Abstract
The CONSTANS (CO)-like gene family has been well studied for its role in the regulation of plant flowering time. However, their role remains poorly understood in cotton. To better understand the possible roles of CO-like in cotton, we performed a comprehensive genome-wide analysis of CO-like genes in cotton. Phylogenetic tree analysis showed that CO-like genes naturally clustered into three groups. Segmental duplication and whole genome duplication (WGD), which occurred before polyploidy, were important contributors to its expansion within the At (“t” indicates tetraploid) and Dt subgenomes, particularly in Group III. Long-terminal repeat retroelements were identified as the main transposable elements accompanying 18 genes. The genotype of GhCOL12_Dt displayed low diversity; it was a candidate involved in domestication. Selection pressure analyses indicated that relaxed purifying selection might have provided the main impetus during the evolution of CO-like genes in upland cotton. In addition, the high expression in the torus and calycle indicated that CO-like genes might affect flowering. The genes from Group II, and those from Group III involved in segmental duplication or WGD, might play important roles in response to drought and salt stress. Overall, this comprehensive genome-wide study of the CO-like gene family would facilitate further detailed studies in cotton.
Keywords: Gossypium, CO-like, segemental duplication, stress response, flowering time
1. Introduction
The CONSTANS (CO) genes encode a small class of transcription factors, characterised by a CCT (CO, CO-like, TOC1 (TIMING OF CAB EXPRESSION1)) domain near the carboxy terminus. The CO-like gene family is subdivided into three groups according to the additional B-box domain near the amino terminus. The genes of Groups I, II, and III have two, one and one zinc-finger B-boxes, respectively; moreover, Group III genes contain a diverged B-box [1]. The CCT domain is involved in nuclear localisation of CO proteins, and the B-box plays vital roles in protein–protein interactions [2,3].
In Arabidopsis, 17 members of the CO-like gene family are divided into three groups. Group I consists of CO and COL1 to COL5, Group II consists of COL6, COL7, COL8, and COL16, and Group III consists of COL9 to COL15 [1]. Increasing numbers of CO-like genes have been identified in several plant species, except Arabidopsis, owing to the increased availability of genome data [4]. Most studies show that CO-like genes play significant roles in the regulation of flowering time in plants. For example, the Arabidopsis CO gene play important roles in the photoperiod pathway and regulation of flowering time [5,6], and its ortholog, Hd1 (Heading date 1), in rice shows a similar biological function and acts as a pivotal regulator in the photoperiod pathway [7,8].
Furthermore, AtCOL5 [9], AtCOL9 [10], and AtCOL12 [11], and the OsCOL9 [12], OsCOL10 [13,14], OsCOL15 [15], and OsCOL16 [16] are involved in regulation of flowering time. In addition to the role in flowering time, recent studies have shown that CO-like genes are associated with plant architecture, abiotic stress response, and auxin homeostasis [17,18,19,20,21]. AtCOL3 is a positive regulator of photomorphogenesis that acts downstream of COP1 (Constitutive photomorphogenic 1) and can promote root growth independent of COP1 [17]. AtCOL4 positively regulates abiotic stress tolerance in plants through abscisic acid (ABA)-dependent pathway. In rice, a CO-like gene, Ghd2 (Grain number, plant height, and heading date2), is a negative regulator for drought tolerance, mediated through regulation of senescence in rice [18]. In Arabidopsis, AtCOL7 is involved in the phytochrome B (phyB)-mediated shade avoidance response and enhanced branching number under high red light:far-red light ratio through phyB-mediated light-quality regulation of auxin homeostasis [19]. AtCOL12 is the substrate of COP1/SPA E3 ligase, and the over-expression of COL12 results in decreased apical dominance and enhanced branching [20]. In rice, OsCOL9 regulates blast resistance through salicylic acid and ethylene signalling pathways [21].
Cotton is the leading fibre crop worldwide and provides large amounts of raw materials for textile fibres and edible oils [22,23,24]. Few studies on CO-like genes in cotton are reported, e.g., ectopic over-expression of cotton COL1 in Arabidopsis rescues the late flowering phenotype of the co-2 mutant [25], but the biological function of most of the CO-like genes are unknown in cotton. Therefore, it is important to understand the biological consequences of the activity of the CO-like gene family in cotton plants. The completion of the cotton genomes makes it possible to perform a comprehensive genome-wide analysis of CO-like genes in cotton [26,27,28,29,30]. Several CO-like genes have been reported in monocotyledonous and dicotyledonous plants, but the evolutionary relationships among these genes are poorly understood.
In the present study, we aimed to identify the CO-like genes from eight different species and use the sequence information for analysing the relationship between monocotyledonous and dicotyledonous plants. Furthermore, we focused on cotton and comprehensively analysed CO-like genes in the allotetraploid upland cotton and two diploid species resembling its wild parents. We performed the identification of gene families, phylogenetic tree analysis, and analysis of segmental duplication, gene structures, domestication selection, chromosome location, and expression patterns.
2. Results
2.1. Gene Identification and Conserved Domain Retrieval
In total, 21, 18, 38, 11, 16, 15, 13, and 16 genes were confirmed as CO-like family members in G. raimondii, G. arboreum, G. hirsutum, cacao, Populus trichocarpa, rice, sorghum, and maize, respectively (Table S1). G. hirsutum contained the largest number of CO-like genes, which was close to that of the two diploid cotton species, G. arboreum and G. raimondii (Table S2). The number of CO-like genes in the diploid cotton G. arboreum exceeded that in maize, rice, sorghum, cacao, and poplar. Because G. raimondii (DD) has a smaller genome size than G. arboreum (AA) and G. hirsutum (DD), it has a higher assembly quality than the two latter species. Therefore, firstly we designated the genes from G. raimondii as COL1 to COL21, and named the genes from G. arboreum (A genome), At subgenome and Dt subgenome of G. hirsutum after their orthologs in D genome. The orthologs from G. raimondii, At subgenome, and Dt subgenome showed good collinearity with homologous chromosomes (Table S2), suggesting that the CO-like genes were conserved in different cotton species during evolution.
2.2. Multiple Sequence Alignment of Conserved Domains
The CO-like was characterised by two conserved domains. The first one was the CCT, which was located near the carboxy terminus, and the second was a zinc-finger B-box domain located near the amino terminus [1]. To better understand the characteristics of the sequences of CO-like genes in cotton, we performed multiple alignments of B-Box and CCT domains. As shown in Figure S1, all genes from upland cotton used in the present study contained the CCT and B-box domain. Nineteen genes contained one CCT domain and two B-boxes (Figure S1A), six genes contained one CCT domain and one B-box domain (Figure S1B), and twelve genes contained one CCT domain, one B-box domain, and one divergent B-box (Figure S1C). Previous reports show that the CO-like genes are divided into three groups based on number and type of B-box. We followed the designation rules and classified the genes in Figure S1A–C into Groups I–III [1]. The sequences of CCT were more conserved within the same groups than among different groups. The sequences of CCT at most of the loci were consensus sequences, and only some loci were divergent. For example, the amino acid residue at 2 (E or V), 9 (R or K), 12 (R or K), 14 (N, K, or T), 18 (E or V), 21 (I or V), 28 (A or S), 32 (M or S), and 41 (A or V) were different. The B-box is a class of zinc fingers, characterised by C-X2-C-X8-C-X7-C-X2-C-X4-H-X8-H, which was identified in Arabidopsis, rice and barley. In the present study, we found that B-box in cotton followed the regular pattern mentioned above, suggesting that the B-boxes are highly conserved during the evolution.
2.3. Phylogenetic Analysis of CO-like Genes among Typical Dicots and Monocots
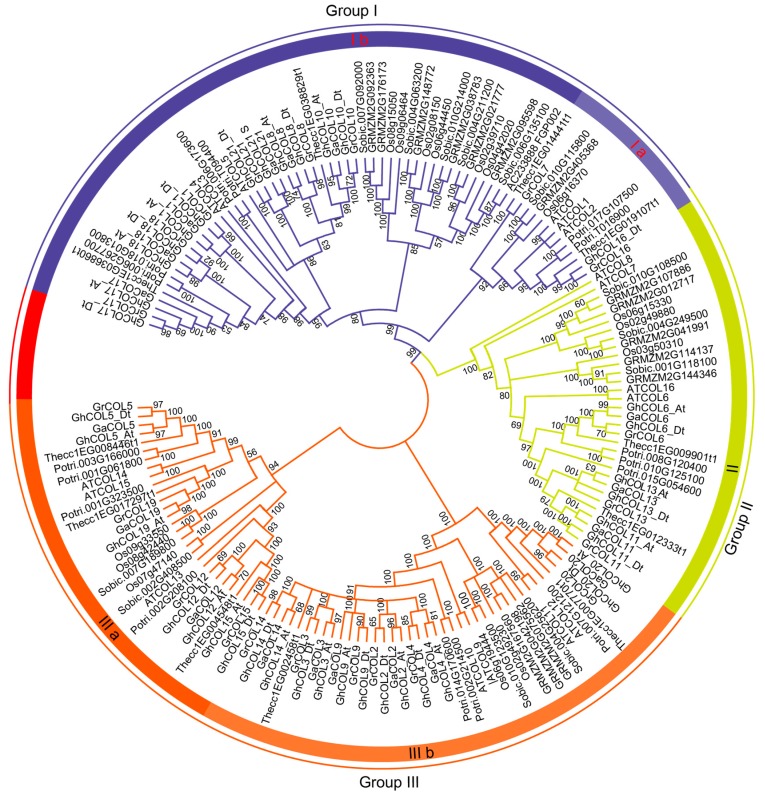
To understand the relationship among the selected species better, we constructed phylogenetic trees. As shown in Figure 1, the CO-like genes were naturally clustered into three groups, designated as I–III. The topology of Maximum likelihood (ML) tree (Figure S2) was similar to that in Figure 1 by neighbour-joining (NJ) method, suggesting that the NJ tree was suitable for further analysis. Group III had the largest number of CO-like genes, whereas Group II had the smallest number of CO-like genes. According to the topology of the tree, Groups I and III have been divided into two sub-groups, termed as a and b, indicating that these two group have experienced expansion, and that their function may have diversified during evolution. The gene numbers from each species in Group II were between two and four, suggesting that this group might not have experienced expansion, and that their biological functions may be very similar.
Figure 1.
Phylogenetic tree of CO-like genes suggesting that CO-like genes were clustered into three groups. The neighbour-joining (NJ) tree was constructed using MEGA 7.0. The outer circle is marked in purple, green, and orange, which represent Groups I, II and III, respectively. The prefixes At, Os, Potri, GRMZM, Sobic, Thecc, Ga, Gr, and Gh, represent A. thaliana, Populus trichocarpa, Oryza sativa, Zea mays, Sorghum bicolor, Theobroma cacao, Gossypium arboreum, G. raimondii, and G. hirsutum, respectively. “At” and “Dt” indicate the A and D subgenomes in upland cotton, respectively. Bootstrapping was used to check the reliability of the tree, and bootstrapping values larger than 50 are displayed near the nodes.
As shown in Figure 1, the genes from cacao always clustered close to cotton, suggesting that the genes of these two species are closely related (Figure 1). The total number of CO-like genes in the diploid G. raimondii was twice of that in cacao, and one cacao gene at the most had three orthologs in diploid cotton. For example, in Group III, Thecc1EG017297t1 only had one ortholog each in the two diploid cotton species, and Thecc1EG002458t1 corresponded to three orthologs in diploid cotton.
Although the orthologs from three cotton species used in the present study display high sequence similarities, the orthologs from the At subgenome and A genome tended to cluster together. This was similar to the tendency observed among the orthologs from the Dt subgenome and D genome. These similarities suggested that the orthologs from At-A or Dt-D might have a common ancestor.
2.4. Gene Expansion and Synteny Analysis
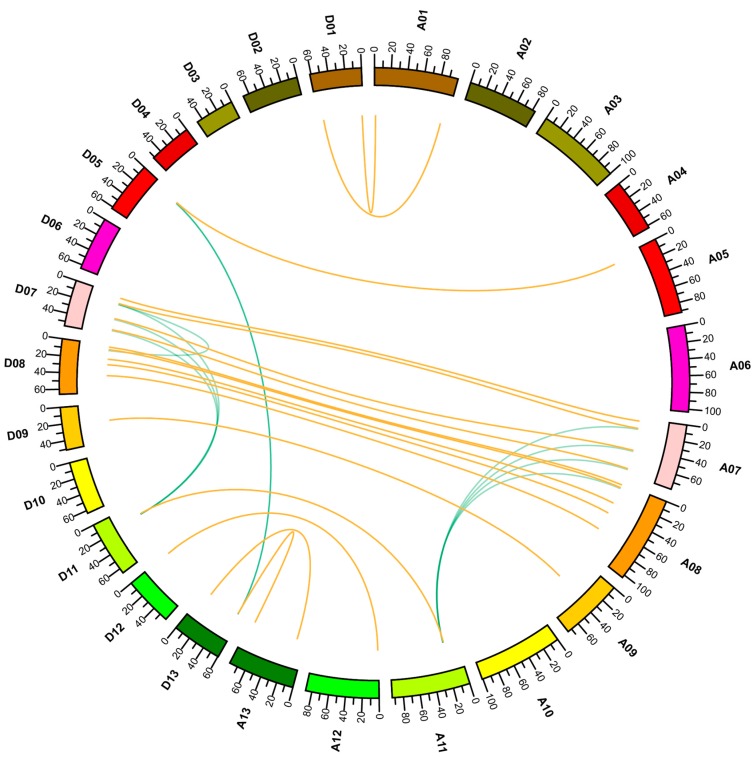
As shown in Figure S3, 36 genes displayed a significant collinearity in the At and Dt subgenomes. Thirty-seven genes were located on 16 chromosomes unevenly, and none of the genes were distributed on 10 chromosomes. Five genes were located on A08 and D08, followed by four on D07/A07, three on D13, two on A01/D01/D13, and one on A05/D05/A09/D09/A12/D12. Only GhCOL21_S was located on scaffold4822 because of incomplete sequencing (Table S2 and Figure S3).
Because most of the orthologs on homologous chromosomes had significant collinearity between At and Dt subgenome, the gene expansion could have occurred before upland cotton formation. Almost half of the CO-like genes were identified as singletons, six genes might have originated from dispersed duplication, and 13 genes were found in collinear blocks, which may have undergone segmental duplication or whole genome duplication (WGD). Except for the GhCOL17–GhCOL18 duplication pair, we noticed that the remaining collinear pairs were from Group III, indicating that segmental duplication or WGD was the major driver of the expansion of Group III. Among the duplicated genes, GhCOL14_At formed four pairs of genes with significant collinearity, and its ortholog GhCOL14_Dt formed three pairs of genes with significant collinearity, indicating that they were active during evolution. There were no tandem and proximal duplications in the chromosomal regions nearby or adjacent to the CO-like genes (Figure 2, Table S3).
Figure 2.
Collinearity analyses within At and Dt subgenomes. The orange lines indicate the ortholog genes from the At and Dt subgenomes. The green lines point towards paralog pairs derived from segmental duplication or whole genome duplication (WGD).
Cotton plants have experienced many duplication events and the gene number has enlarged during evolution. The duplicated genes might have experienced functional divergence, including partial or total loss of the original functions, acquisition of novel functions, or maintenance of a partition of original functions [31]. The CO-like genes may have undergone positive selection, negative selection, and purifying selection during evolution. To inspect the selective pressures exerted on CO-like genes better, we evaluated the ratio of the non-synonymous divergence levels (Ka) versus synonymous divergence levels (Ks) of 27 homologous pairs, including 18 orthologous pairs and nine paralogous pairs derived from segmental duplication/WGD. All the values of Ka/Ks ratios were smaller than 1.0, among which the ratio for five gene pairs was >0.5 and those for five other gene pairs were <0.2 (Table 1), indicating that the CO-like gene family in cotton has experienced relaxed purifying selection pressure, and that their functions may be conserved after gene family expansion.
Table 1.
The Ka and Ks value of homologous gene pairs.
| Paralogous Pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| GhCOL1_At-GhCOL1_Dt | 0.03 | 0.07 | 0.526 |
| GhCOL2_At-GhCOL2_Dt | 0.00 | 0.02 | 0.149 |
| GhCOL3_At-GhCOL3_Dt | 0.01 | 0.04 | 0.199 |
| GhCOL4_At-GhCOL4_Dt | 0.01 | 0.05 | 0.275 |
| GhCOL5_At-GhCOL5_Dt | 0.01 | 0.04 | 0.365 |
| GhCOL6_At_GhCOL6_Dt | 0.01 | 0.03 | 0.550 |
| GhCOL8_At_GhCOL8_Dt | 0.01 | 0.04 | 0.298 |
| GhCOL9_At_GhCOL9_Dt | 0.00 | 0.06 | 0.054 |
| GhCOL10_At-GhCOL10_Dt | 0.01 | 0.06 | 0.159 |
| GhCOL11_At-GhCOL11_Dt | 0.03 | 0.05 | 0.561 |
| GhCOL12_At-GhCOL12_Dt | 0.01 | 0.02 | 0.552 |
| GhCOL13_At-GhCOL13_Dt | 0.01 | 0.03 | 0.365 |
| GhCOL14_At-GhCOL14_Dt | 0.01 | 0.06 | 0.131 |
| GhCOL15_At-GhCOL15_Dt | 0.01 | 0.04 | 0.276 |
| GhCOL17_At-GhCOL17_Dt | 0.01 | 0.03 | 0.205 |
| GhCOL18_At-GhCOL18_Dt | 0.01 | 0.05 | 0.274 |
| GhCOL20_At-GhCOL20_Dt | 0.04 | 0.05 | 0.993 |
| GhCOL21_S-GhCOL21_Dt | 0.04 | 0.07 | 0.515 |
| GhCOL3_Dt-GhCOL14_Dt | 0.09 | 0.41 | 0.213 |
| GhCOL4_Dt-GhCOL14_Dt | 0.09 | 0.40 | 0.223 |
| GhCOL9_Dt-GhCOL2_Dt | 0.11 | 0.53 | 0.211 |
| GhC0L18_Dt-GhCOL17_Dt | 0.09 | 0.73 | 0.119 |
| GhCOL2_Dt-GhCOL14_Dt | 0.09 | 0.53 | 0.175 |
| GhCOL3_At-GhCOL14_At | 0.08 | 0.41 | 0.204 |
| GhCOL2_At-GhCOL14_At | 0.09 | 0.45 | 0.210 |
| GhCOL9_At-GhCOL14_At | 0.09 | 0.45 | 0.210 |
| GhCOL4_At-GhCOL14_At | 0.09 | 0.39 | 0.230 |
TEs are the major drivers of cotton genome expansion; particularly, the long-terminal repeat retrotransposons (LTR) is the major cause for size difference between the A genome (G. arboreum) and the D genome (G. raimondii) [28]. TEs are involved in gene family expansion, in addition to playing important roles in the regulation of gene expression under stress stimulus [32]. The 2000 bp region around the CO-like gene loci contained one DNA transposon and 12 retroelements (Table 2), including one PIF-Harbinger, four L1-type TEs, and eight copia-type TEs (Table 2 and Table S4). By enlarging the scanned region to 10 Kb, many TEs were identified, including that six DNA transposons and 51 retroelements were present.
Table 2.
Transposable elements around the CO-like gene loci.
| Type | Number of Elements | Length Occupied (bp) | Percentage of Sequence (%) | Number of Elements | Length Occupied (bp) | Percentage of Sequence (%) |
|---|---|---|---|---|---|---|
| 2000 bp region | 10,000 bp region | |||||
| DNA transponsons | 1 | 283 | 0.1 | 6 | 109 | 0.04 |
| CMC-EnSpm | 0 | 0 | 0 | 4 | 1172 | 0.42 |
| MULE-MuDR | 0 | 0 | 0 | 0 | 0 | 0 |
| PIF-Harbinger | 1 | 283 | 0.1 | 1 | 283 | 0.1 |
| TcMar-Pogo | 0 | 0 | 0 | 0 | 0 | 0 |
| hAT | 0 | 0 | 0 | 0 | 0 | 0 |
| hAT-Ac | 0 | 0 | 0 | 0 | 0 | 0 |
| hAT-Tag1 | 0 | 0 | 0 | 0 | 0 | 0 |
| hAT-Tip100 | 0 | 0 | 0 | 0 | 0 | 0 |
| hAT-Charlie | 0 | 0 | 0 | 1 | 32 | 0.01 |
| Retroelements | 20 | 6199 | 2.23 | 51 | 25,761 | 9.27 |
| LINE: | 0 | 0 | 0 | 0 | 0 | 0 |
| L1 | 4 | 1085 | 0.39 | 4 | 1085 | 0.39 |
| LTR: | 8 | 2557 | 0.92 | 0 | 0 | 0 |
| Caulimovirus | 0 | 0 | 0 | 0 | 0 | 0 |
| Copia | 8 | 2557 | 0.92 | 36 | 19,874 | 7.15 |
| Gypsy | 0 | 0 | 0 | 11 | 4802 | 1.73 |
| RC: | 0 | 0 | 0 | 0 | 0 | 0 |
| Helitron | 0 | 0 | 0 | 0 | 0 | 0 |
| Low_complexity | 36 | 1880 | 0.68 | 117 | 6419 | 2.31 |
| Simple_repeat | 144 | 6167 | 2.22 | 481 | 20,564 | 7.4 |
| Unspecified | 24 | 3581 | 1.29 | 113 | 38,583 | 13.88 |
The DNA transposons were composed of four MULE-MuDR, one PIF-Harbinger, and one hAT-Charlie, whereas the retroelements were mainly composed of LTRs, including 36 copia-type and 11 Gypsy-type retroelements (Table 2 and Table S5). When the TE distribution was inspected further, we found three L1-type and four copia-type TEs located in the genomic region of GhCOL12_Dt, one PIF-Harbinger located in GhCOL9_Dt, three copia-type TEs located upstream of GhCOL13_Dt, and one L1 and four copia-type TEs inserted in the genomic region of GhCOL12_Dt within a 2000 bp region. Within the 10,000 bp region, we found TEs near the loci of 18 genes. For example, three copia TEs were distributed downstream of GhCOL9_At, two copia-type TEs were located upstream of GhCOL4_At, and one copia-type TE was located downstream of GhCOL4_At. When comparing the origin of the TEs, we found more TEs from the At subgenome than from the Dt subgenome. Simple repeat sequences are more variable and widely spread in the whole genome across the upstream, downstream, and intragenic regions of CO-like genes than TEs are. GhCOL13_At and GhCOL13_Dt shared the same type of TEs belonging to ATCopia29, and GhCOL12_At and GhCOL12_Dt shared the same L1-type TE, indicating that these TEs are ancient and occur in the progenitor genomes of the At and Dt subgenomes.
2.5. The Effect of Domestication on CO-like Genes
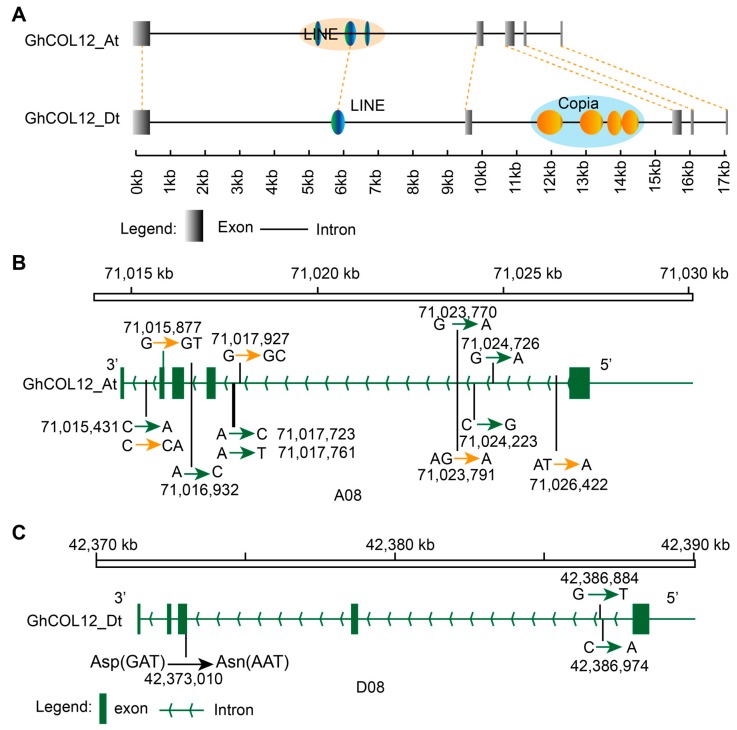
We checked whether the CO-like genes have experienced domestication or not by scanning the CO-like genes in the domestication sweep region and found that GhCOL12 (Gh_D08G1289) was located in approximately 1.6 M region on D08 from 41,400,001–42,600,000 bp. Therefore, we compared the genomic sequences of GhCOL12_At and GhCOL12_Dt, and found that the exons were significantly conserved, whereas the introns were not. Three LINE-type TEs are located in the first intron of GhCOL12_At, whereas only one is located in GhCOL12_Dt. Four copia-type of TEs were inserted in the second intron of GhCOL12_Dt, but none in GhCOL12_At (Figure 3A). There were more sites with single nucleotide polymorphisms (SNPs) in the genomic region of GhCOL12_At than in GhCOL12_Dt (Figure 3B,C), indicating that GhCOL12_At maintained more diversity during evolution than did GhCOL12_Dt. We found seven SNPs and five indels in the GhCOL12_At, among which one indel was present in the fourth exon. As compared with GhCOL12_At, GhCOL12_Dt showed lower diversity in the genomic sequences because only three SNPs were detectable in the genomic region of GhCOL12_Dt. In the third exon of GhCOL12_Dt, a nonsynonymous mutation transformed Asp to Asn.
Figure 3.
Domestication sweep analysis indicating that GhCOL12_Dt was a potential candidate under human-guided evolution: genomic character of GhCOL12_At and GhCOL12_Dt (A); genetic diversity of GhCOL12_At (B); and GhCOL12_Dt (C). The orange and green arrawhead lines indicated SNPs and Indels respectively. The digits closed to the arrawhead lines indicated the SNPs or Indels loci. The black vertical line indicated the SNPs or Indels loci on the chromosomes.
2.6. Gene Structure Analysis
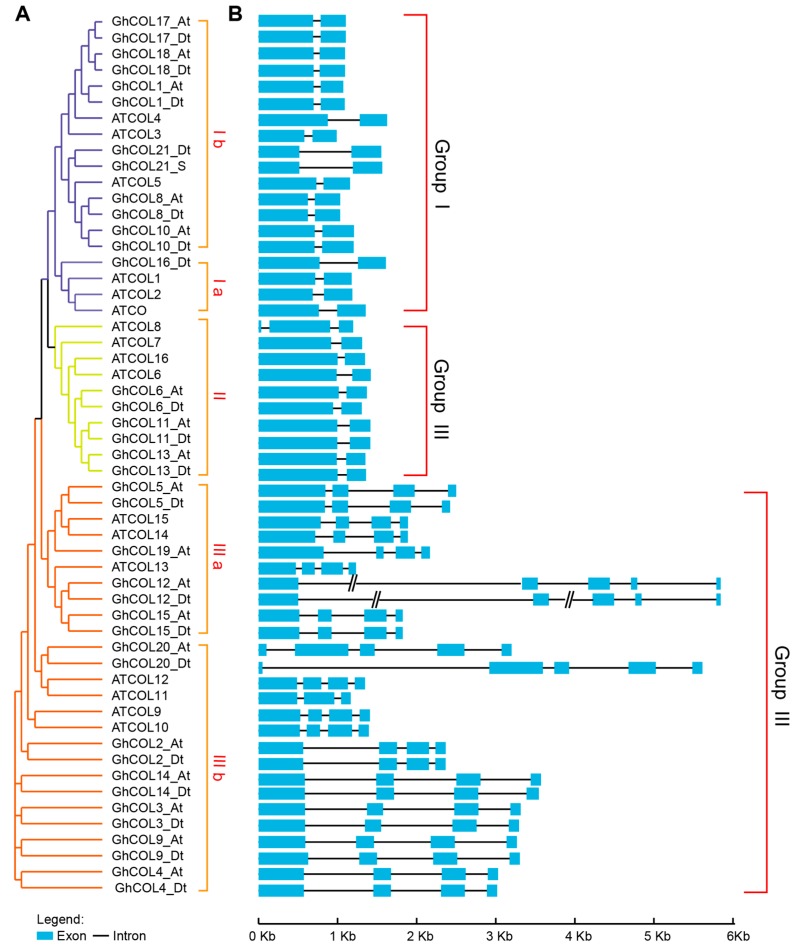
We found that the genes from cotton and Arabidopsis can be divided into three groups as those in Figure 1 (Figure 1 and Figure 4A). Because of the difference in number of genes, the topologies of the two trees displayed some general differences. However, the members within the same clade were the same and the topologies were similar. We found that GhCOL14, GhCOL3, GhCOL4, and GhCOL9 clustered together, displaying a close phylogenetic relationship, which significantly supported the result shown in Figure 2. Figure 2 shows that Group III had undergone gene expansion. The genes shown in Figure 4A were inclined to cluster in cotton- or Arabidopsis-specific manner, suggesting that gene expansion occurred after divergence of cotton and Arabidopsis.
Figure 4.
Comparison of the gene structures between G. hirsutum and A. thaliana. An NJ tree based on the relationship between A. thaliana and G. hirsutum (A); and the number, position, and length of exons and introns within CO-like genes (B). Purple, green, and orange in (A) represent Group I–III, respectively. Boxes and black lines in (B) indicate the exons and introns, respectively.
As shown in Figure 4B, CO-like genes contained multiple exons ranging from two to five. Although the number of exons differed among the groups, the exon patterns within groups were similar. For example, except AtCOL8, the remaining members in Group I and II contained two exons, and the first one was longer than the second one. Except GhCOL12_At/Dt, GhCOL20_At/Dt, and AtCOL11, the other genes of Group III were composed of four exons, among which the first one was the longest. Comparing gene structure of the orthologs from At and Dt subgenomes, the length of exons from the same position were found to be nearly equal, except for GhCOL9_At/Dt, GhCOL16_At/Dt, and GhCOL20_At/Dt gene pairs, indicating that the orthologs between At and Dt were conserved.
2.7. Gene-Expression Patterns in Different Tissues
Gene expression is the bridge between the genomic sequence and plant phenotype. Therefore, we inspected the CO-like gene-expression profiles in different tissues. As shown in Figure S4A, most of the genes were detectable in most of the tissues checked in our study, and we found that the expression levels differed between non-orthologous pairs, indicating that these genes may play diverse roles in different biological processes. As shown in Figure S4A, the genes were clustered according to their expression pattern, and of the genes with similar expression patterns in the heatmap displayed a more positive correlation. Based on the expression pattern, the genes could be roughly divided into a, b, and c clades. All the genes in clade a belonged to Group III, most of those in clade c belonged to Group III, and the genes in clade b were a mixture of the genes from different groups. Most of the genes were highly expressed in the torus and calycle, suggesting that they may be involved in regulation of flowering time. GhCOL3_At/Dt, GhCOL1_At, and GhCOL4_At have relatively high expression level in the early stage of ovule development, suggesting they may be associated with ovule development. Only GhCOL9_Dt was highly expressed during the entire fibre elongation stage, and only six and two genes were highly expressed at 20 days-post-anthesis (DPA) and 5 DPA during the fibre development stage. Ten out of 19 pairs of orthologs showed a highly similar expression pattern, and the remaining orthologous pairs showed some differences, suggesting that these orthologous pairs might have undergone functional diversity. For example, GhCOL12_At and GhCOL12_Dt showed different expression patterns, whereas GhCOL12_Dt displayed a higher expression level than GhCOL12_At in stem and leaves. In addition, we checked the expression level of CO-like genes during seed development. As shown in Figure S4B, most of the genes expressed during seed development, and most of these expressed genes have a low expression levels during the early stage of seed germination. Moreover, the expression level of more than half of the genes showed a tendency of increased expression in the cotyledon. GhCOL4_At/Dt, GhCOL21_At/Dt, and GhCOL5_At showed higher expression levels than that of the other genes in roots.
2.8. Response of CO-like Genes to Photoperiod and Stress
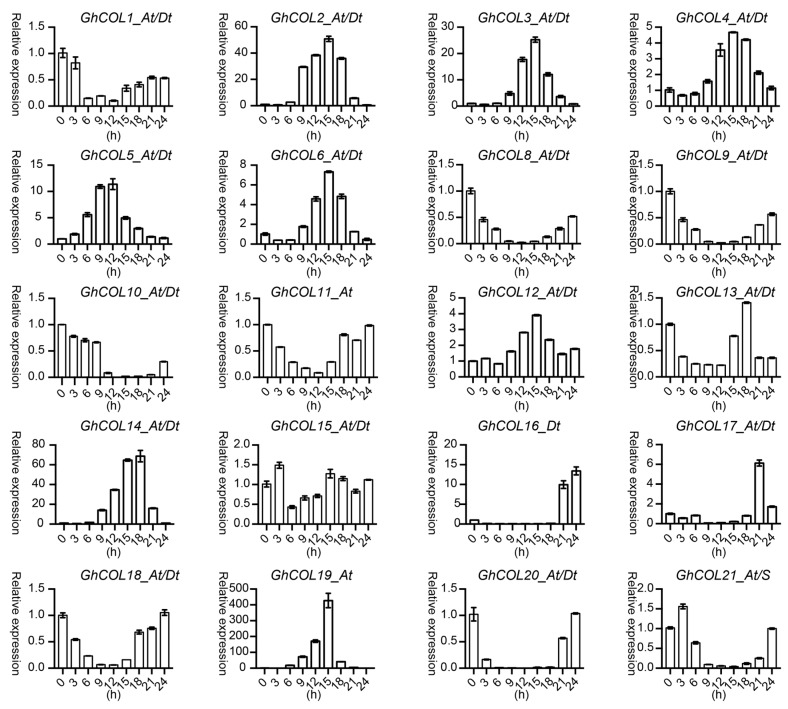
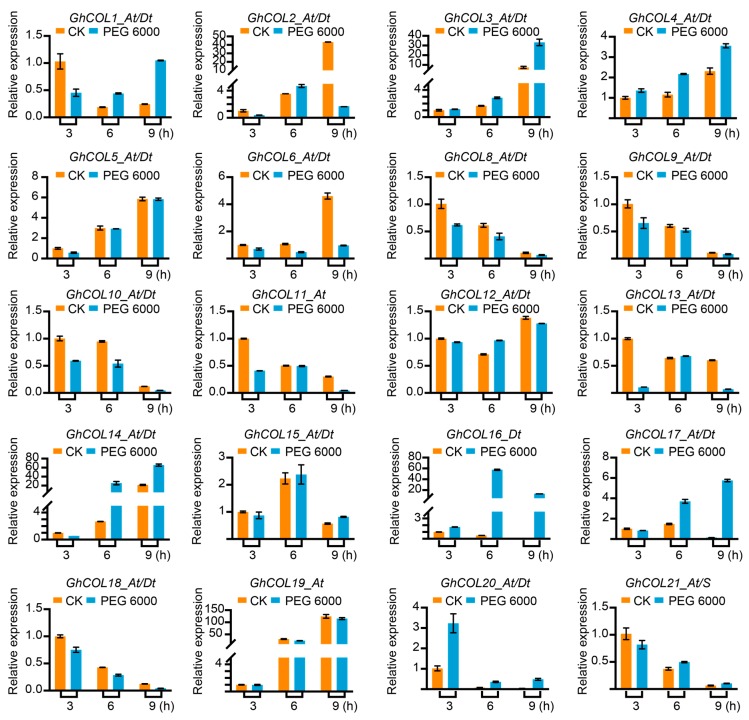
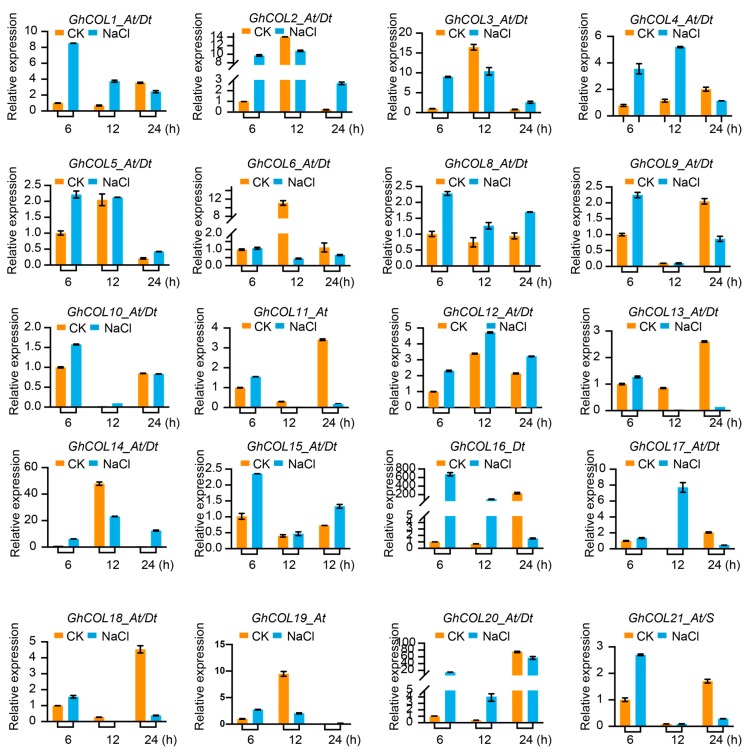
Figure 5 shows that the expression of most genes varied at times, indicating that the CO-like genes responded to the photoperiod. The expression pattern clustered into two groups: in the first, gene expression was initially up-regulated and then down-regulated, while the reverse occurred in the second group. As shown, GhCOL2_At/Dt, GhCOL3_At/Dt, and GhCOL4_At/Dt had relatively low expression levels at 0, 3, and 6 h, were significantly up-regulated from 9 h, peaked at 15 h, and then were down-regulated. However, the expression of GhCOL8_At/Dt, GhCOL11_At/Dt, and GhCOL18_At/Dt was down-regulated from 3 h, decreased to the lowest level at 12 h, and then was up-regulated thereafter. As shown in Figure 6, CO-like genes displayed diverse expression patterns under drought stress. GhCOL3_At/Dt, GhCOL4_At/Dt, GhCOL14_At/Dt, GhCOL17_At/Dt, GhCOL16_At/Dt, and GhCOL20_At/Dt were all up-regulated; the expression of the first eight genes peaked 9 h after treatment, while that of GhCOL20_At/Dt peaked 3 h after treatment. However, GhCOL6_At/Dt, GhCOL8_At/Dt, GhCOL9_At/Dt, GhCOL10_At/Dt, GhCOL11_At/Dt, GhCOL13_At/Dt, GhCOL18_At/Dt, GhCOL19_At, and GhCOL21_At/S were down-regulated when exposed to salt stress. By contrast, the expression of GhCOL1_At/Dt, GhCOL2_At/Dt, GhCOL3_At/Dt, GhCOL4_At/Dt, GhCOL8_At/Dt, GhCOL12_At/Dt, GhCOL14_At/Dt, GhCOL16_At/Dt, and GhCOL17_At/Dt was up-regulated under salt stress (Figure 7). GhCOL3_At/Dt, GhCOL4_At/Dt, GhCOL14_At/Dt, GhCOL16_At/Dt, and GhCOL17_At/Dt were up-regulated under both PEG 6000 and salt treatments, indicating that they may act as positive regulators during stress responses. GhCOL6_At/Dt, GhCOL11_At/Dt, GhCOL13_At/Dt, and GhCOL18_At/Dt were down-regulated under both PEG 6000 and salt treatments, suggesting that they may be negative regulators.
Figure 5.
qPCR analysis showed that CO-like genes responded to the photoperiod. The cotton seedlings were sampled at different times over a 24 h period. The error bar represents the standard deviation of three biological replicates.
Figure 6.
The qPCR analysis showed that CO-like genes might play important roles in drought stress. Four biological replicates were used in this study. The error bar represents the standard deviation of at four biological replicates.
Figure 7.
The qPCR analysis displayed that CO-likes may play important roles in salt stress. Four biological replicates were used in this study for qPCR. The error bar represents the standard deviation of at four biological replicates.
3. Discussion
3.1. CO-like Genes Have Experienced Expansion
CO-like genes encode a small number of transcriptional factors. In the present study, we found that the numbers of CO-like gene family members in each diploid species were different, and that cacao had the fewest genes, which was almost half of that in its close relative G. raimondii, suggesting that the CO-like genes in cotton have experienced expansion. A previous study showed that both cacao and cotton only experienced the paleohexaploidisation event shared by the eudicots, and that cotton has experienced recent duplication events but cacao not, which may be the major reason why the diploid cotton has more CO-like genes [26]. In this study, we found that the number of CO-like genes in G. hirsutum were almost twice of that in G. arboretum and G. raimondii, because G. hirsutum is the typical allotetraploid that was formed by the hybridisation of the A genome and the D genome, followed by chromosome doubling 1–2 million years ago [33]. G. hirsutum was very conserved after polyploidy because the orthologs from At and Dt genome maintained a significant collinearity with each other.
In addition to the WGD, the segmental duplication was a major contributor to gene expansion in the CO-like gene family. Segmental duplication is very common in plants because most plants are diploidised polyploids and maintain large amounts of duplicated chromosomal blocks within their genomes [34]. In our previous study, we found that the segmental duplication was associated with WOX and YABBY genes expansion, and that the genes with segmental duplication showed different expression patterns [22,23]. In the present study, we found nine paralogous pairs were involved in segmental duplication or WGD; however, their expression patterns were similar with a very small value of Ka/Ks between paralogous gene pairs, indicating that these genes may be highly conserved during the evolution.
Cotton is the one of the most important crops in the world and it has been domesticated from the wild-type cotton plants under long-term human selection. Wild cotton species are perennial and are sensitive to photoperiod, whereas the cultivated cotton is photoperiod insensitive, which makes cotton widely cultivated in regions with different sunshine durations [30]. CO-like genes play significant roles in regulation of photoperiod pathway. The rice Hd1, an ortholog of CO, was a possible target of human selection during domestication of rice [35]. Wang et al. found 93 domestication sweeps occupying 178 Mb in the upland cotton genome by comparing the genetic diversity between cultivate cultivar and wild cotton species [36]. Based on comparison of the nucleotide diversity between the wild species and cultivars, we found that GhCOL12_Dt was a candidate gene for domestication selection. The genomic region of GhCOL12_At and GhCOL12_Dt contained different types of TEs and the GhCOL12_At displayed a higher diversity than GhCOL12_Dt did, because GhCOL12_At contained more SNPs than did GhCOL12_Dt. Furthermore, the expression pattern of GhCOL12_Dt and GhCOL12_At differed in the ovule. Overall, it appeared that GhCOL12_Dt and GhCOL12_At might have experienced divergence, and that further experimental validation of this finding is required.
TEs are mobile DNA elements, which are abundant in the genome and distributed in different positions [37]. TE activity and genomic recombination are the two major contributors to gene divergence and play important roles in species divergence. According to the dependence TE mobility on RNA, TEs are classified into retrotransposons and DNA transposons. Retrotransposons are the most abundant TEs in the cotton genome and were divided into LTR and non-LTR types [29]. In the present study, we found that the TEs around the CO-like genes are retrotransposons, and most of them are of the LTR type; moreover, the number of Copia-type TEs was larger than the Gypsy-type TEs, suggesting that Copia may be more active during the cotton evolution. TEs in At may be more active because the At subgenome has more TEs than Dt has. A previous study showed that the TEs may have burst in the progenitor genomes of upland cotton before its formation [30]. However, the distribution of TEs around genes from At and Dt differed, suggesting that these TEs might occur independently in the progenitors of At and Dt. GhCOL13_At and GhCOL13_Dt shared the same type of TEs belonging to ATCopia29, and GhCOL12_At and GhCOL12_Dt shared the same L1-type TE, indicating that these TEs are ancient and occur in the progenitor genomes of the At and Dt subgenomes.
3.2. CO-like Genes Were Highly Conserved during the Evolution
CO-like genes were characterised by the CCT and B-box domain. CCT domains are highly conserved among CO-like genes and B-box domains were more divergent in cotton (Figure 1), and this situation was also found in Arabidopsis, barley, and rice [1], suggesting that B-box may play significant roles in CO-like gene divergence. Although the sequences of B-boxes are diverse in different genes, they shared a higher conserved expression pattern as C-X2-C-X8-C-X7-C-X2-C-X4-H-X8-H, indicating that cysteine residues play important roles in maintaining the stability of the zinc-finger structure. A previous study showed that CO-like genes can be divided into three clades depending on the number and diversity of the B-box [1]. In the present study, we found that CO-like genes from cotton could be divided into three groups (Figure 1 and Figure S1). Members from the same groups showed similar characteristics. Firstly, the genes displayed more similarities in the amino sequences within groups (Figure S1). Secondly, the genes within groups showed a pattern; for example, the genes from Groups II and III mostly consisted of two exons and those from Group I mostly consisted of four exons (Figure 4). Finally, the genes within groups showed a similar expression pattern, particularly in Groups I and III (Figure S4). Most of the genes involved in duplication were from Group I and they displayed a similar expression pattern, indicating that the duplication was a major contributor to the expansion of the genes clustered in Group I, and that functional redundancy may exist among these genes. We found that each group was composed of genes from dicots and monocots, and the genes tended to clustered in dicot- or monocot-specific manner, indicating that the main function has been formed before dicot-monocot divergence.
3.3. CO-like Genes Play Diverse Roles under Stress
CO-like genes have been well studied for their roles in regulation of flowering time [9,10,11,12,13,14,15,16], but their roles in other biological process are poorly understood. Ghd2 (Os02g49880), belonging to Group II, was significantly down-regulated in rice under drought stress [18], which corresponded to GhCOL6_At/Dt, GhCOL11_At/Dt, and GhCOL13_At/Dt in upland cotton. Intriguingly, GhCOL6_At/Dt, GhCOL11_At, and GhCOL13_At/Dt were differentially expressed under artificial drought stress, indicating that the genes from Group II might play significant roles in drought stress response. AtCOL4 was up-regulated under salt, ABA, and osmotic stress, and its ortholog GhCOL1_At/Dt displayed an up-regulated expression pattern under both PEG 6000 and salt stress treatments, indicating that the functions of the genes were conserved during their evolution. Furthermore, GhCOL18_At/Dt displayed a close relationship with GhCOL1_At/Dt, which was down-regulated during the PEG treatment, suggesting that GhCOL18_At/Dt may act as a negative regulator under drought stress conditions. Most of the genes involved in duplication belonged to Group III b, among which GhCOL2_At/Dt, GhCOL3_At/Dt, and GhCOL14_At/Dt were differentially expressed under both drought and salt stress treatments, indicating that genes in Group III b may be involved in stress response and are candidate genes suitable for further study.
4. Materials and Methods
4.1. Gene Identification and Sequence Retrieval
Cotton genome databases for the cotton plants Gossypium arboreum (BJI, version 1.0), G. raimondii, (JGI, version 2.0), and G. hirsutum (NAU, version 1.1) were available from COTTONGEN [38]. The genomic data of rice (version 7.0), sorghum (version 2.1), maize (version 1.1), and cacao (version 1.1) were downloaded from JGI (https://phytozome.jgi.doe.gov/pz/portal.html). The 17 amino acid sequences of CO-like genes from Arabidopsis thaliana were retrieved from The Arabidopsis Information Resource, version 10 (TAIR 10) (http://www.arabidopsis.org). BLASTP and TBLASTN programs were used to search the G. raimondii protein database using all protein sequences of Arabidopsis CO-like genes as queries, and a cut-off e-value of 1e–5. All candidates obtained from BLAST were extracted from the genome database for functional annotation. Interproscan 63.0 was used to annotate the CCT and B-box domain in the candidate sequences. CO-like genes from other species were analysed as described above for G. raimondii.
4.2. Conserved Sequence and Phylogenetic Analysis
For conserved domain analysis, the sequences were extracted from the full-length of CO-like protein sequences according to the domain positions retrieved from Interproscan 63.0, and ClustalX 2.0 was used for multiple-sequence alignments with the default parameters. For phylogenetic analysis, we used the ClustalW program (build-in MEGA 7.0) [39] to perform the multiple sequences alignment using the full-length sequences from rice, sorghum, maize, cacao, Arabidopsis, G. arboreum, G. raimondii, and G. hirsutum. Thereafter, we prepared a phylogenetic tree using the neighbour-joining (NJ) method with 1000 bootstrap replicates. Substitution was evaluated by the Poisson model using the default parameters. To validate the NJ tree, we used the Maximum likelihood method to construct an ML tree. Substitution was evaluated by the Poisson model, and bootstrap method (100 replications) was used to test the tree. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.8524)). All positions with less than 95% site coverage were eliminated.
4.3. Chromosomal Location and Collinearity Analysis
The exon loci were extracted using the method described in our previous study [22]. The chromosomal location was displayed by MapChart [40]. For collinearity analysis, the entire cotton protein sequences of upland cotton were aligned with each other using the basic local alignment search tool (BLAST) with a cut-off e-value of 1 × 10−5. The blastp result was analysed by MCScanx software to produce collinearity blocks across the whole genome. Collinearity pairs within the CO-like family of proteins were extracted to draw a collinearity map with CIRCOS (Version 69.0) software [41]. To analyse the duplicate gene type, the script duplicate_gene_classifier in MCScanx was used to identify the gene type, including singleton, dispersed duplicate, proximal, tandem, segmental duplication/whole genome duplication (WGD), using the blastp result as input with default parameters.
4.4. Calculating Synonymous and Non-Synonymous Substitution Rates
Amino acid sequences from homologous pairs were aligned using Clustal X 2.0 (http://www.ebi.ac.uk/tools/clustalw2) with default parameters. The alignment results were converted to a codon alignment using the online tools PAL2NAL (http://www.bork.embl.de/pal2nal/). The CODEML program was used to calculate the synonymous (Ks) and non-synonymous (Ka) substitution substitution rates with default parameters [42].
4.5. Annotation and Analysis of Transposable Elements
The method for Transposable elements (TEs) were identified in upland cotton according to the method used in our previous study [22]. To analyse the distribution of TEs around CO-like genes, the TEs located 10,000 and 2000 bp upstream and downstream of the CO-like genes were analysed, and we performed statistical analysis of the different types of TEs present.
4.6. Gene Structure Analysis
We used the full-length Arabidopsis and G. hirsutum protein sequences to perform multiple alignments with ClustalW, and used MEGA 7.0 to construct an NJ tree using same method and parameters mentioned above. To draw the gene structure, we extracted the exon positions from the gff3 file using a Perl script, and used the online tool GSDS 2.0 for displaying it [43].
4.7. Transcriptome Data Analysis and Gene-Expression Heatmap
The raw data for RNA-Seq used in this study were downloaded from the national center for biotechnology information center (NCBI) sequence read archive (SRA: PRJNA248163) and analysed using the methods described in our previous study [22]. TopHat (Version 2.0.13) was used for short read mapping with default parameters, and cufflinks (Version 2.2.1) were used to calculate gene-expression levels with default parameters, and fragments per kilobase million (FPKM) values were used to normalise gene-expression levels [44]. The R package “ggplot2” was used to prepare the heatmap.
4.8. Real-Time PCR Analysis
The seeds of upland cotton TM-1 were obtained from the Cotton Research Institute of Chinese Academy of Agricultural Sciences.The seedlings were prepared as our previous report [22]. At the 3-leaf stage, the seedlings were treated with 300 mM, 10% PEG 6000 and water. To study the genes expression pattern in photoperiods, we sampled the roots at 0 (08:00), 3, 6, 9, 12,15,18, 21 and 24 h in normal growth condition (water treatment). For stress analysis, we sampled at 3, 6 and 9 h post PEG 6000 treatment and sampled at 6, 12 and 24 h post NaCl treatment. All samples were immediately frozen in liquid nitrogen and stored at −80 °C. The total RNA was extracted from the roots using RNAprep Pure Plant Kit (TIANGEN, Beijing, China). The first strand cDNA was synthesised using a PrimeScript® RT reagent kit (Takara, Dalian, China) was used to synthesise the first strand of cDNA, and SYBR Premix Ex TaqTM II was used in PCR amplifications. The house-keeping gene, Ghistone3 (GenBank accession no. AF024716), was used as an internal control, and the 2−ΔΔCt method was used to calculate the relative expression level.
5. Conclusions
Previous studies show that members of the CO-like gene family play important roles in the regulation flowering and stress response [9,10,11,12,13,14,15,16,17,18,19,20,21]. The results of the present study suggest that CO-like genes are significantly conserved among cotton and other plant species. Furthermore, whole genome and segmental duplications were two major contributors to the expansion of CO-like genes of cotton. GhCOL12_Dt is a candidate gene involved in selection by humans, which may be an important gene determining the insensitivity of cultivated cotton plants to photoperiod. Moreover, members from Group II and the duplicated genes in Group III might be involved in the mediation of stress responses. Our findings will not only deepen the understanding of the evolution of CO-like genes in cotton, but also facilitate further functional genomic studies on CO-like genes in cotton.
Supplementary Materials
Supplementary Materials can be found at http://www.mdpi.com/1422-0067/19/9/2658/s1.
Author Contributions
Z.Y., F.L. and Q.C. conceived and designed the study; Z.Y., W.Q., Y.J., J.L. and Z.L. prepared figures, and analysed and interpreted the data; Z.Y., W.Q., J.X., H.L., G.Z. and W.H. prepared the manuscript; and X.W. and Y.Y. wrote a part of the manuscript and critically reviewed its intellectual contents. All authors read, edited, and approved the current version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant 31621005) and grants from the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Griffiths S., Dunford R.P., Coupland G., Laurie D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robson F., Costa M.M.R., Hepworth S.R., Vizir I., Pin M., Reeves P.H., Putterill J., Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2002;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 3.Torok M., Etkin L.D. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- 4.Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G. Plant TFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putterill J., Robson F., Lee K., Simon R., Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 6.Putterill J.J., Plummer K.M., Newcomb R.D., Marshall S.D.G. The Carboxylesterase Gene Family from Arabidopsis thaliana. J. Mol. Evol. 2003;57:487–500. doi: 10.1007/s00239-003-2492-8. [DOI] [PubMed] [Google Scholar]
- 7.Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du A., Tian W., Wei M., Yan W., He H., Zhou D., Huang X., Li S., Ouyang X. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant. 2017;10:948–961. doi: 10.1016/j.molp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Hassidim M., Harir Y., Yakir E., Kron I., Green R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta. 2009;230:481–491. doi: 10.1007/s00425-009-0958-7. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X.-F., Wang Z.-Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005;43:758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- 11.Ordoñez-Herrera N., Trimborn L., Menje M., Henschel M., Robers L., Kaufholdt D., Hänsch R., Adrian J., Ponnu J., Hoecker U. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018;176:1327–1340. doi: 10.1104/pp.17.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H., Gu F., Dong S., Liu W., Wang H., Chen Z., Wang J. CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016;479:173–178. doi: 10.1016/j.bbrc.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Tan J., Wu F., Wan J. Flowering time regulation by the CONSTANS-Like gene OsCOL10. Plant Signal. Behav. 2017;12:e1267893. doi: 10.1080/15592324.2016.1267893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan J., Jin M., Wang J., Wu F., Sheng P., Cheng Z., Wang J., Zheng X., Chen L., Wang M., et al. OsCOL10, a CONSTANS-Like Gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell. Physiol. 2016;57:798–812. doi: 10.1093/pcp/pcw025. [DOI] [PubMed] [Google Scholar]
- 15.Wu W., Zhang Y., Zhang M., Zhan X., Shen X., Yu P., Chen D., Liu Q., Sinumporn S., Hussain K., et al. The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 2018;495:1349–1355. doi: 10.1016/j.bbrc.2017.11.095. [DOI] [PubMed] [Google Scholar]
- 16.Wu W., Zheng X.M., Chen D., Zhang Y., Ma W., Zhang H., Sun L., Yang Z., Zhao C., Zhan X., et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017;260:60–69. doi: 10.1016/j.plantsci.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Datta S., Hettiarachchi G.H.C.M., Deng X.-W., Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18:70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Shen J., Xu Y., Li X., Xiao J., Xiong L. Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016;67:5785–5798. doi: 10.1093/jxb/erw344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Ji R., Li H., Zhao T., Liu J., Lin C., Liu B. CONSTANS-LIKE 7 (COL7) is involved in phytochrome B (phyB)-mediated light-quality regulation of auxin homeostasis. Mol. Plant. 2014;7:1429–1440. doi: 10.1093/mp/ssu058. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Zhang Z., Li H., Zhao X., Liu X., Ortiz M., Lin C., Liu B. CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J. Exp. Bot. 2013;64:1017–1024. doi: 10.1093/jxb/ers376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H., Dong S., Sun D., Liu W., Gu F., Liu Y., Guo T., Wang H., Wang J., Chen Z. CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE. 2016;11:e0166249. doi: 10.1371/journal.pone.0166249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z., Gong Q., Qin W., Yang Z., Cheng Y., Lu L., Ge X., Zhang C., Wu Z., Li F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017;17 doi: 10.1186/s12870-017-1065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z., Gong Q., Wang L., Jin Y., Xi J., Li Z., Qin W., Yang Z., Lu L., Chen Q., et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Q., Yang Z., Chen E., Sun G., He S., Butt H.I., Zhang C., Zhang X., Yang Z., Du X., et al. A phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 2018;59:275–289. doi: 10.1093/pcp/pcx180. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Liu Z.-H., Feng L., Zheng Y., Li D.-D., Li X.-B. Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE. 2013;8:e80879. doi: 10.1371/journal.pone.0080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S., et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 27.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 28.Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C., et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 29.Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J., et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., Saski C.A. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 31.Vandepoele K., Simillion C., Van de Peer Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell. 2003;15:2192–2202. doi: 10.1105/tpc.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennetzen J.L., Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014;65:505–530. doi: 10.1146/annurev-arplant-050213-035811. [DOI] [PubMed] [Google Scholar]
- 33.Renny-Byfield S., Page J.T., Udall JA., Sanders W.S., Peterson D.G., Arick M.A., Grover C.E., Wendel J.F. Independent domestication of two old world cotton species. Genome. Biol. Evol. 2016;8:1940–1947. doi: 10.1093/gbe/evw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:4. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi Y., Shimamoto K. Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice. Genes Genet. Syst. 2011;86:175–182. doi: 10.1266/ggs.86.175. [DOI] [PubMed] [Google Scholar]
- 36.Wang M., Tu L., Lin M., Lin Z., Wang P., Yang Q., Ye Z., Shen C., Li J., Zhang L., et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 2017;49:579–587. doi: 10.1038/ng.3807. [DOI] [PubMed] [Google Scholar]
- 37.Arkhipova I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA. 2017;8 doi: 10.1186/s13100-017-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J., Jung S., Cheng C.-H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014;42:D1229–D1236. doi: 10.1093/nar/gkt1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voorrips R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 41.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 43.Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.