-bound DNA sequences of three positions of NTT promoter were detected by PCR and gel electrophoresis; here is a representative graph of three independent experiments, showing binding of C/EBPβ. (C) Knockdown of C/EBPβ in THP-1 cells by si-RNA resulted in lower expression levels of NTT; n = 5, bars represent mean ± SEM. *: p < 0.05 by Wilcoxon signed rank test.
2.2. NTT Regulates Downstream Gene PBOV-1 via HnRNP-U Binding
Next, we investigated how the large lncRNA NTT regulates nearby genes (potentially downstream genes). NTT expression in THP-1 was knocked down by si-RNA and the relative mRNA levels of nearby genes were analyzed (Figure 2A). Transfection of si-NTT in THP-1 led to decreased expression in almost all downstream genes, except L3MBTL3, MYB, and AHI1. NTT knockdown showed the greatest impact on PBOV1 expression (Figure 2A). Since it has been reported that lncRNAs may bind to hnRNP proteins to regulate downstream genes, we further studied if NTT could bind to the PBOV1 promoter via interaction with the hnRNP-U protein. An RNA immunoprecipitation assay showed that NTT could bind to hnRNP-U (Figure 2B). DNA ChIP showed that hnRNP-U binds to two positions of the PBOV1 promoter, and the hnRNP-U binding became undetectable after NTT knockdown (Figure 2C), suggesting NTT might enhance PBOV1 expression by interacting with hnRNP-U binding to the promoter of PBOV1.
Figure 2.
NTT acts in cis on genes at the local proximity. (A) Knockdown of NTT in THP-1 cells resulted in decreased expression levels of downstream genes, with the most prominent effect on PBOV1; n = 3, bars represent mean ± SEM. (B,C) NTT regulates PBOV1 via binding with hnRNP-U to the PBOV1 promoter. (B) NTT RNA binds to hnRNP-U as detected by RNA IP; representative graph from three independent experiments. (C) DNA ChIP assay detecting the promoter sequence of PBOV1 (2 positions) using anti-hnRNP U antibody in THP-1 cells transfected with si-RNA for NTT or scramble control; representative graph from three independent experiments.
2.3. C/EBPβ, NTT, and PBOV1 Expression Levels Were Highly Elevated in Fresh Rheumatoid Arthritis (RA) Patients
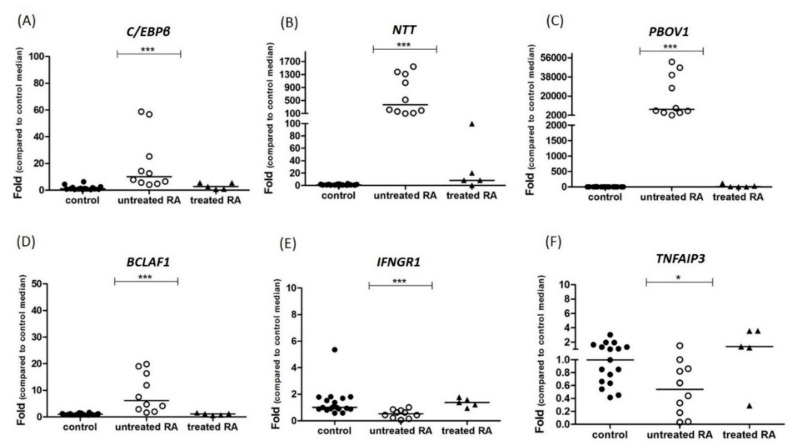
In order to understand the potential roles of NTT in inflammatory diseases, we evaluated the expression levels of C/EBPβ, NTT, and downstream genes in PBMCs derived from first-time diagnosed, untreated RA patients. We were able to follow up with 8 of the 10 patients, and blood was drawn again in 5 of the 8 patients after 2 years of RA treatment. As compared with the levels detected in healthy control PBMCs, C/EBPβ, NTT, and PBOV1 expressions were significantly elevated in freshly diagnosed RA patients (median folds for C/EBPβ = 10.13, NTT = 369.76, PBOV1 = 7455.33), and the levels markedly decreased after 2-year treatment (Figure 3A–C). The expression of another NTT downstream gene, BCLAF1, showed the same trend as PBOV1 but with a much lower fold (Figure 3D). In contrast, as for other NTT–downstream immune-related genes, IFNGR1 and TNFAIP3, the expression levels decreased in freshly diagnosed RA patients (Figure 3E–F).
Figure 3.
C/EBPβ/NTT/PBOV1 expression levels are elevated in peripheral blood mononuclear cells (PBMCs) of untreated rheumatoid arthritis (RA) patients. RNA expression levels of (A) C/EBPβ, (B) NTT, (C) PBOV1, (D) BCLAF1, (E) IFNGR1 and (F) TNFAIP3 in PBMCs derived from healthy controls (n = 17), untreated RA patients (n = 10), and treated RA patients (n = 5, blood drawn after 2 years of follow-up). Lines represent medians; * p < 0.05, *** p < 0.0001 by Kruskal–Wallis tests. RNA expression levels are shown as folds compared with median levels of healthy controls.
2.4. Association of C/EBPβ, NTT, and PBOV1 Expression Levels with RA Disease Severity
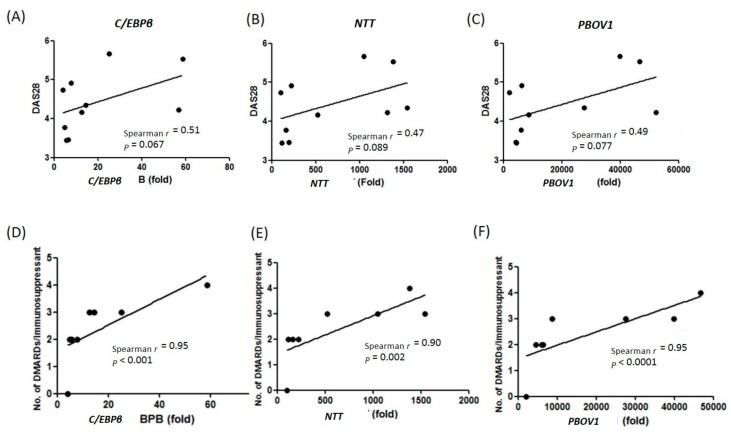
As shown in Table 1, C/EBPβ/NTT/PBOV1 expression levels did not associate with levels of initial laboratory inflammatory markers, including rheumatoid factor (RF), anticyclic citrullinated peptide antibody (anti-CCP), erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP). In addition, C/EBPβ/NTT/PBOV1 expression did not correlate with hemoglobin (Hb), IL-1β, TNFα, and IL-6 levels (Table S1). However, we found a trend of positive correlation between C/EBPβ/NTT/PBOV1 expressions and initial disease activity scores (DAS28) (Table 1, Figure 4A–C). Linear regression analysis showed that C/EBPβ expression level positively correlated with the simplified disease activity index (SDAI) (r2 = 0.54, p = 0.015, Table 1). Furthermore, higher C/EBPβ/NTT/PBOV1 levels positively correlated with the usage of multiple disease-modifying antirheumatic drugs (DMARDs)/immunosuppressants at the 2-year follow-up (Table 1, Figure 4D–F). Only one RA patient (in the initial C/EBPβ/NTT/PBOV1-high group, patient No. 3) required a biological agent (Adalimumab) for treatment. All patients except the one with the highest initial C/EBPβ/NTT/PBOV1 levels (Table 1, patient No. 1) had lower than 2.7 DAS28 scores after 2 years of RA treatment. Collectively, these results suggest that the C/EBPβ/NTT/PBOV1 axis is highly activated in untreated RA patients, and higher expression levels might be associated with a higher disease inflammatory status, which requires more medications to control.
Table 1.
Clinical characteristics of RA (rheumatoid arthritis) patients and their C/EBPβ/NTT/PBOV1 expression levels in PBMCs (peripheral blood mononuclear cells).
| Basic Information | Initial Lab Data | Initial Disease Activity | RNA Expression at RA Diagnosis | Clinical Parameters at 2-Year Follow-Up | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient No. | Sex | Age | RF (IU/mL) | Anti-CCP (U/mL) | ESR (mm/h) | CRP (mg/dL) | DAS28 | SDAI | C/EBPβ (fold) | NTT (fold) | PBOV1 (fold) | Number of Medications Used in Addition to NSAID | DAS28 |
| 1 | F | 57 | 28.2 | 29 | 44 | 1.15 | 5.53 | 19.7 | 58.69 | 1379.57 | 46,663.28 | 4 (Hydroxychloroquine, Sulfasalazine, Cyclosporine, Prednisolone) | 3.86 |
| 2 | F | 56 | 688 | 2.2 | 18 | 0.02 | 4.22 | 15 | 56.69 | 1314.23 | 52,136.28 | NA | NA |
| 3 | F | 55 | 23.4 | 0.8 | 50 | 4.96 | 5.67 | 12.5 | 25.19 | 1045.52 | 39,786.74 | 3 (Hydroxychloroquine, MTX, Adalimumab) | 1.46 |
| 4 | F | 25 | 52.3 | 76 | 106 | 2.57 | 4.35 | 7.8 | 14.27 | 1541.37 | 27,554.49 | 3 (Hydroxychloroquine, MTX, Sulfasalazine) | 1.13 |
| 5 | F | 57 | <20 | 1.4 | 46 | 3.4 | 4.16 | 16 | 12.51 | 522.76 | 8659.09 | 3 (Hydroxychloroquine, Sulfasalazine, Prednisolone) | 2.36 |
| 6 | F | 66 | 413 | 338 | 53 | 1.81 | 4.92 | 11.2 | 7.75 | 216.77 | 6251.56 | 2 (Hydroxychloroquine, Sulfasalazine) | 2.08 |
| 7 | F | 34 | 79.8 | 121 | 8 | 0.03 | 3.45 | 8 | 5.68 | 114.56 | 4420.519 | 2 (Hydroxychloroquine, MTX) | 1.74 |
| 8 | F | 43 | 20 | 3.5 | 28 | 0.68 | 3.46 | 7.1 | 6.52 | 195.36 | 4299.64 | NA | NA |
| 9 | F | 70 | <20 | 0.7 | 50 | 2.52 | 4.74 | 12.8 | 4.13 | 101.83 | 2033.853 | 0 | 2.67 |
| 10 | F | 50 | 137 | 10 | 25 | 0.25 | 3.78 | 8 | 4.77 | 163.14 | 6038.51 | 2 (Hydroxychloroquine, MTX) | 2.20 |
RNA expressions are shown as folds compared to the median of healthy controls. RF: rheumatoid factor; Anti-CCP: anti-cyclic citrullinated peptide antibody; ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; DAS28: disease activity score 28; SDAI: simple disease activity index; NSAID: nonsteroidal anti-inflammatory drug; MTX: methotrexate; NA: not available.
Figure 4.
Correlations of C/EBPβ/NTT/PBOV1 expression levels with RA disease severity score DAS28 (A–C) and usage of multiple disease-modifying antirheumatic drugs (DMARDs)/immunosuppressant (D–F). RNA expression levels are shown as folds compared with median levels of healthy controls.
2.5. Overexpression of PBOV1 in THP-1 Cells Led to Cell Cycle Arrest and Differentiation to Macrophages
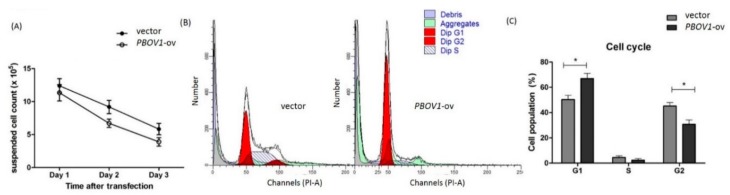
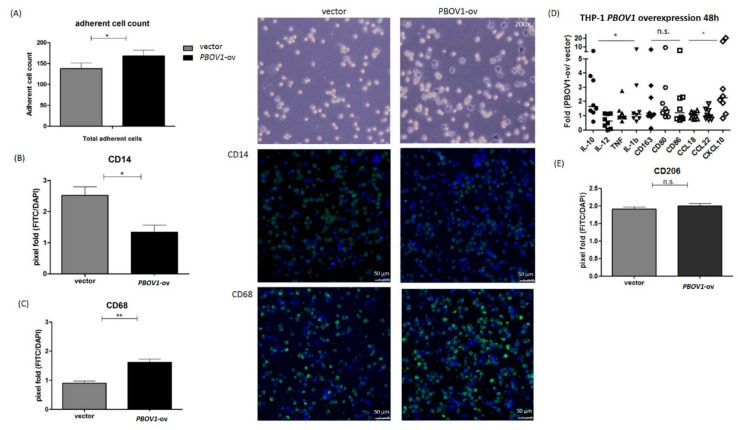
Although marked PBOV1 upregulation was observed in the PBMCs of freshly diagnosed RA patients, the function of PBOV1 in monocytes remains unknown. Therefore, we transfected the PBOV1-overexpression vector to the THP-1 monocytic cell line for 48 h. As compared with cells transfected with the control vector, PBOV1-overexpressing THP-1 cells showed a lower suspended cell count (Figure 5A) but a more adherent cell count (Figure 6A). Furthermore, a higher proportion of cells arrested at the G1 phase of the cell cycle was found in PBOV1-overexpressing THP-1 cells (Figure 5B–C). Further immunohistochemical staining of the adherent cells showed lower monocyte marker CD14 and higher macrophage marker CD68 expressions in PBOV1-overexpressed THP-1 cells as compared with controls, suggesting differentiation toward macrophages (Figure 6B–C).
Figure 5.
Overexpression of PBOV1 in THP-1 cells results in cell cycle arrest. (A) Count of suspended THP-1 cells after transfection of PBOV1-overexpression vectors; n = 6, bars represent mean ± SEM. (B) Cell cycle analysis of THP-1 cells transfected with control vector (left) or with PBOV1-overexpression vector (right) for 48 hours; here are representative graphs of four independent experiments, showing G1 arrest in PBOV1-overexpressing THP-1. (C) Percentage of THP-1 cells in Gap 0/Gap 1 (G0/G1), Synthesis (S), and Gap 2/Mitosis (G2/M) phase; n = 4, bars represent mean ± SEM. * p < 0.05 by Mann Whitney U test.
Figure 6.
Overexpression of PBOV1 in THP-1 cells promotes differentiation into macrophages. (A) Adherent cell counts increased after transfection of PBOV1 overexpression vector for 48 h. Left: Bars represent mean ± SEM, n = 6; Right: images under light microscope after removing suspended cells; * p < 0.05 by Mann–Whiney U test. (B) Decreased CD14 expression in PBOV1-overexpressing THP-1 cells. Left: Columns are CD14-FITC-positive cells counted by Image J tool, bars represent mean ± SEM, n = 5; Right: confocal microscopy images of immunohistochemical staining of CD14 (green) and cell nucleus (blue). * p < 0.05 by Mann Whitney U test (C) Increased CD68 expression in PBOV1-overexpressing THP-1 cells. Left: Columns are CD68-FITC-positive cells counted by Image J tool, bars represent mean ± SEM, n = 5; Right: confocal microscopy images of immunohistochemical staining of CD68 (green) and cell nucleus (blue). ** p < 0.01 by Mann Whitney U test (D) M1/M2 profiling of THP-1 cells transfected with PBOV1-overexpression vector for 48 h. Lines represent medians, n = 8; showing marked elevation in IL-10 and CXCL10 RNA expressions; * p < 0.05 by Kruskal–Wallis test; n.s.: not significant by Kruskal–Wallis test (E) PBOV1-overexpressed THP-1 cells secrete more CXCL10 in the supernatant than control. Bars represent mean ± SEM, n = 6; * p < 0.05 by Mann–Whiney U test. (F) THP-1 cells transfected with control vector or PBOV1-overexpression vector were further differentiated into macrophage (via phorbol 12-myristate 13-acetate (PMA)) and polarized to M2 (via IL-4 and IL-13). Immunohistochemical staining of CD206 shows no difference of expression between THP-1 transfected with control vector or with PBOV1-overexpression vector; n = 5, bars represent mean ± SEM. n.s.: not significant by Mann Whitney U test.
In order to investigate whether PBOV1 also affects macrophage polarization, mRNAs of M1/M2 related cytokines, cell surface markers, and chemokines were analyzed in PBOV1-overexpressed THP-1 cells (Figure 6D). The anti-inflammatory cytokine IL-10 mRNA level was found to be elevated in PBOV1-overexpressed THP-1 cells, while the levels of pro-inflammatory cytokines IL-12, TNF-α, and IL-1β were not (Figure 6D). Conversely, the pro-inflammatory chemokine CXCL10 mRNA level was found to be markedly upregulated in PBOV1-overexpressed THP-1 cells, while the levels of the anti-inflammatory chemokines CCL18 and CCL22 remained unchanged (Figure 6D). The mRNA levels of both M1 (CD80/CD86) and M2 (CD163) related cell markers were increased in cells overexpressing PBOV1 (Figure 6D). We then evaluated the IL-10 and CXCL10 protein levels in supernatants of THP-1 cells transfected with the PBOV1-overexpression vector or control vector for 48 h. However, only elevated secretion of CXCL10 was observed in PBOV1-overexpressed THP-1 cells (Figure 6E). IL-10 protein was undetectable in all supernatants (data not shown). Further polarization of THP-1 cells to M2 macrophages using sequential stimulations with phorbol 12-myristate 13-acetate (PMA), IL-4, and IL-13 revealed no difference of CD206 protein expression between PBOV1-overexpressing cells and cells transfected with the control vector (Figure 6F). Taken together, PBOV1 overexpression in THP-1 cells promoted differentiation to macrophages and increased CXCL10 secretion. CXCL10 could potentially mobilize more immune cells, which then may be trafficked to the site of inflammation together with the differentiated macrophages. The graphical summary showing the proposed model of the roles of C/EBPβ/NTT/PBOV1 upregulation in RA pathogenesis is shown in Figure 7.
Figure 7.
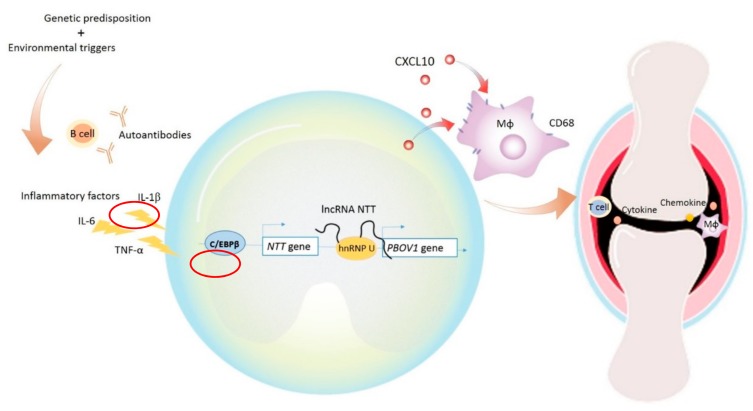
Graphical summary showing the proposed model of the roles of C/EBPβ/NTT/PBOV1 upregulation in early RA pathogenesis. RA pathology could be induced by environmental triggers in genetically predisposed individuals. Autoantibodies produced by B cells and inflammatory cytokines such as IL-6, IL-1β, and TNF-α appear in the disease process. Perhaps under the stimulation of these factors, the transcription factor C/EBPβ binds to the promoter of NTT, leading to the transcription of NTT. NTT binds to the promoter of PBOV1 via hnRNP-U, then the enhanced expression of PBOV1 promotes monocyte/macrophage differentiation and the secretion of cytokines and chemokines including CXCL10. CXCL10 can promote the migration of immune cells to the site of inflammation, such as the synovial joints in RA.
3. Discussion
As the central player in orchestrating complex immune responses, monocytes/macrophages are equipped with the ability to react to different stimuli dynamically [2]. Although data are still limited, lncRNAs have been reported to be the link connecting signals transduced by transcription factors and the activation or repression of downstream genes in macrophages [8,14,17,28]. Here, we report that NTT is expressed in human monocytes/macrophages and is highly upregulated (around 100- to 1000-fold of normal control) in the PBMCs of untreated RA. While being regulated by the transcription factor C/EBPβ, NTT acts in cis on gnomically closely located genes related to differential immune responses and macrophage differentiation, adding a layer of complexity to the fine tuning of monocyte functions.
Previous researchers have identified differentially expressed lncRNAs in PBMCs and synovial fibroblasts in patients with RA, a chronic inflammatory joint disease [29,30,31,32]. While the functions of most of these lncRNAs remains unknown, Hotair has been reported to enhance the migration of activated macrophages, thereby participating in RA pathogenesis [31]. In this study, NTT was found to be highly elevated in the PBMCs of untreated RA and correlated with the requirement of multiple DMARDs/immunosuppressants during 2 years of treatment. In early RA, NTT is possibly upregulated by the transcription factor C/EBPβ, which is known to drive myeloid-derived suppressor cell (MDSC) differentiation and the polarization of the more regulatory M2 macrophages [2,33,34,35]. The increased amount of NTT bound to the promoter of the downstream gene PBOV1 via hnRNP-U, and it upregulated PBOV1 expression to an average of 18,000-fold as compared with healthy controls. These results are consistent with the report that lncRNAs could elevate downstream gene expression via increasing mRNA stability by interacting with hnRNP-U; however, we could not rule out the possibility that NTT might also exert its cis-action on PBOV1 via other mechanisms, such as binding to histone modifying complexes [36,37].
Prostate and breast cancer overexpressed 1 (PBOV1) was first described to be upregulated in certain cancers involving tumor proliferation and is variably associated with patient survival [24,25,27]. In prostate cancer cell lines, PBOV1 overexpression was found to promote tumor proliferation and cell cycle progression [25]. However, the role of PBOV1 in autoimmune diseases is still unknown. We are the first to investigate the function of PBOV1 in monocytes. As observed in untreated RA PBMCs, high levels of PBOV1 might be downstream of C/EBPβ/NTT hyperactivations. Although C/EBPβ is known to drive macrophages towards an anti-inflammatory phenotype [33,34], we found that macrophages differentiated from PBOV1-overexpressed THP-1 cells upregulated the RNA expressions of IL-10 (mostly between 1- and 2-fold) and the pro-inflammatory chemokine CXCL10 (mostly more than 2-fold). We could not detect IL-10 protein in the supernatant at 48 or 64 h after transfection, suggesting the detection of IL-10 RNA upregulation might be nonspecific and transient. As for CXCL10, markedly elevated protein levels were also detected in PBOV1-overexpressed THP-1 supernatants. The pro-inflammatory chemokine CXCL10 has been reported to attract activated macrophages, T cells, and NK cells to the site of inflammation and is upregulated in autoimmune diseases, including RA [38,39,40,41,42]. It is suggested that CXCL10 increases the migration of inflammatory cells via CXCR3-mediated ERK activation and stimulates the production of osteoclastogenic cytokines in CD4+ T cells, resulting in bone destruction in RA [43]. We therefore speculate that NTT-induced monocyte/macrophage PBOV1 expression in genetically predisposed individuals plays a pathogenic role in RA via enhancing the secretion of the chemokine CXCL10, which could lead to synovial inflammation and joint destruction (Figure 7). Studies on the expressions of NTT and PBOV1 in blood monocytes and synovial macrophages of different disease phases might help to clarify their pathogenic roles in RA. Since NTT is also expressed in T cells, the function of NTT in regulating macrophage—T cell interactions in RA development warrant further research.
4. Materials and Methods
4.1. Patient Subjects
Peripheral blood mononuclear cells (PBMCs) from freshly diagnosed rheumatoid arthritis patients and sex- and age-matched healthy controls were collected. RNA was extracted by the TRIzol method, further reverse transcribed to cDNA, and tested for the relative expression levels of NTT and upstream and downstream genes. This study was approved by China Medical University Hospital Research Ethics Committee on 05 March 2016 (CMUH105-REC2-006, for project “Exploring the regulation and function of a long non-coding RNA NTT in inflammation”).
4.2. THP-1 Cell Culture, Differentiation, and Polarization Assays
THP-1 cells were grown in LPS-free complete RPMI medium containing 10% fetal bovine serum at 37 °C in an incubator with 5% CO2. In M2-polarization experiments, THP-1 cells were differentiated into macrophages using 50 µM PMA for 5 days; THP-1-derived macrophages were then polarized to M2 via treatment of 20 ng/mL IL-4 and 20 ng/mL IL-13 (R&D Systems, Minneapolis, MN, USA) for 2 days.
4.3. Isolation of Primary Immune Cells
Human primary T cells and monocytes were isolated from healthy subjects’ peripheral blood by using RosetteSep human T cell enrichment cocktail and RosetteSep human monocyte enrichment cocktail (Stemcell technologies, Vancouver, BC, Canada), followed by Ficoll Paque gradient centrifugation. Macrophages were further derived from monocytes via replacing half of the culture medium each day for 7 days.
4.4. Chromatin Immunoprecipitation Assay (ChIP)
THP-1 cells were crosslinked with 0.4% formaldehyde and the reaction was stopped by adding glycine. Fixed cells were lysed according to magnetic chromatin immunoprecipitation kit protocol. The nuclear fractions were sonicated to reduce DNA length to 200–500 bp. The chromatin extract was incubated with magnetic beads and either anti-C/EBPβ/anti-hnRNP-U/anti-ATF3 (all from Abcam, Cambridge, UK) or anti-IgG (negative control) antibody. Another portion of sheared DNA were prepared for making an input control. After immunoprecipitation, the immune complex was eluted, the protein was unlinked, and the associated DNA fragments were purified according to the manufacturer’s instructions (Active Motif, Carlsbad, CA, USA). Finally, PCR of the purified DNA using primers for the respective promoter sequences were performed.
4.5. Electroporation for Knockdown and Overexpression Assays
THP-1 was transfected with 10 nM si-RNAs for C/EBPβ, si-RNA for NTT, or negative control-siRNA (all purchased from MDBio, Taipei, Taiwan) by using electroporation. Briefly, 5 × 106 cells were resuspended in 500 µL serum-free RPMI medium and incubated with 150 nM siRNA in electroporation cuvettes for 5 min on ice. Cells were then electroporated at 220 mV and 950 µF using Bio-rad Gene Pulser (Midland, ON, Canada) according to manufacturer’s instructions. For PBOV1-overexpression assays, THP-1 cells were transfected with 10 µg/mL pCMV6-Entry vector cloned with PBOV1 or the control vector (both vectors were purchased from OriGene, Rockville, MD, USA) for 48 h. Subsequently, RNAs were extracted for real-time PCR analysis, and culture supernatants were collected for IL-10 (R&D Systems) and CXCL10 (R&D Systems) ELISA analyses.
4.6. Reverse-Transcriptase PCR (RT-PCR)
Nuclear and cytoplasmic cell lysates were extracted using the Nuclear/Cytoplasmic Extraction kit (Thermo Fisher Scientific, Waltham, MA, USA) and followed by TRIzol RNA extraction. Two micrograms of RNA were reverse-transcribed using reverse transcriptase and buffers. The resulting cDNA underwent quantitative real-time PCR analysis. The house-keeping gene GAPDH was used as the internal control for total and cytoplasmic cell fraction, while U2 snRNA gene was used as the internal control for nuclear cell fraction.
NTT promoter primer sequences:
Position 9501–9746: forward 5′-TGGTATGAACAAGGCAGCAG-3′, reverse 5′-CCTTGTTTGTGCCCATCTCT-3′; position 6846–7059: forward 5′-GGGTGAAAGCAGCCTGTG-3′, reverse 5′-CAGAACAAAAAGAACCCCTGA-3′; position 5825–6095: forward 5′-TCTCCAATCCCCTCTGTGAC-3′, reverse 5′-GAAGGCTATGGGCACTTTCA-3′; position 8204–8425: forward 5′-TCAACATATGCCTTACCTCACA-3′, reverse: 5′-TACCAGGAGCTTGGGATGTT-3′
Gene primer sequences:
NTT forward 5′-CTTGGCCTAAAAGGGGATG-3′, reverse 5′-GCACCTTTGGTCTCCTTCAC-3′; IFNGR1 forward 5′-CATGCAGGGTGTGAGCAG-3′, reverse 5′-AACATTAGTTGGTGTAGGCACTGA-3′; TNFAIP3 forward 5′-TGCACACTGTGTTTCATCGAG-3′, reverse 5′-ACGCTGTGGGACTGACTTTC-3′; HIVEP2 forward 5′-CGGCAAGCTTACATCATCAA-3′, reverse: 5′-AGGACGCATCAGGTTTCATC-3′; PBOV1 forward 5′-GAAAAAGATTCTCATCACTCAAC-3′, reverse 5′-GGTTCTCAAACAGCCTTCC-3′; C/EBPβ forward 5′-CGGGCTCAGGAGAAACTTTA-3′, reverse 5′-GGGGTGGCCGCTATTAGT-3′; GAPDH: forward 5′-AGCCACATCGCTCAGACAC-3′, reverse 5′-GCCCAATACGACCAAATCC-3′; U2 snRNA: forward 5′-TTTGGCTAAGATCAAGTGTAGTATCTGTTC-3′, reverse 5′-AATCCATTTAATATATTGTCCTCGATAGA-3′
4.7. Cell Cycle Analysis
Transfected THP-1 cells were collected and fixed in 70% ethanol at −20 °C overnight. Fixed cells were washed and labelled with propidium iodide (Sigma-Aldrich, St. Louis, MO, USA) in the presence of RNase A (Sigma-Aldrich) and Triton X-100 for 30 min in the dark. FACSCanto flow cytometer and the ModFit LT program were used to analyze the percentages of cells within each phase of the cell cycle.
4.8. Immunofluorescence Staining
Monolayer of THP-1 cells was attached to slides using the Cytospin centrifugation method. Cells were fixed by 100% methanol and stained with primary antibodies (anti-CD14, anti-CD68, anti-CD206, all from Abcam, Cambridge, UK) at 4 °C overnight after blocking with PBS (phosphate buffered saline) containing 1% (wt/vol) fetal bovine serum (FBS). Cells were then washed and stained with fluorochrome-conjugated secondary antibodies. DAPI (4′,6-diamidino-2-phenylindole) nuclear stain was applied. Slides were further analyzed under a confocal laser scanning microscope. Image J software was used to quantify the relative amount of positive staining.
4.9. Statistical Analyses
Mann–Whitney U test was applied to compare the percentage of macrophage marker expression between THP-1 cells with or without transfection of PBOV1-overexpression vector. Kruskal–Wallis test was used to compare the expression levels of NTT and related genes in healthy subjects, fresh RA patients, and treated RA patients. Spearman’s test was performed to analyze the correlation between NTT expression level and RA disease activity. All statistical tests were performed on GraphPad Prism version 5 (La Jolla, CA, USA).
5. Conclusions
This study suggests that the lncRNA NTT is a regulator of inflammation in monocytes, and its activation participates in monocyte/macrophage differentiation via PBOV1 upregulation. Hyperactivation of the C/EBPβ/NTT/PBOV1 axis might play a role in the pathogenesis of RA via enhanced secretion of the proinflammatory chemokine CXCL10.
Acknowledgments
We thank Shih-Ya Hung (China Medical University, Taichung, Taiwan) for technical assistance; Der-Yuan Chen (China Medical University Hospital, Taichung, Taiwan) for comments on the manuscript; Ms. Yu-Hsuan Juan for graphical assistance.
Abbreviations
| lncRNA | Long noncoding RNA |
| PBMC | Peripheral blood mononuclear cells |
| RA | Peripheral blood mononuclear cells |
| NTT | Noncoding transcript in T cells |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/9/2806/s1.
Author Contributions
Conceptualization, C.-A.Y. and J.-G.C.; Data curation, C.-A.Y. and J.-C.Y.; Formal analysis, C.-A.Y. and J.-C.Y.; Funding acquisition, C.-A.Y.; Investigation, C.-A.Y., J.-P.L., J.-C.Y., I.-L.L., and Y.-C.H.; Methodology, C.-A.Y., J.-P.L., J.-C.Y., I.-L.L., Y.-C.C., and J.-G.C.; Project administration, J.-P.L. and J.-L.L.; Resources, J.-P.L., J.-L.L., and J.-G.C.; Supervision, J.-L.L. and J.-G.C.; Validation, Y.-C.H.; Visualization, Ch.-A.Y.; Writing—original draft, C.-A.Y.; Writing—review & editing, J.-G.C.
Funding
The project was supported by funds from China Medical University, grant number CMU105-N-07, from China Medical University Hospital, Taiwan, grant DMR-107-205, and from the Ministry of Science and Technology (MOST), Taiwan, grant 106-2314-B-039-047-MY3.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shi C., Pamer E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 3.Roberts C.A., Dickinson A.K., Taams L.S. The interplay between monocytes/macrophages and CD4(+) T cell subsets in rheumatoid arthritis. Front. Immunol. 2015;6:571. doi: 10.3389/fimmu.2015.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy S. miRNA in macrophage development and function. Antioxid. Redox Signal. 2016;25:795–804. doi: 10.1089/ars.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Self-Fordham J.B., Naqvi A.R., Uttamani J.R., Kulkarni V., Nares S. MicroRNA: Dynamic regulators of macrophage polarization and plasticity. Front. Immunol. 2017;8:1062. doi: 10.3389/fimmu.2017.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Xiao J., Wu N., Liu C., Xu J., Liu F., Wu L. MicroRNA-223 regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPβ. Cell Rep. 2015;13:1149–1160. doi: 10.1016/j.celrep.2015.09.073. [DOI] [PubMed] [Google Scholar]
- 7.Simpson L.J., Ansel K.M. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J. Clin. Investig. 2015;125:2242–2249. doi: 10.1172/JCI78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y.G., Satpathy A.T., Chang H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heward J.A., Lindsay M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang R., Tang J., Chen Y., Deng L., Ji J., Xie Y., Wang K., Jia W., Chu W.M., Sun B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathy N.W., Chen X.M. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J. Biol. Chem. 2017;292:12375–12382. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy M.B., Medvedev A.E. Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity. J. Leukoc. Biol. 2016;99:839–850. doi: 10.1189/jlb.2RU1215-575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Cao X. Long noncoding RNAs in innate immunity. Cell Mol. Immunol. 2016;13:138–147. doi: 10.1038/cmi.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satpathy A.T., Chang H.Y. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 16.Wei S., Zhao M., Wang X., Li Y., Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J. Hematol. Oncol. 2016;9:44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B., et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Chao T.C., Chang K.Y., Lin N., Patil V.S., Shimizu C., Head S.R., Burns J.C., Rana T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amarante M.K., de Lucca F.L., de Oliveira C.E., Pelegrinelli Fungaro M.H., Reiche E.M., Muxel S.M., Ehara Watanabe M.A. Expression of noncoding mRNA in human blood cells activated with synthetic peptide of HIV. Blood Cells Mol. Dis. 2005;35:286–290. doi: 10.1016/j.bcmd.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Liu A.Y., Torchia B.S., Migeon B.R., Siliciano R.F. The human NTT gene: Identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997;39:171–184. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 21.Eshleman E.M., Delgado C., Kearney S.J., Friedman R.S., Lenz L.L. Down regulation of macrophage IFNGR1 exacerbates systemic L. monocytogenes infection. PLoS Pathog. 2017;13:e1006388. doi: 10.1371/journal.ppat.1006388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vande Walle L., Van Opdenbosch N., Jacques P., Fossoul A., Verheugen E., Vogel P., Beyaert R., Elewaut D., Kanneganti T.D., Van Loo G., et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattabiraman D.R., Gonda T.J. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 24.Samusik N., Krukovskaya L., Meln I., Shilov E., Kozlov A.P. PBOV1 is a human de novo gene with tumor-specific expression that is associated with a positive clinical outcome of cancer. PLoS ONE. 2013;8:e56162. doi: 10.1371/journal.pone.0056162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan T., Wu R., Liu B., Wen H., Tu Z., Guo J., Yang J., Shen G. PBOV1 promotes prostate cancer proliferation by promoting G1/S transition. Onco Targets Ther. 2016;9:787–795. doi: 10.2147/OTT.S92682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An G., Ng A.Y., Meka C.S., Luo G., Bright S.P., Cazares L., Wright G.L.J., Veltri R.W. Cloning and characterization of UROC28, a novel gene overexpressed in prostate, breast, and bladder cancers. Cancer Res. 2000;60:7014–7020. [PubMed] [Google Scholar]
- 27.Wang L., Niu C.H., Wu S., Wu H.M., Ouyang F., He M., He S.Y. PBOV1 correlates with progression of ovarian cancer and inhibits proliferation of ovarian cancer cells. Oncol. Rep. 2016;35:488–496. doi: 10.3892/or.2015.4396. [DOI] [PubMed] [Google Scholar]
- 28.Xin J., Li J., Feng Y., Wang L., Zhang Y., Yang R. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. Onco Targets Ther. 2017;10:1307–1315. doi: 10.2147/OTT.S124201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Li X., Jiang C., Qian W., Tse G., Chan M.T.V., Wu W.K.K. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. 2018;51:51. doi: 10.1111/cpr.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigdel K.R., Cheng A., Wang Y., Duan L., Zhang Y. The emerging functions of long noncoding rna in immune cells: Autoimmune diseases. J. Immunol. Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J., Kim D., Han J., Kim Y., Lee M., Jin E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015;15:121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 32.Wu G.C., Pan H.F., Leng R.X., Wang D.G., Li X.P., Li X.M., Ye D.Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015;4:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R.G., Rosenthal N., Nerlov C. A CREB-C/EBPβ cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber R., Pietsch D., Panterodt T., Brand K. Regulation of C/EBPβ and resulting functions in cells of the monocytic lineage. Cell Signal. 2012;24:1287–1296. doi: 10.1016/j.cellsig.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Pham T.H., Langmann S., Schwarzfischer L., El Chartouni C., Lichtinger M., Klug M., Krause S.W., Rehli M. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J. Biol. Chem. 2007;282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 36.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Y., Liu X., Xie M., Liu M., Ye M., Li M., Chen X.M., Li X., Zhou R. The NF-κβ-responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 2017;199:3571–3582. doi: 10.4049/jimmunol.1700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrovic-Djergovic D., Popovic M., Chittiprol S., Cortado H., Ransom R.F., Partida-Sanchez S. CXCL10 induces the recruitment of monocyte-derived macrophages into kidney, which aggravate puromycin aminonucleoside nephrosis. Clin. Exp. Immunol. 2015;180:305–315. doi: 10.1111/cei.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu B., Li J., Wu C., Liu C., Yan X., Chang X. CXCL10 and TRAIL are upregulated by TXNDC5 in rheumatoid arthritis fibroblast-like synoviocytes. J. Rheumatol. 2018;45:335–340. doi: 10.3899/jrheum.170170. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Q., Kim T., Pang J., Sun W., Yang X., Wang J., Song Y., Zhang H., Sun H., Rangan V., et al. A novel function of CXCL10 in mediating monocyte production of proinflammatory cytokines. J. Leukoc. Biol. 2017;102:1271–1280. doi: 10.1189/jlb.5A0717-302. [DOI] [PubMed] [Google Scholar]
- 41.Karin N., Razon H. Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine. 2018;109:24–28. doi: 10.1016/j.cyto.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Carvalheiro T., Horta S., Van Roon J.A.G., Santiago M., Salvador M.J., Trindade H., Radstake T., da Silva J.A.P., Paiva A. Increased frequencies of circulating CXCL10-, CXCL8- and CCL4-producing monocytes and Siglec-3-expressing myeloid dendritic cells in systemic sclerosis patients. Inflamm. Res. 2018;67:169–177. doi: 10.1007/s00011-017-1106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.H., Kim B., Jin W.J., Kim H.H., Ha H., Lee Z.H. Pathogenic roles of CXCL10 signaling through CXCR3 and TLR4 in macrophages and T cells: Relevance for arthritis. Arthritis Res. Ther. 2017;19:63. doi: 10.1186/s13075-017-1353-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.