Abstract
Mechanistic/mammalian target of rapamycin (mTOR), an atypical serine/threonine kinase of the phosphoinositide 3-kinase- (PI3K-) related kinase family, elicits a vital role in diverse cellular processes, including cellular growth, proliferation, survival, protein synthesis, autophagy, and metabolism. In the cardiovascular system, the mTOR signaling pathway integrates both intracellular and extracellular signals and serves as a central regulator of both physiological and pathological processes. MicroRNAs (miRs), a class of short noncoding RNA, are an emerging intricate posttranscriptional modulator of critical gene expression for the development and maintenance of homeostasis across a wide array of tissues, including the cardiovascular system. Over the last decade, numerous studies have revealed an interplay between miRNAs and the mTOR signaling circuit in the different cardiovascular pathophysiology, like myocardial infarction, hypertrophy, fibrosis, heart failure, arrhythmia, inflammation, and atherosclerosis. In this review, we provide a comprehensive state of the current knowledge regarding the mechanisms of interactions between the mTOR signaling pathway and miRs. We have also highlighted the latest advances on mTOR-targeted therapy in clinical trials and the new perspective therapeutic strategies with mTOR-targeting miRs in cardiovascular diseases.
1. Introduction
Cardiovascular disease (CVD) is one of the leading cause of mortality and morbidity in the world and is a global pandemic threat to human health [1, 2]. Coronary artery diseases (CAD) such as ischemia reperfusion injury (I/R) and acute myocardial infarction (AMI) are the primary forms of CVD that account for the majority of the deaths. Apart from this, additional comorbid factors like diabetes [3, 4], obesity [5], inflammation [6], and atherosclerosis [7] escalate the complication associated with heart disease and increase the incidence of death. The underlying mechanisms involved in cardiovascular complication are complex and multifactorial. The existence of several metabolic perturbations in diseases like diabetes and inflammation further pose a tough challenge in understanding the mechanism and pathology of CVD. These obstacles largely impede our goal to develop an effective treatment against progression of CVD and its prevention. However, current therapies for heart diseases have been substantially improved using integrated genome-based evidences and molecular clues. Our recent understanding of genomics and the regulation of gene expression by noncoding RNAs (ncRNA) during both normal and pathological conditions encourage us in exploring the novel therapies for heart disease with a unique perspective.
In the cardiovascular system, the mechanistic target of rapamycin (mTOR) pathway regulates both physiological and pathological processes in the heart [8]. mTOR is an evolutionarily conserved signaling pathway found in various species including yeast [9, 10], Caenorhabditis elegans [11, 12], drosophila [13, 14], and mammals [15–18]. mTOR is a master regulator of cell metabolism and plays a central role in integrating various signaling network [19]. mTOR participates in the fundamental aspect of cell function and is therefore indispensable for cellular life. It governs several key cellular processes such as nutrient sensing [20–22], protein synthesis [23, 24], cell proliferation [14], and apoptosis [25, 26]. mTOR is also actively involved in the epigenetic regulation of gene expression and control process like aging [27] and autophagy [28]. However, aberrant regulation of mTOR is known to play a significant role in various maladies including cancer [29], diabetes [30], aging [31], and cardiovascular diseases [32]. mTOR plays an important role in normal cardiac development [33–36] and during cardiac pathophysiologic condition [37, 38].
Recent studies have demonstrated that mTOR signaling pathway is profoundly influenced by small noncoding RNAs, and an interplay between these two molecules define a synergistic regulation of gene expression [39–41]. The unique combination and cross talk between mTOR and miRs have opened up research interest from a distinct perspective and to revisit mTOR signaling in the light of miR. The intention of this review article is to highlight our recent understanding on mTOR pathway in cardiovascular system and its coordinated interaction with miRs to fine-tune the regulation of gene expression under both normal and pathological conditions.
1.1. Structure, Mechanism, and Function of mTOR Complexes
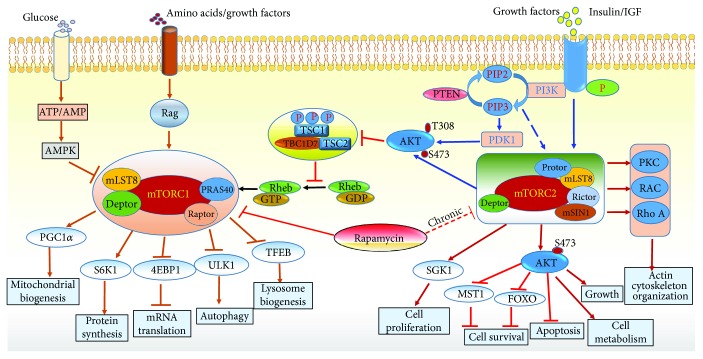
The mTOR macromolecular complex is a serine/threonine protein kinase of 289 kDa that belongs to phosphatidylinositol-3-kinase (PI3K) family of proteins and governs several cellular processes including protein synthesis and metabolic regulation [42]. Hall and colleagues first identified target of rapamycin 1 (TOR1) and TOR2 in yeast Saccharomyces cerevisiae [43, 44], which was subsequently characterized in mammalian cells and hence called mTOR [42, 45, 46]. Discovered in the year 1970 for its antifungal property [47], rapamycin played a bigger role in elucidating the cellular function of mTOR [42, 48]. The mTOR consists of two major distinct complexes termed as mTORC1 and mTORC2 (Figure 1) and have different sensitivity towards its inhibitor rapamycin [49]. mTORC1 and mTORC2 are similarly large, weighing in at ~1.2 and ~1.4 MDa, respectively [50]. The central core catalytic subunit, mTOR, is common to both complexes and characterized by their own unique subunits. TOR proteins contain ∼2500 amino acids and comprise several distinct domains [51], including 32 tandem HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, and Tor1) repeats towards their N-termini, followed by FAT (FRAP, ATM, and TRRAP) domain consist of multiple antiparallel α-helical features, termed tetratricopeptide repeats (TPRs) [51].
Figure 1.
Schematic representation of various subunits of mTORC1 and mTORC2 complex and its upstream signaling regulators and cellular function. The mammalian target of rapamycin (mTOR); insulin growth factor (IGF); adenosine monophosphate activated protein kinase (AMPK); eukaryotic translation initiation factor 4E- (eIF4E-) binding protein 1 (4EBP1); proline-rich AKT substrate 40 (PRAS40); tuberous sclerosis protein ½ (TSC1/2); Ras homolog enriched in brain (Rheb); phosphoinositide 3 kinase (PI3K); Unc-51 like autophagy activating kinase (ULK); ribosomal protein S6 kinase beta-1 (S6K1); forkhead box O transcription factor (FOXO); serum/glucocorticoid-regulated kinase 1 (SGK1); peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α).
mTORC1 consist of five components (1) mTOR (mammalian target of rapamycin), the central catalytic subunit, (2) raptor (regulatory-associated protein of mTOR) [52, 53], (3) mLST8 or GβL (mammalian lethal with Sec13 protein 8) [54, 55]. (4) PRAS40 (proline-rich AKT substrate 40 kDa) [56–58], and (5) Deptor (DEP-domain-containing mTOR-interacting protein) [59, 60]. The cryoelectron microscopy structure of human mTORC1 revealed that mTORC1 maintains an obligate dimer with an overall rhomboid shape and a central cavity by interlocking mTOR-raptor interactions [61]. The principal interaction between raptor and mTOR consists of an α-solenoid stack formed between the horn and bridge domains of mTOR and the raptor armadillo domain [62].
The precise function of all mTOR-interacting proteins in mTORC1 complex remains to be understood. It was shown that rapamycin forms complex with FKBP12 and interacts with mTOR subunit and inhibits mTORC1 activity [53, 63, 64]. Experimental evidences also suggest that raptor might affect mTORC1 activity by regulating assembly of the complex and by recruiting substrates for mTOR [11, 21, 60]. The role of mLST8 in mTORC1 function is partially understood. Studies using raptor-, rictor-, or mLST8-deficient mice demonstrate that mLST8 is required for mTORC1 activity and is necessary to maintain the rictor-mTOR, but not the raptor-mTOR, interaction [55]. The interaction among different proteins in mTORC1 complexes is crucial and determines its active state. For example, the subunits of mTORC1, PRAS40 and Deptor, are known to interact with each other and docked to the core complex, resulting in the inhibition of mTORC1. PRAS40 and Deptor are phosphorylated by mTORC1, which blocks their interaction leading to the activation of mTORC1 [56–59].
Relatively, much less is known about the regulation of mTORC2 compared to mTORC1. Essentially, mTORC2 contains six different proteins, many of which are components of mTORC1: (1) mTOR; (2) rictor (rapamycin-insensitive companion of mTOR), unique to mTORC2; (3) mammalian stress-activated protein kinase-interacting protein (mSIN1) [65]; (4) protein observed with Rictor-1 (Protor-1); (5) mLST8; and (6) Deptor, an important interacting protein in mTORC1. mTORC2 contains its unique subunit rictor, which is insensitive to rapamycin. This is due to the fact that rapamycin when forms a complex with FKBP12 subunit, it does not bind to mTORC2 and losses it ability to block its activity [66–68]. The N-terminal region of rictor is composed of helical repeat clusters, which binds to mTOR as well as makes multiple contacts with mSin1. The FRB domain of mTOR also shows multiple cross-links with mSin1 and C-terminal regions of rictor. Rictor and mSin1 together generate a steric hindrance to inhibit binding of FKBP12-rapamycin to mTOR, revealing the mechanism for rapamycin insensitivity of mTORC2 [50, 69].
Recent evidences suggest that rapamycin inhibits mTORC1 at low concentration, and a prolonged chronic inhibition leads to inactivation of mTORC2 [70, 71]. Moreover, interaction between rictor and mSIN1 is essential for their own stability and to form the mTORC2 complex, since the deletion of SIN1 blocked the phosphorylation of AKT at serine 473 residue leading to the disruption of rictor-mTOR [72]. Deptor is common to both mTORC1 and C2 and acts as an endogenous inhibitor of mTORC2 [59]. mLST8 is also shown to be a crucial element in the mTORC2 complex formation since the ablation of this protein destabilize mTORC2. Interestingly, several of the subunits in mTORC1 and 2 are common to each other but they interact in an exclusive mechanism that characterize the individual complex. Even though they are unique in many aspects, mTORC1 and mTORC2 phosphorylate entirely different substrates and consequently have distinct function [55].
Several external stimuli such as nutrient, insulin, growth factors, leptin, and stress signals regulate mTOR complexes. However, mTORC1 and C2 respond to these factors differently and have exclusive downstream effects. The primary effector pathway of mTORC1 is through activation of ribosomal proteins S6 kinase 1 and 2 (S6K1/2) by phosphorylating their hydrophobic motif (HM), on Thr389 and Thr 388, respectively, which promotes mRNA biogenesis as well as translational initiation and elongation of protein synthesis. Other substrates of mTORC1 includes 4E- (eIF4E-) binding protein1 and (4EBP1), which are also involved in the activation of gene expression and protein translation. mTORC1 complex is also very sensitive to nutrients, particularly amino acids and glucose level. Deprivation of amino acids especially leucine results in rapid dephosphorylation of S6K1 and 4EBP1 and results in the inactivation of mTORC1 [13, 20]. The energy status of the cell is also sensed by mTORC1 through AMP-activated protein kinase (AMPK). AMPK is phosphorylated in response to low cellular energy status indicated by the high AMP/ATP ratio. The activated AMPK in turn inhibits cell growth via TSC2-dependent suppression of mTORC1 activity and blocks the phosphorylation of S6K1 and 4EBP1 mediated by mTORC1 [22, 73, 74]. Apart from its role in protein synthesis, mTORC1 is also involved in catabolic processes such as apoptosis and autophagy. Under starvation, mTORC1 phosphorylates ULK1 (Unc-51-like autophagy activating kinase), thereby preventing its activation by AMPK, an important activator of autophagy [75, 76].
mTORC2 is widely recognized to play an important role in cell proliferation and response to growth factors such as insulin. Unlike mTORC1, which acts through various downstream effectors, mTORC2 mainly acts through insulin/PI3K pathway via phosphorylation of AKT at serine 473, Thr 308, and Thr 450 residues upon stimulation by insulin [77, 78]. Recent evidence also suggest that mTORC2 can phosphorylate AKT at S377/T479 residues in the C-terminal end and can regulate apoptosis [79]. The mTORC2 subunit mSin1 contains a phosphoinositide-binding PH domain that is critical for the insulin-dependent regulation of mTORC2 activity [65, 80, 81]. Insulin binding to its tyrosine kinase receptor activates IRS and recruits activated PI3K. [82] The PI3-PDK pathway phosphorylates AKT in a mTORC2-dependent manner [83]. mTORC2 phosphorylates several protein kinases including PKA, B, C, G, SGK1 (serum/glucocorticoid-induced kinase 1), and Rho1 (GDP-GTP exchange protein-2), resulting in their stabilization and activation [84–90]. Rictor enables mTORC2 to directly phosphorylate AKT at its Ser473 and facilitates Thr308 phosphorylation by PDK1 (phosphoinositide-dependent kinase 1) as part of the insulin-signaling cascade [91].
Interestingly, there is a cross talk between mTORC1 and mTORC2 and they are functionally interconnected. Apparently, mTORC1 inhibits mTORC2 through phosphorylation of rictor and mTORC2 regulates mTORC1 through phosphorylation of AKT, which controls both the activity and abundance of AKT [92]. Rictor subunit of mTORC2 complex can be phosphorylated by S6K1, a downstream effector of mTORC1, and this phosphorylation negatively regulates the mTORC2-dependent phosphorylation of AKT-S473 [93]. In contrast, upon stimulation with growth factors, mTORC2 activates AKT, which in turn enhances mTORC1 activity through the inactivation of TSC1/2 (tuberous sclerosis complex). The TSC2 is inactivated by AKT-dependent phosphorylation, which destabilizes TSC2 and disrupts its interaction with TSC1 [94, 95].
1.2. MicroRNA Biogenesis and Mode of Action
MicroRNAs (miRs) are small noncoding RNAs consisting of approximately 22 nucleotide in size and function as gene suppressors [96, 97]. They bind to the 3′ untranslated region (UTR) of mRNA and regulate their expression via either degradation of mRNA transcript or interfere in the translation process [98, 99]. The regulation of gene expression mediated by miR has now been widely recognized as a major molecular mechanism employed by cells to control various function and signaling pathway [100–103], including AKT, AMPK, JNK, and TGF-β [104–114].
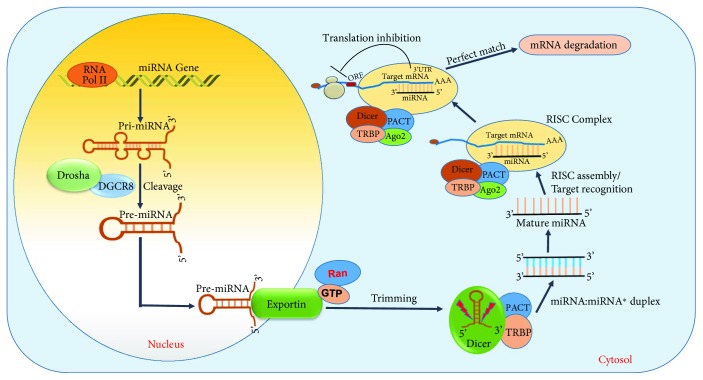
miRs are encoded across genomic locations including introns and intergenic. Once synthesized and matured through several steps these miRs bind to the complementary 3′UTR of their target mRNA and either degrade or silence them [115–117]. miRs undergo a series of maturation process before they develop into a mature miR (Figure 2). They are initially synthesized from their respective genomic region by the enzyme RNA polymerase II into a hairpin structure of approximately ~400–500 bps nucleotide into a primary miR transcript [118, 119]. They are further cleaved by the enzyme Drosha into a 70 nt length nucleotide, named pre-miR [120, 121]. The pre-miR then binds with the protein exportin-5 which transports them out of the nucleus for further trimming [122]. Once in the cytoplasm, they undergo further cleavage by the enzyme ribonuclease III (RNase III) and dicer in to a 22 nt mature miR [123–125]. The mature miR, depending on the complementary nucleotide sequence of its seed region (2–7) at its 5′ end, forms complex with its target mRNA. The double stranded miR-mRNA complex induces the RNA-induced silencing complex (RISC) and targets them for degradation or gene suppression [126–128]. A near perfect match between the seed region (~8 nt) of miR and mRNA leads to a complete degradation of the mRNA, while a partial complementarity results in the suppression of the gene expression [129–133]. miRs are transcribed either as an individual miRNA (e.g., miR-1) or as a family of clusters (e.g., miR-17~92) [134]. The coding region for miRNA can arise from either strand of the DNA and can have multiple mRNA targets [135–137].
Figure 2.
Illustration of miRNA biogenesis pathway and mode of gene regulation. RNA polymerase (RNA-Pol); Drosha; DiGeorge syndrome critical region-8 (DGCR8); guanosine triphosphate (GTP); RAs-related nuclear protein (Ran); human immunodeficiency virus trans-activating response RNA-binding protein (TRBP); argonaute 2 (Ago2); RNA-induced silencing complex (RISC); protein kinase RNA activator (PACT); open reading frame (ORF); 3′ untranslated region (3′UTR).
2. mTOR in Cardiovascular Diseases
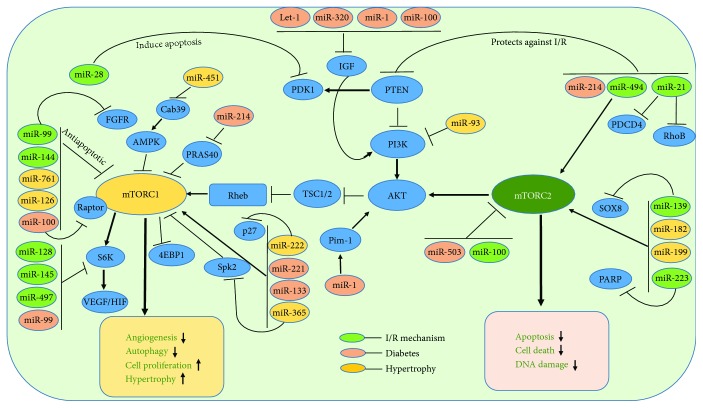
The role of mTOR in cancer and aging is well recognized and documented with numerous scientific publications [35, 138–144]. However, their role in cardiovascular system is still in the early stages and yet to be elucidated. mTOR plays an important role in the normal development of cardiovascular system and is crucial during pathophysiological conditions [35, 145–150]. Nevertheless, studies have revealed a novel role for mTOR in CVDs like ischemia reperfusion (I/R) injury [151–153], heart failure [154–156], and its associated risk factors including diabetes [146, 157, 158] and aging [31, 159, 160]. Recent research findings also indicate that the components involved in mTOR pathway are regulated by miRs in cancer and other diseases [39, 40, 161, 162]. A comprehensive role of miRs regulating mTOR signaling in cardiovascular diseases is depicted in Figure 3.
Figure 3.
Diagram depicting network of coordinated interaction between miRNA and mTOR pathway in the regulation of cardiovascular diseases. Fibroblast growth factor receptors (FGFR); insulin-like growth factor 1 (IGF-1) sphingosine kinase 2 (Spk2); regulation of proline-rich AKT substrate 40 kDa (PRAS40); phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K); phosphoinositide-dependent kinase l (PDK1); calcium-binding protein 39 (Cab39); poly (ADP-ribose) polymerase (PARP); SRY-related HMG-box 8 (SOX8); programmed cell death protein 4 (PDCD4); Ras homolog gene family, member B (RhoB); phosphatase and tensin homolog (PTEN); vascular endothelial growth factor (VEGF); hypoxia-inducible factors (HIF); tuberous sclerosis protein (TSC1/2); ras homolog enriched in brain (Rheb); ribosomal protein S6 kinase beta-1 (S6K1); proviral integration site for Moloney murine leukemia virus-1 (Pim-1).
2.1. miRNA-Dependent Regulation of mTOR in Ischemia Reperfusion Injury
Oxidative stress induced by ROS generation is a major mechanism of cell injury during myocardial I/R injury [163]. Deprivation of oxygen during I/R stress activates mTORC1 [146, 151, 158, 164] and controls several downstream kinases and leads to cellular effects such as apoptosis [165], autophagy [166, 167], and proliferation [22]. Accumulating evidences point out several different miRs are involved in the modulation of mTOR signaling via targeting mTOR-interacting partners [40, 168–171].
mTOR mediates cardiomyocyte response during ischemia and is an important determinant of cell survival [172, 173]. Inhibition of mTORC1 was shown to be beneficial for the survival of cardiomyocytes via induction of autophagy [174]. Sciarretta et al. demonstrated that selective and direct mTORC1 activation is detrimental during acute cardiac energy deprivation, whereas both pharmacological and genetic mTORC1 inhibition are protective [175]. Pretreatment with rapamycin, the mTOR inhibitor, reduced myocardial infarct size after I/R injury by attenuating necrosis and apoptosis in cardiomyocytes [158, 176]. Reperfusion therapy with rapamycin also attenuated myocardial infarction and apoptosis by activation of PI3K and ERK [177]. Activation of autophagy prevents unwanted expenditure of cellular energy to damaged cells, especially mitochondria, which results in an increased ROS generation [174, 178]. Cardiac-specific overexpression of Rheb leads to the inhibition of Atg7, a key effector protein in the autophagy cascade, and enhances cardiomyocyte cell death through activation of Rheb/mTORC1 signaling pathway [175]. Interestingly, mTOR inhibition with rapamycin promotes the survival of oxygen-deprived cardiomyocytes through activation of autophagy via inhibition of Ras homolog enriched in brain (Rheb) protein [175]. These results indicate that Rheb is a main regulator of mTORC1 during cardiomyocyte energy stress, and Rheb/mTORC1 inhibition promotes cell survival through activation of autophagy [175]. Moreover, obesity and metabolic syndrome, which are characterized by increased myocardial susceptibility to ischemic injury and cardiovascular mortality, are associated with inadvertent activation of the Rheb/mTORC1 pathway and reduction of autophagy [175, 179–181]. Mice with high fat diet- (HFD-) induced obesity and metabolic syndrome exhibit deregulated cardiac activation of Rheb/mTORC1 and inhibition of cardiac autophagy, which lead to increased ischemic injury [175]. Rapamycin treatment before prolong ischemia (3 hours) increases autophagy and significantly reduces the myocardial infarction of both HFD-treated mice [175].
Autophagy is a delicate process that involves a closely-knit transcriptional and epigenetic regulation through miRs [182, 183]. Overexpression of microRNA-99a (miR-99a) through intramyocardial delivery improved cardiac function after MI stress and prevented cell death via enhancing autophagy in mTOR/P70/S6K-dependent signaling pathway [184]. Notably, overexpression of miR-99a in the border zone of infarct area prevented cell apoptosis, but increased autophagy via inhibiting mTOR/P70/S6K [184]. Furthermore, the expression level of miR-99a was reduced in neonatal mice ventricular myocytes (NMVMs) subjected to hypoxia. Similarly, intramyocardial delivery of lenti-miR-99a in mice showed a significant improvement in both left ventricular (LV) function and cell survival post four weeks of MI compared to sham groups [184]. Even though the study illustrated that miR-99a is cardioprotective through decreasing mTOR activity, it did not show a direct target of miR-99a. However, further evidence from the same group also showed that fibroblast growth factor receptor 3 (FGFR3) to be a direct target of miR-99a and hinted a possible role for FGFR-mTOR pathway in MI-induced hypertrophy [185]. Interestingly, multiple findings established a link between miR-99a and FGFR in cell proliferation via mTOR signaling [186–188].
Prevention of the loss of cardiomyocyte during I/R injury is the primary focal point and strategic approach to avoid cardiac dysfunction post-MI, which can potentially be achieved by regulating apoptosis and autophagy [189]. Experiments using primary neonatal cultured mouse cardiomyocytes identified a direct link between miR-28 and PDK1, an immediate upstream regulator of mTOR in the PI3K pathway [190]. Induction of oxidative stress in cardiomyocyte using hydrogen peroxide elevated the expression of miR-28 and increased apoptosis-mediated cell death [190]. Interestingly, overexpression of miR-28 downregulated p-AKT, p-p70, and p-mTOR suggesting a direct interference of mTOR signaling by miR-28 [190]. Mechanistically, PDK1 was found to be a direct target of miR-28 and target-binding assay using luciferase activity and PDK protein expression after transfection with miR-28 confirmed the prediction [190]. Similarly, miR-223 has been reported to play an important role in cell survival by regulation of autophagy and apoptosis [191]. The miR-223 was upregulated in the border zone of infarct area in rats subjected to LAD occlusion [191]. Moreover, overexpression of miR-223 protected H9c2 cells and neonatal rat cardiomyocytes (NRCMs) against hypoxia-induced apoptosis by directly targeting PARP-1 [191]. Decisively, this study showed that H9c2 and NRCMs cells treated with miR-223 mimic increased p-AKT and p-mTOR expression under hypoxic conditions and the protective effect of miR-223 was abolished upon treatment with miR-223 inhibitor [191]. Although, earlier study by van Rooij et al. reported an upregulation of miR-223 in human failing heart tissues [192], but did not explore on the mechanism. The same study showed that miR-29 family are downregulated in the region of the fibrotic scar after MI.
Phosphatase and tensin homology deleted from chromosome 10 (PTEN) are an important determinant for the activation of AKT through PI3 kinase-mTOR pathway. It enhances cardiomyocyte cell death and increases cardiac dysfunction during MI and I/R injury [193–196]. Genetic ablation or pharmacological inhibition of PTEN has been shown to be cardioprotective against MI and vascular remodeling [196, 197]. Our laboratory demonstrated that miR-21 had a powerful cardio protective effect against I/R injury [198–200]. miR-21 expression is induced in the border zone of the infarcted hearts, but it is significantly decreased in the infarcted area [181, 192]. Overexpression of miR-21 protects against I/R injury by reducing myocardial infarct size and apoptosis, by its target genes, PTEN, and programmed cell death 4 (PDCD4) [201–203]. Recent studies suggest that the therapeutic effects seen with miR-21 may be mediated through PTEN/AKT/mTOR signaling pathway [204]. It was observed that miR-21 expression was downregulated, and autophagy was remarkably increased in H9c2 cells during H/R injury. Simultaneously, increased apoptosis after H/R injury was associated with reduction of Bcl2-Bax ratio. Such an effect was abolished by overexpression of miR-21 with a miR-21 precursor, which also inhibited autophagic activity and decreased apoptosis accompanied by the activation of the AKT/mTOR pathway [204]. Thus, it appears that miR-21 plays an active role in disrupting the PTEN-AKT-mTOR pathway. Similar result was also observed in cardiac stem cells, where miR-21 reduced hydrogen peroxide- (H2O2-) induced apoptosis, as evidenced by the downregulation of caspase-3 and Bax and upregulation of the antiapoptotic Bcl-2 [205]. Overexpression of miR-21 suppressed the expression of PTEN, a direct target of miR-21, with simultaneous increased in the phosphorylation state of AKT. Moreover, the antiapoptotic effect of miR-21 was abolished in cells treated with miR-21 inhibitor and PI3 inhibitor, LY294002, suggesting an involvement of PTEN/PI3K/AKT signaling in miR-21-mediated antiapoptotic effect [205]. Although miR-21 has been considered to promote cellular proliferation, invasion, and migration in various types of tumors [206, 207], rapamycin treatment induced the expression of miR-21 in human umbilical vein endothelial cells (HUVECs) but attenuated endothelial cell proliferation and migration [208]. RhoB, an important partner in AKT-mTOR pathway, is a direct target of miR-21, and silencing of RhoB impairs endothelial cell migration and tubulogenesis, thus providing a possible mechanism for miR-21 to inhibit angiogenesis after rapamycin treatment [208, 209]. However, raptor knockdown, but not rictor silencing, upregulates miR-21 expression, and inhibition of miR-21 blunted the antiproliferative and antimigration effects of rapamycin treatment [208].
Interestingly, miR-21 was shown to be upregulated with rapamycin treatment in angiomyolipoma-derived cells isolated from patient with lymphangioleiomyomatosis (LAM) [210]. Conceptually, LAM is induced by mutation in the tuberous sclerosis complex genes (TSC1 or TSC2) [211] and results in hyperactivation of mTOR signaling, which is characterized by proliferation of smooth muscle-like cells and leads to the malfunction of the lungs. The study demonstrated that 19 miRNAs were differently regulated by rapamycin, and miR-21 was shown to be robustly upregulated in TSC2-deficient 621–101 cells (Renal angiomyolipoma cells). This study also suggested that rapamycin-mediated upregulation of miR-21 is independent of AKT signaling, but rather dependent on mTOR. More importantly, the study also demonstrated that rapamycin potentiates the Drosha-mediated posttranscriptional processing of pri-miR-21 to pre-miR-21. Moreover, rapamycin was clinically shown to improve pulmonary function in LAM patients. Surprisingly, a recent another study demonstrated a significant upregulation of miR-21 in Tsc2-deficient cells compared to wild type controls, which was further induced by rapamycin [212]. Experimental evidences suggest that miR-21 induced proliferation, tumor growth, and offered resistance to apoptosis in TSC2-deficient cells. Moreover, data analysis of RNA Seq implicated that miR-21 promoted mitochondrial adaptation and homeostasis in Tsc-2-deficient cells. Inhibition of miR-21 using LNA reduced the tumor size and mitochondrial function. In addition, rapamycin cotreatment with miR-21 inhibition more efficiently reduced tumorigenic growth of Tsc2-deficient cells in vivo xenograft model. Importantly, this study showed that rapamycin increased mitochondrial content and polarization, and these effects of rapamycin were miR-21-dependent. The study also proposed that the unexpected upregulation of miR-21 in TSC2-deficient cells was partly due to miR-21 regulation of mTOR in a noncanonical pathway via either STAT3 or Rheb [212]. Recently, mTORC1 was also reported to regulate the miRNA biogenesis pathway itself [213]. Extensive expression analysis of 752 miRs in TSC2-deficient cells, treated with Torin1 (inhibitor of both mTORC1 and C2), demonstrated an upregulation of majority of miRs in consistent with the increased activity of microprocessor (the multiprotein complex that includes Drosha (a type III RNAse) and DGCR8) in TSC2-deficient cells. Microprocessor activity is regulated in part by GSK3β, which is phosphorylated at S9 and subsequently inhibited by mTORC2 via AKT. Inhibition of mTORC1 impaired the microprocessor activity through regulation of Drosha and GSK3β-dependent pathways via mTORC2 [213].
Numerous studies reported that miR-21 is involved in a variety of disorders and is highly upregulated during cardiac remodeling [201, 214–216]. However, genetic deletion of miR-21 or acute inhibition of miR-21 did not alter the pathological responses of the heart to pressure overload or other stresses, which suggests that miR-21 is not required for cardiac hypertrophy, fibrosis, or loss of contractile function in response to acute or chronic injury in mice [217]. However, the precise role of miR-21 regulating mTOR signaling in cardiovascular system is still not well evolved, and the effect of miR-21 on proliferative cells like endothelial, smooth muscle cell, and nonproliferative cells like cardiomyocytes may be different. However, several reports in cancer biology strongly suggest an active role for miR-21 in regulating mTOR signaling largely through PTEN/PI3K/AKT pathway [218–222].
Angiogenesis is an important process that restores blood supply to the infarct area post-MI and I/R injury and improves cardiac function [223, 224]. In this context, miR-100 was reported to be an antiangiogenic miR and functioned through repressing mTOR signaling after induction of hind-limb ischemia in mice [162]. miR-100 modulated proliferation, tube formation, and sprouting activity of endothelial cells and migration of vascular smooth muscle cells and functions as an endogenous repressor of mTOR. Inhibition of miR-100 by specific antagomirs stimulated angiogenesis with functional improvement of perfusion after femoral artery occlusion in mice. Moreover, the stimulatory effect of antagomir therapy was abolished by simultaneous rapamycin treatment, demonstrating that the angiogenic effect of miR-100 inhibition in hind-limb ischemia was dependent on its target gene mTOR [162]. Nevertheless, this study did not address the specific role of mTORC1 or C2 in blocking the angiogenic response and did not use a long-term treatment with rapamycin [162].
In tumor glioma, miR-128 is downregulated and acts as a tumor suppressor by directly targeting p70S6K1 [225]. Overexpression of miR-128 attenuated cell proliferation, tumor growth, and angiogenesis by suppressing p70S6K1 and its downstream signaling molecules such as HIF-1 and VEGF expression. Similarly, the expression of miR-145 is downregulated in colon and ovarian cancer, and overexpression of miR-145 inhibits tumor growth and angiogenesis by targeting p70S6K1 and suppressing its downstream angiogenic factors HIF-1 and VEGF [226]. Another miR, miR-497 is downregulated in breast, cervical, head-and-neck, colorectal, and prostate cancers, and overexpression of miR-497 sensitizes the resistant ovarian tumor to cisplatin treatment by targeting mTOR and p70S6K1 [227]. Among the three miRs previously reported to target p70S6k1, only miR-128-3p is downregulated in human cardiomyocytes during H/R by Tongxinluo (TXL, a traditional Chinese medicine, widely used to treat cardiovascular and cerebrovascular diseases). Interestingly, TXL restored p70S6K1 but had no effects on miR-145-5p and miR-497-5p [228]. Inhibition of miR-128-3p activated mTOR via increasing the phosphorylation and abundance of p70s6k1.
PI3K/AKT/mTOR pathway has been shown to be suppressed by miR-139 in I/R injury in H9c2 cell line [229]. The overexpression of SOX8, a target of miR-139, alleviates hypoxia-induced cell injury via activation of PI3K/AKT/mTOR and MAPK pathway [229]. Recently, several miR profiling studies revealed that miR-494 was downregulated in human failing hearts as well as ischemic/hypertrophic hearts of animals [230–232]. The cardiac-specific overexpression of miR-494 in mice protected hearts against I/R-triggered injury; conversely, knockdown of endogenous miR-494 by antagomir sensitized hearts to I/R-induced injury [233]. The overexpression of miR-494 suppressed the levels of proapoptotic proteins (PTEN, ROCK1, and CaMKIIδ) after I/R injury, which also induced AKT signaling in concert, a critical survival pathway in the myocardium mediated through mTORC2 activation [233]. Also, the inhibition of miR-494 using antagomir elevated the level of PTEN while simultaneously suppressing the level of pAKT (S473) after I/R injury [233].
Apart from the intracellular regulation of mTOR pathway by miRs, miRs packed in exosomes can affect cardiac function. Based on the miRNA array data, remote ischemic preconditioning (rIPC) altered the myocardial expression of miR-144 in mice [234]. Initially, it was shown that rIPC increased the myocardial expression of miR-144, whereas I/R injury alone significantly reduced the level of miR-144. Intriguingly, the exosomes isolated from tissue samples of rIPC hearts were rich with the expression of miR-144 upon rIPC. Moreover, intravenous administration of miR-144 via tail vein injection induced early and delayed cardioprotection in Langendorff isolated perfused model of I/R injury [234]. This study also showed that miR-144 directly targeted mTOR as evident with the decreased p-mTOR and increased autophagy signaling upon miR-144 administration. More precisely p-AKT (S473), a marker for mTORC2 activation, was increased in the heart upon miR-144 injection in mice [234]. These finding suggest that miR-144 acts via suppressing mTORC1 while simultaneously activating mTORC2 complex [234]. In silico analysis of miRNA-target mRNA prediction algorithm (TargetScan 6.0) revealed two specific miR-144 binding sites in the mTOR 3′UTR region with perfect Watson–Crick matches at miRNA positions 1–7 and 2–8 [235]. The interaction of miR-144 and mTOR and its clinical significance have been evaluated in human cancer biology. Specifically, the downregulation of miR-144 leads to poor prognosis of cancer patients via activation of the mTOR signaling pathway [235].
2.2. Regulation of mTOR through miRNA in Diabetes and Obesity
Diabetes is a major risk factor for CVD and is characterized by elevated blood glucose, insulin resistance/deficiency, and metabolic abnormalities [236, 237]. Since mTOR is sensitive to nutrient, excessive glucose level in the blood stream activates mTOR [146, 151, 158, 238]. Prolonged activation of mTORC1 induces insulin resistance in adipose tissue through the S6K1-mediated inhibition of insulin signaling that disrupts the recruitment and activation of PI3K via phosphorylation of insulin receptor substrate-1 (IRS-1) [239, 240]. Similar aberrant mechanism in cardiovascular tissues, in conditions like diabetic and obesity, leads to cardiac abnormalities through S6K1-IRS-1 [241] and its effector kinases like MAPK [242], AMPK [241], and glycogen synthase kinase-3β (GSK3β) [148, 243, 244]. Several miRNAs were identified to play a role in diabetes by regulating insulin signaling and glucose metabolism [238, 245]; [246, 247]. Some of the prominent miRs that regulate mTOR pathway are miR-133a, miR-100, miR-221, miR-483-3p, miR-133a, miR-503, miR-214, microRNA-99a, miR-143, miR-126, and miR-181a-5p.
Inhibition of Let-7 family of miR was shown to be beneficial and promoted cardiac function against I/R injury in diabetic rats [248]. I/R injury in diabetic rat significantly increased let-7 miR as well as infarct size, while antagomir let-7-treated diabetic group offered protection against I/R [248]. Moreover, the myocardial expression of IGF1 and GLUT4 as well as p-AKT (S473) were significantly lower with activation of mTOR in diabetic group. Notably, blocking of let-7 expression or treatment with rapamycin effectively increased AKT phosphorylation at S473 residue, while simultaneously blocked mTOR phosphorylation [248]. IGF plays an important role in glucose metabolism and in the development of insulin resistance, which are crucial events in diabetic cardiomyopathy. miR-1 has been shown to directly targeted IGF-1 [249] and regulated PI3-AKT pathway [250]. In support of this notion, it was shown that miR-1 increased during diabetic cardiomyopathy, which led cardiomyocyte apoptosis through targeting Pim-1 (proviral integration site for Moloney murine leukemia virus-1) [251]. Inhibition of miR-1-dependent downregulation of Pim-1 using miR-1 antagomir resulted in the elevation of phosphorylated AKT and abrogation of diabetic-induced cardiac apoptosis [251]. Similarly, miR-320 is also identified to directly target IGF-1 and VEGF and impairs angiogenesis in myocardial microvascular endothelial cells (MMVEC) isolated from Goto-Kakizaki (GK) diabetic rats [252]. Published studies also demonstrated that miR-99a suppressed the expression of IGF-1 and inactivated mTOR in vascular smooth muscle cells (VSMC) [253]. The hyperinsulin-mediated proliferation and migration of VSMC were reversed by overexpression of miR-99 or inhibition of mTOR. Moreover, overexpression of miR-99a reduced AKT and ERK1/2 activity while suppressing p70S6K, a downstream target of mTORC1 [253].
miR-133a is one of the predominantly expressed miRs in the cardiac tissue, which plays a protective role against pathological remodeling by inhibiting cardiac hypertrophy and cardiac fibrosis in diabetes [254, 255]. Studies in the murine model show that diabetes attenuates miR-133a in hearts [256, 257]. Additionally, a diabetic heart failure (DHF) patient population study showed that the attenuation in the level of miR-133a in diabetic hearts was associated with the exacerbation of autophagy and hypertrophy and suppression of mTOR [258]. In contrast, another interesting study conducted to evaluate the cardiac dysfunction in the offspring of maternal diet-induced obesity revealed a role for miR-133a in cardiac hypertrophy [259]. The results showed that the level of miR-133 is significantly increased in ventricular tissue of the Mat-Ob group and cardiac hypertrophy in the offspring [259]. Most notably, AKT1-Ser473 phosphorylation as well as levels of phospho-ERK1/2, phospho-mTOR, and phospho-p38MAPK were significantly elevated in the Mat-Ob group [259], suggesting an active role of mTOR in the development of cardiac hypertrophy upon diet-induced maternal obesity [259].
Elevated levels of fatty acids and glucose observed in obesity and diabetes mellitus (DM) contribute to systematic inflammation [260, 261]. Blood miRNAs signatures in patients with diabetes with/without obesity revealed a significant reduction of circulating miR-100 in obese normoglycemic subjects and subjects with T2D compared to healthy and lean individuals [262]. Visceral adipose miR-100 was also lower in obese patients with T2D compared to those without. Reduced miR-100 levels were associated with adverse metabolic indices, which may lead to the differentiation of fat tissues and subsequent lipid accumulation, potentially contributing to increased obesity. miR-100 led to the differentiation of adipocytes by modulating its direct target IGFR (insulin growth factor receptor), mTOR, and vLDLR signaling.
A recent study characterized the function of the endothelial-enriched miR-100 during vascular inflammation and atherogenesis [263]. It was reported that miR-100 directly repressed several components of mTORC1-signaling, including mTOR and raptor, which led to the stimulation of endothelial autophagy and diminished activity of the proinflammatory transcription factor NF-κB. In a low-density lipoprotein receptor-deficient atherosclerotic mouse model, inhibition of miR-100 enhanced atherosclerotic plaque formation and a higher macrophage content of the plaque, whereas miR-100 mimic attenuated atherogenesis in the aortic root and in the abdominal aorta. Moreover, miR-100 mimic suppressed mTOR and the transcription factor SREBP-2, which subsequently controlled lipid metabolism in hepatocytes. mTOR inhibition with rapamycin showed anti-inflammatory effects through decreasing the expression of E-Selectin, intracellular adhesion molecule 1 (ICAM-1), and vascular cell adhesion molecule-1 (VCAM-1) in response to endothelial cell activation with TNF-α. In addition, rapamycin abolished the effects of miR-100 inhibition with TNF-α on endothelial adhesion molecule protein expression, confirming the essential role of intact mTOR signaling in the anti-inflammatory effects of miR-100 [263].
Vascular remodeling and cardiac hypertrophy is one of the adverse effect of diabetes and results in end-stage heart failure [264, 265]. To address this phenomenon, cardiac hypertrophy was induced by angiotensin II (Ang II) treatment in diabetic OVE26 mice, and the role of miR-221 on autophagy was investigated [266]. The results demonstrated that Ang II treatment increased the phosphorylation of c-Jun, JNK, mTOR, and miR-221, while decreasing the level of p27, a direct target of miR-221 and regulator of p-mTOR [266]. Direct downregulation of p27 by miR-221 led to mTOR activation and diminished cardiac autophagy of diabetic OVE26 and/or Ang II-treated mice, resulting in cardiac hypertrophy [266].
mTOR plays a contrasting role in type I DM, where there is an insufficient insulin secretion due to deficient pancreatic β-cells. In gestational diabetes mellitus (GDM), it was shown that knockdown of miR-503 enhanced insulin secretion of pancreatic β-cells, promoted cell proliferation, and protected cells from apoptosis [267]. mTOR has been identified as a direct target of miR-503, and suppression of miR-503 improves insulin secretion and pancreatic β-cells proliferation [267]. The regulation of mTOR pathway by miR is also evident in renal cortex of type 1 diabetic mice [268]. Elevation of miR-214 under high glucose conditions decreased the levels of its target PTEN and increased AKT activity (p-S473) and led to phosphorylation of its substrates glycogen synthase kinase-3β and phosphorylation of PRAS40. In contrast, antimiR-214 blocked the phosphorylation of both AKT and PRAS40 and attenuated renal cell hypertrophy, suggesting that inactivation of both mTORC1 and C2 is beneficial [268]. Consistent with this finding, studies using placental tissue from women with GDM demonstrated a robust activation of both mTORC1 and C2 as evident with the increased phosphorylation of AKT (S473), (4EBP1), and p70 S6 kinase (S6K) [269]. Data also showed that miR-143 was significantly high using placental tissue and trophoblast cells, and it impaired mitochondrial respiration via targeting hexokinase (HK), a rate-limiting enzyme in glycolysis [269]. Similarly, miR-99a has been shown to be involved in insulin-dependent glucose consumption in human liver cells (HLL7702) via directly targeting mTOR [270]. Cells treated with insulin suppressed the level of miR-99a while increasing glucose consumption and activation of mTOR. In contrast, the overexpression of miR-99a or rapamycin treatment reversed insulin-mediated glucose utilization [270].
2.3. Interaction of mTOR and miRNA in Vascular Remodeling and Hypertrophy
Given the role of mTOR in regulating protein synthesis through S6K [271] and cell cycle control [272, 273], it is well established that mTOR play a key role in cardiac hypertrophy [274–276]. In fact, several reports support this notion as mTOR inhibitors have antihypertrophic property [277–279]. Due to its antiproliferative properties, mTOR inhibitors have also been approved as anticancer drugs [280–282]. Intriguingly, the identification of miRNAs as novel emerging regulators of mTOR signaling has provided new insights into a multitude of biological processes, especially in tissue remodeling and hypertrophy, which has been appreciated by the scientific community in cardiac physiology [103, 169, 185]. Hypertrophic stimuli such as phenylephrine [283], angiotensin II (Ang II) [37, 284, 285], and endothelin-1 [286] are known to activate mTORC1 in the heart and result in robust vascular remodeling leading to heart failure [274]. However, mTORC2 is essential for the preservation of cardiac function and attenuation of pressure overload-induced cardiac hypertrophy [287]. It is increasingly apparent that mTOR [156, 283] and miR [216, 254] have a critical role in the development of cardiac hypertrophy and it is becoming important to understand the mechanism by which these two major regulators communicate with each other.
Cardiomyocyte-specific miR-199a overexpression inhibited autophagy and induced cardiac hypertrophy via targeting glycogen synthase kinase 3β (GSK3β) involving mTOR signaling [39]. The mTOR signaling was activated in miR-199a transgenic hearts [39]. In addition, treatment with rapamycin blocked the activation of p-mTOR and p-S6 in miR-199 transgenic mice and attenuated hypertrophy with induction of autophagy [39]. Data also indicated that miR-761 expression was reduced during Ang II-induced proliferation of VSMCs, and exogenous miR-761 delivery effectively inhibited the Ang II-induced VSMC proliferation. [288]. Experimental evidence showed that miR-761 directly targets mTOR and reduced its abundance [288]. Similarly, miR-99a was shown to negatively regulate hypertrophy through mTOR signaling pathway [185]. Interestingly, mice displayed an increase in mTOR activity starting at first week through 8 weeks following TAC- (transverse aortic constriction-) induced cardiac hypertrophy [185]. Overexpression of miR-99a suppressed mTOR and attenuated cardiac hypertrophy and cell death in TAC mouse model. Overexpression of miR-99a attenuated cardiac hypertrophy in TAC mice and cellular hypertrophy in cardiomyocytes subjected to Ang II or isoprenaline (ISO) through suppression of expression of mTOR [185].
In contrast, it has been shown that cardiac-specific overexpression of miR-222 induced pathological cardiac remodeling and heart failure in mice [289]. Transgenic mice with cardiac-specific expression of miR-222 (Tg-miR-222 mice) developed severe cardiac fibrosis and apoptosis, which led pathological cardiac remodeling and heart failure. The autophagy was inhibited in the hearts of Tg-miR-222 mice with activation of mTOR, but the expression of p27 was downregulated in the hearts of Tg-miR-222 mice [289]. It was suggested that miR-222 induced autophagy through activation of both mTORC1 and C2 complexes as shown with a substantial increase in both p-mTOR and p-S6 (Ser240/244) in transgenic Tg-miR-222 mice [289]. In the context of these findings, Su et al. also reported a role for p27-mTOR in the development of cardiac hypertrophy [290]. Cardiac-specific overexpression of miR-221, driven by the α-myosin heavy chain, resulted in hypertrophic hearts at 4 weeks of age with increased expression levels of ANP and BNP [290]. Moreover, miR-221 also inhibited autophagy, as demonstrated by downregulation of LC3-1/LC3-II ratio and an increase in p62 expression level [290]. Further, miR-221 overexpression in H9C2 cells and in primary cardiomyocytes showed decreased autophagosome formation as demonstrated with low number of EGFP-LC3 puncta [290]. More importantly, phosphorylation levels of mTOR and its substrates phospho-mTOR (S2448), phospho-4EBP1 (T37/46), and phospho-S6 (S235/236) levels were all significantly increased in Tg-miR-221 hearts at 4 weeks of age compared with those in the nontransgenic controls [290]. Conversely, silencing miR-221 in H9C2 cells and cardiomyocytes decreased the levels of phospho-mTOR, phospho-S6K, and phospho-S6, thereby establishing a link between miR-221 and mTOR signaling in the induction of cardiac hypertrophy [290]. Similarly, miR-365 was shown to promote cardiac hypertrophy through inhibition of autophagy by suppressing S-phase kinase-associated protein 2 (SPK2), an important activator of autophagy [291]. Conceptually, it was demonstrated that Spk2 induces autophagy through inhibition of mTORC1 and reverses adverse effect of cardiac hypertrophy [291]. Notably, Ang II treatment of cardiomyocytes increased the phosphorylation of the mTORC1 downstream effectors S6K and 4EBP1 and decreased the level of Spk2. Inhibition of mTOR activation, using rapamycin, completely abolished the Ang II-mediated inhibition of autophagy via miR-365-Spk2-dependent mechanism [291].
Recent studies have suggested that a long noncoding RNA (lncRNA), myocardial infarction-associated transcript (MIAT), plays a role in vascular remodeling and cardiac hypertrophy [41]. In this study, the authors demonstrated a three way link between MIAT, miR-93, and mTOR network. The upregulation of MIAT was associated with the decrease in miR-93 in Ang II-induced cardiac hypertrophy in rat [41]. Furthermore, it was shown that MIAT positively regulated TLR4 expression by acting as a sponge for miR-93 expression [41]. Overexpression of miR-93 attenuated MIAT-induced increase of TLR4 level in cardiomyocytes and attenuated Ang II-induced cardiac hypertrophy. In contrast, MIAT knockdown or miR-93 overexpression led to a significant inhibition on the protein levels of PI3K, p-AKT, and p-mTOR and blunted Ang II-mediated cardiac hypertrophy [41]. This study also suggested a strong corelation between miR-93, TLR4, and mTOR signaling, since overexpression of TLR4 enhanced the expression of miR-93 and blocked the protection observed with p-mTOR inhibition [41].
High fat diet (HFD) consumption for a prolonged time induces cardiac hypertrophy [292], and mTOR being a nutrition sensor plays an active role in mediating this effect in the heart [293]. Microarray analyses of the heart tissue of mice on HFD for 8 and 20 weeks identified a role for miR-451 in the development of cardiac hypertrophy [103]. Calcium-binding protein 39 (Cab39) is a direct target of miR-451 and an upstream kinase of AMP-activated protein kinase (AMPK). Suppression of miR-451 protected neonatal rat cardiac myocytes against palmitate-induced lipotoxicity through a mechanism that involves Cab39 [103]. In addition, cardiomyocyte-specific miR-451 knockout mice were resistant to HFD-induced cardiac hypertrophy. Protein levels of Cab39 and phosphorylated AMPK were increased, and phosphorylated mTOR and S6 phosphorylation were significantly suppressed in cardiomyocyte of the HFD-fed miR-451 cKO mice compared with control mouse hearts [103]. These findings elucidated an interesting aspect of AMPK-miR-451 and mTOR cross talk in cardiac hypertrophy.
Angiogenesis is an important process that plays a detrimental role in post-MI, and its abnormal regulation leads to cardiac hypertrophy [294]. mTOR and its downstream target AKT have been involved in the control of angiogenesis process during I/R injury [295–298]. Placental growth factor (P1GF), a member of vascular endothelial growth factor (VEGF) family, has been shown to induce cardiac angiogenesis and leads to hypertrophic heart [299]. Cardiac-specific overexpression of P1GF induced cardiac angiogenesis with increased expression of miR-182 at 6 weeks onset of angiogenesis process [299]. The study also found blunting of miR-182 upregulation in PlGF-induced eNOS−/− mice, suggesting that miR-182 acts through NO-independent pathway to regulate angiogenesis [299]. Since NO exerts its function through AKT, it was further shown that mTORC1 was involved in the induction of angiogenesis and cardiac hypertrophy. Suppression of miR-182 using antimiR-182 decreased the phosphorylation of AKTSer473 and p70-S6KThr389, thus indicating an important regulatory effect of miR-182 on the AKT/mTORC1 pathway [299].
Endothelial cell dysfunction contributes to coronary vascular tone and results in atherosclerosis by affecting various growth factors, such as vascular endothelial growth factor, fibroblast growth factors (FGFs), and platelet-derived growth factors [300, 301]. PI3K/AKT/mTOR pathway plays a role in endothelial function and in the development of atherosclerosis [302]. miR-126 has been shown to play a role in alleviating oxidized low-density lipoprotein (ox-LDL) induced HUVEC injury through suppression of AKT-mTOR pathway [303]. The overexpression of miR-126 reversed ox-LDL-induced cell injury and apoptosis in HUVECs [303]. Conceptually, treatment of HUVECs with ox-LDL increased the phosphorylation of mTOR through activation of PI3K and AKT, and miR-126 mimics restored the impaired autophagic flux via inhibition of PI3K/AKT/mTOR pathway [303].
A recent study by Bera et al. revealed a significant role of miR-214 in the activation of mTORC1 that contributed to high-glucose-induced mesangial and proximal tubular cell hypertrophy and fibronectin expression [268]. miR-214 expression is increased in the renal cortex of type 1 diabetic mice. High glucose treatment induced the expression of miR-214 and decreased its target, PTEN, in mesangial and proximal tubular epithelial cells [268]. Suppression of PTEN subsequently increased the AKT-dependent mTORC1 activation to induce mesangial and proximal tubular cell hypertrophy and fibronectin expression. Quenching of miR-214 expression inhibited high-glucose-stimulated cell hypertrophy and expression of the matrix protein fibronectin. In contrast, overexpression of miR-214 suppressed PTEN and increased AKT activity similar to high glucose and led to phosphorylation of two mTORC1 inhibitors, PRAS40 and tuberin, which contributes to high-glucose-stimulated mTORC1 activation [268].
Interestingly, a recent study demonstrated that overexpression of lncRNA Plscr4 alleviated pressure overload-induced cardiac hypertrophy in mice and attenuated the increased cell surface area of cultured neonatal mouse cardiomyocytes treated with Ang II [304]. The study identified that Plscr4 elicits the antihypertrophic effects by repressing the prohypertrophy gene miR-214. Mitofusin 2 (Mfn2), which is located at the mitochondrial outer membrane, plays a negative regulator of cardiac hypertrophy by modulating mitochondrial fusion [305, 306]. Mfn2 is a direct target of miR-214 in the hypertrophic heart [307]. The interaction between Plscr4 and miR-214 attenuated the inhibitory effects of miR-214 on Mfn2. The overexpression of Plscr4 rescued the decreased expression of Mfn2 by sponging miR-214 in response to hypertrophic stress and, therefore, resisted mitochondrial dysfunction to alleviate hypertrophic growth [307]. However, the exclusive interplay between lncRNA Plascr4 and mTOR in regulation mediated by miR-214 of cardiac hypertrophy is yet to be identified.
3. Therapeutic Potential of miRNA and mTOR Inhibitors in CVD
Rapamycin, received the FDA approval in 1999, has been successfully used as an effective immunosuppressant post-organ transplantation to prevent allograft rejection [308]. The rapamycin-eluting coronary stent received first FDA approval in 2003 for use in coronary-artery stents to prevent restenosis [309–311]. Rapamycin is also used clinically for some rare forms of cancer (pediatric and adult patients with subependymal giant cell astrocytoma (SEGA), progressive neuroendocrine tumors of pancreatic origin (PNET), and SEGA associated with tuberous sclerosis (TS)) (http://www.cancer.gov/cancertopics/druginfo/fda-everolimus) [312, 313]. Multiple clinical trials of rapamycin are currently underway for several other disease conditions including lymphangioleiomyomatosis (LAM) [314], other metabolism modulating interventions on the elderly (NCT02874924), ALS (amyotrophic lateral sclerosis) (NCT03359538), Sturge-Weber syndrome (SWS) (NCT03047980), and type 1 diabetes (NCT01060605; NCT00014911-both are completed) [315, 316].
Rapalogs, the modified form of rapamycin, are widely considered in clinical trials for its anticancer property. In fact, the National Cancer Institute has registered more than 200 clinical trials involving either rapamycin or modified form of rapamycin both as monotherapy and as combination treatment cancer (NCT01698918; NCT00337376; NCT00930930) [317–320]. Due to the successful outcome of rapamycin in the clinical trials, several drugs analogs of rapamycin with modified chemical structure such as sirolimus, temsirolimus (CCI-779), everolimus (RAD001), and ridaforolimus (AP-23573) are being evaluated for enhanced treatment of several diseases [321–323]. In 2009, everolimus received approval from the FDA for HER2-negative breast cancer (advanced HR+ BC) patients in combination with exemestane after failure of a nonsteroidal aromatase inhibitor (Afinitor: Highlights of Prescribing Information). (http://www.accessdata.fda.gov/drugsatfda_docs/label/2012/022334s016lbl.pdf).
Current mTOR inhibitors available in the market are not complex-specific and can either partially suppress mTORC1 or completely block mTORC1 as well as C2. Therefore, several pharmaceutical companies ventured to develop second generation of mTOR inhibitor that can block both mTORC1 and C2. These inhibitors are designed to completely block the core catalytic activity of mTOR by acting as an ATP-competitive agents to mTOR subunit. On the contrary, diseases like cancer and cell cycle irregularities need specific inhibition of mTORC1 without interfering the activity of mTORC2. Since mTORC1 is vital for basic cellular process, it is indispensable, and its complete inhibition may lead to unwanted side effects. To overcome these obstacles, scientists are also in pursuit of developing inhibitor that target rictor or raptor to silence either mTORC1 or C2. Although mTOR inhibitors are promising drug for cancer treatment and immunosuppressant, an unmet clinical trial is essential for their therapeutic use in cardiovascular diseases. Substantially, evidences from laboratory models and preclinical trials suggest that mTOR inhibition in the heart is beneficial and prevents cell apoptosis [146, 158] and autophagy [324–327]. Interestingly, inhibition of mTOR by rapamycin or other rapalogs are shown to alter the expression pattern of miRs in the cardiovascular system [162, 199, 208, 228]. Especially, alteration of miRs through mTOR inhibition that changes the expression level of PTEN and other downstream targets can offer new treatment strategies [328]. Many miRNA-based therapies for cancer are in clinical trial and have shown efficiency in reducing tumor malignancy [312]. Mimic of miR-34 are currently being tested in phase I clinical trials (NCT01829971) for its anticancer properties [329]. It was demonstrated that low level of miR-34 is an indicator of poor prognosis in osteosarcoma (OS) patients. Sirolimus increases the sensitivity of human OS cells to anticancer drugs in vitro by upregulating miR-34b and suppressing its target p21-activated protein kinase 1 (PAK1) and ATP-binding cassette subfamily B member 1 (ABCB1) [329]. In contrast, the miR-34 family (miR-34a, -34b, and -34c) is upregulated in the heart in response to stress, including myocardial infarction or pressure overload via TAC [330]. Diabetes also increases the expression of miR-34a both in the heart and in circulation [331]. miRNA Therapeutics Inc. is developing an LNA-modified antimiR against miR-34a, which attenuates MI-induced remodeling and dysfunction, and also improves cardiac function and increase angiogenesis with activation of AKT in a model of pressure overload-induced pathological hypertrophy and dysfunction [330]. Mechanistically, miR-34 can directly target protein phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP2), a negative regulator of the PI3K/AKT/mTOR pathway.
Upregulation of miR-92a was shown to activate PI3K/AKT/mTOR pathway and inhibit cell apoptosis induced by chemotherapy in mantle cell lymphoma (MCL) cells [332]. Downregulation of miR-92a could inhibit the growth of tumors in a xenograft MCL mouse model [332]. Interestingly, pharmaceutical company Miragen developed MRG-110, an inhibitor of miR-92a, to enhance the revascularization process in ischemic heart disease. However, inhibition of angiogenesis is the goal for cancer therapy, and it should be assumed that miR-92a acts differently in cardiovascular system [333].
MGN-1374, a miR-15 inhibitor, is under the developmental stage by miRagen Therapeutics for treating myocardial infarction [334, 335]. Studies conducted in MDA-MB-231 breast cancer cells demonstrated overexpression of miR-15b/16 led to inhibition of cell proliferation causing G1 cell cycle arrest as well as caspase-3-dependent apoptosis by directly suppressing mRNA levels of RPS6KB1 and mTOR [336]. In addition, miR-15 was shown to regulate CD4+ regulatory T cells (Tregs) expression, which is essential for preventing autoimmunity. Overexpression of miR-15b/16 significantly enhanced the induction of Tregs in Dicer−/− CD4+ T cells and suppressed the mTOR expression as evident with the decrease in phosphorylation of its downstream target, ribosomal protein S6 [337].
Cardiac expression of miR-208 was upregulated upon Ang II treatment and induced obesity through upregulation of mTORC1 in Zucker obese (ZO) rats [338]. Whereas, rapamycin treatment attenuated weight gain despite leptin resistance by attenuating the expression of miR-208 and increasing the expression of cardiac mediator complex subunit 13 (MED13), a suppresser of obesity, in ZO rats [338]. In addition, therapeutic inhibition of miR-208 prevents pathological cardiac remodeling, which coincides with a significant improvement in survival and cardiac function during heart disease [339]. MED13 is negatively regulated by a heart-specific miR-208a [340]. In this context, MGN-9103 (a LNA-modified antisense oligonucleotide against a cardiac-specific miR-208LNA) is a novel potential therapeutic candidate developed by miRagen Therapeutics for the treatment of obesity, diabetes, and metabolic syndrome and to improve cardiac function and survival rates during heart failure (http://drugprofiles.informa.com/drug_profiles/18925-mgn-9103). These research findings and clinical trials described above highlight the potential of miRNA-based therapies with an emphasis on mTOR signaling. Although several studies established a clear synergistic effect of miRNA and mTOR in the treatment of cancer, there are scarce reports of clinical trial in cardiovascular field. Nevertheless, conceptual treatments in laboratory models describing mTOR inhibition mediated miR changes and vice versa are encouraging and may lead to novel treatments in cardiovascular diseases in the future.
4. Conclusion
The role of mTOR in controlling the cellular dynamics in cardiovascular system provides confidence to consider mTOR and its related kinases as targets for therapeutic intervention. Most remarkably, changes in epigenetic signature of miRs upon mTOR inhibition can lead to identify novel miRNA-based treatment for cardiovascular diseases. Moreover, antagomir-based treatment options can specifically target individual mTOR complex and eliminate common side effects seen with dual mTOR inhibitors. Further understanding of the interfunctional relationship between mTORC1 and C2 complexes and its association with miRNA is warranted to develop an efficient miRNA-based therapeutics and diagnostics in cardiovascular system.
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1HL134366) (Anindita Das and Rakesh C. Kukreja).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Benjamin E. J., Virani S. S., Callaway C. W., et al. Heart disease and stroke Statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137(12):e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Writing Group Members, Mozaffarian D., Benjamin E. J., et al. Executive summary: heart disease and stroke statistics--2016 update: a report from the American Heart Association. Circulation. 2016;133(4):447–454. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 3.Jia G., Hill M. A., Sowers J. R. Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circulation Research. 2018;122(4):624–638. doi: 10.1161/CIRCRESAHA.117.311586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vistisen D., Witte D. R., Brunner E. J., et al. Risk of cardiovascular disease and death in individuals with prediabetes defined by different criteria: the Whitehall II study. Diabetes Care. 2018;41(4):899–906. doi: 10.2337/dc17-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krittanawong C., Tunhasiriwet A., Wang Z., et al. Meta-analysis comparing frequency of overweight versus normal weight in patients with new-onset heart failure. The American Journal of Cardiology. 2018;121(7):836–843. doi: 10.1016/j.amjcard.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Ruparelia N., Chai J. T., Fisher E. A., Choudhury R. P. Inflammatory processes in cardiovascular disease: a route to targeted therapies. Nature Reviews Cardiology. 2017;14(3):133–144. doi: 10.1038/nrcardio.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low Wang C. C., Hess C. N., Hiatt W. R., Goldfine A. B. Clinical update: cardiovascular disease in diabetes mellitus: atherosclerotic cardiovascular disease and heart failure in type 2 diabetes mellitus-mechanisms, management, and clinical considerations. Circulation. 2016;133(24):2459–2502. doi: 10.1161/CIRCULATIONAHA.116.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sciarretta S., Forte M., Frati G., Sadoshima J. New insights into the role of mTOR signaling in the cardiovascular system. Circulation Research. 2018;122(3):489–505. doi: 10.1161/CIRCRESAHA.117.311147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. Regulation of longevity and stress resistance by Sch 9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 10.Kaeberlein M., Powers R. W., 3rd, Steffen K. K., et al. Regulation of yeast replicative life span by TOR and Sch 9 in response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 11.Hara K., Maruki Y., Long X., et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 12.Jia K., Chen D., Riddle D. L. The TOR pathway interacts with the insulin signaling pathway to regulate C. Elegans larval development, metabolism and life span. Development. 2004;131(16):3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 13.Gao X., Zhang Y., Arrazola P., et al. Tsc tumour suppressor proteins antagonize amino-acid–TOR signalling. Nature Cell Biology. 2002;4(9):699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., Stallock J. P., Ng J. C., Reinhard C., Neufeld T. P. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes & Development. 2000;14(21):2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunn G. J., Williams J., Sabers C., Wiederrecht G., Lawrence J. C., Jr, Abraham R. T. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. The EMBO Journal. 1996;15(19):5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison D. E., Strong R., Sharp Z. D., et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaleghpour K., Pyronnet S., Gingras A. C., Sonenberg N. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding. Protein 1 and p 70 S6 kinase activities. Molecular and Cellular Biology. 1999;19(6):4302–4310. doi: 10.1128/MCB.19.6.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott P. H., Brunn G. J., Kohn A. D., Roth R. A., Lawrence J. C. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proceedings of the National Academy of Sciences. 1998;95(13):7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes & Development. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 21.Kim D. H., Sarbassov D. D., Ali S. M., et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 22.Inoki K., Zhu T., Guan K. L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. Regulation of elongation factor 2 kinase by p 90 (RSK1) and p 70 S6 kinase. The EMBO Journal. 2001;20(16):4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holz M. K., Ballif B. A., Gygi S. P., Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 25.Castedo M., Roumier T., Blanco J., et al. Sequential involvement of Cdk 1, mTOR and p 53 in apoptosis induced by the HIV-1 envelope. The EMBO Journal. 2002;21(15):4070–4080. doi: 10.1093/emboj/cdf391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumder P. K., Febbo P. G., Bikoff R., et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nature Medicine. 2004;10(6):594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 27.Schoeftner S., Blanco R., de Silanes I. L., et al. Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proceedings of the National Academy of Sciences. 2009;106(46):19393–19398. doi: 10.1073/pnas.0909265106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei F. Z., Cao Z., Wang X., et al. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11(12):2309–2322. doi: 10.1080/15548627.2015.1117734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guertin D. A., Sabatini D. M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Chong Z. Z., Maiese K. Mammalian target of rapamycin signaling in diabetic cardiovascular disease. Cardiovascular Diabetology. 2012;11(1):p. 45. doi: 10.1186/1475-2840-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy B. K., Lamming D. W. The mechanistic target of rapamycin: the grand conducTOR of metabolism and aging. Cell Metabolism. 2016;23(6):990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciarretta S., Volpe M., Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circulation Research. 2014;114(3):549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P., Shan T., Liang X., Deng C., Kuang S. Mammalian target of rapamycin is essential for cardiomyocyte survival and heart development in mice. Biochemical and Biophysical Research Communications. 2014;452(1):53–59. doi: 10.1016/j.bbrc.2014.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentges K. E., Sirry B., Gingeras A. C., et al. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proceedings of the National Academy of Sciences. 2001;98(24):13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangloff Y. G., Mueller M., Dann S. G., et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Molecular and Cellular Biology. 2004;24(21):9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y., Pires K. M. P., Whitehead K. J., et al. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS One. 2013;8(1, article e54221) doi: 10.1371/journal.pone.0054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Teran B., Lopez J. A., Rodriguez E., et al. p38γ and δ promote heart hypertrophy by targeting the mTOR-inhibitory protein DEPTOR for degradation. Nature Communications. 2016;7, article 10477 doi: 10.1038/ncomms10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Cicchetti G., Onda H., et al. Loss of Tsc 1/Tsc 2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. Journal of Clinical Investigation. 2003;112(8):1223–1233. doi: 10.1172/JCI200317222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Song Y., Liu L., et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death and Differentiation. 2017;24(7):1205–1213. doi: 10.1038/cdd.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Huang B., Wang H. Y., Chang A., Zheng X. F. S. Emerging role of microRNAs in mTOR signaling. Cellular and Molecular Life Sciences. 2017;74(14):2613–2625. doi: 10.1007/s00018-017-2485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Wang J., Sun L., Zhu S. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. European Journal of Pharmacology. 2018;818:508–517. doi: 10.1016/j.ejphar.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 42.Brown E. J., Albers M. W., Bum Shin T., et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 43.Heitman J., Movva N. R., Hall M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 44.Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. TOR controls translation initiation and early G1 progression in yeast. Molecular Biology of the Cell. 1996;7(1):25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu M. I., Katz H., Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proceedings of the National Academy of Sciences. 1994;91(26):12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiederrecht G. J., Sabers C. J., Brunn G. J., Martin M. M., Dumont F. J., Abraham R. T. Mechanism of action of rapamycin: new insights into the regulation of G1-phase progression in eukaryotic cells. Progress in Cell Cycle Research. 1995;1:53–71. doi: 10.1007/978-1-4615-1809-9_5. [DOI] [PubMed] [Google Scholar]
- 47.Sehgal S. N., Baker H., Vezina C. Rapamycin (AY-22, 989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. The Journal of Antibiotics. 1975;28(10):727–732. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- 48.Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., Hall M. N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73(3):585–596. doi: 10.1016/0092-8674(93)90144-F. [DOI] [PubMed] [Google Scholar]
- 49.Lorenz M. C., Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. Journal of Biological Chemistry. 1995;270(46):27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 50.Gaubitz C., Prouteau M., Kusmider B., Loewith R. TORC2 structure and function. Trends in Biochemical Sciences. 2016;41(6):532–545. doi: 10.1016/j.tibs.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Knutson B. A. Insights into the domain and repeat architecture of target of rapamycin. Journal of Structural Biology. 2010;170(2):354–363. doi: 10.1016/j.jsb.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navé B. T., Margriet Ouwens D., Withers D. J., Alessi D. R., Shepherd P. R. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochemical Journal. 1999;344(2):427–431. doi: 10.1042/bj3440427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D. H., Sabatini D. M. Raptor and mTOR: subunits of a nutrient-sensitive complex. Current Topics in Microbiology and Immunology. 2004;279:259–270. doi: 10.1007/978-3-642-18930-2_15. [DOI] [PubMed] [Google Scholar]
- 54.Jacinto E., Loewith R., Schmidt A., et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature Cell Biology. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 55.Guertin D. A., Stevens D. M., Thoreen C. C., et al. Ablation in Mice of the mTORC Components raptor, rictor, or mLST8 Reveals that mTORC2 Is Required for Signaling to Akt-FOXO and PKCα, but Not S6K1. Developmental Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Wang L., Harris T. E., Roth R. A., Lawrence J. C., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. Journal of Biological Chemistry. 2007;282(27):20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 57.Huang B., Porter G. Expression of proline-rich Akt-substrate PRAS40 in cell survival pathway and carcinogenesis. Acta Pharmacologica Sinica. 2005;26(10):1253–1258. doi: 10.1111/j.1745-7254.2005.00184.x. [DOI] [PubMed] [Google Scholar]
- 58.Sancak Y., Thoreen C. C., Peterson T. R., et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Peterson T. R., Laplante M., Thoreen C. C., et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu M., Wilk S. A., Wang A., et al. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. Journal of Biological Chemistry. 2010;285(47):36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Molecular Cell. 2010;38(5):768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aylett C. H. S., Sauer E., Imseng S., et al. Architecture of human mTOR complex 1. Science. 2016;351(6268):48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 63.Koser P. L., Eng W.-K., Bossard M. J., et al. The tyrosine 89 residue of yeast FKBP12 is required for rapamycin binding. Gene. 1993;129(2):159–165. doi: 10.1016/0378-1119(93)90264-4. [DOI] [PubMed] [Google Scholar]
- 64.Bossard M. J., Bergsma D. J., Brandt M., et al. Catalytic and ligand binding properties of the FK506 binding protein FKBP12: effects of the single amino acid substitution of Tyr82to Leu. Biochemical Journal. 1994;297(2):365–372. doi: 10.1042/bj2970365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frias M. A., Thoreen C. C., Jaffe J. D., et al. mSin1 is necessary for Akt/PKB phosphorylation, and Its isoforms define three distinct mTORC2s. Current Biology. 2006;16(18):1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 66.Thedieck K., Polak P., Kim M. L., et al. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2(11, article e1217) doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loewith R., Jacinto E., Wullschleger S., et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular Cell. 2002;10(3):457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 68.Sarbassov D. D., Ali S. M., Kim D.-H., et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current Biology. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 69.Chen X., Liu M., Tian Y., et al. Cryo-EM structure of human mTOR complex 2. Cell Research. 2018;28(5):518–528. doi: 10.1038/s41422-018-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarbassov D. D., Ali S. M., Sengupta S., et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 71.Zeng Z., Sarbassov D. D., Samudio I. J., et al. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood. 2007;109(8):3509–3512. doi: 10.1182/blood-2006-06-030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jacinto E., Facchinetti V., Liu D., et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127(1):125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 73.Hardie D. G. AMPK and raptor: matching cell growth to energy supply. Molecular Cell. 2008;30(3):263–265. doi: 10.1016/j.molcel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 74.Tzatsos A., Tsichlis P. N. Energy depletion inhibits phosphatidylinositol 3-kinase/Akt signaling and induces apoptosis via AMP-activated protein kinase-dependent phosphorylation of IRS-1 at Ser-794. Journal of Biological Chemistry. 2007;282(25):18069–18082. doi: 10.1074/jbc.M610101200. [DOI] [PubMed] [Google Scholar]
- 75.Kim J., Kundu M., Viollet B., Guan K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk 1. Nature Cell Biology. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gwinn D. M., Shackelford D. B., Egan D. F., et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Breuleux M., Klopfenstein M., Stephan C., et al. Increased AKT S473 phosphorylation after mTORC1 inhibition is rictor dependent and does not predict tumor cell response to PI3K/mTOR inhibition. Molecular Cancer Therapeutics. 2009;8(4):742–753. doi: 10.1158/1535-7163.MCT-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Celton-Morizur S., Merlen G., Couton D., Margall-Ducos G., Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. Journal of Clinical Investigation. 2009;119(7):1880–1887. doi: 10.1172/jci38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu P., Begley M., Michowski W., et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508(7497):541–545. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao C. A., Ortiz-Vega S., Sun Y. Y., Chien C. T., Chuang J. H., Lin Y. Association of mSin1 with mTORC2 Ras and Akt reveals a crucial domain on mSin1 involved in Akt phosphorylation. Oncotarget. 2017;8(38):63392–63404. doi: 10.18632/oncotarget.18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ebner M., Sinkovics B., Szczygiel M., Ribeiro D. W., Yudushkin I. Localization of mTORC2 activity inside cells. The Journal of Cell Biology. 2017;216(2):343–353. doi: 10.1083/jcb.201610060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Myers M. G., Jr., White M. F. Insulin signal transduction and the IRS proteins. Annual Review of Pharmacology and Toxicology. 1996;36(1):615–658. doi: 10.1146/annurev.pa.36.040196.003151. [DOI] [PubMed] [Google Scholar]
- 83.Alessi D. R., Andjelkovic M., Caudwell B., et al. Mechanism of activation of protein kinase B by insulin and IGF-1. The EMBO Journal. 1996;15(23):6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 84.Xie J., Herbert T. P. The role of mammalian target of rapamycin (mTOR) in the regulation of pancreatic β-cell mass: implications in the development of type-2 diabetes. Cellular and Molecular Life Sciences. 2012;69(8):1289–1304. doi: 10.1007/s00018-011-0874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alam H., Maizels E. T., Park Y., et al. Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/AKT/Ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. Journal of Biological Chemistry. 2004;279(19):19431–19440. doi: 10.1074/jbc.M401235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mavrakis M., Lippincott-Schwartz J., Stratakis C. A., Bossis I. mTOR kinase and the regulatory subunit of protein kinase A (PRKAR1A) spatially and functionally interact during autophagosome maturation. Autophagy. 2006;3(2):151–153. doi: 10.4161/auto.3632. [DOI] [PubMed] [Google Scholar]
- 87.Doronzo G., Viretto M., Russo I., et al. Nitric oxide activates PI3-K and MAPK signalling pathways in human and rat vascular smooth muscle cells: influence of insulin resistance and oxidative stress. Atherosclerosis. 2011;216(1):44–53. doi: 10.1016/j.atherosclerosis.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 88.Lu M., Wang J., Jones K. T., et al. mTOR complex-2 activates ENaC by phosphorylating SGK1. Journal of the American Society of Nephrology. 2010;21(5):811–818. doi: 10.1681/ASN.2009111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao D., Wan L., Wei W. Phosphorylation of rictor at Thr 1135 impairs the rictor/cullin-1 complex to ubiquitinate SGK1. Protein & Cell. 2010;1(10):881–885. doi: 10.1007/s13238-010-0123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gordon B. S., Kazi A. A., Coleman C. S., et al. RhoA modulates signaling through the mechanistic target of rapamycin complex 1 (mTORC1) in mammalian cells. Cellular Signalling. 2014;26(3):461–467. doi: 10.1016/j.cellsig.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. The EMBO Journal. 2008;27(14):1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hagiwara A., Cornu M., Cybulski N., et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metabolism. 2012;15(5):725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 93.Dibble C. C., Asara J. M., Manning B. D. Characterization of rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Molecular and Cellular Biology. 2009;29(21):5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature Cell Biology. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 95.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular Cell. 2002;10(1):151–162. doi: 10.1016/S1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 96.Lee Y., Jeon K., Lee J. T., Kim S., Kim V. N. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO Journal. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stark A., Brennecke J., Russell R. B., Cohen S. M. Identification of Drosophila microRNA targets. PLoS Biology. 2003;1(3):p. E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lai E. C. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genetics. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 99.Zeng Y., Wagner E. J., Cullen B. R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Molecular Cell. 2002;9(6):1327–1333. doi: 10.1016/S1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 100.McManus D. D., Freedman J. E. MicroRNAs in platelet function and cardiovascular disease. Nature Reviews Cardiology. 2015;12(12):711–717. doi: 10.1038/nrcardio.2015.101. [DOI] [PubMed] [Google Scholar]
- 101.Quiat D., Olson E. N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. Journal of Clinical Investigation. 2013;123(1):11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu M., Mo Y. Y. The Akt-associated microRNAs. Cellular and Molecular Life Sciences. 2012;69(21):3601–3612. doi: 10.1007/s00018-012-1129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuwabara Y., Horie T., Baba O., et al. MicroRNA-451 exacerbates lipotoxicity in cardiac myocytes and high-fat diet-induced cardiac hypertrophy in mice through suppression of the LKB1/AMPK pathway. Circulation Research. 2015;116(2):279–288. doi: 10.1161/CIRCRESAHA.116.304707. [DOI] [PubMed] [Google Scholar]
- 104.Yang Z., Han Y., Cheng K., Zhang G., Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Proliferation. 2014;47(6):587–595. doi: 10.1111/cpr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kato M., Putta S., Wang M., et al. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature Cell Biology. 2009;11(7):881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sayed D., Abdellatif M. AKT-ing via microRNA. Cell Cycle. 2010;9(16):3233–3237. doi: 10.4161/cc.9.16.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu H., Buchan R. J., Cook S. A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovascular Research. 2010;86(3):410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 108.Chen M. B., Wei M. X., Han J. Y., et al. MicroRNA-451 regulates AMPK/mTORC1 signaling and fascin1 expression in HT-29 colorectal cancer. Cellular Signalling. 2014;26(1):102–109. doi: 10.1016/j.cellsig.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 109.Li Y., Cai X., Guan Y., et al. Adiponectin upregulates MiR-133a in cardiac hypertrophy through AMPK activation and reduced ERK1/2 phosphorylation. PLoS One. 2016;11(2, article e0148482) doi: 10.1371/journal.pone.0148482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao M., Luo R., Liu Y., et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR–p-PI3K/AKT-c-JUN. Nature Communications. 2016;7, article 11309 doi: 10.1038/ncomms11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sui H., Cai G. X., Pan S. F., et al. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Molecular Cancer Therapeutics. 2014;13(12):3137–3151. doi: 10.1158/1535-7163.MCT-14-0167. [DOI] [PubMed] [Google Scholar]
- 112.Li R., Yan G., Zhang Q., et al. miR-145 inhibits isoproterenol-induced cardiomyocyte hypertrophy by targeting the expression and localization of GATA6. FEBS Letters. 2013;587(12):1754–1761. doi: 10.1016/j.febslet.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeng X., Huang C., Senavirathna L., Wang P., Liu L. miR-27b inhibits fibroblast activation via targeting TGFβ signaling pathway. BMC Cell Biology. 2017;18(1):p. 9. doi: 10.1186/s12860-016-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nagalingam R. S., Sundaresan N. R., Noor M., Gupta M. P., Solaro R. J., Gupta M. Deficiency of cardiomyocyte-specific microRNA-378 contributes to the development of cardiac fibrosis involving a transforming growth factor β (TGFβ1)-dependent paracrine mechanism. Journal of Biological Chemistry. 2014;289(39):27199–27214. doi: 10.1074/jbc.M114.580977. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Vella M. C., Choi E. Y., Lin S. Y., Reinert K., Slack F. J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes & Development. 2004;18(2):132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Moor C. H., Meijer H., Lissenden S. Mechanisms of translational control by the 3' UTR in development and differentiation. Seminars in Cell & Developmental Biology. 2005;16(1):49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 117.Brennecke J., Stark A., Russell R. B., Cohen S. M. Principles of microRNA–target recognition. PLoS Biology. 2005;3(3, article e85) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee Y., Kim M., Han J., et al. MicroRNA genes are transcribed by RNA polymerase II. The EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou H., Xia X. G., Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Research. 2005;33(6):p. e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tijsterman M., Plasterk R. H. A. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117(1):1–3. doi: 10.1016/S0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 121.Lee Y., Ahn C., Han J., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 122.Lund E., Guttinger S., Calado A., Dahlberg J. E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 123.Carmell M. A., Hannon G. J. RNase III enzymes and the initiation of gene silencing. Nature Structural & Molecular Biology. 2004;11(3):214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 124.Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Han J., Lee Y., Yeom K. H., Kim Y. K., Jin H., Kim V. N. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Development. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hutvagner G., Zamore P. D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 127.Han M. H., Goud S., Song L., Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proceedings of the National Academy of Sciences. 2004;101(4):1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krol J., Sobczak K., Wilczynska U., et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. Journal of Biological Chemistry. 2004;279(40):42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 129.Grimson A., Farh K. K.-H., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lewis B. P., Burge C. B., Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 131.Zeng Y., Cullen B. R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9(1):112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lai E. C., Wiel C., Rubin G. M. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10(2):171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 134.Tanzer A., Stadler P. F. Molecular evolution of a microRNA cluster. Journal of Molecular Biology. 2004;339(2):327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 135.Khvorova A., Reynolds A., Jayasena S. D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 136.Matranga C., Tomari Y., Shin C., Bartel D. P., Zamore P. D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 137.Wang B., Li S., Qi H. H., Chowdhury D., Shi Y., Novina C. D. Distinct passenger strand and mRNA cleavage activities of human argonaute proteins. Nature Structural & Molecular Biology. 2009;16(12):1259–1266. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 138.Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proceedings of the National Academy of Sciences. 2002;99(21):13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jaeschke A., Hartkamp J., Saitoh M., et al. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. The Journal of Cell Biology. 2002;159(2):217–224. doi: 10.1083/jcb.jcb.200206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pene F., Claessens Y. E., Muller O., et al. Role of the phosphatidylinositol 3-kinase/Akt and mTOR/P70S6-kinase pathways in the proliferation and apoptosis in multiple myeloma. Oncogene. 2002;21(43):6587–6597. doi: 10.1038/sj.onc.1205923. [DOI] [PubMed] [Google Scholar]
- 141.Brown J. R., Hamadani M., Hayslip J., et al. Voxtalisib (XL765) in patients with relapsed or refractory non-Hodgkin lymphoma or chronic lymphocytic leukaemia: an open-label, phase 2 trial. The Lancet Haematology. 2018;5(4):e170–e180. doi: 10.1016/S2352-3026(18)30030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xie K., Ryan D. P., Pearson B. L., et al. Epigenetic alterations in longevity regulators, reduced life span, and exacerbated aging-related pathology in old father offspring mice. Proceedings of the National Academy of Sciences. 2018;115(10):E2348–E2357. doi: 10.1073/pnas.1707337115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & Development. 2009;23(4):496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lamming D. W., Ye L., Sabatini D. M., Baur J. A. Rapalogs and mTOR inhibitors as anti-aging therapeutics. The Journal of Clinical Investigation. 2013;123(3):980–989. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shioi T., McMullen J. R., Tarnavski O., et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107(12):1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 146.Das A., Salloum F. N., Filippone S. M., et al. Inhibition of mammalian target of rapamycin protects against reperfusion injury in diabetic heart through STAT3 signaling. Basic Research in Cardiology. 2015;110(3):p. 31. doi: 10.1007/s00395-015-0486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xia Y., Wen H. Y., Young M. E., Guthrie P. H., Taegtmeyer H., Kellems R. E. Mammalian target of rapamycin and protein kinase a signaling mediate the cardiac transcriptional response to glutamine. Journal of Biological Chemistry. 2003;278(15):13143–13150. doi: 10.1074/jbc.M208500200. [DOI] [PubMed] [Google Scholar]
- 148.Juhaszova M., Zorov D. B., Kim S. H., et al. Glycogen synthase kinase-3β mediates convergence of protection signaling to inhibit the mitochondrial permeability transition pore. Journal of Clinical Investigation. 2004;113(11):1535–1549. doi: 10.1172/JCI19906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao X., Lu S., Nie J., et al. Phosphoinositide-dependent kinase 1 and mTORC2 synergistically maintain postnatal heart growth and heart function in mice. Molecular and Cellular Biology. 2014;34(11):1966–1975. doi: 10.1128/MCB.00144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Park I. H., Erbay E., Nuzzi P., Chen J. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Experimental Cell Research. 2005;309(1):211–219. doi: 10.1016/j.yexcr.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 151.Samidurai A., Salloum F. N., Durrant D., Chernova O. B., Kukreja R. C., Das A. Chronic treatment with novel nanoformulated micelles of rapamycin, Rapatar, protects diabetic heart against ischaemia/reperfusion injury. British Journal of Pharmacology. 2017;174(24):4771–4784. doi: 10.1111/bph.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kong Q., Dai L., Wang Y., et al. HSPA12B attenuated acute myocardial ischemia/reperfusion injury via maintaining endothelial integrity in a PI3K/Akt/mTOR-dependent mechanism. Scientific Reports. 2016;6(1, article 33636) doi: 10.1038/srep33636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oka S. I., Hirata T., Suzuki W., et al. Thioredoxin-1 maintains mechanistic target of rapamycin (mTOR) function during oxidative stress in cardiomyocytes. Journal of Biological Chemistry. 2017;292(46):18988–19000. doi: 10.1074/jbc.M117.807735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Benard L., Oh J. G., Cacheux M., et al. Cardiac stim 1 silencing impairs adaptive hypertrophy and promotes heart failure through inactivation of mTORC2/Akt signaling. Circulation. 2016;133(15):1458–1471. doi: 10.1161/CIRCULATIONAHA.115.020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sciarretta S., Zhai P., Maejima Y., et al. mTORC2 regulates cardiac response to stress by inhibiting MST1. Cell Reports. 2015;11(1):125–136. doi: 10.1016/j.celrep.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shende P., Plaisance I., Morandi C., et al. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123(10):1073–1082. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 157.Zhao D., Yang J., Yang L. Insights for oxidative stress and mTOR signaling in myocardial ischemia/reperfusion injury under diabetes. Oxidative Medicine and Cellular Longevity. 2017;2017:12. doi: 10.1155/2017/6437467.6437467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Das A., Durrant D., Koka S., Salloum F. N., Xi L., Kukreja R. C. Mammalian target of rapamycin (mTOR) inhibition with rapamycin improves cardiac function in type 2 diabetic mice: potential role of attenuated oxidative stress and altered contractile protein expression. Journal of Biological Chemistry. 2014;289(7):4145–4160. doi: 10.1074/jbc.M113.521062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lesniewski L. A., Seals D. R., Walker A. E., et al. Dietary rapamycin supplementation reverses age-related vascular dysfunction and oxidative stress, while modulating nutrient-sensing, cell cycle, and senescence pathways. Aging Cell. 2017;16(1):17–26. doi: 10.1111/acel.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Flynn J. M., O'Leary M. N., Zambataro C. A., et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12(5):851–862. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang M., Sun D., Li S., et al. Lin28a protects against cardiac ischaemia/reperfusion injury in diabetic mice through the insulin-PI3K-mTOR pathway. Journal of Cellular and Molecular Medicine. 2015;19(6):1174–1182. doi: 10.1111/jcmm.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grundmann S., Hans F. P., Kinniry S., et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123(9):999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 163.Zweier J., Talukder M. The role of oxidants and free radicals in reperfusion injury. Cardiovascular Research. 2006;70(2):181–190. doi: 10.1016/j.cardiores.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 164.Zhang J., Kim J., Alexander A., et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nature Cell Biology. 2013;15(10):1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Goorden S. M. I., Hoogeveen-Westerveld M., Cheng C., et al. Rheb is essential for murine development. Molecular and Cellular Biology. 2011;31(8):1672–1678. doi: 10.1128/MCB.00985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. mTOR regulation of autophagy. FEBS Letters. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim Y. C., Guan K. L. mTOR: a pharmacologic target for autophagy regulation. Journal of Clinical Investigation. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Totary-Jain H., Sanoudou D., Ben-Dov I. Z., et al. Reprogramming of the microRNA transcriptome mediates resistance to rapamycin. Journal of Biological Chemistry. 2013;288(9):6034–6044. doi: 10.1074/jbc.M112.416446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang P., Liu X. M., Ding L., Zhang X. J., Ma Z. L. mTOR signaling-related microRNAs and cancer involvement. Journal of Cancer. 2018;9(4):667–673. doi: 10.7150/jca.22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sato T., Nakashima A., Guo L., Coffman K., Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29(18):2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Volinia S., Calin G. A., Liu C. G., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Efeyan A., Zoncu R., Chang S., et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493(7434):679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang D., Contu R., Latronico M. V. G., et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. Journal of Clinical Investigation. 2010;120(8):2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dutta D., Xu J., Kim J. S., Dunn W. A., Jr., Leeuwenburgh C. Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity. Autophagy. 2013;9(3):328–344. doi: 10.4161/auto.22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sciarretta S., Zhai P., Shao D., et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125(9):1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khan S. A., Salloum F., Das A., Xi L., Vetrovec G. W., Kukreja R. C. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. Journal of Molecular and Cellular Cardiology. 2006;41(2):256–264. doi: 10.1016/j.yjmcc.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 177.Filippone S. M., Samidurai A., Roh S. K., et al. Reperfusion therapy with rapamycin attenuates myocardial infarction through activation of AKT and ERK. Oxidative Medicine and Cellular Longevity. 2017;2017:16. doi: 10.1155/2017/4619720.4619720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li Y., Chen C., Yao F., et al. AMPK inhibits cardiac hypertrophy by promoting autophagy via mTORC1. Archives of Biochemistry and Biophysics. 2014;558:79–86. doi: 10.1016/j.abb.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 179.Flegal K. M., Carroll M. D., Ogden C. L., Curtin L. R. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 180.Abdulla J., Kober L., Abildstrom S. Z., Christensen E., James W. P. T., Torp-Pedersen C. Impact of obesity as a mortality predictor in high-risk patients with myocardial infarction or chronic heart failure: a pooled analysis of five registries. European Heart Journal. 2008;29(5):594–601. doi: 10.1093/eurheartj/ehn010. [DOI] [PubMed] [Google Scholar]
- 181.du Toit E. F., Smith W., Muller C., et al. Myocardial susceptibility to ischemic-reperfusion injury in a prediabetic model of dietary-induced obesity. American Journal of Physiology. Heart and Circulatory Physiology. 2008;294(5):H2336–H2343. doi: 10.1152/ajpheart.00481.2007. [DOI] [PubMed] [Google Scholar]
- 182.Wang X. T., Wu X. D., Lu Y. X., et al. Potential involvement of MiR-30e-3p in myocardial injury induced by coronary microembolization via autophagy activation. Cellular Physiology and Biochemistry. 2017;44(5):1995–2004. doi: 10.1159/000485905. [DOI] [PubMed] [Google Scholar]
- 183.Sermersheim M. A., Park K. H., Gumpper K., et al. Micro RNA regulation of autophagy in cardiovascular disease. Frontiers in Bioscience. 2017;22:p. 48. doi: 10.2741/4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Q., Xie J., Li R., et al. Overexpression of microRNA-99a attenuates heart remodelling and improves cardiac performance after myocardial infarction. Journal of Cellular and Molecular Medicine. 2014;18(5):919–928. doi: 10.1111/jcmm.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li Q., Xie J., Wang B., et al. Overexpression of microRNA-99a attenuates cardiac hypertrophy. PLoS One. 2016;11(2, article e0148480) doi: 10.1371/journal.pone.0148480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen C., Zhao Z., Liu Y., Mu D. microRNA-99a is downregulated and promotes proliferation, migration and invasion in non-small cell lung cancer A549 and H1299 cells. Oncology Letters. 2015;9(3):1128–1134. doi: 10.3892/ol.2015.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Oneyama C., Ikeda J., Okuzaki D., et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011;30(32):3489–3501. doi: 10.1038/onc.2011.63. [DOI] [PubMed] [Google Scholar]
- 188.Sun M., Hong S., Li W., et al. MiR-99a regulates ROS-mediated invasion and migration of lung adenocarcinoma cells by targeting NOX4. Oncology Reports. 2016;35(5):2755–2766. doi: 10.3892/or.2016.4672. [DOI] [PubMed] [Google Scholar]
- 189.Saito T., Sadoshima J. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circulation Research. 2015;116(8):1477–1490. doi: 10.1161/CIRCRESAHA.116.303790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhu R. Y., Zhang D., Zou H. D., Zuo X. S., Zhou Q. S., Huang H. MiR-28 inhibits cardiomyocyte survival through suppressing PDK1/Akt/mTOR signaling. In Vitro Cellular & Developmental Biology - Animal. 2016;52(10):1020–1025. doi: 10.1007/s11626-016-0065-6. [DOI] [PubMed] [Google Scholar]
- 191.Liu X., Deng Y., Xu Y., Jin W., Li H. MicroRNA-223 protects neonatal rat cardiomyocytes and H9c2 cells from hypoxia-induced apoptosis and excessive autophagy via the Akt/mTOR pathway by targeting PARP-1. Journal of Molecular and Cellular Cardiology. 2018;118:133–146. doi: 10.1016/j.yjmcc.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 192.van Rooij E., Sutherland L. B., Thatcher J. E., et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proceedings of the National Academy of Sciences. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Crackower M. A., Oudit G. Y., Kozieradzki I., et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110(6):737–749. doi: 10.1016/S0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 194.Mocanu M. M., Yellon D. M. PTEN, the Achilles' heel of myocardial ischaemia/reperfusion injury? British Journal of Pharmacology. 2007;150(7):833–838. doi: 10.1038/sj.bjp.0707155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ruan H., Li J., Ren S., et al. Inducible and cardiac specific PTEN inactivation protects ischemia/reperfusion injury. Journal of Molecular and Cellular Cardiology. 2009;46(2):193–200. doi: 10.1016/j.yjmcc.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 196.Oudit G. Y., Kassiri Z., Zhou J., et al. Loss of PTEN attenuates the development of pathological hypertrophy and heart failure in response to biomechanical stress. Cardiovascular Research. 2008;78(3):505–514. doi: 10.1093/cvr/cvn041. [DOI] [PubMed] [Google Scholar]
- 197.Keyes K. T., Xu J., Long B., Zhang C., Hu Z., Ye Y. Pharmacological inhibition of PTEN limits myocardial infarct size and improves left ventricular function postinfarction. American Journal of Physiology-Heart and Circulatory Physiology. 2010;298(4):H1198–H1208. doi: 10.1152/ajpheart.00915.2009. [DOI] [PubMed] [Google Scholar]
- 198.Toldo S., Das A., Mezzaroma E., et al. Induction of microRNA-21 with exogenous hydrogen sulfide attenuates myocardial ischemic and inflammatory injury in mice. Circulation Cardiovascular Genetics. 2014;7(3):311–320. doi: 10.1161/circgenetics.113.000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Huang W., Tian S. S., Hang P. Z., Sun C., Guo J., Du Z. M. Combination of microRNA-21 and microRNA-146a attenuates cardiac dysfunction and apoptosis during acute myocardial infarction in mice. Molecular Therapy - Nucleic Acids. 2016;5, article e296 doi: 10.1038/mtna.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sayed D., He M., Hong C., et al. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. Journal of Biological Chemistry. 2010;285(26):20281–20290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cheng Y., Zhang C. MicroRNA-21 in cardiovascular disease. Journal of Cardiovascular Translational Research. 2010;3(3):251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dong S., Cheng Y., Yang J., et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. The Journal of Biological Chemistry. 2009;284(43):29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tu Y., Wan L., Fan Y., et al. Ischemic postconditioning-mediated miRNA-21 protects against cardiac ischemia/reperfusion injury via PTEN/Akt pathway. PLoS One. 2013;8(10, article e75872) doi: 10.1371/journal.pone.0075872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang Z., Wu S., Kong F., et al. MicroRNA-21 protects against cardiac hypoxia/reoxygenation injury by inhibiting excessive autophagy in H9c2 cellsviathe Akt/mTOR pathway. Journal of Cellular and Molecular Medicine. 2017;21(3):467–474. doi: 10.1111/jcmm.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Deng W., Wang Y., Long X., et al. miR-21 reduces hydrogen peroxide-induced apoptosis in c-kit+cardiac stem cells in vitro through PTEN/PI3K/Akt signaling. Oxidative Medicine and Cellular Longevity. 2016;2016:14. doi: 10.1155/2016/5389181.5389181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gabriely G., Wurdinger T., Kesari S., et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and Cellular Biology. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Moriyama T., Ohuchida K., Mizumoto K., et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Molecular Cancer Therapeutics. 2009;8(5):1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 208.Jin C., Zhao Y., Yu L., Xu S., Fu G. MicroRNA-21 mediates the rapamycin-induced suppression of endothelial proliferation and migration. FEBS Letters. 2013;587(4):378–385. doi: 10.1016/j.febslet.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 209.Sabatel C., Malvaux L., Bovy N., et al. MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One. 2011;6(2, article e16979) doi: 10.1371/journal.pone.0016979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Trindade A. J., Medvetz D. A., Neuman N. A., et al. MicroRNA-21 is induced by rapamycin in a model of tuberous sclerosis (TSC) and lymphangioleiomyomatosis (LAM) PLoS One. 2013;8(3, article e60014) doi: 10.1371/journal.pone.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Carsillo T., Astrinidis A., Henske E. P. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proceedings of the National Academy of Sciences. 2000;97(11):6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lam H. C., Liu H. J., Baglini C. V., et al. Rapamycin-induced miR-21 promotes mitochondrial homeostasis and adaptation in mTORC1 activated cells. Oncotarget. 2017;8(39):64714–64727. doi: 10.18632/oncotarget.19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ogorek B., Lam H. C., Khabibullin D., et al. TSC2 regulates microRNA biogenesis via mTORC1 and GSK3β. Human Molecular Genetics. 2018;27(9):1654–1663. doi: 10.1093/hmg/ddy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Small E. M., Frost R. J. A., Olson E. N. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121(8):1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thum T., Gross C., Fiedler J., et al. Micro RNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 216.van Rooij E., Sutherland L. B., Liu N., et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Patrick D. M., Montgomery R. L., Qi X., et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. Journal of Clinical Investigation. 2010;120(11):3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Darido C., Georgy S. R., Cullinane C., et al. Stage-dependent therapeutic efficacy in PI3K/mTOR-driven squamous cell carcinoma of the skin. Cell Death & Differentiation. 2018;25(6):1146–1159. doi: 10.1038/s41418-017-0032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang P., Guan Q., Zhou D., Yu Z., Song Y., Qiu W. miR-21 inhibitors modulate biological functions of gastric cancer cells via PTEN/PI3K/mTOR pathway. DNA and Cell Biology. 2018;37(1):38–45. doi: 10.1089/dna.2017.3922. [DOI] [PubMed] [Google Scholar]
- 220.Song L., Liu S., Zhang L., et al. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumour Biology. 2016;37(9):12161–12168. doi: 10.1007/s13277-016-5073-3. [DOI] [PubMed] [Google Scholar]
- 221.Yu X., Li R., Shi W., et al. Silencing of microRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomedicine & Pharmacotherapy. 2016;77:37–44. doi: 10.1016/j.biopha.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 222.Ng R., Song G., Roll G. R., Frandsen N. M., Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. Journal of Clinical Investigation. 2012;122(3):1097–1108. doi: 10.1172/JCI46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cochain C., Channon K. M., Silvestre J. S. Angiogenesis in the infarcted myocardium. Antioxidants & Redox Signaling. 2013;18(9):1100–1113. doi: 10.1089/ars.2012.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.van der Laan A. M., Piek J. J., van Royen N. Targeting angiogenesis to restore the microcirculation after reperfused MI. Nature Reviews Cardiology. 2009;6(8):515–523. doi: 10.1038/nrcardio.2009.103. [DOI] [PubMed] [Google Scholar]
- 225.Shi Z. M., Wang J., Yan Z., et al. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 2012;7(3, article e32709) doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Xu Q., Liu L. Z., Qian X., et al. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Research. 2012;40(2):761–774. doi: 10.1093/nar/gkr730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Xu S., Fu G. B., Tao Z., et al. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6(28):26457–26471. doi: 10.18632/oncotarget.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen G. H., Xu C. S., Zhang J., et al. Inhibition of miR-128-3p by Tongxinluo protects human cardiomyocytes from ischemia/reperfusion injury via upregulation of p70s6k1/p-p70s6k1. Frontiers in Pharmacology. 2017;8 doi: 10.3389/fphar.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gong L. C., Xu H. M., Guo G. L., Zhang T., Shi J. W., Chang C. Long non-coding RNA H19 protects H9c2 cells against hypoxia-induced injury by targeting microRNA-139. Cellular Physiology and Biochemistry. 2017;44(3):857–869. doi: 10.1159/000485354. [DOI] [PubMed] [Google Scholar]
- 230.Thum T., Catalucci D., Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovascular Research. 2008;79(4):562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 231.Yang B., Lu Y., Wang Z. Control of cardiac excitability by microRNAs. Cardiovascular Research. 2008;79(4):571–580. doi: 10.1093/cvr/cvn181. [DOI] [PubMed] [Google Scholar]
- 232.Thum T., Galuppo P., Wolf C., et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116(3):258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 233.Wang X., Zhang X., Ren X. P., et al. MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation. 2010;122(13):1308–1318. doi: 10.1161/CIRCULATIONAHA.110.964684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li J., Rohailla S., Gelber N., et al. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Research in Cardiology. 2014;109(5):p. 423. doi: 10.1007/s00395-014-0423-z. [DOI] [PubMed] [Google Scholar]
- 235.Iwaya T., Yokobori T., Nishida N., et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 236.Atlas Writing Group, Timmis A., Townsend N., et al. European Society of Cardiology: cardiovascular disease. Statistics 2017. European Heart Journal. 2018;39(7):508–579. doi: 10.1093/eurheartj/ehx628. [DOI] [PubMed] [Google Scholar]
- 237.Newman J. D., Schwartzbard A. Z., Weintraub H. S., Goldberg I. J., Berger J. S. Primary prevention of cardiovascular disease in diabetes mellitus. Journal of the American College of Cardiology. 2017;70(7):883–893. doi: 10.1016/j.jacc.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang Y., Sun X., Icli B., Feinberg M. W. Emerging roles for microRNAs in diabetic microvascular disease: novel targets for therapy. Endocrine Reviews. 2017;38(2):145–168. doi: 10.1210/er.2016-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Um S. H., D'Alessio D., Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metabolism. 2006;3(6):393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 240.Werner E. D., Lee J., Hansen L., Yuan M., Shoelson S. E. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. Journal of Biological Chemistry. 2004;279(34):35298–35305. doi: 10.1074/jbc.M405203200. [DOI] [PubMed] [Google Scholar]
- 241.Kim J.-a., Jang H.-J., Martinez-Lemus L. A., Sowers J. R. Activation of mTOR/p70S6 kinase by ANG II inhibits insulin-stimulated endothelial nitric oxide synthase and vasodilation. American Journal of Physiology-Endocrinology and Metabolism. 2012;302(2):E201–E208. doi: 10.1152/ajpendo.00497.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Simm A., Schlüter K.-D., Diez C., Piper H. M., Hoppe J. Activation of p70S6Kinase byβ-adrenoceptor agonists on adult cardiomyocytes. Journal of Molecular and Cellular Cardiology. 1998;30(10):2059–2067. doi: 10.1006/jmcc.1998.0768. [DOI] [PubMed] [Google Scholar]
- 243.Zhang Y., Welzig C. M., Picard K. L., et al. Glycogen synthase kinase-3β inhibition ameliorates cardiac parasympathetic dysfunction in type 1 diabetic Akita mice. Diabetes. 2014;63(6):2097–2113. doi: 10.2337/db12-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang L., Wang X., Proud C. G. Activation of mRNA translation in rat cardiac myocytes by insulin involves multiple rapamycin-sensitive steps. American Journal of Physiology-Heart and Circulatory Physiology. 2000;278(4):H1056–H1068. doi: 10.1152/ajpheart.2000.278.4.H1056. [DOI] [PubMed] [Google Scholar]
- 245.Sun X., Lin J., Zhang Y., et al. MicroRNA-181b improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue. Circulation Research. 2016;118(5):810–821. doi: 10.1161/CIRCRESAHA.115.308166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jordan S. D., Kruger M., Willmes D. M., et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature Cell Biology. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 247.Trajkovski M., Hausser J., Soutschek J., et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 248.Li J., Ren Y., Shi E., et al. Inhibition of the Let-7 family microRNAs induces cardioprotection against ischemia-reperfusion injury in diabetic rats. The Annals of Thoracic Surgery. 2016;102(3):829–835. doi: 10.1016/j.athoracsur.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 249.Shan Z. X., Lin Q. X., Fu Y. H., et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochemical and Biophysical Research Communications. 2009;381(4):597–601. doi: 10.1016/j.bbrc.2009.02.097. [DOI] [PubMed] [Google Scholar]
- 250.Rozengurt E. Mechanistic target of rapamycin (mTOR): a point of convergence in the action of insulin/IGF-1 and G protein-coupled receptor agonists in pancreatic cancer cells. Frontiers in Physiology. 2014;5 doi: 10.3389/fphys.2014.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Katare R., Caporali A., Zentilin L., et al. Intravenous gene therapy with PIM-1 via a cardiotropic viral vector halts the progression of diabetic cardiomyopathy through promotion of prosurvival signaling. Circulation Research. 2011;108(10):1238–1251. doi: 10.1161/CIRCRESAHA.110.239111. [DOI] [PubMed] [Google Scholar]
- 252.Wang X. H., Qian R. Z., Zhang W., Chen S. F., Jin H. M., Hu R. M. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clinical and Experimental Pharmacology and Physiology. 2009;36(2):181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 253.Zhang Z. W., Guo R. W., Lv J. L., et al. MicroRNA-99a inhibits insulin-induced proliferation, migration, dedifferentiation, and rapamycin resistance of vascular smooth muscle cells by inhibiting insulin-like growth factor-1 receptor and mammalian target of rapamycin. Biochemical and Biophysical Research Communications. 2017;486(2):414–422. doi: 10.1016/j.bbrc.2017.03.056. [DOI] [PubMed] [Google Scholar]
- 254.Carè A., Catalucci D., Felicetti F., et al. MicroRNA-133 controls cardiac hypertrophy. Nature Medicine. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 255.Chen S., Puthanveetil P., Feng B., Matkovich S. J., Dorn G. W., II, Chakrabarti S. Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. Journal of Cellular and Molecular Medicine. 2014;18(3):415–421. doi: 10.1111/jcmm.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chavali V., Tyagi S. C., Mishra P. K. Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/− Akita hearts. Cell Biochemistry and Biophysics. 2014;68(1):25–35. doi: 10.1007/s12013-013-9679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chavali V., Tyagi S. C., Mishra P. K. MicroRNA-133a regulates DNA methylation in diabetic cardiomyocytes. Biochemical and Biophysical Research Communications. 2012;425(3):668–672. doi: 10.1016/j.bbrc.2012.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nandi S. S., Duryee M. J., Shahshahan H. R., Thiele G. M., Anderson D. R., Mishra P. K. Induction of autophagy markers is associated with attenuation of miR-133a in diabetic heart failure patients undergoing mechanical unloading. American Journal of Translational Research. 2015;7(4):683–696. [PMC free article] [PubMed] [Google Scholar]
- 259.Fernandez-Twinn D. S., Blackmore H. L., Siggens L., et al. The programming of cardiac hypertrophy in the offspring by maternal obesity is associated with hyperinsulinemia, AKT, ERK, and mTOR activation. Endocrinology. 2012;153(12):5961–5971. doi: 10.1210/en.2012-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Donath M. Y. Targeting inflammation in the treatment of type 2 diabetes: time to start. Nature Reviews Drug Discovery. 2014;13(6):465–476. doi: 10.1038/nrd4275. [DOI] [PubMed] [Google Scholar]
- 261.Dregan A., Charlton J., Chowienczyk P., Gulliford M. C. Chronic inflammatory disorders and risk of type 2 diabetes mellitus, coronary heart disease, and stroke: a population-based cohort study. Circulation. 2014;130(10):837–844. doi: 10.1161/CIRCULATIONAHA.114.009990. [DOI] [PubMed] [Google Scholar]
- 262.Pek S. L. T., Sum C. F., Lin M. X., et al. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Molecular and Cellular Endocrinology. 2016;427:112–123. doi: 10.1016/j.mce.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 263.Pankratz F., Hohnloser C., Bemtgen X., et al. MicroRNA-100 suppresses chronic vascular inflammation by stimulation of endothelial autophagy. Circulation Research. 2018;122(3):417–432. doi: 10.1161/CIRCRESAHA.117.311428. [DOI] [PubMed] [Google Scholar]
- 264.Suhara T., Baba Y., Shimada B. K., Higa J. K., Matsui T. The mTOR signaling pathway in myocardial dysfunction in type 2 diabetes mellitus. Current Diabetes Reports. 2017;17(6):p. 38. doi: 10.1007/s11892-017-0865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Seferovic J. P., Tesic M., Seferovic P. M., et al. Increased left ventricular mass index is present in patients with type 2 diabetes without ischemic heart disease. Scientific Reports. 2018;8(1):p. 926. doi: 10.1038/s41598-018-19229-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Qian L. B., Jiang S. Z., Tang X. Q., et al. Exacerbation of diabetic cardiac hypertrophy in OVE26 mice by angiotensin II is associated with JNK/c-Jun/miR-221-mediated autophagy inhibition. Oncotarget. 2017;8(63):106661–106671. doi: 10.18632/oncotarget.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Xu K., Bian D., Hao L., et al. microRNA-503 contribute to pancreatic beta cell dysfunction by targeting the mTOR pathway in gestational diabetes mellitus. EXCLI Journal. 2017;16:1177–1187. doi: 10.17179/excli2017-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Bera A., Das F., Ghosh-Choudhury N., Mariappan M. M., Kasinath B. S., Ghosh Choudhury G. Reciprocal regulation of miR-214 and PTEN by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. American Journal of Physiology Cell Physiology. 2017;313(4):C430–C447. doi: 10.1152/ajpcell.00081.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Muralimanoharan S., Maloyan A., Myatt L. Mitochondrial function and glucose metabolism in the placenta with gestational diabetes mellitus: role of miR-143. Clinical Science. 2016;130(11):931–941. doi: 10.1042/CS20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Li W., Wang J., Chen Q. D., et al. Insulin promotes glucose consumption via regulation of miR-99a/mTOR/PKM2 pathway. PLoS One. 2013;8(6, article e64924) doi: 10.1371/journal.pone.0064924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.von Walden F., Liu C., Aurigemma N., Nader G. A. mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription, and chromatin remodeling. American Journal of Physiology-Cell Physiology. 2016;311(4):C663–C672. doi: 10.1152/ajpcell.00144.2016. [DOI] [PubMed] [Google Scholar]
- 272.Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Molecular and Cellular Biology. 2004;24(1):200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Laplante M., Sabatini D. M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.McMullen J. R., Sherwood M. C., Tarnavski O., et al. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109(24):3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 275.Bodine S. C., Stitt T. N., Gonzalez M., et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell Biology. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 276.Volkers M., Sussman M. mTOR/PRAS40 interaction: hypertrophy or proliferation. Cell Cycle. 2013;12(23):3579–3580. doi: 10.4161/cc.26822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kushwaha S. S., Raichlin E., Sheinin Y., et al. Sirolimus affects cardiomyocytes to reduce left ventricular mass in heart transplant recipients. European Heart Journal. 2008;29(22):2742–2750. doi: 10.1093/eurheartj/ehn407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Crespo-Leiro M. G., Hermida-Prieto M. Sirolimus treatment of left ventricular hypertrophy: who, and when? European Heart Journal. 2008;29(22):2703–2704. doi: 10.1093/eurheartj/ehn470. [DOI] [PubMed] [Google Scholar]
- 279.Kuzman J. A., O’Connell T. D., Gerdes A. M. Rapamycin prevents thyroid hormone-induced cardiac hypertrophy. Endocrinology. 2007;148(7):3477–3484. doi: 10.1210/en.2007-0099. [DOI] [PubMed] [Google Scholar]
- 280.Rao R. D., Buckner J. C., Sarkaria J. N. Mammalian target of rapamycin (mTOR) inhibitors as anti-cancer agents. Current Cancer Drug Targets. 2004;4(8):621–635. doi: 10.2174/1568009043332718. [DOI] [PubMed] [Google Scholar]
- 281.Rodrik-Outmezguine V. S., Okaniwa M., Yao Z., et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534(7606):272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ilagan E., Manning B. D. Emerging role of mTOR in the response to cancer therapeutics. Trends in Cancer. 2016;2(5):241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Boluyt M. O., Zheng J. S., Younes A., et al. Rapamycin inhibits alpha 1-adrenergic receptor-stimulated cardiac myocyte hypertrophy but not activation of hypertrophy-associated genes. Evidence for involvement of p 70 S6 kinase. Circulation Research. 1997;81(2):176–186. doi: 10.1161/01.RES.81.2.176. [DOI] [PubMed] [Google Scholar]
- 284.Hafizi S., Wang X., Chester A. H., Yacoub M. H., Proud C. G. ANG II activates effectors of mTOR via PI3-K signaling in human coronary smooth muscle cells. American Journal of Physiology-Heart and Circulatory Physiology. 2004;287(3):H1232–H1238. doi: 10.1152/ajpheart.00040.2004. [DOI] [PubMed] [Google Scholar]
- 285.Sadoshima J., Izumo S. Rapamycin selectively inhibits angiotensin II-induced increase in protein synthesis in cardiac myocytes in vitro. Potential role of 70-kD S6 kinase in angiotensin II-induced cardiac hypertrophy. Circulation Research. 1995;77(6):1040–1052. doi: 10.1161/01.RES.77.6.1040. [DOI] [PubMed] [Google Scholar]
- 286.Huang H., Zhang P., Wang Z., Tang F., Jiang Z. Activation of endothelin-1 receptor signaling pathways is associated with neointima formation, neoangiogenesis and irreversible pulmonary artery hypertension in patients with congenital heart disease. Circulation Journal. 2011;75(6):1463–1471. doi: 10.1253/circj.CJ-10-0670. [DOI] [PubMed] [Google Scholar]
- 287.Shende P., Xu L., Morandi C., et al. Cardiac mTOR. Complex 2 preserves ventricular function in pressure-overload hypertrophy. Cardiovascular Research. 2016;109(1):103–114. doi: 10.1093/cvr/cvv252. [DOI] [PubMed] [Google Scholar]
- 288.Cho J. R., Lee C. Y., Lee J., et al. MicroRNA-761 inhibits angiotensin II-induced vascular smooth muscle cell proliferation and migration by targeting mammalian target of rapamycin. Clinical Hemorheology and Microcirculation. 2015;63(1):45–56. doi: 10.3233/CH-151981. [DOI] [PubMed] [Google Scholar]
- 289.Su M., Chen Z., Wang C., et al. Cardiac-specific overexpression of miR-222 induces heart failure and inhibits autophagy in mice. Cellular Physiology and Biochemistry. 2016;39(4):1503–1511. doi: 10.1159/000447853. [DOI] [PubMed] [Google Scholar]
- 290.Su M., Wang J., Wang C., et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death and Differentiation. 2015;22(6):986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wu H., Wang Y., Wang X., Li R., Yin D. MicroRNA-365 accelerates cardiac hypertrophy by inhibiting autophagy via the modulation of Skp2 expression. Biochemical and Biophysical Research Communications. 2017;484(2):304–310. doi: 10.1016/j.bbrc.2017.01.108. [DOI] [PubMed] [Google Scholar]
- 292.Fang C. X., Dong F., Thomas D. P., Ma H., He L., Ren J. Hypertrophic cardiomyopathy in high-fat diet-induced obesity: role of suppression of forkhead transcription factor and atrophy gene transcription. American Journal of Physiology-Heart and Circulatory Physiology. 2008;295(3):H1206–H1215. doi: 10.1152/ajpheart.00319.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sung M. M. Y., Koonen D. P. Y., Soltys C.-L. M., Jacobs R. L., Febbraio M., Dyck J. R. B. Increased CD36 expression in middle-aged mice contributes to obesity-related cardiac hypertrophy in the absence of cardiac dysfunction. Journal of Molecular Medicine. 2011;89(5):459–469. doi: 10.1007/s00109-010-0720-4. [DOI] [PubMed] [Google Scholar]
- 294.Oka T., Akazawa H., Naito A. T., Komuro I. Angiogenesis and cardiac hypertrophy: maintenance of cardiac function and causative roles in heart failure. Circulation Research. 2014;114(3):565–571. doi: 10.1161/CIRCRESAHA.114.300507. [DOI] [PubMed] [Google Scholar]
- 295.Jaba I. M., Zhuang Z. W., Li N., et al. NO triggers RGS4 degradation to coordinate angiogenesis and cardiomyocyte growth. Journal of Clinical Investigation. 2013;123(4):1718–1731. doi: 10.1172/JCI65112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kemi O. J., Ceci M., Wisloff U., et al. Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. Journal of Cellular Physiology. 2008;214(2):316–321. doi: 10.1002/jcp.21197. [DOI] [PubMed] [Google Scholar]
- 297.Tirziu D., Chorianopoulos E., Moodie K. L., et al. Myocardial hypertrophy in the absence of external stimuli is induced by angiogenesis in mice. Journal of Clinical Investigation. 2007;117(11):3188–3197. doi: 10.1172/JCI32024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Jabs M., Rose A. J., Lehmann L. H., et al. Inhibition of endothelial notch signaling impairs fatty acid transport and leads to metabolic and vascular remodeling of the adult heart. Circulation. 2018;137(24):2592–2608. doi: 10.1161/CIRCULATIONAHA.117.029733. [DOI] [PubMed] [Google Scholar]
- 299.Li N., Hwangbo C., Jaba I. M., et al. miR-182 modulates myocardial hypertrophic response induced by angiogenesis in heart. Scientific Reports. 2016;6(1):p. 21228. doi: 10.1038/srep21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Gimbrone M. A., Jr., Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circulation Research. 2016;118(4):620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Tabas I., Garcia-Cardena G., Owens G. K. Recent insights into the cellular biology of atherosclerosis. The Journal of Cell Biology. 2015;209(1):13–22. doi: 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhai C., Cheng J., Mujahid H., et al. Selective inhibition of PI3K/Akt/mTOR signaling pathway regulates autophagy of macrophage and vulnerability of atherosclerotic plaque. PLoS One. 2014;9(3, article e90563) doi: 10.1371/journal.pone.0090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tang F., Yang T. L. MicroRNA-126 alleviates endothelial cells injury in atherosclerosis by restoring autophagic flux via inhibiting of PI3K/Akt/mTOR pathway. Biochemical and Biophysical Research Communications. 2018;495(1):1482–1489. doi: 10.1016/j.bbrc.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 304.Lv L., Li T., Li X., et al. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Molecular Therapy - Nucleic Acids. 2018;10:387–397. doi: 10.1016/j.omtn.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yu H., Guo Y., Mi L., Wang X., Li L., Gao W. Mitofusin 2 inhibits angiotensin II-induced myocardial hypertrophy. Journal of Cardiovascular Pharmacology and Therapeutics. 2011;16(2):205–211. doi: 10.1177/1074248410385683. [DOI] [PubMed] [Google Scholar]
- 306.Papanicolaou K. N., Khairallah R. J., Ngoh G. A., et al. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Molecular and Cellular Biology. 2011;31(6):1309–1328. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sun M., Yu H., Zhang Y., Li Z., Gao W. MicroRNA-214 mediates isoproterenol-induced proliferation and collagen synthesis in cardiac fibroblasts. Scientific Reports. 2015;5(1, article 18351) doi: 10.1038/srep18351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Calne R. Y., Lim S., Samaan A., et al. Rapamycin for immunosuppression in organ allografting. The Lancet. 1989;334(8656):p. 227. doi: 10.1016/S0140-6736(89)90417-0. [DOI] [PubMed] [Google Scholar]
- 309.Moses J. W., Leon M. B., Popma J. J., et al. Sirolimus-eluting stents versus standard stents in patients with stenosis in a native coronary artery. New England Journal of Medicine. 2003;349(14):1315–1323. doi: 10.1056/NEJMoa035071. [DOI] [PubMed] [Google Scholar]
- 310.Munoz J. S., Abizaid A., Mintz G. S., et al. Intravascular ultrasound study of effects of overlapping sirolimus-eluting stents. The American Journal of Cardiology. 2004;93(4):470–473. doi: 10.1016/j.amjcard.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 311.Schofer J., Schluter M., Gershlick A. H., et al. Sirolimus-eluting stents for treatment of patients with long atherosclerotic lesions in small coronary arteries: double-blind, randomised controlled trial (E-SIRIUS) The Lancet. 2003;362(9390):1093–1099. doi: 10.1016/S0140-6736(03)14462-5. [DOI] [PubMed] [Google Scholar]
- 312.Xie J., Wang X., Proud C. G. mTOR inhibitors in cancer therapy. F1000Res. 2016;5 doi: 10.12688/f1000research.9207.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Meng L.-h., Zheng X. F. S. Toward rapamycin analog (rapalog)-based precision cancer therapy. Acta Pharmacologica Sinica. 2015;36(10):1163–1169. doi: 10.1038/aps.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Glasgow C. G., Steagall W. K., Taveira-Dasilva A., et al. Lymphangioleiomyomatosis (LAM): molecular insights lead to targeted therapies. Respiratory Medicine. 2010;104(Supplement 1):S45–S58. doi: 10.1016/j.rmed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Benedini S., Ermetici F., Briganti S., et al. Insulin-mimetic effects of short-term rapamycin in type 1 diabetic patients prior to islet transplantation. Acta Diabetologica. 2018;55(7):715–722. doi: 10.1007/s00592-018-1141-z. [DOI] [PubMed] [Google Scholar]
- 316.Gala-Lopez B., Kin T., O'Gorman D., et al. Microbial contamination of clinical islet transplant preparations is associated with very low risk of infection. Diabetes Technology & Therapeutics. 2013;15(4):323–327. doi: 10.1089/dia.2012.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kwitkowski V. E., Prowell T. M., Ibrahim A., et al. FDA approval summary: temsirolimus as treatment for advanced renal cell carcinoma. The Oncologist. 2010;15(4):428–435. doi: 10.1634/theoncologist.2009-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Motzer R. J., Escudier B., Oudard S., et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. The Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 319.Zaytseva Y. Y., Valentino J. D., Gulhati P., Mark Evers B. mTOR inhibitors in cancer therapy. Cancer Letters. 2012;319(1):1–7. doi: 10.1016/j.canlet.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 320.Royce M., Bachelot T., Villanueva C., et al. Everolimus plus endocrine therapy for postmenopausal women with estrogen receptor-positive, human epidermal growth factor. Receptor 2-negative advanced breast Cancer: a clinical trial. JAMA Oncology. 2018;4(7):977–984. doi: 10.1001/jamaoncol.2018.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Zheng Y., Jiang Y. mTOR inhibitors at a glance. Molecular and Cellular Pharmacology. 2015;7(2):15–20. [PMC free article] [PubMed] [Google Scholar]
- 322.Munakata W., Tobinai K. Clinical development of voxtalisib: a pan-PI3K/mTOR inhibitor. The Lancet Haematology. 2018;5(4):e134–e135. doi: 10.1016/S2352-3026(18)30034-6. [DOI] [PubMed] [Google Scholar]
- 323.Lai Z. W., Kelly R., Winans T., et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. The Lancet. 2018;391(10126):1186–1196. doi: 10.1016/S0140-6736(18)30485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Bishu K., Ogut O., Kushwaha S., et al. Anti-remodeling effects of rapamycin in experimental heart failure: dose response and interaction with angiotensin receptor blockade. PLoS One. 2013;8(12, article e81325) doi: 10.1371/journal.pone.0081325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Kobashigawa J. A., Pauly D. F., Starling R. C., et al. Cardiac allograft vasculopathy by intravascular ultrasound in heart transplant patients: substudy from the everolimus versus mycophenolate mofetil randomized, multicenter trial. JACC: Heart Failure. 2013;1(5):389–399. doi: 10.1016/j.jchf.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 326.De Rosa R., Silverio A., Varricchio A., et al. Meta-analysis comparing outcomes after everolimus-eluting bioresorbable vascular scaffolds versus everolimus-eluting metallic stents in patients with acute coronary syndromes. The American Journal of Cardiology. 2018;122(1):61–68. doi: 10.1016/j.amjcard.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 327.El-Chemaly S., Taveira-Dasilva A., Goldberg H. J., et al. Sirolimus and autophagy inhibition in lymphangioleiomyomatosis: results of a phase I clinical trial. Chest. 2017;151(6):1302–1310. doi: 10.1016/j.chest.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Jamaspishvili T., Berman D. M., Ross A. E., et al. Clinical implications of PTEN loss in prostate cancer. Nature Reviews Urology. 2018;15(4):222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhou Y., Zhao R. H., Tseng K. F., et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacologica Sinica. 2016;37(4):519–529. doi: 10.1038/aps.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Bernardo B. C., Gao X.-M., Winbanks C. E., et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proceedings of the National Academy of Sciences. 2012;109(43):17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Fomison-Nurse I., Saw E. E. L., Gandhi S., et al. Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death and Differentiation. 2018;25(7):1336–1349. doi: 10.1038/s41418-017-0047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Rao E., Jiang C., Ji M., et al. The miRNA-17∼92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26(5):1064–1072. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 333.Zhang L., Zhou M., Qin G., Weintraub N. L., Tang Y. MiR-92a regulates viability and angiogenesis of endothelial cells under oxidative stress. Biochemical and Biophysical Research Communications. 2014;446(4):952–958. doi: 10.1016/j.bbrc.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Hullinger T. G., Montgomery R. L., Seto A. G., et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circulation Research. 2012;110(1):71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Hennessy E. J., Moore K. J. Using microRNA as an alternative treatment for hyperlipidemia and cardiovascular disease. Journal of Cardiovascular Pharmacology. 2013;62(3):247–254. doi: 10.1097/FJC.0b013e31829d48bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Janaki Ramaiah M., Lavanya A., Honarpisheh M., Zarea M., Bhadra U., Bhadra M. P. MiR-15/16 complex targets p70S6 kinase 1 and controls cell proliferation in MDA-MB-231 breast cancer cells. Gene. 2014;552(2):255–264. doi: 10.1016/j.gene.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 337.Singh Y., Garden O. A., Lang F., Cobb B. S. MicroRNA-15b/16 enhances the induction of regulatory T cells by regulating the expression of rictor and mTOR. Journal of Immunology. 2015;195(12):5667–5677. doi: 10.4049/jimmunol.1401875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Gul R., Mahmood A., Luck C., et al. Regulation of cardiac miR-208a, an inducer of obesity, by rapamycin and nebivolol. Obesity. 2015;23(11):2251–2259. doi: 10.1002/oby.21227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Montgomery R. L., Hullinger T. G., Semus H. M., et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Grueter C. E., van Rooij E., Johnson B. A., et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149(3):671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]