Abstract
In immunosuppressed patients, BKPyV-variants emerge carrying rearranged non-coding control-regions (rr-NCCRs) that increase early viral gene region (EVGR) expression and replication capacity. BKPyV also encodes microRNAs, which have been reported to downregulate EVGR-encoded large T-antigen transcripts, to decrease viral replication in infected cells and to be secreted in exosomes. To investigate the interplay of NCCR and microRNAs, we compared archetype- and rr-NCCR-BKPyV infection in cell culture. We found that laboratory and clinical rr-NCCR-BKPyV-strains show higher replication rates but significantly lower microRNA levels than archetype virus intracellularly and in exosomes. To investigate whether rr-NCCR or increased EVGR activity modulated microRNA levels, we examined the (sp1-4)NCCR-BKPyV, which has an archetype NCCR-architecture but shows increased EVGR expression due to point mutations inactivating one Sp1 binding site. We found that microRNA levels following (sp1-4)NCCR-BKPyV infection were as low as in rr-NCCR-variants. Thus, NCCR rearrangements are not required for lower miRNA levels. Accordingly, Sp1 siRNA knock-down decreased microRNA levels in archetype BKPyV infection but had no effect on (sp1-4)- or rr-NCCR-BKPyV. However, rr-NCCR-BKPyV replication was downregulated by exosome preparations carrying BKPyV-microRNA prior to infection. To explore the potential relevance in humans, urine samples from 12 natalizumab-treated multiple sclerosis patients were analysed. In 7 patients, rr-NCCR-BKPyV were detected showing high urine BKPyV loads but low microRNAs levels, whereas the opposite was seen in 5 patients with archetype BKPyV. We discuss the results in a dynamic model of BKPyV replication according to NCCR activity and exosome regulation, which integrates immune selection pressure, spread to new host cells and rr-NCCR emergence.
Keywords: polyomavirus, non-coding control region, microRNA, exosomes, persistence, immunosuppression, BK virus
1. Introduction
BK polyomavirus (BKPyV) is one of more than 10 human polyomaviruses (HPyV) and it infects >90% of the general human population without ill effects [1,2,3]. BKPyV causes nephropathy or haemorrhagic cystitis in immunocompromised patients, particularly after kidney or allogeneic hematopoietic cell transplantation [4,5]. Seroprevalence data indicate that BKPyV transmission occurs in early childhood [6] via the oral or respiratory route [7]. Subsequently, BKPyV reaches the renourinary tract, presumably by primary viremia [7], where the virus establishes a latent infection [8]. Asymptomatic low-level BKPyV shedding has been demonstrated in 10% of BKPyV IgG-seropositive healthy blood donors [9] providing evidence of immune escape in adult immunocompetent hosts. Antibody levels decline during adult life [9,10] unless significant re-exposure occurs which includes immunocompromised patients [11,12,13,14]. In immunocompromised individuals, however, urinary BKPyV shedding is more frequent, often with urine viral loads of exceeding 7 log10 copies/mL that become apparent as “decoy cell” shedding [2,15,16,17,18]. In kidney transplant or allogeneic hematopoietic cell transplant patients, high-level BKPyV replication precedes nephropathy or haemorrhagic cystitis, respectively [4,5]. Increased BKPyV reactivation rates have also been observed in other solid organ transplant recipients and in HIV/AIDS patients [19,20,21]. However, the molecular details of regulating BKPyV replication in immunocompetent individuals and the relevance for disease progression are not well understood.
BKPyV has a double-stranded DNA genome of approximately 5 kb, which can be divided into: (i) the early viral gene region (EVGR) encoding the small and the large T-antigen; (ii) the late viral gene region (LVGR) encoding the capsid proteins Vp1, Vp2, Vp3, agnoprotein and microRNAs; and (iii) the non-coding control region (NCCR) [2,22]. The NCCR harbours the origin of genome replication ori and promoter/enhancer with DNA-binding sites for transcription factors mediating the secondary host cell specificity [23], as well as the timing and course of EVGR-expression, viral DNA-replication and LVGR-expression [2].
BKPyV sequences commonly found in the urine of healthy persons have an archetype NCCR architecture of sequence blocks arbitrarily denoted O-P-Q-R-S [2,9]. In immunocompromised patients with BKPyV-disease, viral variants with rearranged NCCR (rr-NCCR) architecture have been shown to emerge as majority species and are associated with disease severity [24]. In these patients, archetype NCCR-BKPyV is still found in urine but molecular cloning has demonstrated the presence of a viral quasi-species with co-existing rr-NCCR minority species [25], which may be an indicator of imminent pathology [26]. In vitro studies and in vivo observations support the view that rr-NCCRs confer a higher replicative activity in vitro but which depends on the lack of cellular immune functions in vivo [24]. A similar link between rr-NCCR and disease was also observed for JCPyV in HIV/AIDS patients with progressive multifocal leukoencephalopathy [27,28], or for HPyV-7 and HPyV-9 [23]. Together, the data suggest that HPyV-NCCR rearrangements arising in immunocompromised patients are not only a surrogate marker of long-standing immunologically uncontrolled replication but also represent a virulence determinant of activated EVGR expression and increased replication capacity causing disease.
Given the complex diversity of NCCR rearrangements that affect various transcription factor binding sites as well as the overall architecture, a systematic study of inactivating specific transcription factor binding sites by point mutation was conducted, which maintained the linear archetype architecture of BKPyV-NCCR [29]. Of note, mutations inactivating the Sp1 site located proximal to the LVGR promoter termed SP1-4 resulted in a phenotype functionally equivalent to NCCR rearrangements (group 1) and which had been identified in patients with BKPyV disease [29]. Intriguingly, a similar, albeit low-affinity Sp1 binding site SP1-2 has been located upstream of the EVGR promoter, the inactivation of which (e.g., sp1-2) decreased EVGR expression and replication (group 3) [29]. Further mutational dissection of the BKPyV-NCCR as well as electrophoretic mobility shift assays and chromatin immunoprecipitation analysis revealed that EVGR-expression involves a classic inducible TATA-box promoter. Upstream, the EVGR promoter partially overlaps with a constitutive housekeeping gene-type LVGR promoter using a TATA-like box in the opposite orientation, in which a high-affinity SP1-4 sites acts as a central switch of bidirectional gene expression [30]. sp1-4 point mutations inactivating Sp1 binding cause constitutive activation of EVGR expression and increased viral replication without NCCR rearrangements [30].
Besides the NCCR, another layer of regulating EVGR expression has been described at the posttranscriptional level mediated by microRNAs (miRNAs) miRNA-B1-3p and miRNA-B1-5p encoded in the distal LVGR [22,31,32,33]. Similar to other PyVs [31,33,34,35], BKPyV miRNAs are short noncoding RNAs, which target large T-antigen transcripts and thereby down-regulate viral replication [32,36,37]. This posttranscriptional safeguard has been implicated in the escape from large T-antigen-specific cytotoxic T-cells [34], which have recently been linked to the effective curtailing of BKPyV replication in kidney transplant patients [38,39]. The regulation of miRNAs expression is subject of ongoing studies, which may involve sequences close to the miRNA gene as well as the NCCR [22,40,41]. Moreover, miRNAs of BKPyV and the closely related JCPyV miRNAs have been detected in blood, urine and cerebrospinal fluid samples, often together with the corresponding viral loads with few cases reporting the nature of the NCCR structures [42,43]. It has been suggested that urinary exosomes associated BKPyV-miRNA may be a surrogate marker for BKPyV pathology [44]. Thus, the association between miRNAs and exosomes has also raised questions about their regulatory potential in non-infected neighbouring cells [43,45,46].
Since the NCCR and the miRNA represent two different, formally independent modalities of regulating BKPyV replication at the transcriptional and post-transcriptional level, respectively, we examined the interplay of NCCR architecture and miRNAs expression following BKPyV infection. Imperiale and colleagues reported that BKPyV miRNA levels were low following infection with laboratory strains carrying rr-NCCR. In a first step, we therefore compared miRNA levels in archetype and different laboratory- and patient-derived rr-NCCR-BKPyV variants in cell culture. We also examined the miRNA levels seen with the (sp1-4)NCCR-BKPyV and (sp1-2)NCCR-BKPyV, which, as outlined above, have an archetype architecture but show increased and decreased EVGR expression and viral replication, respectively. The role of Sp1 was further evaluated by siRNA-SP1 known-down. To investigate whether or not rr-NCCR-BKPyV replication could still be down-regulated by mircoRNA, we transferred exosomes carrying BKPyV-microRNA cargo onto uninfected cells prior to infection. Finally, we explore the potential relationship of BKPyV load, NCCR architecture and miRNA levels in vivo in urines from multiple sclerosis patients treated with natalizumab. We then attempt to integrate the results in a dynamic model of transcriptional and posttranscriptional regulation and discuss the potential implications in immunocompetent and immunosuppressed hosts, favouring archetype and rearranged NCCR-BKPyV, respectively.
2. Materials and Methods
2.1. Urine Sampling
The urine samples used in this study were obtained from 12 relapsing-remitting multiple sclerosis patients undergoing intravenous natalizumab treatment at the Multiple Sclerosis Center, Neurological Institute Mondino, Pavia, and had enrolled for a previous study [47]. None of them developed symptoms or signs of polyomavirus-associated diseases at presentation or during the follow-up. The study was approved by the local institutional review board, “Fondazione Istituto Neurologico Mondino”, Pavia, Italy (n. 101MS326). Human samples were taken after obtaining the informed consent from the patients or control subjects in accordance with the tenets of the Declaration of Helsinki.
2.2. Cell Cultures and BkPyV Molecular Clones
African green monkey kidney, SV40-transformed Cos-7 cells line (CRL1651, ATCC, Manassas, VA, USA [48]) were grown in Dulbecco modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum (FBS, SIGMA, Milan, Italy). Primary human renal proximal tubule epithelial cells (RPTECs, PCS-400-010, ATCC, Manassas, VA, USA) were grown in an epithelial cell medium supplemented with epithelial cell growth supplement and 2% FBS (ScienceCell Research Laboratory, Carlsbad, CA, USA). BKPyV strains were derived from molecular clones carrying archetype NCCR (clone WW1.4) [24], a rr-NCCR laboratory strain (Dunlop, Akron, OH, USA) [49], clinical rr-NCCR variants having deletions in the Q- and the R-block (clone del5.3, del15.10) [24,29] and the archetype NCCR variants carrying point mutations of Sp1 binding sites in the Q-block (clone sp1-4) or in the P-block (clone sp1-2) [29,30].
2.3. BKPyV Infection of Cos-7 and RPTECs
Transfection of BKPyV genomic DNA into Cos-7 cells was performed at 90–95% confluence in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. At 6 h posttransfection, the medium was replaced by DMEM containing 10% FBS. At 14 days posttransfection, Cos-7 cells were harvested by scraping off cells in 1/10 of the cell culture supernatant. Virus was released by 3 cycles of freeze-thawing of the cells and centrifugation at 800 g for 5 min. For the infection experiments, Cos-7 and RPTECs were seeded at 7.5 × 105 cells and 3.12 × 105 cells per well in 6-well plate with 2 mL of DMEM (10% FBS) or supplemented EpiCM medium (2% FBS). After 24 h at a confluence of approximately 70%, Cos-7 and RPTECs were exposed to 500 microliters of the corresponding virus preparations (MOI 1) obtained from Cos-7 cells after transfection at 37 °C for 2 h followed by removal and replacement with DMEM (10% FBS) or supplemented EpiCM medium (0.5% FBS). Cells and supernatant were harvested at 12, 24, 48 and 72 h post infection. Titration revealed that approximately 5–10 × 106 genome copies of BKPyV-Dunlop genomes on 50,000 RPTECs or Cos-7 cells at infection typically elicits a multiplicity of infection of 1 after 48 h according to fluorescent focus forming units using staining for large T-antigen; H.H. Hirsch and M. Wernli, unpublished results and see References [50,51].
2.4. Plasmid Exosomes BKPyV miRNA Expression in Cos-7 Cells
BKPyV-miRNAs were inserted into the pcDNA 6.2GW/EmGFP-miR (Invitrogen, Carlsbad, CA, USA) vectors and cloned according to the manufacturer’s instructions. To this end, specific oligonucleotide pairs were designed on the BKPyV miRNA mature sequence (5′TGCTGATCTGAGACTTGGGAAGAGCATTTTTGGCCACTGACTGAATGCTCTTCCCATCTCAGAT3′ forward oligonucleotide and 5′CCTGATCTGAGATGGGAAGAGCATTCAGTCAGTGGCCAAAAATGCTCTTCCCAAGTCTCAGATC3′ reverse oligonucleotide; BKPyV-miRNA mature sequence is underlined below the sequences shown). Next, the corresponding expression vectors pcDNA6-BK-miRNA (10 µg) were transfected into Cos-7 cells (7.5 × 105 cells per well in 6-well plate) using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). BKPyV-miRNA expression vectors system (encodes for the green fluorescence protein GFP) efficiency was assessed at 24h after transfection based on the number of GFP-positive cells, as determined by flow cytometry. In subsequently experiments exosomes carrying BKPyV-miRNA were purified at 24 h, 48 h and 72 h from transfected Cos-7 cells and were quantified. The sequence of the miRNA inserts was confirmed by sequencing.
2.5. Exosome Enriched Vesicles Extraction
The exosome-enriched vesicles were isolated starting from 250 µL of cells supernatant collected by prior centrifugation at 14,000 g for 20 min, using the exosome-specific extraction kit (Norgen, Thorold, ON, Canada), following the manufacturer’s protocols. Characterization of the exosome-preparation was done by demonstrating the presence of 112 nm vesicles present at a density of a mean total particles concentration of 107/mL. Further characterization was achieved by western blot, detecting three markers of exosome tetraspanin/CD63, CD81 and Annexin II (Supplement Figure S1). Before analysis, exosome-enriched vesicles were treated with RNase and DNase to remove miRNA and DNA not protected inside the exosomes.
2.6. Exosome Addition and Anti BKPyV-miRNA Inhibition in Cos-7 Cells
Exosomes carrying BKPyV-miRNA (103 copies) were added to the Cos-7 cells (7.5 × 105 cells per well in 6-well plate) and used to test the effect on subsequent viral infection. Where indicated, the exosomes were added to Cos-7 cells in the absence or presence of inhibitory specific antago-BK-miRNA-5p (Inh-BK-miRNA, a phosphorothioate oligonucleotide with the sequence complementary to the BK-miRNA) synthetic molecule, which had been transfected into Cos-7 cells using a 5 µM final concentration, together with lipofectamine 2000 (Life Technologies, Foster City, CA, USA). Unrelated miRNA (Unrel-miRNA, a phosphorothioate oligonucleotide with an unrelated sequence) synthetic molecule, was used as control.
2.7. BKPyV DNA Quantification
Viral DNA was extracted from 0.15 mL of urine, from 2 × 104 cells and cell supernatant of tissue cultures with the High Pure PCR Template Preparation kit (Roche, Basel, Switzerland). DNA extracted was subjected to quantitative real-time PCR (qPCR) assays using primer and probe targeting Vp1 gene (BKVPf forward primer 5′-AGTGGATGGGCAGCCTATGTA-3′, BKVPr reverse primer 5′-TCATATCTGGGTCCCCTGGA-3 and BKVPp TaqMan MGB probe labelled with VIC VIC-5′ AGGTAGAAGAGGTTAGGGTGTTTGATGGCACAG 3′) (Life Technologies, Foster City, CA, USA). Each reaction was carried out with negative controls (no template) and DNA standards (diluted to contain 101–106 copies per millilitre) of a plasmid containing the BKPyV molecular clone. The lowest limit of detection of the assay was 10 copies per millilitre of urine and micrograms of total DNA [52].
2.8. BKPyV Pre-miRNA and Mature miRNA Quantification
Total RNA was isolated from 2.0 × 106 Cos-7 and RPTECs cells using the mirVana miRNA isolation Kit (Ambion, Foster City, CA, USA), and from exosomes contained in 250 µL of cell-free supernatant that had previously been centrifuged at 14,000 g for 20 min using an RNA exosome-specific circulating extraction kit (Norgen). The miRNA expression was measured and quantified with a specific pre-miRNA and mature bk-miRNA-5p quantitative stem-loop RT-PCR MiRNA assay whose primers were designed on the specific region of the BKPyV WW clone (Life Technologies, Foster City, CA, USA) according to the manufacturer’s protocol. Each reaction was performed in triplicate using 10 ng of extracted RNA, including negative controls (no template) and synthesized oligonucleotides as standards (diluted to contain 101–106 copies). The lowest detection limit of the assay was 10 copies/ng of RNA. The assay was specific and reproducible, as demonstrated in preliminary experiments using a BKPyV oligonucleotide standard (with <0.5 Ct value inter-assay variation) and observing no amplification of unrelated oligonucleotide targets.
2.9. siRNA knock-Down of Sp1 and Immunoblotting
Cos-7 cells were transfected in a 6-well plate using per well 2 µg of siRNA targeting Sp1 (SP1-siRNA, Mission esiRNA SIGMA), or 2 µg of siRNA scrambled form (Scr-siRNA), or mock-treated without siRNA added (mock). At 24 h post-transfection, Cos-7 cells were infected at multiplicity of infection (MOI) of 1, using BKPyV-infectious supernatant obtained after transfection of Cos-7 cells. BK-miRNA-5p expression in cells and presence in exosomes enriched vesicles were measured at 48 h post infection. Immunoblots were used to compare Sp1 expression and cytochrome P450 as control. Briefly, cell extracts were prepared from Cos-7 cells at 48 h after transfection of SP1-siRNA, Scr-siRNA and mock-treatment and 15 µg of extract were analysed by 10% SDS-PAGE, transferred onto nitrocellulose membrane (BIO-RAD), block the membrane with 5% bovine serum albumin and then incubated with the rabbit anti-Sp1 (PLA0044, SIGMA) and rabbit anti-cytochrome-P450 polyclonal antibody (PA1-343, SIGMA) followed by a peroxidase-conjugated anti-rabbit IgG antibody (A0545, SIGMA).
2.10. NCCR Sequencing
A nested-PCR was used to obtain the NCCR product for sequencing. Briefly, 100 ng of total DNA was amplified using two pairs of primers: first pair of primers, BKTT1 forward 5′ AAG GTC CAT GAG CTC CAT GGA TTC TTC C 3′ and BKTT2 reverse 5′ CTA GGT CCC CCA AAA GTG CTA GAG CAG C 3′, generating a 684 bp DNA fragment; the second pair of primers, BK-1 forward 5′ GGCCTCAGAAAAAGCTTCCACACCCTTACTACTTGA 3′ and BK-2 reverse 5′ CTTGTCGTGACAGCTGGCGCAGAA 3′, that amplified a portion of the first amplicon generating a fragment of 354 bp. The PCR products were purified using the PCR purification Kit (Qiagen, Hilden, Germany) and sequenced using the BigDye Terminator Cycle-Sequencing Ready Reaction (Applied Biosystems, Foster City, CA, USA). The sequences were analysed and edited using Bioedit 5.0.9 (Tom Hall of Ibis Therapeutics, Carlsbad, CA, USA).
2.11. Statistical Tests
The data were analysed using Student’s t-tests. p-values less than 0.05 were considered statistically significant.
3. Results
3.1. Rearranged NCCR-BKPyV Variants Show Higher Viral Loads But 10-Fold Lower miRNA Expression than Archetype NCCR BKPyV
To address the role of NCCR and microRNAs, we compared archetype and rearranged NCCR-BKPyV in cell culture. Accordingly, cloned genomes of the archetype NCCR-BKPyV-WW(1.4) and the laboratory stain BKPyV-Dunlop, which carries a rr-NCCR (Figure 1) were transfected into Cos-7 cells and infectious supernatants were prepared. Following infection, BKPyV-loads rapidly increased in Dunlop-infected cells but only slowly in archetype WW(1.4) infected cultures over 72h post infection (Figure 2A; left top panel). As shown previously [24,50,53], only one viral replication cycle is complete for the Dunlop strain under these conditions and only few secondary infection events have started as evidenced by large T-antigen positive/Vp1-negative immunofluorescence. BKPyV miRNA also increased during this time course but the levels seen in the archetype-infected cells (Figure 2A; left middle panel) and in the corresponding exosome preparations (Figure 2A; left bottom panel) were almost as high as in the Dunlop-infected cultures and therefore much higher than expected from the replication kinetics and the available genome copy number in particular.
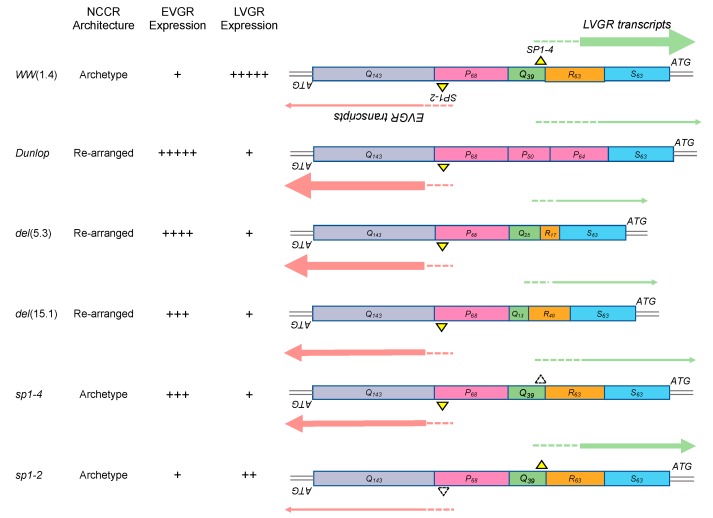
Figure 1.
Schematic representation of NCCR architecture and EVGR and LVGR expression pattern of BKPyV strains. BKPyV NCCR architecture, Sp1 binding sites and expression levels are schematically summarized for the archetype (WW1.4), the laboratory-derived rr-NCCR (Dunlop), isolates from kidney transplant patients carrying rearranged NCCRs: BKPyV rr-NCCR del(5.3) and del(15.10) genome, as well as the BKPyV archetype NCCR carrying point mutations inactivating Sp1 binding to LVGR-proximal (sp1-4) and EVGR-proximal binding sites (sp1-2). Sp1 binding to the directional (non-palindromic) sequences is indicated as yellow triangle, dashed-line triangles indicate point mutations. The sequence blocks O, P, Q, R and S are indicated by coloured boxes and their respective length in base pairs in subscripts. The relative expression levels of the EVGR and the LVGR is represented by the thickness of the red and green arrows, whereby +++++, very strong; to + weak (see references [24,29,30]).
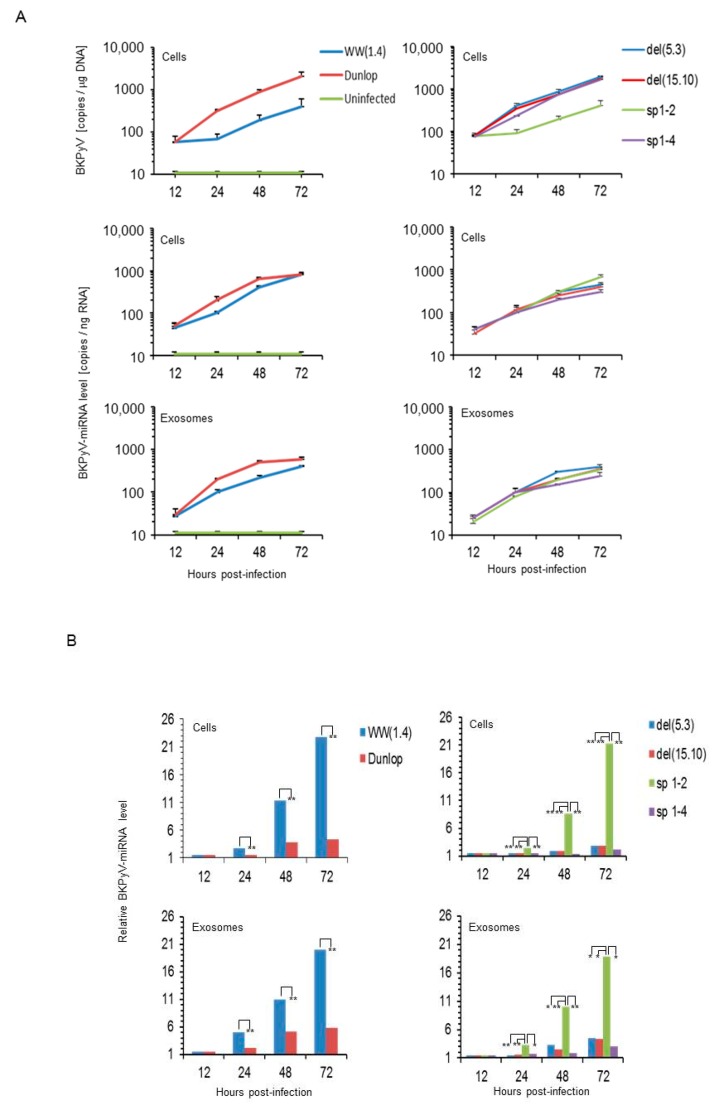
Figure 2.
Viral replication and miRNA levels of archetype and different laboratory- or patient-derived BKPyV variants in cell culture. (A) Cos-7 cells were infected with the indicated strains (Figure 1) and viral load and BK-miRNA-5p expression were measured in cells and exosome preparations at 12, 24, 48 and 72 h post infection. BKPyV genome load was expressed per total µg DNA; miRNA given per total ng RNA (mean ± standard deviation of 3 independent experiments). (B) Time course comparing of miRNA expression levels in cells or exosome preparations following infection with archetype or indicated NCCR variant strains as described in Figure 1. Results are expressed as BKPyV-miRNA relative levels to DNA genome copy number normalized to archetype WW(1.4) clone values obtained at 12 h post-infection set to 1 (-fold; *, p < 0.05; ** p < 0.01).
Direct comparison of the miRNA levels expressed per BKPyV genome copy number revealed significantly higher expression in cells and in exosome preparations generated during archetype BKPyV infection than observed for the rr-NCCR-Dunlop strain (Figure 2B; left panels).
Given the difference in NCCR architecture between archetype WW(1.4) and rearranged laboratory Dunlop virus, we were interested to examine the patient-derived rr-NCCRs del(5.3) and del(15.10) variants that had emerged in kidney transplant recipients and had been shown to increase EVGR expression and replication rates in several previous studies [24,29]. The results demonstrated that both of patient-derived rr-NCCR variants behaved similar to the Dunlop-strain showing high replication levels and low miRNA expression in cells and exosome preparations (Figure 2A,B; right panels).
To investigate whether NCCR rearrangements or increased EVGR activity played a role in modulating the miRNA levels, we examined the (sp1-4)NCCR-BKPyV, which has an archetype architecture but shows increased EVGR expression and viral replication due to point mutations inactivating the SP1-4 binding site upstream of the LVGR transcription start site [29,30] (Figure 1). As a control, we chose the (sp1-2)NCCR-BKPyV, in which the SP1-2 site located upstream of the EVGR transcription start site was inactivated by point mutations and which showed only low EVGR expression and replication [29,30] (Figure 1). We observed high replication rates and low miRNA expression in cells and exosome-preparations for the (sp1-4)NCCR-BKPyV, whereas the opposite was true for (sp1-2)NCCR-BKPyV (Figure 2A,B; right panels). The results indicated lower miRNA expression was associated with increased EVGR expression and replication rates and that rearrangements of the NCCR were not necessary for the observed reduction in miRNA levels.
3.2. Sp1 Expression Levels Modulate BKPyV miRNA Levels in Archetype NCCR Virus And Variants With Low EVGR Expression But Not in Viral Variants Having an Activated EVGR Expression
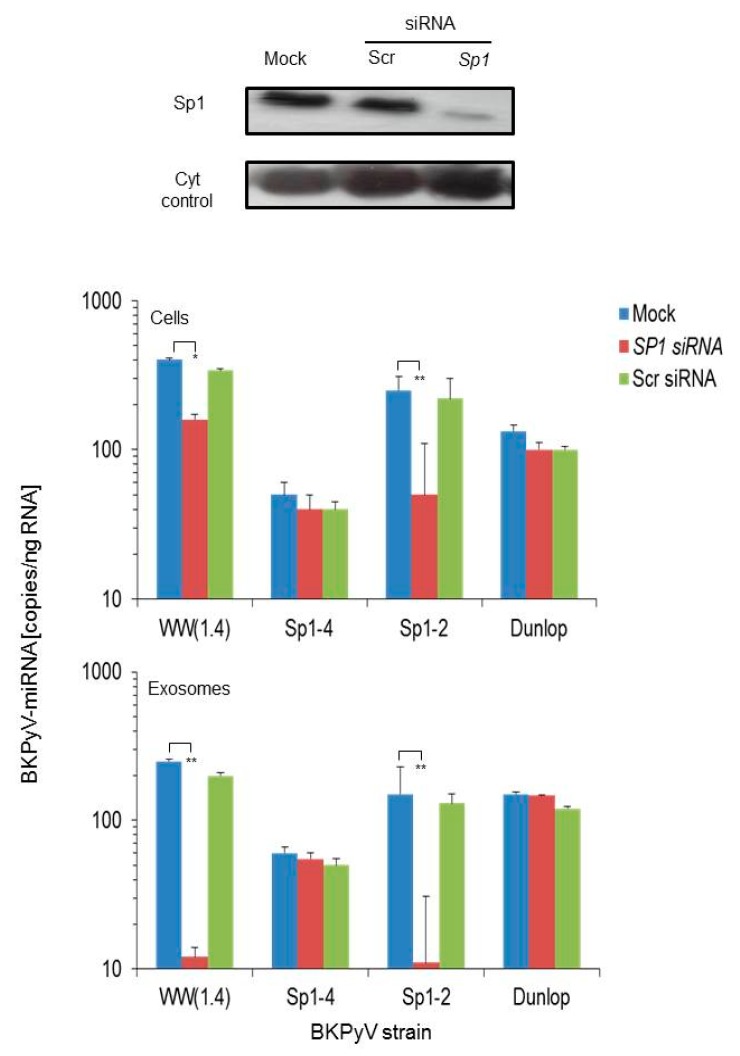
Since the bi-directional expression of the archetype NCCR is critically modulated by Sp1 binding sites with high (SP1-4) and low affinity (SP1-2) binding sites in the EVGR and LVGR promoter, respectively [29,30], we wondered if knock-down of Sp1 had any influence on miRNA expression levels. Accordingly, Cos-7 cells were transfected with siRNA targeting SP1, with scrambled siRNA, or only mock-treated and miRNA expression levels were analysed after infection with archetype, the two point-mutant derivatives sp1-2 and sp1-4; and the Dunlop strain (Figure 1).
Sp1 knock-down was verified by immunoblotting demonstrating a significant reduction in the siRNA-SP1- but not in the scrambled siRNA- or the mock-treated control cells (Figure 3; top panel). In the mock and scrambled siRNA controls, the original observation was confirmed showing high levels of BKPyV-miRNA following archetype- and (sp1-2)NCCR-BKPyV infection, whereas low levels were seen following (sp1-4)NCCR-BKPyV and Dunlop infection, both in cells and corresponding exosome preparations (Figure 3; blue and green bars). In the siRNA-SP1 treated cells, no change was seen in BKPyV-miRNA levels in Dunlop- or (sp1-4)NCCR-BKPyV-infected cells, whereas a significant reduction was seen in archetype and (sp1-2)NCCR-BKPyV-infected cells (Figure 3; red bars). The results indicated that knock-down of Sp1 protein by siRNA was associated with a significant reduction in cellular and exosomal miRNA levels in BKPyV strains having an intact SP1-4 binding site, whereas there was little change in cells infected with virus strains showing constitutively activated EVGR expression. Thus, Sp1 availability for binding on the SP1-4 site appeared to play a key role in BKPyV-miRNA expression levels.
Figure 3.
Effect of Sp1 knock-down on BKPyV miRNA-5p expression. Cos-7 cells were transfected with SP1-siRNA, scrambled Scr-siRNA or mock-treated and at 24 h posttransfection, infected with the indicated viral strains. Top panel shows the immunoblot of Sp1 and cytochrome P450 protein levels at 48 h post-transfection as described in Materials & Methods. Middle and lower panel shows the BK-miRNA-5p expression measured in cells and in exosome preparations at 48 h post infection, respectively. The values result from 3 independent experiments (mean ± standard deviation; Student’s t test, ** p < 0.01).
3.3. BKPyV miRNA Levels in Primary Human Renal Tubular Epithelial Cells (RPTECs) Inversely Correlate with the Evgr-Activity and Replication Rate of Archetype and rr-NCCR-Viruses
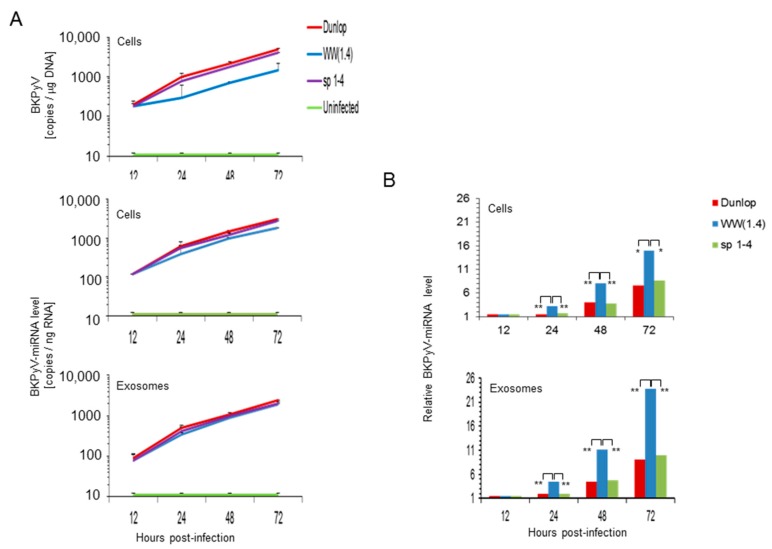
To extend these studies to RPTECs, the natural target of BKPyV infection in the human host, these cells were infected with archetype WW(1.4)-, (sp1-4)- and Dunlop strains and BKPyV-DNA load and miRNA-5p expression was measured at 12, 24, 48 and 72 h post-infection. As described previously [29], BKPyV-Dunlop and the point mutant sp1-4 replicated significantly faster than the archetype BKPyV-WW(1.4) but there was little difference in miRNA per total RNA in cells or exosome preparations (Figure 4A). However, the BKPyV-miRNA-5p levels per viral genome were significantly higher in the archetype-infected RPTECs and exosomes than in the corresponding preparations following Dunlop or (sp1-4)-infection (Figure 4B).
Figure 4.
RPTECs infected with BKPyV variants having increased EVGR activity are associated with lower miRNA levels than archetype BKPyV. (A) Time course of BKPyV DNA replication and miRNA-5p expression in RPTECs and corresponding exosome- preparations. RPTECs were infected and viral DNA genome and BKPyV-miRNA-5p expression were measured at 12, 24, 48 and 72 h post infection as detailed in Materials & Methods. BKPyV genome load was expressed per total µg DNA; miRNA was quantified per total ng RNA (mean ± standard deviation of 3 independent experiments). (B) Comparison of relative miRNA expression levels per BKPyV genome copy (template) in cells. Results are expressed as BKPyV-miRNA levels normalized to BKPyV genome copy number, archetype WW(1.4) values at 12 h post-infection are set to 1 (-fold; * p < 0.05; ** p < 0.01).
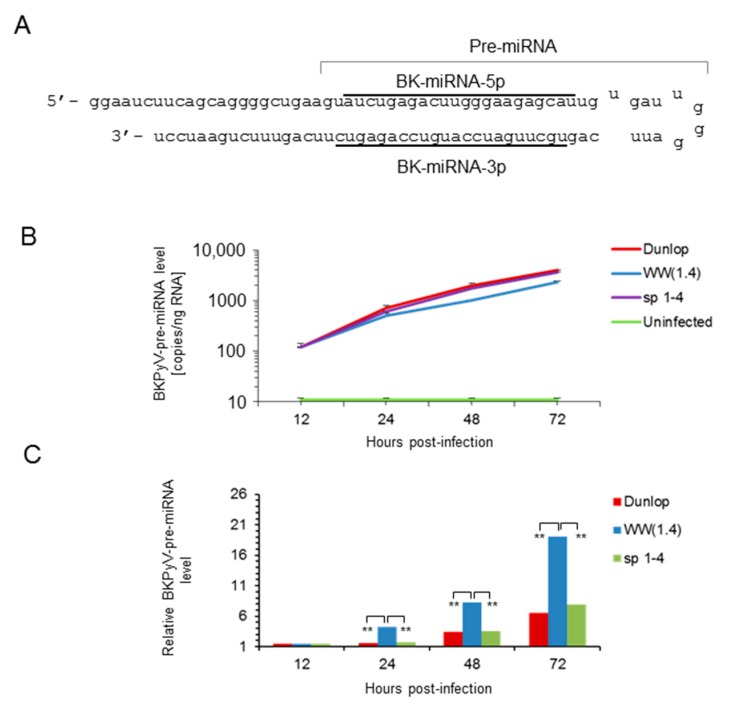
To address the question, whether or not the differences in mature BKPyV-miRNA-5p were related to the expression levels of the miRNA gene or differences in maturation or degradation, we examined the amount of precursor miRNA (pre-miRNA) which consists of the 5′- and 3′-miRNA linked by a hairpin loop in RPTECs (Figure 5). The results indicated that the BKPyV pre-miRNA levels behaved similar to what has been observed for the mature BKPyV-miRNA-5p, namely being high in archetype virus but low in the variants (Dunlop and sp1-4 mutant) having increased EVGR activity.
Figure 5.
Time course of BKPyV pre-miRNA expression in infected RPTECs. (A) Schematic representation of the BKPyV pre-miRNA hairpin and the corresponding BKPyV-miRNA-5p and miRNA-3p sequence (underlined). (B) Time course of BKPyV pre-miRNA expression levels after infection of RPTECs with the indicated strains. Mean values ± standard deviation of 3 independent experiments are shown. (C) Comparison of relative BKPyV pre-miRNA expression levels per BKPyV genome copy (template) in cells. Results are expressed as BKPyV-miRNA levels normalized to BKPyV genome copy number, archetype WW(1.4) values at 12 h post-infection are set to 1 (-fold; ** p < 0.01).
3.4. Pre-Infection Addition of BKPyV-Exosome Preparations Containing BKPyV miRNA Inhibit BKPyV Replication of Dunlop or sp1-4 Mutant But Have No Effect On Archetype BKPyV or sp1-2 Mutant Strains
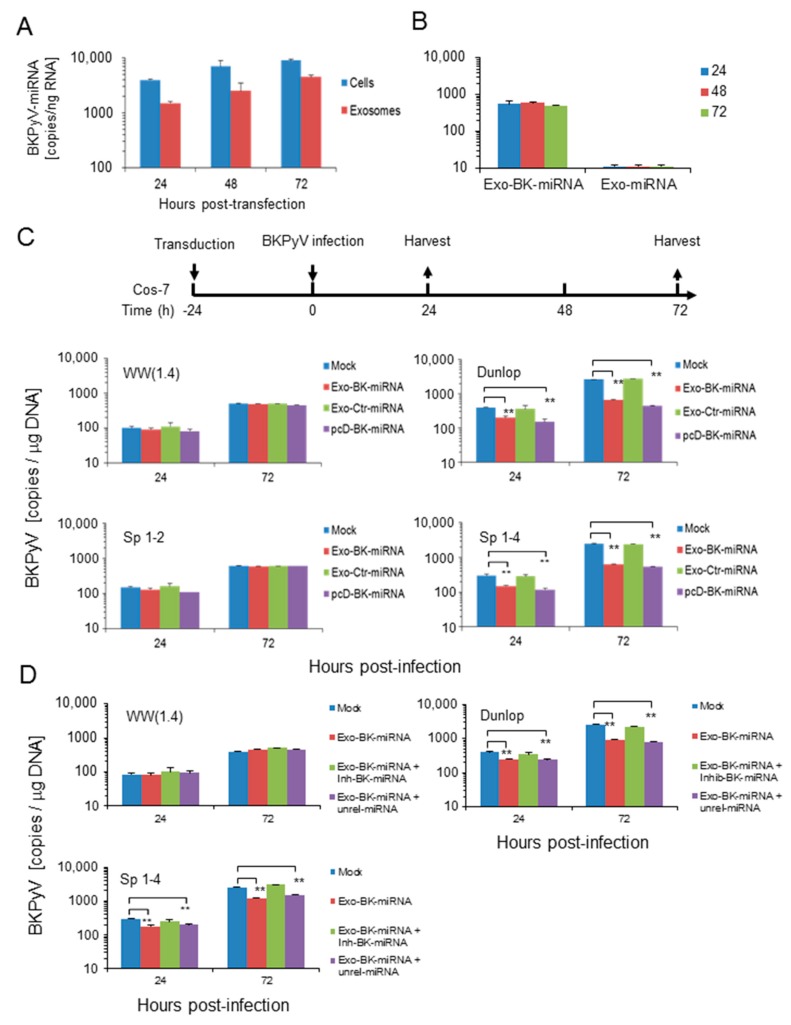
Given the rapid replication of BKPyV-variants with activated EVGR expression having only low levels of BKPyV-miRNA per genome template, we wondered if rr-NCCR-BKPyV replication could still be down-regulated by miRNA. To this end, we transferred exosome preparations carrying BKPyV-microRNA cargo onto uninfected cells prior to BKPyV infection. To avoid potential interference or confounding factors associated with exosome preparations from BKPyV-infected cells, we prepared exosomes from Cos-7 cells transfected with the plasmid vector pcDNA6-BK-miRNA expressing BKPyV miRNA-5p and the pcDNA6-miRNA vector expressing an unrelated miRNA-5p as control (see Materials & Methods).
The time course up to 72 h post-transfection showed increasing BKPyV-miRNA-5p levels in cells and in corresponding exosome preparations (Figure 6A). Exosome uptake and internalization after addition on Cos-7 cells was verified by the positive fusion assay performed based on octadecyl-rhodamine lipid probe dequenching as previously described (Supplement Figure S2). The amount of detectable BKPyV-miRNA-5p after exosome addition did not change up to 72 h post-addition, whereas no BKPyV-miRNA was detected when using exosomes from control-transfected cells (Figure 6B).
Figure 6.
Addition of BKPyV-exosome preparations containing BKPyV-miRNA-5p before infection inhibits replication of BKPyV variants with activated EVGR expression (Dunlop and sp1-4 mutant) but not archetype or (sp1-2) BKPyV replication. (A) Cos-7 cells were transfected with pcDNA6-BK-miRNA (pcD-BK-miRNA). Then, BK-miRNA expression in Cos-7 cells and in exosomes enriched vesicles derived from supernatant was measured after indicated time post transfection. (B) Detection of BKPyV-miRNA-5p in Cos-7 cells after addition of exosome preparations from pcDNA6-BK-miRNA-5p and control transfected or mock-treated cells. (C) BKPyV loads after exosome addition prepared from cells transfected with the indicated expression vectors (Exo-BK-miRNA, unrelated miRNA, or mock), or after direct transfection of the pcDNA6-BK-miRNA expression vector (see flow-chart). (D) BKPyV loads after exosome addition prepared from cells transfected with the indicated expression vectors (Exo-BK-miRNA, unrelated miRNA, in the presence or absence of a specific antago BK-miRNA called Inh-BK-miRNA), or after direct transfection of the pcDNA6-BK-miRNA expression vector (see flow-chart). The corresponding exosome preparations are indicated according to the transfection vector (e.g., BKPyV-miRNA-5p; unrelated miRNA; mock; in the presence of Inh-BK-miRNA, a synthetic phosphoro-thioate oligonucleotide with the sequence complementary to the BK-miRNA or a corresponding unrelated miRNA. The values represent the mean ± standard, ** p < 0.01.
We next tested the effect of exosome preparation on subsequent viral infection. The results indicate that replication of the BKPyV-Dunlop and the point mutant (sp1-4)-NCCR-BKPyV was significantly inhibited, if infection was performed after addition of exosomes prepared from BKPyV-miRNA-5p transfected cells. Instead, exosomes prepared from cells transfected with an unrelated miRNA expression vector or from untransfected cells (mock) had no effect (Figure 6C; top panels). Under these conditions, archetype BKPyV and the (sp1-2)-point mutant replicated slowly but without significant inhibition by exogenous exosome addition. Of note, no inhibition of BKPyV-Dunlop or sp1-4 mutant replication was observed, when exosome preparations carrying BKPyV-miRNA-5p and a specific antago-BK-miRNA (Inh-BK-miRNA) molecules, whereas unrelated synthetic miRNA (Unrel-miRNA) had no effect (Figure 6D; bottom panels).
3.5. Analysis of the BKPyV NCCR Architecture and Exosomes Content in Urine Samples of Immunocompromised Patients
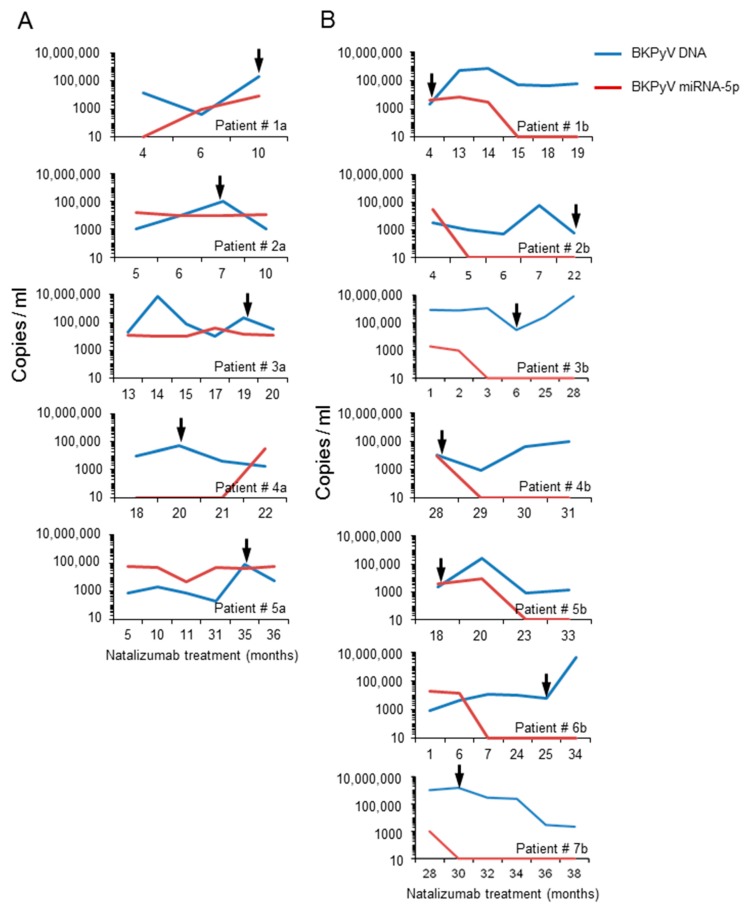
To explore the potential relevance in humans, urine samples from 12 natalizumab-treated multiple sclerosis patients were analysed. Twelve out of 25 BKPyV-DNA positive patients were identified among 42 natalizumab-treated multiple sclerosis patients enrolled in previous study [47]. The sequencing of the BKPyV NCCRs identified the presence of predominantly rr-NCCR in the urine samples collected from 7 patients, subjected to long term natalizumab treatment. Archetype NCCR BKPyV strains were detected in the urine samples from the remaining 5 patients. Overall, the urine BKPyV load in patients with predominantly rr-NCCR BKPyV was higher than in patients with archetype BKPyV. The opposite was true for BKPyV miRNA-5p expression levels, being low in the 7 patients carrying rr-NCCR variants and in the 5 patients shedding archetype NCCR BKPyV (Figure 7).
Figure 7.
BKPyV DNA viral load and miRNA-5p expression in urine samples from multiple sclerosis patients. Urine sample were analysed over indicated time of follow-up and at the indicated time points, urine BKPyV loads and miRNA levels in exosome preparations were determined. Arrows indicate the time at which NCCR sequence was obtained from urine samples. Panels on the left show the results obtained for 5 patients with BKPyV carrying archetype NCCRs (patient ID 1a–5a); Panels on right show those with rearranged NCCR-BKPyV (patient ID 1b–7b).
4. Discussion
The regulation of BKPyV persistence and reactivation is receiving increasing attention [8] in the light of the almost universal but well controlled BKPyV infection of the general human population and the increasing BKPyV diseases in patients under potent immunosuppressive regimens [4,5,54,55]. So far, two major mechanisms of regulating BKPyV replication have emerged, which involve transcriptional, NCCR-based and post-transcriptional miRNA-based mechanisms targeting EVGR expression [24,41]. Pioneering work from Imperiale and co-workers has demonstrated that both layers of regulation appear to interact since low BKPyV miRNA-5p levels were seen in infection by rapidly replicating BKPyV carrying rr-NCCRs, whereas high miRNA-5p levels were seen in archetype BKPyV [41].
In this study, we extend these observations from laboratory strains to patient-derived isolates and show that BKPyV miRNA-5p levels are decreased following infection of patient BKPyV carrying diverse NCCR rearrangements (Figure 1). Moreover, we demonstrate that point mutations inactivating a single Sp1 binding site (sp1-4) in an otherwise archetype NCCR have a similar effect. This (sp1-4)-mutation has been shown to confer a phenotype of increased EVGR expression and more rapid viral replication (group 1) similar to rr-NCCR BKPyV variants [29]. Thus, our results indicate that NCCR rearrangements are not required for the lowered miRNA levels but suggest that the activation of EVGR expression is important. The latter was supported by another Sp1 point mutant (sp1-2) showing which permits only modest EVGR expression, while LVGR expression is also reduced (group 3) [29] and which showed high-levels of BKPyV miRNA similar to the archetype WW(1.4)-NCCR-BKPyV. Of note, the BKPyV miRNA-5p expression of archetype and (sp1-2)-mutated NCCR-BKPyV could be significantly diminished by reduced Sp1 levels following siRNA-SP1 knock-down. Conversely, variants carrying rearranged or (sp1-4)-mutated NCCRs were not affected. Parallel experiments in human RPTECs generated similar results, suggesting that the observations were also relevant in the human situation. The data support the notion that BKPyV miRNA-5p levels and the NCCR activity are critically linked by Sp1 binding to the SP1-4 site. Independently, a principle role of Sp1 for BKPyV replication has recently emerged from a genome-wide interference study [56].
While our study provides a detailed accounting of the inverse association of EVGR activity and miRNA-5p expression, the exact molecular mechanisms need to be addressed further. Inverting the orientation of the rr-NCCR has been shown to confer high LVGR expression at the expense of EVGR [23,24] and corresponding recombinant viruses show increased miRNA expression [41]. Whether such LVGR transcription opens the access to the miRNA promoter region, provides transcription factors and enzymatic complexes, or extended transcripts is presently discussed. Conversely, activated EVGR transcription might simply outcompete the available miRNA transcripts, or confer resistance to downregulation by another as yet unknown mechanism including antisense stealth transcripts or processing from pre-miRNA to miRNA. In our study, we observed that BKPyV pre-miRNA were similarly affected by the NCCR activity as the mature miRNA-5p suggesting that transcript generation rather than miRNA processing, maturation or degradation was affected. No evidence for a difference in cellular miRNA by packaging and secretion as exosomes was obtained, since both cellular and exosomal levels mirror-imaged the activity of the NCCR in both compartments. The possibility that BKPyV miRNAs were no longer able to downregulate replication of variants with activated EVGR was refuted in time course experiments adding BKPyV-miRNA-5p loaded exosome preparations to host cells prior to infection, which demonstrated a significant reduction of the BKPyV-Dunlop and the (sp1-4)-point mutant. This interpretation was supported by inclusion of an antagonist synthetic phosphorothioate oligonucleotide reversing replication inhibition.
The polyomavirus miRNA-5p has been proposed to act as an important safeguard silencing residual large-T-antigen expression of the archetype BKPyV during viral persistence [34,41]. Efficient transcriptional and posttranscriptional synergy in downregulation of large T-antigen expression may permit escape from cytotoxic CD8 T-cell effectors, which we have been shown to preferentially target T-antigen epitopes [38,39]. Conversely, a bi-directional link between NCCR activity and miRNA-expression appears biologically plausible, when signals of activating EVGR expression are sensed in the latently infected host cell, for example by displacing Sp1 from the SP1-4 binding site in the LVGR promoter [30]. Downregulating the posttranscriptional miRNA then permits for an efficient progression through the viral life cycle. In immunosuppressed patients lacking sufficient CD8 T-cell activity, the high and prolonged viral replication allows for the emergence of rr-NCCR variants conferring an activated EVGR expression and high-replication capacity [24], while reducing posttranscriptional interference through BK-miRNA-5p downregulation
Given the potential clinical relevance for immunocompromised patients, we explored BKPyV shedding, NCCR architecture and miRNA levels in multiple sclerosis patients treated with natalizumab. We found that patients shedding BKPyV with rr-NCCRs had on average higher urine viral loads but lower miRNA-5p levels in the exosome-enriched vesicles. Conversely, patients shedding BKPyV with the archetype NCCR architecture showed lower viral loads and typically higher miRNA-5p levels in urinary exosome preparations. Thus, these preliminary data, if confirmed, seem to be consistent in their almost dichotomous nature and also provide an incentive for further work, which may be of relevance for clinical diagnostic and therapeutic approaches [42,44].
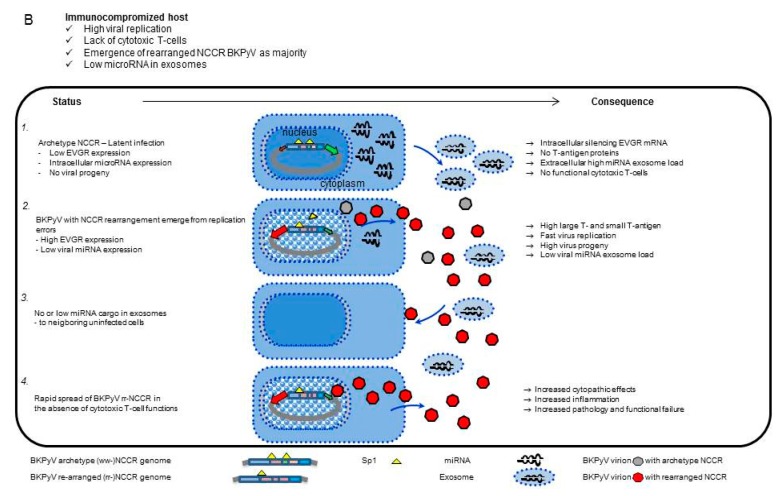
Finally, there is increasing evidence reported that other viruses including those that have the propensity to establish latent/persistent infections such as herpesviruses use miRNA regulation not only intracellularly but also in exosomes [57]. Thus, besides a principle role in virus biology, our data, together with those of other researchers, strongly suggest that viral miRNA should be explored further in a virological and clinical context. To integrate our results into the work of other researchers and to stimulate the corresponding projects, we present a model (see Figure 8) in which host cell signals, viral NCCR activity and miRNA expression permit fine tuning of persistence, reactivation of replication, spread to neighbouring host cells, for example, in the epithelial monolayer of the renal tubules and hiding from cytotoxic large T-antigen-specific CD8 T-cells, unless the host is immunocompromised. The potential role of exosomes and miRNA cargo in cell to cell communication offers an interesting possibility that could potentially be harnessed for antiviral therapy.
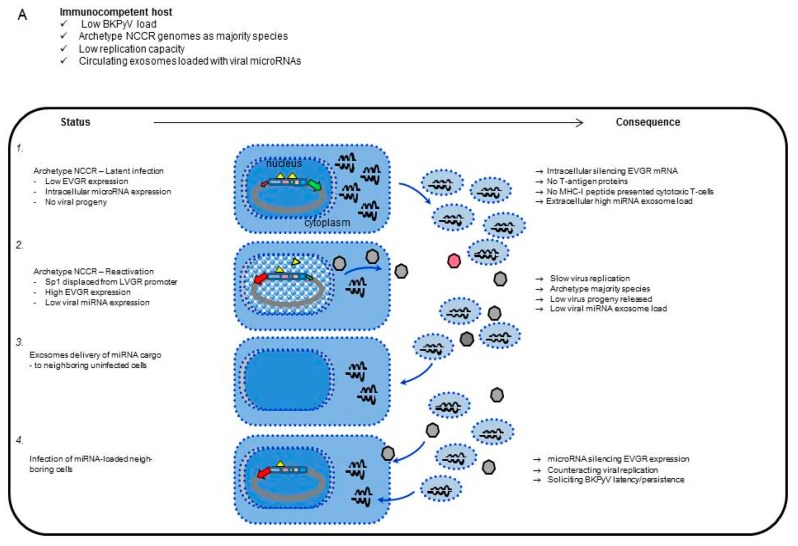
Figure 8.
Model of BKPyV miRNAs expression and exosome traffic. (A) Immunocompetent host:1- Archetype NCCR-BKPyV establishes latent infection with low EVGR expression and strong silencing by high-level miRNA expression; 2- Displacement of Sp1 from SP1-4 through host cell activation permits activation of EVGR expression and lowering of miRNA expression; 3- Exosomes deliver BKPyV miRNA cargo to uninfected cells; 4- Subsequent infection with archetype BKPyV counteract new rounds of viral replication, thereby avoid recognition by cytotoxic T-cells. (B) Immunocompromised host: 1- Status and consequences during BKPyV archetype persistent infection; 2- BKPyV with rearranged NCCR emerge in immunocompromised patients as the immune selection pressure fades; 3- No or low miRNA cargo in exosome delivery to uninfected neighbouring cells; 4- Rapid spread, increased cytopathology and functional failure.
Acknowledgments
The work was supported by grants from the Fondazione “Istituto di Ricerca Virologica Oretta Bartolomei Corsi” Florence, Italy to S.G.; and an appointment grant by the University of Basel to H.H.H.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4915/10/9/466/s1, Figure S1: Size, concentration and markers of Cos-7 exosome enriched vesicles., Figure S2: Cells adsorption and penetration of exosomes enriched vesicles containing BKPyV miRNAs.
Author Contributions
“F.M., S.G., H.H.H. and Z.W. conceived and designed the experiments; F.M., Z.W., M.C.G. and F.H.W., performed the experiments; S.G., H.H.H., S.D. and P.F. analyzed the data; F.M., Z.W., F.H.W., M.C.G., S.D., and P.F. contributed reagents/materials/analysis tools; S.G. and H.H.H. wrote the paper.”
Funding
Istituto di Ricerca Virologica Oretta Bartolomei Corsi, 2016.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.DeCaprio J.A., Imperiale M.J., Major E.O. Polyomaviruses. In: Knipe D.M., Howley P., editors. Fields Virology. 6th ed. LWW; Philadelphia, PA, USA: 2013. pp. 1633–1661. [Google Scholar]
- 2.Greenlee J.E., Hirsch H.H. Polyomaviruses. In: Microbiology A.A.S.F., editor. Clinical Virology. 4th ed. John Wiley & Sons Ltd.; Chichester, UK: 2017. pp. 599–623. [Google Scholar]
- 3.Rinaldo C.H., Hirsch H.H. The human polyomaviruses: From orphans and mutants to patchwork family. APMIS. 2013;121:681–684. doi: 10.1111/apm.12125. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch H.H., Babel N., Comoli P., Friman V., Ginevri F., Jardine A., Lautenschlager I., Legendre C., Midtvedt K., Munoz P., et al. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clin. Microbiol. Infect. 2014;20(Suppl. 7):74–88. doi: 10.1111/1469-0691.12538. [DOI] [PubMed] [Google Scholar]
- 5.Cesaro S., Dalianis T., Hanssen Rinaldo C., Koskenvuo M., Pegoraro A., Einsele H., Cordonnier C., Hirsch H.H., Group E. ECIL guidelines for the prevention, diagnosis and treatment of BK polyomavirus-associated haemorrhagic cystitis in haematopoietic stem cell transplant recipients. J. Antimicrob. Chemother. 2018;73:12–21. doi: 10.1093/jac/dkx324. [DOI] [PubMed] [Google Scholar]
- 6.Knowles W.A. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV) Adv. Exp. Med. Biol. 2006;577:19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch H.H., Steiger J. Polyomavirus BK. Lancet Infect. Dis. 2003;3:611–623. doi: 10.1016/S1473-3099(03)00770-9. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale M.J., Jiang M. Polyomavirus Persistence. Annu. Rev. Virol. 2016;3:517–532. doi: 10.1146/annurev-virology-110615-042226. [DOI] [PubMed] [Google Scholar]
- 9.Egli A., Infanti L., Dumoulin A., Buser A., Samaridis J., Stebler C., Gosert R., Hirsch H.H. Prevalence of Polyomavirus BK and JC Infection and Replication in 400 Healthy Blood Donors. J. Infect. Dis. 2009;199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 10.Kean J.M., Rao S., Wang M., Garcea R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leuenberger D., Andresen P.A., Gosert R., Binggeli S., Strom E.H., Bodaghi S., Rinaldo C.H., Hirsch H.H. Human polyomavirus type 1 (BK virus) agnoprotein is abundantly expressed but immunologically ignored. Clin. Vaccine Immunol. 2007;14:959–968. doi: 10.1128/CVI.00123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodaghi S., Comoli P., Boesch R., Azzi A., Gosert R., Leuenberger D., Ginevri F., Hirsch H.H. Antibody Responses to Recombinant Polyomavirus BK Large T and VP1 Proteins in Pediatric Kidney Transplant Patients. J. Clin. Micro. 2009;47:2577–2585. doi: 10.1128/JCM.00030-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastrana D.V., Brennan D.C., Cuburu N., Storch G.A., Viscidi R.P., Randhawa P.S., Buck C.B. Neutralization serotyping of BK polyomavirus infection in kidney transplant recipients. PLoS Pathog. 2012;8:e1002650. doi: 10.1371/journal.ppat.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt T., Adam C., Hirsch H.H., Janssen M.W., Wolf M., Dirks J., Kardas P., Ahlenstiel-Grunow T., Pape L., Rohrer T., et al. BK Polyomavirus-Specific Cellular Immune Responses Are Age-Dependent and Strongly Correlate With Phases of Virus Replication. Am. J. Transplant. 2014;14:1334–1345. doi: 10.1111/ajt.12689. [DOI] [PubMed] [Google Scholar]
- 15.Funk G.A., Gosert R., Comoli P., Ginevri F., Hirsch H.H. Polyomavirus BK replication dynamics in vivo and in silico to predict cytopathology and viral clearance in kidney transplants. Am. J. Transplant. 2008;8:2368–2377. doi: 10.1111/j.1600-6143.2008.02402.x. [DOI] [PubMed] [Google Scholar]
- 16.Randhawa P., Vats A., Shapiro R. Monitoring for polyomavirus BK and JC in urine: Comparison of quantitative polymerase chain reaction with urine cytology. Transplantation. 2005;79:984–986. doi: 10.1097/01.TP.0000157573.90090.FD. [DOI] [PubMed] [Google Scholar]
- 17.Drachenberg C.B., Papadimitriou J.C., Hirsch H.H., Wali R., Crowder C., Nogueira J., Cangro C.B., Mendley S., Mian A., Ramos E. Histological patterns of polyomavirus nephropathy: Correlation with graft outcome and viral load. Am. J. Transplant. 2004;4:2082–2092. doi: 10.1046/j.1600-6143.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch H.H., Knowles W., Dickenmann M., Passweg J., Klimkait T., Mihatsch M.J., Steiger J. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 2002;347:488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- 19.Randhawa P., Uhrmacher J., Pasculle W., Vats A., Shapiro R., Eghtsead B., Weck K. A comparative study of BK and JC virus infections in organ transplant recipients. J. Med. Virol. 2005;77:238–243. doi: 10.1002/jmv.20442. [DOI] [PubMed] [Google Scholar]
- 20.Jiang M., Abend J.R., Johnson S.F., Imperiale M.J. The role of polyomaviruses in human disease. Virology. 2009;384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egli A., Helmersen D.S., Taub K., Hirsch H.H., Johnson A. Renal Failure Five Years After Lung Transplantation Due to Polyomavirus BK-Associated Nephropathy. Am. J. Transplant. 2010;10:2324–2330. doi: 10.1111/j.1600-6143.2010.03265.x. [DOI] [PubMed] [Google Scholar]
- 22.Imperiale M.J. Polyomavirus miRNAs: The beginning. Curr. Opin. Virol. 2014;7:29–32. doi: 10.1016/j.coviro.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ajuh E., Wu Z., Kraus E., Weissbach F.H., Bethge T., Gosert R., Fischer N., Hirsch H.H. Novel Human Polyomavirus non-coding control regions differ in bi-directional gene expression according to host cell, large T-antigen expression and clinically occurring rearrangements. J. Virol. 2018;92:e02231-17. doi: 10.1128/JVI.02231-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosert R., Rinaldo C.H., Funk G.A., Egli A., Ramos E., Drachenberg C.B., Hirsch H.H. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen G.H., Hirsch H.H., Rinaldo C.H. Functional analysis of polyomavirus BK non-coding control region quasispecies from kidney transplant recipients. J. Med. Virol. 2009;81:1959–1967. doi: 10.1002/jmv.21605. [DOI] [PubMed] [Google Scholar]
- 26.Sharma P.M., Gupta G., Vats A., Shapiro R., Randhawa P.S. Polyomavirus BK non-coding control region rearrangements in health and disease. J. Med. Virol. 2007;79:1199–11207. doi: 10.1002/jmv.20909. [DOI] [PubMed] [Google Scholar]
- 27.Gosert R., Kardas P., Major E.O., Hirsch H.H. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. J. Virol. 2010;84:10448–10456. doi: 10.1128/JVI.00614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khanna N., Wolbers M., Mueller N.J., Garzoni C., Du Pasquier R.A., Fux C.A., Vernazza P., Bernasconi E., Viscidi R., Battegay M., et al. JC virus-specific immune responses in human immunodeficiency virus type 1 patients with progressive multifocal leukoencephalopathy. J. Virol. 2009;83:4404–4411. doi: 10.1128/JVI.02657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bethge T., Hachemi H.A., Manzetti J., Gosert R., Schaffner W., Hirsch H.H. Sp1 sites in the noncoding control region of BK polyomavirus are key regulators of bidirectional viral early and late gene expression. J. Virol. 2015;89:3396–33411. doi: 10.1128/JVI.03625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bethge T., Ajuh E., Hirsch H.H. Imperfect Symmetry of Sp1 and Core Promoter Sequences Regulates Early and Late Virus Gene Expression of the Bidirectional BK Polyomavirus Noncoding Control Region. J. Virol. 2016;90:10083–10101. doi: 10.1128/JVI.01008-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan C.S., Grundhoff A., Tevethia S., Treisman R., Pipas J.M., Ganem D. Expression and function of microRNAs in viruses great and small. Cold Spring Harb. Symp. Quant. Biol. 2006;71:351–356. doi: 10.1101/sqb.2006.71.065. [DOI] [PubMed] [Google Scholar]
- 32.Lagatie O., Tritsmans L., Stuyver L.J. The miRNA world of polyomaviruses. Virol. J. 2013;10:268–288. doi: 10.1186/1743-422X-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo G.J., Fink L.H., O’Hara B., Atwood W.J., Sullivan C.S. Evolutionarily conserved function of a viral microRNA. J. Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan C.S., Grundhoff A.T., Tevethia S., Pipas J.M., Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 35.Cantalupo P., Doering A., Sullivan C.S., Pal A., Peden K.W., Lewis A.M., Pipas J.M. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 2005;79:13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cullen B.R. Viruses and microRNAs. Nat. Genet. 2006;38:25S–30S. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 37.Bauman Y., Mandelboim O. MicroRNA based immunoevasion mechanism of human polyomaviruses. RNA Biol. 2011;8:591–594. doi: 10.4161/rna.8.4.15587. [DOI] [PubMed] [Google Scholar]
- 38.Cioni M., Leboeuf C., Comoli P., Ginevri F., Hirsch H.H. Characterization of Immunodominant BK Polyomavirus 9mer Epitope T Cell Responses. Am. J. Transplant. 2016;16:1193–1206. doi: 10.1111/ajt.13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leboeuf C., Wilk S., Achermann R., Binet I., Golshayan D., Hadaya K., Hirzel C., Hoffmann M., Huynh-Do U., Koller M.T., et al. BK Polyomavirus-Specific 9mer CD8 T Cell Responses Correlate With Clearance of BK Viremia in Kidney Transplant Recipients: First Report From the Swiss Transplant Cohort Study. Am. J. Transplant. 2017;17:2591–2600. doi: 10.1111/ajt.14282. [DOI] [PubMed] [Google Scholar]
- 40.White M.K., Safak M., Khalili K. Regulation of gene expression in primate polyomaviruses. J. Virol. 2009;83:10846–10856. doi: 10.1128/JVI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broekema N.M., Imperiale M.J. miRNA regulation of BK polyomavirus replication during early infection. Proc. Natl. Acad. Sci. USA. 2013;110:8200–8205. doi: 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martelli F., Giannecchini S. Polyomavirus microRNAs circulating in biological fluids during viral persistence. Rev. Med. Virol. 2017:e1927. doi: 10.1002/rmv.1927. [DOI] [PubMed] [Google Scholar]
- 43.Rocca A., Martelli F., Delbue S., Ferrante P., Bartolozzi D., Azzi A., Giannecchini S. The JCPYV DNA load inversely correlates with the viral microrna expression in blood and cerebrospinal fluid of patients at risk of PML. J. Clin. Virol. 2015;70:1–6. doi: 10.1016/j.jcv.2015.06.104. [DOI] [PubMed] [Google Scholar]
- 44.Kim M.H., Lee Y.H., Seo J.W., Moon H., Kim J.S., Kim Y.G., Jeong K.H., Moon J.Y., Lee T.W., Ihm C.G., et al. Urinary exosomal viral microRNA as a marker of BK virus nephropathy in kidney transplant recipients. PLoS ONE. 2017;12:e0190068. doi: 10.1371/journal.pone.0190068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giovannelli I., Clausi V., Nukuzuma S., Della Malva N., Nosi D., Giannecchini S. Polyomavirus JC microRNA expression after infection in vitro. Virus Res. 2016;213:269–273. doi: 10.1016/j.virusres.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 46.Giovannelli I., Martelli F., Repice A., Massacesi L., Azzi A., Giannecchini S. Detection of JCPyV microRNA in blood and urine samples of multiple sclerosis patients under natalizumab therapy. J. Neurovirol. 2015;21:666–670. doi: 10.1007/s13365-015-0325-3. [DOI] [PubMed] [Google Scholar]
- 47.Delbue S., Elia F., Carloni C., Pecchenini V., Franciotta D., Gastaldi M., Colombo E., Signorini L., Carluccio S., Bellizzi A., et al. JC virus urinary excretion and seroprevalence in natalizumab-treated multiple sclerosis patients. J. Neurovirol. 2015;21:645–652. doi: 10.1007/s13365-014-0268-0. [DOI] [PubMed] [Google Scholar]
- 48.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 49.Henriksen S., Mittelholzer C., Gosert R., Hirsch H.H., Rinaldo C.H. Human BK Polyomavirus Plasmid pBKV (34-2) (Dunlop) Contains Mutations Not Found in the Originally Published Sequences. Genome Announc. 2015;3:e00046-15. doi: 10.1128/genomeA.00046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirsch H.H., Yakhontova K., Lu M., Manzetti J. BK Polyomavirus Replication in Renal Tubular Epithelial Cells Is Inhibited by Sirolimus but Activated by Tacrolimus Through a Pathway Involving FKBP-12. Am. J. Transplant. 2016;16:821–832. doi: 10.1111/ajt.13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li R., Sharma B.N., Linder S., Gutteberg T.J., Hirsch H.H., Rinaldo C.H. Characteristics of polyomavirus BK (BKPyV) infection in primary human urothelial cells. Virology. 2013;440:41–50. doi: 10.1016/j.virol.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Tremolada S., Delbue S., Castagnoli L., Allegrini S., Miglio U., Boldorini R., Elia F., Gordon J., Ferrante P. Mutations in the external loops of BK virus VP1 and urine viral load in renal transplant recipients. J. Cell Physiol. 2010;222:195–199. doi: 10.1002/jcp.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernhoff E., Gutteberg T.J., Sandvik K., Hirsch H.H., Rinaldo C.H. Cidofovir Inhibits Polyomavirus BK Replication in Human Renal Tubular Cells Downstream of Viral Early Gene Expression. Am. J. Transplant. 2008;8:1413–1422. doi: 10.1111/j.1600-6143.2008.02269.x. [DOI] [PubMed] [Google Scholar]
- 54.Binet I., Nickeleit V., Hirsch H.H., Prince O., Dalquen P., Gudat F., Mihatsch M.J., Thiel G. Polyomavirus disease under new immunosuppressive drugs: A cause of renal graft dysfunction and graft loss. Transplantation. 1999;67:918–922. doi: 10.1097/00007890-199903270-00022. [DOI] [PubMed] [Google Scholar]
- 55.Hirsch H.H., Steiger J., Mihatsch M.J. Immunosuppression and BKV Nephropathy. N. Engl. J. Med. 2002;347:2079–2080. doi: 10.1056/NEJM200212193472517. [DOI] [PubMed] [Google Scholar]
- 56.Zhao L., Imperiale M.J. Identification of Rab18 as an Essential Host Factor for BK Polyomavirus Infection Using a Whole-Genome RNA Interference Screen. mSphere. 2017;2:e00291-174. doi: 10.1128/mSphereDirect.00291-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meckes D.G., Jr. Exosomal communication goes viral. J. Virol. 2015;89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.