Abstract
Sonic Hedgehog (Shh) signaling is characterized by non-cell autonomy; cells expressing Shh do not respond to the ligand. Here, we identify several Shh mutations that can activate the Hedgehog (Hh) pathway cell-autonomously. Cell-autonomous pathway activation requires the extracellular cysteine rich domain of Smoothened, but is otherwise independent of the Shh receptors Patched1 and −2. Many of the Shh mutants that gain activity fail to undergo auto processing resulting in the perdurance of the Shh pro-peptide, a form of Shh that is sufficient to activate the Hh response cell-autonomously. Our results demonstrate that Shh is capable of activating the Hh pathway via Smoothened, independently of Patched1/2, and that it harbors an intrinsic mechanism that prevents cell-autonomous activation of the Shh response.
Introduction
Sonic Hedgehog (Shh) is a signaling molecule indispensable for vertebrate embryonic development and adult stem cell maintenance. Impaired regulation of the Hedgehog (Hh) pathway can cause of various birth defects and diseases, including Holoprosencephaly (HPE) and a several tumor types (Bale, 2002). Shh is synthesized as a 45 kD pro-peptide encompassing the signaling Shh N-terminal domain (ShhN) and an intein-like domain C-terminal domain (ShhC). The ShhC domain mediates an autocatalytic cleavage of Shh (Hall et al., 1997), resolved by the addition of a cholesterol moiety on the C-terminus of the 19 kD ShhN domain (ShhNp) (J. J. Lee et al., 1994). ShhNp becomes further modified with the attachment of a palmitoyl group to its N-terminus (Buglino and Resh, 2008). Lipid modified ShhNp is then secreted from expressing cells through a mechanism involving Dispatched1 (Disp1), Scube2, and ADAM metalloproteases. The release of ShhNp from cell membranes might require the removal of its cholesterol modification, which results in soluble and biologically active ligands capable of non-cell autonomous signaling (Jakobs et al., 2014; Ohlig et al., 2012).
In the absence of Shh ligand, the receptors Patched1 (Ptch1) and Patched2 (Ptch2) inhibit the signal transducer Smoothened (Smo) through a non-stoichiometric mechanism (Alfaro et al., 2014; Taipale et al., 2002; J. Zhang et al., 2001). Ptch1 and Ptch2 are extracellular Shh receptors that regulate the Hh responses in vivo (Allen et al., 2011; Goodrich et al., 1997; Izzi et al., 2011). Binding between Shh and Ptch1 appears to be largely mediated via the palmitoylated first 22 amino acid residues (Qi et al., 2018) that suffice for Hh pathway activation (Tukachinsky et al., 2016). Binding of Shh to the antagonist Hhip (Bosanac et al., 2009), encompasses the Zn2+ ion coordinated within Shh that is part of a larger and highly conserved motif resembling a zinc peptidase catalytic domain (Hall et al., 1995). Structural analysis of interactions between Shh and Hhip showed that they resemble molecular interactions between matrix metalloproteases (MMPs) and the tissue inhibitor of metalloproteinase (TIMP) (Bosanac et al., 2009). This observation is consistent with the notion that Shh is a peptidase (Roelink, 2018). In Zn2+ peptidases, the Shh-E177 equivalent abstracts a proton from the catalytic water at the Zn2+ coordination domain, which is followed by a nucleophilic attack of the OH− on the peptide backbone. Shh-E177A is, therefore, predicted to be impaired for the intrinsic Zn2+ peptidase activity. Analysis of this mutant has revealed two interesting properties. First, Shh-E177A is unable to mediate signaling from the notochord to the overlying neural tube (non-cell autonomously), but is more capable than Shh of inducing the Hh response when expressed in the developing neural tube (likely cell-autonomously) (Himmelstein et al., 2017). Second, purified ShhN-E177A is more stable in solution than ShhN, indicating a cannibalistic peptidase activity that is intrinsic to ShhN. Nevertheless, The conserved catalytic site, and consequently any putative enzymatic activity, has been shown not to be required for signaling (Fuse et al., 1999), and is thus it is often referred to as a “pseudo active” site in Shh. As these experiments were performed with the truncated Shh-C199* (ShhN) mutant, a role for the peptidase activity might be associated with the processing of the Shh pre-protein or the function of ShhNp. The Zn2+ coordinating residues H141 and D148 (mouse numbering) have been found mutated in HPE, indicating that these residues are important for Shh signaling (Roessler et al., 2009). SHH-H140P was shown to be unable to undergo auto-processing, a characteristic shared with several other mutations both in the N-terminal and C-terminal domains (Traiffort et al., 2004). Still, the resulting Shh pro-protein is active and is internalized by Ptch1 (Tokhunts et al., 2010).
ShhN can act as a cellular chemoattractant (Angot et al., 2008; Bijlsma et al., 2007). The chemotactic response to Shh is directional and does not require de novo transcription or translation, nor does it require the function of Gli proteins (Bijlsma et al., 2007; 2008; Chinchilla et al., 2010; Lipinski et al., 2008). Chemotaxis towards Shh is mediated by Smo (Charron et al., 2003); however, it does not require the localization of Smo to the primary cilium, a prerequisite of the transcriptional response. Chemotaxis can be mediated by forms of Smo unable to activate the transcriptional response to Shh (Bijlsma et al., 2012), indicating fundamental differences between these two activities of Smo. Translocation of Smo to the primary cilium is negatively affected by Ptch1 function (Rohatgi et al., 2007), and we have previously shown that cells lacking Ptch1 retain the ability to migrate towards ShhN (Alfaro et al., 2014). Because Ptch1 and Ptch2 have overlapping functions (Alfaro et al., 2014; Y. Lee et al., 2006; Roberts et al., 2016), it is not clear whether the chemotactic response in Ptch1LacZ/LacZ cells is mediated by Ptch2 or if it is a Ptch1/2-independent signaling event.
Smo is required for the responses to Shh. Smo is a Class F G-protein coupled receptor (GPCR) and belongs to a receptor superfamily predominantly defined by Frizzleds (Frz), the canonical receptors of the Wnt pathway (Bhanot et al., 1996; Kristiansen, 2004). Smo and Frzs share over 25% sequence identity; both contain a characteristic heptahelical domain as well as a conserved extracellular Cysteine Rich Domain (CRD). Wnts bind to the CRD of Frz receptors through two distinct binding sites, one of which is a protein-lipid interface, to initiate signal transduction (Janda et al., 2012). Although Ptch1-mediated inhibition of Smo is thought to target a specific hydrophobic binding pocket within the heptahelical domain of Smo, the CRD of Smo can also regulate Hh pathway activity by binding to a variety of sterols- most of which cause an activation of the response (Myers et al., 2013; Nachtergaele et al., 2013; Nedelcu et al., 2013; Corcoran and Scott, 2006; Huang et al., 2016; Xiao et al., 2017; Huang et al., 2018). The regulatory interactions involving the CRD of Smo demonstrate that activation of the Hh pathway is multifaceted, and that Ptch1-mediated inhibition is only part of a more complex regulation of Smo activity. In many Shh-induced tumors Shh signaling is predominantly non-cell autonomous (García-Zaragoza et al., 2012; Shaw et al., 2009; Yauch et al., 2008), the non-Shh-expressing cells are sensitive to Smo inhibitors, whereas cells expressing Shh are not. This notion of non-autonomy in Shh signaling is further supported by the observation that in embryos, cells that express Shh (e.g. cells in the notochord and floor plate) only have low levels of Ptch1, a gene that is universally upregulated in cells responding to Shh. A gradient of Ptch1 expression adjacent to, and away from the floor plate reflects the graded Shh response in the developing neural tube, a pattern that appears to be associated with all sites of Shh/Ptch1 expression in the embryo (Sagai et al., 2009).
Here we assess if the negative correlation between Shh expression and Ptch1 expression affects the apparent inability of Shh-expressing cells to respond to the ligand. We find that expression of Shh and several Shh mutants activates the Hh response cell-autonomously in cells lacking all Ptch1 and Ptch2 activity. Ptch1LacZ/LacZ;Ptch2−/− cells also retain the ability to migrate towards a source of ShhN, a non-canonical Hh response mediated by Smo. We demonstrate that this Shh-mediated Ptch1/2-independent activation requires the CRD of Smo. We find several Shh mutants that activate the Hh response better than wild type Shh, and many of these mutants exhibit defects in processing resulting in the perdurance of Shh pro-protein, further supporting the notion that not ShhNp, but instead the Shh pro-protein induces Smo activation cell autonomously. An obligatory Shh pro-protein retains its activity when expressed in the developing neural tube, and the amount of Shh pro-protein, but not ShhNp correlates with the level of cell-autonomous Smo activation, indicating that auto-processing of Shh turns the active Shh pro-protein into the cell-autonomously inactive ShhNp.
Results
The Shh pro-protein and ShhN can activate the Hh response cell-autonomously independent of Patched1 and −2
We set out to test if the absence or scarcity of Ptch1 causes the inability of cells to respond to Shh. To assure lack of all Ptch activity, we used Ptch1LacZ/LacZ;Ptch2−/− cells. These Ptch1/2-deficient cells do not induce the transcriptional Hh response when exposed to exogenously supplied ShhN (Roberts et al., 2016), which also ensures that any Hh pathway activity we measure is strictly cell-autonomous.
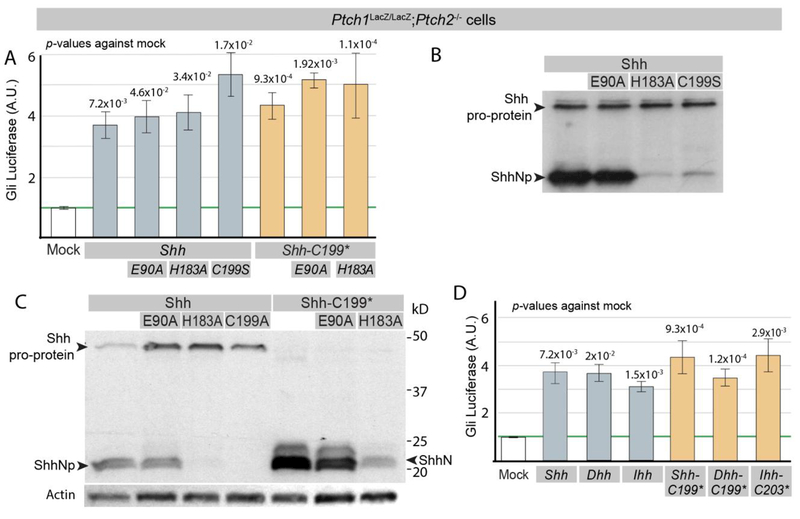
To similarly assess the requirement of Shh (co-)receptors to activate the Hh response, we used Shh-E90A (a mutant unable to bind to Boc, Cdo, and Gas1 (Allen et al., 2011; Izzi et al., 2011)) and Shh-H183A a mutant that breaks the Zn2+ triad, which interferes with Hhip binding (Gong et al., 2018). Both Shh and its mutant forms effectively induced the transcriptional Hh response after transfection in Ptch1LacZ/LacZ;Ptch2−/− cells (Figure 1A). Shh protein was readily detected in transfected Ptch1LacZ/LacZ;Ptch2−/− cells (Figure 1B,C). In these cells, Shh is normally processed into a cholesterol-associated form (ShhNp) (19 kD), but full-length Shh pro-protein (45 kD) can be detected as well. Cells expressing Shh-H183A had little N-terminal processed Shh (ShhNp), and Shh was primarily present in its full length form (Figure 1B,C). It thus appears that the ability to induce the transcriptional Hh response cell-autonomously correlates with the presence of the full length form of Shh, and not with ShhNp. To determine if the Shh pro-protein could mediate the activation of the Hh response cell-autonomously, we tested Shh-C199S, a form of Shh largely unable to undergo autoproteolysis (Figure 1B) and found that this mutant was an equally potent inducer of the Hh response (Figure 1A). This indicates that ShhNp is not active in this assay. As cholesterol-modified ShhNp is thought to undergo activation upon shedding after removal of its lipid moieties (Ohlig et al., 2011), we tested a form of Shh that due to truncation at G198 does not have the cholesterol moiety (Shh-C199*). We found that Shh-C199* (Figure 1C) retains the ability to induce the Hh response cell-autonomously (Figure 1A). The mechanism by which Shh-E90A, -H183A and Shh-C199* allow activation of the Hh pathway cell-autonomously appears to be related, as the double mutants (Shh-C199*/E90A and Shh-C199*/H183A) have the same activities as the single mutants (Figure 1A), and do not produce additive effects.
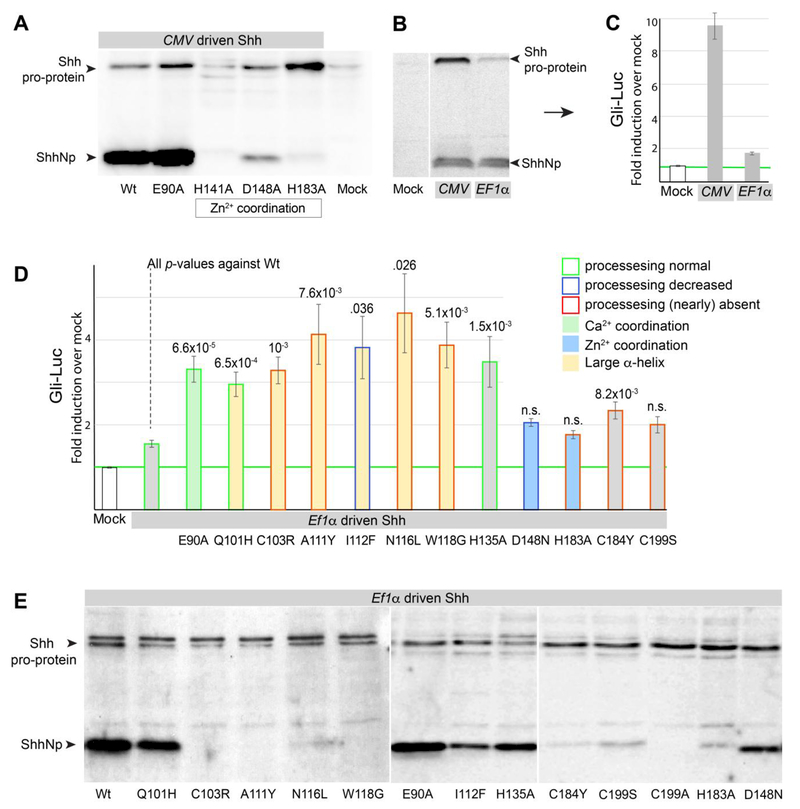
Figure 1. The Shh pro-protein can induce Gli-luciferase independent of Ptch1/2.
(A) Ptch1LacZ/LacZ;Ptch2−/− cells were transfected with Gli-Luciferase (Luc) alone (Mock) or cotransfected with Gli-Luc and Shh, Shh-E90A, Shh-H183A, Shh-C199S, Shh-C199*, Shh-C199*/E90A, or Shh-C199*/183A. Luciferase levels in mock transfected Ptch1LacZ/LacZ;Ptch2−/− cells were set at “1”. (B) Western blot analysis of Shh, Shh-E90A, Shh-H183A, Shh-C199S, all driven by the CMV promoter. (C) Western blot analysis of Shh, Shh-E90A, Shh-H183A, Shh-C199A Shh-C199*, Shh-C199*/E90A, and Shh-C199*/183A protein expression in Ptch1LacZ/LacZ;Ptch2−/− cells, using an antibody directed against the N-terminal domain of Shh. all driven by the CMV promoter, except Shh, which was driven by the EF1α promoter. (D) Ptch1LacZ/LacZ;Ptch2−/− cells were transfected with Gli-Luc alone (Mock) or co-transfected with Gli-Luc and Shh, Dhh, Ihh, Shh-C199*, Dhh-C199*, or Ihh-C203*. Luciferase levels in Mock transfected cells were normalized to 1. All error bars are s.e.m., p values (Student t-test, 2 tailed) against mock are indicated were relevant, n=4 (A), n≥3 (C), independent biological replicates, of triple or quadruple parallel experiments.
Together these results demonstrate that presumed loss-of-function mutations (Shh-H183A, Shh-E90A, Shh-C199S, Shh-C199*/E90A, and Shh-C199*/H183A), all have the ability to signal cell-autonomously in cells lacking Ptch1/2 receptors. Cell-autonomous activation of the Hh response by Shh does not require processing of the pro-protein, nor does it require it require correct Zn2+ and Ca2+ coordination. In this assay Dhh, Ihh as well as their N-terminally truncated forms (Dhh-C199*, Ihh-C203*) were found to be active (Figure 1 D).
Shh mutants can activate the Hh response in the developing neural tube
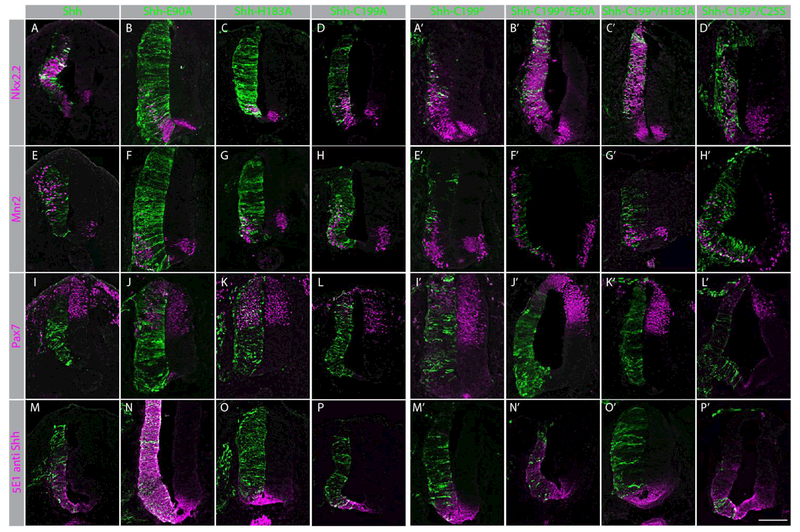
Mis-expression of the Shh mutants in the developing neural tube, allows the assessment of their ability to activate the Hh response in vivo. Along with Shh-E90A and Shh-H183A, we tested Shh-199A, a mutant form that cannot undergo autoproteolysis (Figure 1C), and ShhN-C25S a mutant form lacking the N-terminal acyl group thought to be central for the binding to Ptch1 (Qi et al., 2018; Tukachinsky et al., 2016). We assessed the activity of Shh mutants in vivo by coelectroporating Shh, Shh-E90A, Shh-H183A, or Shh-199A, like Shh-C199S coding for a persistent pro-protein (Roelink et al., 1995), together with GFP in developing chick neural tubes. Expression of Shh, Shh-E90A, and Shh-C199A caused an expansion of Nkx2.2 and Mnr2 domains, as well as repression of Pax7 expression (Figure 2, panels A-H), indicating that these forms of Shh are active. Electroporation of Shh-H183A does not result in activation of the pathway (Figure 2 C, G, K). However, we found that Shh-C199*/H183A strongly induced the Hh response after electroporation (Figure 2 C’,G’,K’), thus indicating that correct Zn2+ coordination is not required per se to activate the Hh response in vivo, but appears to be required for in vivo signaling by the pro-protein. Consistent with the activity of Shh-E90A, we found that Shh-C199*/E90A was active in this assay, as was Shh-C199*/C25S, indicating that association with Ptch1 is not required for activation of the Hh response in vivo. Altogether, the in vivo results support our findings in vitro indicating that the binding of Shh to Ptch1/2, Cdo, Boc or Gas1 is not necessary to activate the Hh response in vivo, although there appears to be a requirement for the correct Zn2+ coordination for signaling by the Shh prop-protein. Nevertheless, these results demonstrate that also in vivo Hh pathway activation can be mediated by the Shh pro-protein.
Figure 2. Shh mutants activate the Hh pathway in vivo.
Panels A-P: Cross-sections of stage 20 HH chicken embryo neural tubes co-electroporated at stage 10 HH with GFP and Shh (A, E, I, M), Shh-E90A (B, F, J, N), Shh-H183A (C, G, K, O), or Shh-C199A (D, H, L, P). Cells expressing GFP, and thus probably Shh (mutants) are labeled in green. Sections are stained with antibodies to Nkx2.2 (A-D), Mnr2 (E-H), Pax7 (I-L), and Shh (using 5E1, M-P) labeled in magenta. Panels A’-P’: Cross-sections of stage 20 HH chicken embryo neural tubes co-electroporated at stage 10 HH with GFP and Shh-C199* (A’,E’,I’,M’), Shh-C199*/E90A (B’,F’,J’,N’), or Shh-C199*/H183A (C’,G’,K’,O’). Shh-C199*/C25S (D’,H’,L’,P’). Cells expressing GFP, and thus probably the Shh mutants are labeled in green. Sections are stained with antibodies to Nkx2.2 (A-D, A’-D’), Mnr2 (E-H, E’-H’), Pax7 (I-L, I’-L’), and Shh (using 5E1, M’-P’) labeled in magenta. The scale bar is 100μm.
Cell-autonomous activation of the Hh response by ShhN requires Smo
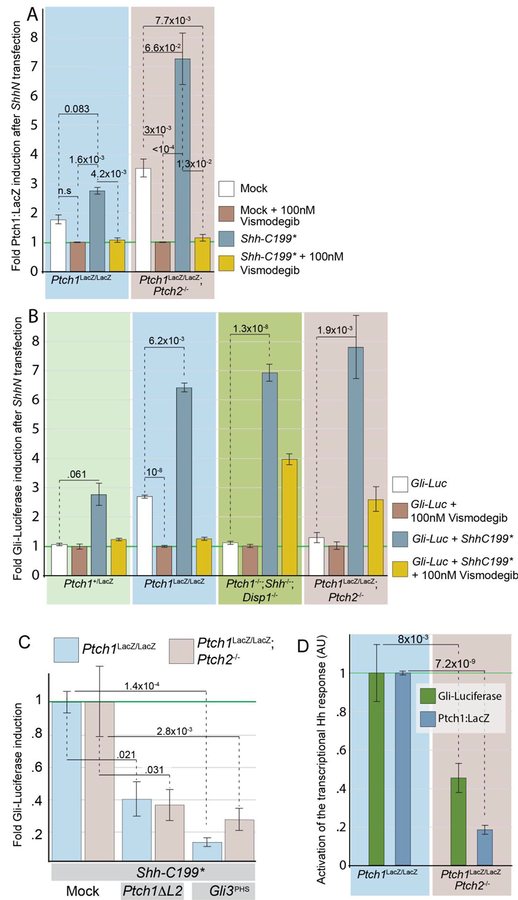
Both the in vitro and in vivo experiments Show that the non-cholesterol-modified form of Shh (Shh-C199*) is very active, and we used this commonly used form of Shh to assess if the observed activation of the Shh response independent of Ptch1/2 function is mediated by Smo. The Ptch1/2 null fibroblast line used in this study is derived from Ptch1LacZ/LacZ;Ptch2−/− mESCs (Alfaro et al., 2014; Roberts et al., 2016), which in turn are derived from Ptch1LacZ/LacZ mESCs (Goodrich et al., 1997), and consequently, these cells carry the Ptch1:LacZ null allele. As Ptch1 itself is invariably induced in cells responding to Shh, the Ptch1:LacZ allele has found widespread use as a dependable readout of Hh pathway activation. To further assess the nature of cell-autonomous pathway activation, we also used Ptch1LacZ/LacZ fibroblasts, used to study the of role of Ptch1 (Taipale et al., 2002), as a control. Transfection of Shh-C199* into either cell line resulted in an activation of the transcriptional Hh response, i.e. an increase of transcriptional activity over the mock transfected cells. This activation could be blocked with the addition of vismodegib, a small molecule inhibitor of Smo (Sharpe et al., 2015) (Figure 3A). The ability of vismodegib to block Ptch1/2-independent activation of the Hh response, as a consequence of Shh-C199* expression, demonstrates that this activity is mediated by Smo. This experiment was repeated using the Gli-Luciferase reporter as an independent readout in four independent cell lines of varying genotypes. The Gli-Luciferase reporter was invariably induced upon transfection with Shh-C199*, regardless of whether the cells were Ptch1/2 proficient, Ptch1 null, or lacked both Ptch1 and Ptch2 function (Figure 3B). The resulting Hh pathway activation could be lowered by vismodegib in all cell lines, further demonstrating the involvement of Smo in cell-autonomous signaling. Gli-Luciferase induction after Shh-C199* expression confirms the results obtained using the Ptch1:LacZ allele for quantification (Figure 3A). As shown before, Ptch1LacZ/LacZ fibroblasts that are derived from a Ptch1LacZ/LacZ have a moderate intrinsically upregulated Hh response after serum withdrawal (Taipale et al., 2002) as measured using Gli-Luciferase. This appears to be a unique characteristic of this particular cell line, as neither Ptch1LacZ/LacZ;Ptch2−/− fibroblasts, nor Ptch1−/−;Disp1−/−;Shh−/− fibroblasts exhibit this property (Figure 3 B,D).
Figure 3. Activation of the Hh response by Shh-C199* does not require Ptch1/2 activity.
(A) Ptch1LacZ/LacZ cells and Ptch1LacZ/LacZ;Ptch2−/− (indicated) were assessed for Ptch1:LacZ expression after mock transfection (white and brown bars) or Shh-C199* transfection (blue and yellow bars). Each condition was treated with either a Smo inhibitor, 100 nM Vismodegib (brown and yellow bars), or a DMSO vehicle control (white and blue bars). β-Gal levels in mock transfected cells treated with 100 nM Vismodegib for each cell line were normalized to 1 to visualize intrinsic upregulation of the Hh response. Error bars in A are s.e.m., p-values (Student t-test, 2 tailed) are indicated were relevant, n≥3 of independent biological replicates of triple or quadruple independent parallel experiments. (B) Ptch1+/LacZ cells, Ptch1LacZ/LacZ cells, Ptch1−/−;Shh−/−;Disp1−/−, and Ptch1LacZ/LacZ;Ptch2−/− cells (indicated) were co-transfected with Gli-Luc and GFP (Mock; white/brown bars) or co-transfected with Gli-Luc and Shh-C199* (blue and yellow bars). Each condition was treated with either a Smo inhibitor, 100 nM Vismodegib (brown and yellow bars), or a DMSO vehicle control (white and blue bars). Luciferase levels in mock transfected cells treated with 100 nM Vismodegib for each cell line were normalized to 1. (C) Gli-luc levels were assayed in Ptch1LacZ/LacZ (blue bars) and Ptch1LacZ/LacZ;Ptch2−/− (pink bars) transfected with Ptch1ΔL2,or Gli3phs together with Gli-luc and Shh-C199*. Luciferase levels in Shh-C199*, mock cotransfected cells were set at “1” for each cell line. Error bars in B, C are s.e.m., p-values (Student t-test, 2 tailed) are indicated were relevant, n≥3, independent biological replicates of triple or quadruple independent parallel experiments. (D) Hh pathway activation in Ptch1LacZ/LacZ and Ptch1LacZ/LacZ;Ptch2−/− was measured in parallel using Gli-luc and Ptch1:LacZ. Using both methods we find higher Hh pathway activation in the Ptch1LacZ/LacZ cells than the Ptch1LacZ/LacZ;Ptch2−/− cells. Luciferase and LacZ levels in Ptch1LacZ/LacZ cells were normalized to 1. Error bars in D are s.e.m., p values (Student t-test, 2 tailed) are indicated were relevant, n≥6.
Cell-autonomous activation of the Hh response by Shh-C199*N in cells lacking Ptch1/2 activity could be inhibited by a dominant inhibitory form of Ptch1 and by the downstream transcription factor Gli3. Expression of Ptch1ΔL2 (Briscoe et al., 2001) or Gli3PHS (Meyer and Roelink, 2003) prevented upregulation of the Hh response by Shh-C199* in both Ptch1LacZ/LacZ and Ptch1LacZ/LacZ;Ptch2−/− cells (Figure 3C). Ptch1 catalytically acts upon Smo (Taipale et al., 2002), which in turn acts via the Gli transcription factors (Ruiz i Altaba, 1998; Stamataki et al., 2005), thus these results indicate that ShhN expression activates Smo downstream of Ptch1/2 function and upstream of Gli function.
The extracellular Cysteine Rich Domain of Smo is required for its activation by Shh
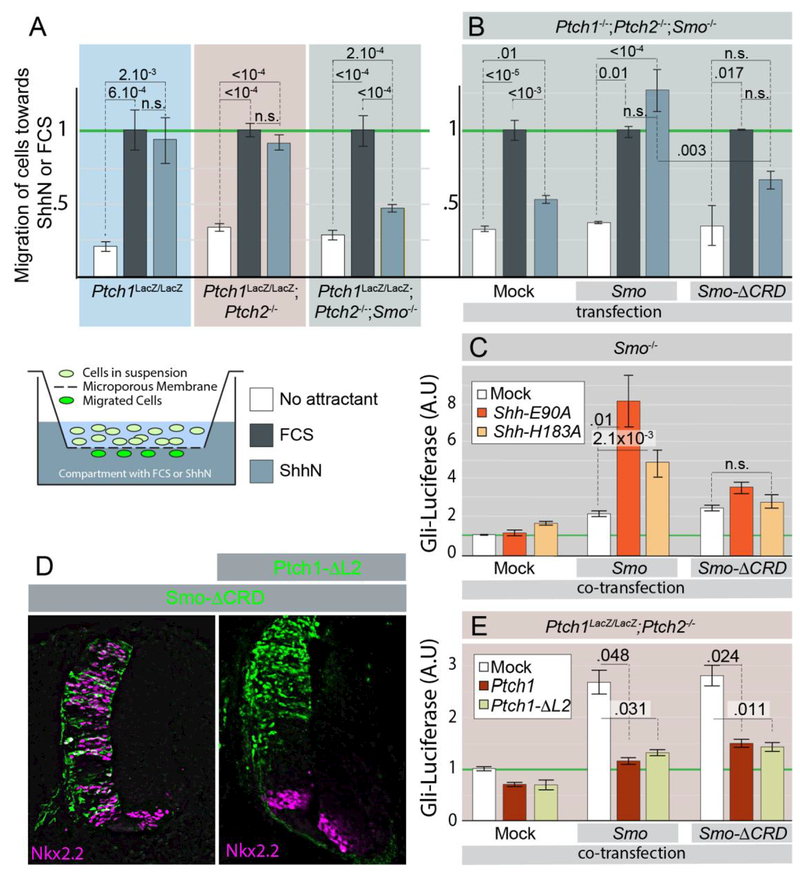
We tested the ability of Ptch1LacZ/LacZ;Ptch2−/− fibroblasts to migrate towards a localized source of ShhN using a modified Boyden chamber (H. C. Chen, 2005). We found that Ptch1LacZ/LacZ;Ptch2−/− cells were indistinguishable from Ptch1LacZ/LacZ cells in their ability to migrate towards ShhN (Figure 4A). Shh chemotaxis was largely abolished in Ptch1LacZ/LacZ;Ptch2−/−;Smo−/− cells; however, the ability to migrate towards FCS, the positive control attractant, was unaffected (Figure 4A).
Figure 4. SmoΔCRD retains sensitivity to Ptch1-mediated inhibition in vitro and in vivo, but cannot be activated by Shh in a Ptch1/2-independent manner.
(A) Cells with the indicated genotypes were assayed in a modified Boyden chamber (diagram) for their ability to migrate towards FCS or ShhN. Migration was normalized to FCS for each condition. (B) Ptch1−/−;Ptch2−/−;Smo−/− cells were mock transfected, or transfected with Smo or Smo-ΔCRD. The transfected cells were assayed in a modified Boyden chamber for their ability to migrate towards FCS or ShhN. Error bars are s.e.m., p values (Student t-test, 2 tailed), n≥4. (C) Smo−/− cells were transfected with Gli-Luc, and co-transfected with Smo or Smo-ΔCRD and Shh-E90A or Shh-H183A as indicated. Gli-Luciferase levels were quantified, and the levels in double mock transfected cells were normalized to 1. (D) Stage 10 chicken embryos were electroporated with Smo-ΔCRD (green) or co-electroporated with Smo-ΔCRD and Ptch1-ΔL2 (Green). The ventral marker NKX2.2 was visualized (purple). (E) Ptch1LacZ/LacZ;Ptch2−/− cells were transfected with Gli-Luc, and co-transfected with Smo or Smo-ΔCRD, and Ptch1 or Ptch1-ΔL2 as indicated. Gli-Luciferase levels were quantified, and the levels in mock/mock/Gli-Luc transfected cells were normalized to 1. Error bars for C and E are s.e.m., p values (Student t-test, 2 tailed) against mock are indicated were relevant, n≥4 (C), n≥3 (E) independent biological replicates of triple or quadruple independent parallel experiments.
We could restore the ability of Ptch1LacZ/LacZ;Ptch2−/−;Smo−/− cells to migrate towards ShhN upon transfection with wild type Smo, demonstrating the sufficiency of Smo to mediate Shh chemotaxis independent of Ptch1/2 function. (Figure 4A). However, transfection of a form of Smo lacking its CRD (Smo-ΔCRD) did not restore the chemotactic response in Ptch1LacZ/LacZ;Ptch2−/−;Smo−/− cells (Figure 4B). These results demonstrate that Smo can respond to ShhN in the extracellular space to mediate the chemotactic response in the absence of Ptch1/2, and the CRD of Smo is required for this signaling event.
We used a Smo−/− fibroblast cell instead of the preferred Ptch1LacZ/LacZ;Ptch2−/−;Smo−/− cell line to assess the requirement of the CRD of Smo to mediate the cell-autonomous transcriptional response. This was necessitated by our experience that Ptch1LacZ/LacZ;Ptch2−/−;Smo−/− fibroblasts do not survive the serum deprivation necessary for the transcriptional assays. To ensure that we strictly measured cell-autonomous signaling, we used the Shh-E90A and Shh-H183A mutants that cannot signal non-cell autonomously. As expected, transfection of either Shh-H183A or Shh-E90A alone in Smo−/− cells did not result in the induction of the Hh response (Figure 4C). Transfection of Smo or Smo-ΔCRD alone caused a small upregulation of the Hh response. Consistent with earlier results (Figure 1A), we found that co-transfection of Shh-E90A or Shh-H183A together with Smo caused a strong induction of the Hh response. In contrast, neither Shh-H183A nor Shh-E90A was capable of synergizing with Smo-ΔCRD to induce the Hh response, further demonstrating that the activation of Smo by Shh requires the CRD of Smo.
Despite its inability to respond to Shh, Smo-ΔCRD remains subject to inhibition by Ptch1. Mis-expression of Smo-ΔCRD in the developing chick neural tube after electroporation causes an upregulation of the Hh response (Figure 4D), consistent with earlier reports that Smo-ΔCRD retains a relatively high level of basal activity in vivo (Kwong et al., 2014). The activity of Smo-ΔCRD was suppressed by co-expression of Ptch1ΔL2, coding for a dominant-inhibitory form of Ptch1 (Briscoe et al., 2001) (Figure 4D). Similarly, expression of either Smo or Smo-ΔCRD in Ptch1LacZ/LacZ;Ptch2−/− cells results in transcriptional Hh activity, which can be inhibited by co-expression with Ptch1 or Ptch1ΔL2 (Figure 4E). Together these results indicate that the CRD of Smo is required for its activation by ShhN, but not for its repression by Ptch1.
Holoprosencephaly-causing mutants can activate the Hh response cell-autonomously
Shh-H183A cannot undergo autoproteolytic processing (Figure 1B), and to further assess the role of the Zn2+ coordination triad is we also tested Shh-H141A and Shh-D148A. Shh-H141A did not auto-process, while Shh-D148A processed very poorly (Figure 5A). Both H141 and D148 are found mutated in HPE, a congenital syndrome that can be caused by aberrant SHH signaling (Hehr et al., 2010; Odent et al., 1999; Roessler et al., 2009). It was observed before that several HPE mutants prevent processing of the SHH pro-protein (Traiffort et al., 2004), indicating that the perdurance of the SHH pro-protein might contribute HPE. As we found that the Shh pro-protein (Shh-C199A) was able to induce the Hh response in vivo, we tested if failure of Shh processing could alter the Hh response. The autocatalytic processing of the Shh pro-protein is relatively slow, and to minimize potential overexpression artefacts we tested Shh expressed using the human Elongation Factor α (EF1α) promoter, which is much weaker than the CMV promoter used in the experiments described above. Shh driven by the EF1α promoter is a poor inducer of the Hh response in cells lacking Ptch1/2, in contrast to Shh driven by the CMV promoter (Figure 1A, 5C). Analysis of Shh expressed in the same transfected cells (Figure 5B) revealed that the amount of observed Shh pro-protein correlates with the level of Hh pathway induction. In contrast, the amount ShhNp, was not correlated with pathway activity (Figure 5 C,D), consistent with the observations above (Figure 1 A,B). This further advances the notion that ShhNp is unable to induce the Hh response cell-autonomously and that the Shh pro-protein can activate the Hh response.
Figure 5. Shh processing is not required for the cell-autonomous activation of the Hh response in Ptch1LacZ/LacZ;Ptch2−/− cells.
(A) Shh western blot showing that mutations in the Zn2+ coordination triad (H141A, D148A, H183A) severely compromise, or prevent Shh processing. E90A (affecting Ca2+ coordination) undergoes normal processing. (B) Shh western blot showing expression levels (high for the CMV and low for the EF1α promoter) affect the level of Shh pro-protein more so than ShhNp. (C) The induction of the Gli-Luciferase was measured after transfection of EF1α- or CMV-driven Shh. The induction of the response correlates with the amount of Shh pro-protein, not ShhNp. (D) Gli-Luciferase induction by Shh mutants driven off the EF1α promoter. Several HPE mutants located in the large α-helix (Q101H, C103R, A111Y, I112F, N116L, W118G), mutants involved in Zn2+ coordination (D148N, H183A), E90A (Ca2+ coordination), H135 (near the catalytic domain), C184Y (commonly mutated in HPE), and C199S (cleavage site for auto-processing, mutated in HPE) were assessed. Error bars for E are s.e.m., p values (Student t-test, 2 tailed) are indicated were relevant, n=3 of independent biological replicates of triple or quadruple independent parallel experiments. (E) Western blot analysis of EF1α-driven Shh mutants assessed in F. Note that the ability to auto-process is often impaired in α-helix and Zn2+ coordination mutants.
The low level of Hh pathway induction observed in cells expressing EF1α-driven Shh allowed us to assess if Shh mutations can result in a gain-of-function phenotype, as measured by activation of the transcriptional Hh response cell-autonomously, as would be predicted by perdurance of the full-length Shh pro-protein. We tested mutations affecting the Zn2+ coordination domain, the Ca2+ coordination domain, and a series of HPE mutants, in particular those in the large α-helix that dominates the face of Shh opposite to the Zn2+ coordination domain. All these mutants were directly introduced into the EF1α–driven Shh clone that had little activity after transfection. We found that Zn2+ coordination triad mutants D148N (found in HPE), and H183A, (blue bars, Figure 5E), caused minimal activation of the Hh response as compared to wild-type Shh in Ptch1LacZ/LacZ;Ptch2−/− cells. Traiffort demonstrated that W118G does not undergo autoproteolysis (Traiffort et al., 2004). Tryptophan118 is at the C-terminal end of the large α-helix that is a common site for mutations found in holoprosencephaly patients (Figure 6C), and we tested Q101H, C103R, A111Y I112F, N116L and W118G. All these of Shh point mutations are gain-of-function mutants in their ability to induce the Hh response cell-autonomously as compared to wild type Shh. This activation does not require processing of the Shh pro-protein, as a majority of the tested HPE mutants in the large α-helix do not give rise to ShhNp (Figure 5F). This suggests that at least some cases of HPE could be caused by the acquisition of Ptch1/2-independent Hh signaling via the accumulating SHH pro-protein, while leaving non-autonomous signaling via the wild type allele intact.
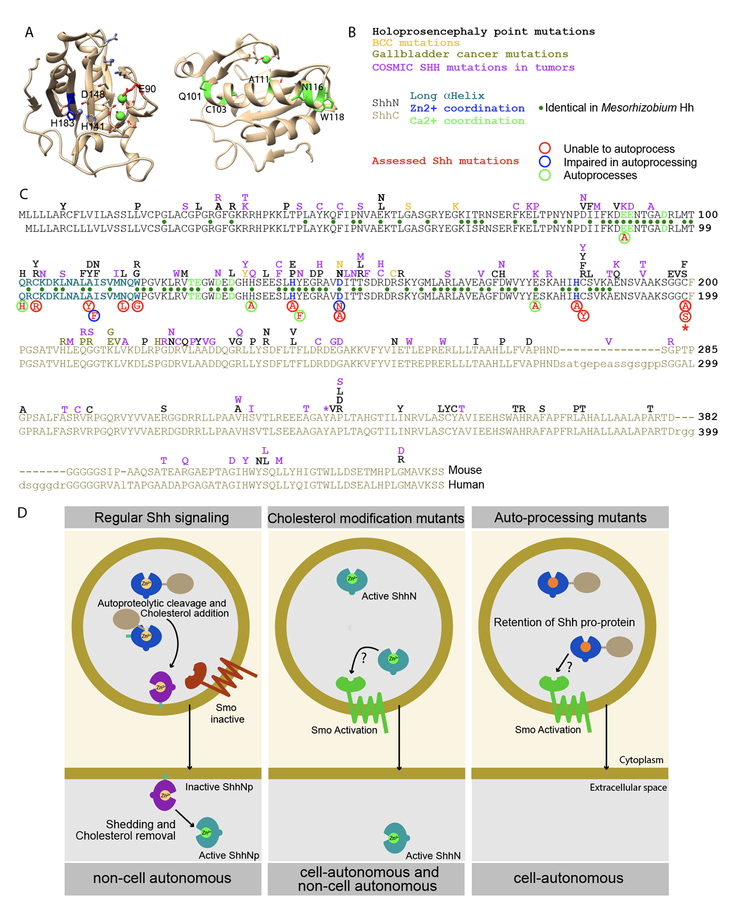
Figure 6. Overview and model.
Representation of the ShhN crystal structure with salient residues indicated. On the left is a view into the Zn2+ (grey)/Ca2+ (green) coordination domains, on the right is the opposite view with the large α-helix in front, with mutated residues indicated in green. (B, C) Lineup of the mouse (top) and human Shh. All tested mutants are indicated below the lineup, with their activity and ability to process indicated. Above the line are Shh point mutations described for holoprosencephaly, BCCs, Gallbladder Cancers, and those curated in the COSMIC database. Identical residues found in Mesorhizobium Hh are indicated with green dots between the lines. (D) During regular Hh signaling, the potentially active pro-protein auto-cleaves to yield the inactive ShhNp form. Further processing mediates activation and release, thus allowing signaling in trans. Mutants that do not receive the cholesterol modification always exist in an active form, thus mediating both cell-autonomous and non-cell autonomous signaling. Repressed auto-processing of the Shh pro-protein results in perdurance of this active form, thus causing cell-autonomous activation of the Hh response.
Discussion
Ptch1/2-independent activation of Smo by Shh
Our observations challenge the canonical model that Smo activation is solely mediated by the Shh-induced release of Ptch1 inhibition. Despite the ample evidence that Ptch1 is an efficient inhibitor of Smo (Goodrich et al., 1997), we find that: 1) the loss of Ptch1/2 does not inevitably result in maximal Smo activation, and 2) several forms of Shh can activate the migrational and transcriptional Hh responses in the absence of Ptch1/2 function. Nevertheless, Ptch1 function is required in those same cells to transcriptionally respond to ShhN provided extracellularly (Roberts et al., 2016). A central role of Ptch1 function is to regulate Smo entry into the primary cilium (Rohatgi et al., 2007), which is required for the transcriptional Hh response (Goetz and Anderson, 2010) but dispensable for the chemotactic response (Bijlsma et al., 2012). Our observations are consistent with a model in which Shh can both bind to Ptch1/2 (Rohatgi et al., 2007), resulting in the localization of Smo to the primary cilium where it mediates the transcriptional response, and activate Smo independent of Ptch1/2 function, resulting in fast changes in cell shape that underlie chemotaxis. This model would require the function of Ptch1/2 to couple extracellular Shh to the Gli-mediated transcriptional response, something that might be circumvented by some of Shh mutants we described.
Whether cells lacking Ptch1 or Ptch2 retain sensitivity to Shh in vivo is unclear. However, some observations indicate that they might. The spectrum of tumors observed in Ptch1 null and in Ptch1/2 heterozygous mice (Y. Lee et al., 2006) are commonly found at sites where Shh is expressed; and Shh-induced tumors often arise at the same locations (Beachy et al., 2004). The positional overlap between tumors induced by the loss of Ptch function and Shh-induced tumors is compatible with our model that cells lacking Ptch1/2 function nevertheless retain some ability to respond to Shh. Our model is further supported by the observation that the establishment of left/right asymmetry in mouse embryos, determined by Pitx2 expression, requires Smo, Shh, and Ihh, but not Ptch1 (X. M. Zhang et al., 2001). Ptch1−/− mutant embryos establish normal asymmetric expression of Pitx2, indicating that this Hh/Smo-mediated symmetry-breaking event occurs independently of Ptch1. Furthermore, the upregulation of pathway activity observed in Ptch1LacZ/LacZ mice coincides with a widespread expression of Shh in the developing neural tube, leaving open the possibility that this activation remains Shh-dependent.
The ability of Shh to activate Smo in a Ptch1/2-independent manner is supported by the observations that ShhN activates the Hh pathway in cells lacking these receptors and that forms of Shh unable to bind to the extracellular receptors activate the pathway in a Smo-dependent manner. Furthermore, we demonstrate that the CRD of Smo is required for both Shh-mediated transcriptional and migrational responses. The notion that the CRD of Smo is a target of Shh-mediated activation is supported by previous observations that Smo-ΔCRD has a decreased sensitivity to Shh in vitro and in vivo (Aanstad et al., 2009; Myers et al., 2013; Nachtergaele et al., 2013). The regulation of Smo facilitated by Ptch1/2-mediated inhibition likely targets the heptahelical domain of Smo, a notion that has previously been suggested based on the action of many small molecule Smo inhibitors, such as Cyclopamine (J. K. Chen et al., 2002) and vismodegib (Byrne et al., 2016). This indicates that the mechanism by which Smo is inhibited and activated are not the same, consistent with the findings that: 1) Ptch1 can inhibit forms of Smo lacking the CRD (Myers et al., 2013), and 2) the binding of oxysterols and cholesterol to the CRD can activate Smo (Huang et al., 2016; Nachtergaele et al., 2013; 2012; Nedelcu et al., 2013).
Shh auto-processing is not required for Smo activation
Our results demonstrate that a wide variety of Shh mutants, several previously characterized as “dead”, have the ability to activate Smo cell-autonomously when expressed in cells lacking the canonical receptors in vitro and in wild-type cells in vivo. Shh-C199A, which remains unprocessed as a full length precursor poorly signals non-cell autonomously (Pettigrew et al., 2014; Roelink et al., 1995), but can induce the Hh response cell-autonomously consistent with earlier observations (Tokhunts et al., 2010). Several Shh mutations cause persistence of the pro-proteins (C103R, A111Y, I112F, N116L, W118G, and C184Y) and these mutants can activate Hh response cell-autonomously better than Shh when expressed at low levels. These results indicate that one important function of the unusual processing and modification of Shh is to prevent activation of the Hh response in cells that express the ligand, thus reinforcing the predominant non-cell autonomy that characterizes Shh signaling (García-Zaragoza et al., 2012; Shaw et al., 2009; Yauch et al., 2008).
The Zn2+ coordination site is involved in the binding to Hhip (Bosanac et al., 2009) 5E1 (Maun et al., 2010) and possibly Ptch1 (Gong et al., 2018). The resemblance to the active site of Zn2+ peptidases was recognized early (Hall et al., 1995), and this site was thought be not catalytically active, but instead was proposed to mediate binding to Ptch1 (Fuse et al., 1999). Although widely accepted, this observation is not supported by a structure of Shh bound to Ptch1 which leaves the putative catalytic domain unobscured by Ptch1 (Qi et al., 2018). The Zn2+ coordination domain is required for autoproteolytic processing, consistent with the notion that the Zn2+ triad is catalytically active. At low expression levels the Zn2+ triad mutants are poor inducers of the Hh response at best. However, several other mutants outside the triad prevent autoproteolytic processing, and gain the ability to induce the Hh response cell-autonomously. This indicates that the Zn2+ coordination domain, even in the pro-protein, appears to be involved in the activation of the Hh response in cells lacking Ptch1/2, and might thus be involved in the Ptch1/2-independent activation of Smo. We think this is part of the explanation why Shh-H183A is inactive in vivo and might indicate that the Shh-mediated activation of Smo involves proteolytic cleavage of an unknown target.
The requirement for correct Zn2+ coordination must act in concert with the proposed proteolysis of the pre-protein via the C-terminal domain. This mechanism requires C199 to allow a thiol-ester intermediate to be resolved by cholesterol (Porter et al., 1996). Our observation that C199S undergoes some processing (Figure 1B, 5E) cannot be explained by the Beachy model but is consistent with a function of Shh a peptidase that auto-processes via a mechanism that resembles that of bacterial proteases (Wetmore et al., 1994).
Shh mutants found in tumors and in individuals with holoprosencephaly
It is striking that several mutations found in SHH-induced Basal Cell Carcinomas (BCCs) (Couvé-Privat et al., 2004) (SHH-D147N and SHH-R155C) are defective in auto-processing (Figure 5 and not shown). It is unlikely that these mutations are inhibitory to Hh signaling and consistent with the idea that such mutants are gain-of-function as measured by the cell-autonomous activation of the Hh response. SHH Point mutations curated in the COSMIC database tend to be more prevalent in the SHH N-domain, but are present in the SHH C-domain, in particular right after the autoproteolytic cleavage site. Asymmetric distribution indicates that at least some of the mutations are ‘drivers’ in tumor formation. Several of the N-domain mutations involve residues that activate the cell autonomous response when mutated (H134Q and C183Y, human numbering). Several point mutations in gall bladder tumors are found in SHHC (Dixit et al., 2017), and at least one of these (Shh-K213E) fails to process. These findings are consistent with a model in which SHH mutations permit cell-autonomous activation of the Hh response, thereby contributing to tumor progression.
In humans, heterozygous SHH mutants causing single amino acid substitutions can cause HPE (Hehr et al., 2010), although mice heterozygous for a null Shh allele are normal. It is thus possible that some of these HPE-causing point mutations affect the function of the wild type SHH as dominant-negatives or are gain-of-function mutations. The analysis of point mutations that cause HPE (Roessler et al., 2009) reveal an interesting pattern. Residues involved the Zn2+ coordination are targets for point mutations in HPE (H140, and D147, human numbering), but also heterozygous mutations in the C-terminal domain of SHH can cause HPE (Hehr et al., 2010). Mutations in the C-terminal domain likely interfere with auto-processing of full length Shh and thus, like several mutations in the SHHN domain result in accumulation of the pro-protein. It is, therefore, possible that unprocessed SHH functions as a dominant-negative form of Shh as proposed by Singh et al. (Singh et al., 2009). Alternatively, a cell-autonomous activation of the Hh response (a gain-of-function mutation) in cells heterozygous for this mutation could contribute to HPE. We think the latter explanation is plausible given the gain-of-function phenotype we find associated with Shh mutants that fail to process.
Model
Based on our results in cells lacking Ptch1/2 and those of others, we propose the following model to account for the non-cell autonomy of Shh signaling (Figure 6 D). Although the Shh pro-protein has the ability to activate the Hh response, this is prevented by the processing into the inactive ShhNp form. This form is acted upon by Disp1, Scube2, and metalloproteases to be released in an active form (Jakobs et al., 2014) that signals non-cell autonomously. Mutations that prevent efficient processing of the Shh pro-protein or prevent the addition of the cholesterol moiety allow cell autonomous activation of the Hh response. The activation of this response does not require, but likely is antagonized by Ptch1/2, It does, however, require the CRD of Smo, perhaps indicating that Smo is a receptor for Shh, a role for which becomes apparent in the absence of Ptch1/2 function.
Materials and Methods
Materials:
Vismodegib was a gift from Dr. Fred de Sauvage (Genentech). SAG was from EMD Biochemicals. Recombinant ShhN protein was from R&D Systems. Cell Tracker Green CMFDA was from Invitrogen.
Electroporations:
Hamburger-Hamilton (HH) stage 10 Gallus gallus embryos were electroporated caudally in the developing neural tube using standard procedures (Meyer and Roelink, 2003). Embryos were incubated for another 48 h following electroporation to about HH stage 20, dissected, fixed in 4% PFA, mounted in Tissue-Tek OCT Compound (Sakura) and sectioned.
Immunofluorescence:
Antibodies for mouse Pax7 (1:10), Mnr2 (1:100), Nkx2.2 (1:10), Shh (5E1, 1:20) were from the Developmental Studies Hybridoma Bank. The Rabbit α-GFP (1:1000) antibody was from Invitrogen, and the Goat α-hOlig2 (1:100) antibody was from R&D Systems. The mouse α-acetylated tubulin (1:200) was from Sigma Aldrich. Alexa488 and Alexa568 secondary antibodies (1:1000) were from Invitrogen. Nuclei were stained with DAPI (Invitrogen).
DNA Constructs:
The Gli-Luciferase reporter and the Renilla control were gifts from Dr. H. Sasaki (Sasaki et al., 1997). Ptch1 was a gift from Dr. Scott (Stanford University, CA, USA). Ptch1-Δloop2 was a gift from Dr. Thomas Jessell (Columbia University, NY, USA). Ptch1 channel mutants were previously described (Alfaro et al., 2014). SmoΔCRD was a gift from J. Reiter (Aanstad et. al., 2009). SmoM2 was from Genentech (F. de Sauvage). The following mutations were created using Quikchange mutagenesis (Stratagene): Shh-E90A, Shh-H183A, Shh-C199*/E90A, Shh-C199*/H183A, Smo-ΔCRD-CLD, Smo-L112D/W113Y. Dhh and Ihh were gifts from Charles Emerson Jr. (University of Massachusetts Medical School, MA, USA). The N-terminally truncated forms were made by site directed mutagenesis of C199 (Dhh) and C203 (Ihh) to a stop codon. Shh-C199A was previously described (Roelink et al., 1995). Gli3PHS was previously described (Meyer et. al., 2003).
Cell Culture:
Ptch1LacZ/LacZ;Ptch2−/−, Ptch1LacZ/LacZ;Ptch2−/−;Smo−/−, and Ptch1−/−;Disp1−/−;Shh−/− fibroblasts were obtained by plating mESCs at a density of 8.0×105 cells in 6-well plates and transfected with the large T antigen from the SV40 virus (Gökhan et al., 1998) in ES medium. Cells were then switched to DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) without LIF. Ptch1+/LacZ and Ptch1LacZ/LacZ fibroblasts (gifts from Dr. Scott) were cultured in DMEM supplemented with 10% FBS (Invitrogen) and maintained under standard conditions. Identity of these lines was confirmed by the presence of the LacZ recombination in the Ptch1 locus, the presence of 40 chromosomes per cell, and mouse-specific DNA sequences of the edited genes. Cells were routinely tested for Mycoplasma by Hoechst stain, and grown in the presence of tetracycline and gentamycin at regular intervals. Cultures with visible extra-nuclear staining, likely infected with Mycoplasma, were discarded. None of the cell lines used in this study is listed in the Database of Cross-Contaminated or Misidentified Cell Lines. Fibroblast-like lines derived from the mESCs were re-sequenced at the edited loci to confirm their identity.
Transfection:
Cells were transiently transfected for 24h at 80–90% confluency using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
Modified Boyden Chamber Migration Assay:
Cell migration assays were performed as previously described (Bijlsma et. al. 2007). Cells were labeled with 10 μM CellTracker Green (Invitrogen) in DMEM for one hour. The well compartments were set up with the specified chemoattractant (ShhN (R&D Systems) .75 μg/ml, about 30nM, (re-suspended in 0.1%BSA in PBS), 10% FCS, or no attractant (plus 0.1%BSA in PBS) and pre-warmed at 37°C. Cells were then detached with 5mM EDTA and re-suspended in DMEM without phenol red and supplemented with 50mM HEPES. Cells were transferred into FluoroBlok Transwell inserts (BD Falcon) at 5.0×104 cells per insert. GFP-spectrum fluorescence in the bottom compartment was measured every 2 min for 99 cycles (approximately 3 hours), after which background fluorescence (medium without cells) and a no-attractant control was subtracted from each time point. Starting points of migration were set to 0.
Gli-Luciferase Assay:
Fibroblasts were plated at a density of 3×104 in 24 well plates and transfected with Gli-Luciferase, CMV-Renilla (control plasmid), and specified plasmids 24 hours after plating. Cells were grown to confluency and then switched to low serum medium (0.5% FBS) alone or with specified concentrations of Vismodegib. After 24 hours, cells were lysed and the luciferase and renilla activity in lysates was measured using the Dual Luciferase Reporter Assay System (Promega). Raw Luciferase values were normalized against Renilla values for each biological replicate to control against variation in transfection efficiency. Individual luciferase/renilla values were then normalized against the mock control for each experiment.
LacZ Assay:
Fibroblasts were plated at a density of 3×104 in 24 well plates and transfected with plasmids 24 hours after plating. Fibroblasts were grown to confluency and then switched to a low serum medium (0.5% FBS) alone or with specified concentrations of Vismodegib or SAG. After 24 hours, cells were lysed and lysates were analyzed using the Galacto-Light™ chemiluminescence kit (Applied Biosciences) for level of LacZ expression. Raw chemiluminescence values were normalized against total protein for each biological replicate. Protein concentration was determined with a Bradford assay using the Bio-Rad Protein Assay Dye Reagent.
Western Blots:
Ptch1LacZ/LacZ;Ptch2−/− cells were transfected with Shh mutants as indicated. 48 hours after transfection, Ptch1LacZ/LacZ;Ptch2−/− cells were rinsed with PBS and lysed with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 1% Igepal, 0.5% Sodium Deoxycholate, and protease inhibitors) for 30 min on ice. Protein lysate was cleared by centrifugation at 13,000g for 30 min at 4 °C. 20 μg of each sample was run on a 12% SDS-PAGE gel and transferred to a 0.45 μ nitrocellulose membrane. Membranes were blocked with 5% milk in Tris-buffered saline with 0.1% Tween-20 (TBS-T) and incubated with a rabbit polyclonal anti-Shh antibody (H160; Santa Cruz Biotechnology) at 1:250. A goat anti-rabbit HRP-conjugated secondary antibody (BioRad) was used at 1:10000.
Highlights:
Cells that lack Ptch1 and Ptch2 upregulate the Hh response after transfection with ShhN
Ptch1/2 null cells are capable of Shh chemotaxis
Ptch1/2-independent responses to Shh require the N-terminal extracellular CRD of Smo
Zn2+ coordination domain mutations prevent auto-processing of the Shh pro-protein
SHH mutations found in holoprosencephaly prevent auto-processing of the pro-protein
The Shh pro-protein retains the Ptch1/2-independent activity to activate the Hh response
Shh mutants unable to auto-process or bind Zn2+ are active in vivo
Acknowledgements:
This work was supported by NIH grant 1R01GM117090 to HR. Vismodegib was a gift from Dr. de Sauvage (Genentech), Dhh and Ihh were a gift from Dr. Charles P. Emerson III (U. Mass. Med. School). Ptch1LacZ/LacZ fibroblasts were a gift of Dr. M. Scott (Carnegie). Dr. A. Alfaro cloned the Shh binding mutants (E90A, H183A) and made the initial observation that they activate the Hh pathway in vivo. We thank Dr. M. Barro for her help with Western blotting, Dr. M. F. Bijlsma (AMC Amsterdam) for his help with the migration assays, and Dr. B. Roberts (Allen Institute for Cell Science) for his comments and advice. We also thank Fatma Ozguc for help with genome editing and Michelle Boisvert for her help with generating Shh mutants, and Carina Jägers and Wei Guo for commenting on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aanstad P, Santos N, Corbit KC, Scherz PJ, Trinh le A, Salvenmoser W, Huisken J, Reiter JF, Stainier DY, 2009. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol 19, 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro AC, Roberts B, Kwong L, Bijlsma MF, Roelink H, 2014. Ptch2 mediates the Shh response in Ptch1−/− cells. Development (Cambridge, England) 141, 3331–3339. doi: 10.1242/dev.110056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP, 2011. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Developmental cell 20, 775–787. doi: 10.1016/j.devcel.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot E, Loulier K, Nguyen-Ba-Charvet KT, Gadeau AP, Ruat M, Traiffort E, 2008. Chemoattractive activity of sonic hedgehog in the adult subventricular zone modulates the number of neural precursors reaching the olfactory bulb. Stem cells (Dayton, Ohio) 26, 2311–2320. [DOI] [PubMed] [Google Scholar]
- Bale AE, 2002. Hedgehog signaling and human disease. Annu Rev Genomics Hum Genet 3, 47–65. doi: 10.1146/annurev.genom.3.022502.103031 [DOI] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM, 2004. Tissue repair and stem cell renewal in carcinogenesis. Nature 432, 324–331. doi: 10.1038/nature03100 [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R, 1996. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382, 225–230. [DOI] [PubMed] [Google Scholar]
- Bijlsma MF, Borensztajn KS, Roelink H, Peppelenbosch MP, Spek CA, 2007. Sonic hedgehog induces transcription-independent cytoskeletal rearrangement and migration regulated by arachidonate metabolites. Cellular signalling 19, 2596–2604. [DOI] [PubMed] [Google Scholar]
- Bijlsma MF, Damhofer H, Roelink H, 2012. Hedgehog-stimulated chemotaxis is mediated by smoothened located outside the primary cilium. Sci Signal 5, ra60. doi: 10.1126/scisignal.2002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma MF, Peppelenbosch MP, Spek CA, Roelink H, 2008. Leukotriene synthesis is required for hedgehog-dependent neurite projection in neuralized embryoid bodies but not for motor neuron differentiation. Stem cells (Dayton, Ohio) 26, 1138–1145. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Maun HR, Scales SJ, Wen X, Lingel A, Bazan JF, de Sauvage FJ, Hymowitz SG, Lazarus RA, 2009. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nature structural & molecular biology 16, 691–697. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Chen Y, Jessell TM, Struhl G, 2001. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Molecular cell 7, 1279–1291. [DOI] [PubMed] [Google Scholar]
- Buglino JA, Resh MD, 2008. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. The Journal of biological chemistry 283, 22076–22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MSP, Newstead S, Rohatgi R, Siebold C, 2016. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522. doi: 10.1038/nature18934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M, 2003. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell 113, 11–23. [DOI] [PubMed] [Google Scholar]
- Chen HC, 2005. Boyden chamber assay. Methods in molecular biology (Clifton, N.J 294, 15–22. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA, 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & development 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla P, Xiao L, Kazanietz MG, Riobo NA, 2010. Hedgehog proteins activate proangiogenic responses in endothelial cells through non-canonical signaling pathways. Cell cycle (Georgetown, Tex 9, 570–579. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Scott MP, 2006. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proceedings of the National Academy of Sciences of the United States of America 103, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvé-Privat S, Le Bret M, Traiffort E, Queille S, Coulombe J, Bouadjar B, Avril MF, Ruat M, Sarasin A, Daya-Grosjean L, 2004. Functional analysis of novel sonic hedgehog gene mutations identified in basal cell carcinomas from xeroderma pigmentosum patients. Cancer research 64, 3559–3565. doi: 10.1158/0008-5472.CAN-03-4040 [DOI] [PubMed] [Google Scholar]
- Dixit R, Pandey M, Tripathi SK, Dwivedi AND, Shukla VK, 2017. Comparative Analysis of Mutational Profile of Sonic hedgehog Gene in Gallbladder Cancer. Dig. Dis. Sci 62, 708–714. doi: 10.1007/s10620-016-4438-1 [DOI] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA, 1999. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. U.S.A. 96, 10992–10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Zaragoza E, Pérez-Tavarez R, Ballester A, Lafarga V, Jiménez-Reinoso A, Ramírez A, Murillas R, Gallego MI, 2012. Intraepithelial paracrine Hedgehog signaling induces the expansion of ciliated cells that express diverse progenitor cell markers in the basal epithelium of the mouse mammary gland. Developmental biology 372, 28–44. doi: 10.1016/j.ydbio.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV, 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet 11, 331–344. doi: 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, Yan N, 2018. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science (New York, N.Y 112, eaas8935. doi: 10.1126/science.aas8935 [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP, 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science (New York, N.Y 277, 1109–1113. [DOI] [PubMed] [Google Scholar]
- Gökhan S, Song Q, Mehler MF, 1998. Generation and regulation of developing immortalized neural cell lines. Methods 16, 345–358. doi: 10.1006/meth.1998.0689 [DOI] [PubMed] [Google Scholar]
- Hall TM, Porter JA, Beachy PA, Leahy DJ, 1995. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 378, 212–216. doi: 10.1038/378212a0 [DOI] [PubMed] [Google Scholar]
- Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ, 1997. Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell 91, 85–97. [DOI] [PubMed] [Google Scholar]
- Hehr U, Pineda-Alvarez DE, Uyanik G, Hu P, Zhou N, Hehr A, Schell-Apacik C, Altus C, Daumer-Haas C, Meiner A, Steuernagel P, Roessler E, Winkler J, Muenke M, 2010. Heterozygous mutations in SIX3 and SHH are associated with schizencephaly and further expand the clinical spectrum of holoprosencephaly. Hum. Genet 127, 555–561. doi: 10.1007/s00439-010-0797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelstein DS, Cajigas I, Bi C, Clark BS, Van Der Voort G, Kohtz JD, 2017. SHH E176/E177-Zn(2+) conformation is required for signaling at endogenous sites. Developmental biology 424, 221–235. doi: 10.1016/j.ydbio.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, Salic A, 2016. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 166, 1176–1187.e14. doi: 10.1016/j.cell.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, Liu J, Kruse AC, Salic A, 2018. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell. doi: 10.1016/j.cell.2018.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi L, Lévesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F, 2011. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Developmental cell 20, 788–801. doi: 10.1016/j.devcel.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs P, Exner S, Schürmann S, Pickhinke U, Bandari S, Ortmann C, Kupich S, Schulz P, Hansen U, Seidler DG, Grobe K, 2014. Scube2 enhances proteolytic Shh processing from the surface of Shh-producing cells. Journal of Cell Science 127, 1726–1737. doi: 10.1242/jcs.137695 [DOI] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC, 2012. Structural basis of Wnt recognition by Frizzled. Science (New York, N.Y 337, 59–64. doi: 10.1126/science.1222879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen K, 2004. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther 103, 21–80. [DOI] [PubMed] [Google Scholar]
- Kwong L, Bijlsma MF, Roelink H, 2014. Shh-mediated degradation of Hhip allows cell autonomous and non-cell autonomous Shh signalling. Nat Commun 5, 4849. doi: 10.1038/ncomms5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von KD, Porter JA, Sun BI, Beachy PA, 1994. Autoproteolysis in hedgehog protein biogenesis. Science (New York, N.Y 266, 1528–1537. [DOI] [PubMed] [Google Scholar]
- Lee Y, Miller HL, Russell HR, Boyd K, Curran T, McKinnon PJ, 2006. Patched2 modulates tumorigenesis in patched1 heterozygous mice. Cancer research 66, 6964–6971. doi: 10.1158/0008-5472.CAN-06-0505 [DOI] [PubMed] [Google Scholar]
- Lipinski RJ, Bijlsma MF, Gipp JJ, Podhaizer DJ, Bushman W, 2008. Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC cell biology 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maun HR, Wen X, Lingel A, de Sauvage FJ, Lazarus RA, Scales SJ, Hymowitz SG, 2010. Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. The Journal of biological chemistry 285, 26570–26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer NP, Roelink H, 2003. The amino-terminal region of Gli3 antagonizes the Shh response and acts in dorsoventral fate specification in the developing spinal cord. Developmental biology 257, 343–355. [DOI] [PubMed] [Google Scholar]
- Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, Bazan JF, Beachy PA, 2013. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Developmental cell 26, 346–357. doi: 10.1016/j.devcel.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, Rohatgi R, 2012. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nature chemical biology 8, 211–220. doi: 10.1038/nchembio.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, Whalen DM, Mydock LK, Zhao Z, Malinauskas T, Krishnan K, Ingham PW, Covey DF, Siebold C, Rohatgi R, 2013. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife 2, e01340. doi: 10.7554/eLife.01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu D, Liu J, Xu Y, Jao C, Salic A, 2013. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nature chemical biology. doi: 10.1038/nchembio.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odent S, Atti Bitach T, Blayau M, Mathieu M, Aug J, Delezo de AL, Gall JY, Le Marec B, Munnich A, David V, Vekemans M, 1999. Expression of the Sonic hedgehog (SHH ) gene during early human development and phenotypic expression of new mutations causing holoprosencephaly. Hum. Mol. Genet 8, 1683–1689. [DOI] [PubMed] [Google Scholar]
- Ohlig S, Farshi P, Pickhinke U, van den Boom J, Höing S, Jakuschev S, Hoffmann D, Dreier R, Schöler HR, Dierker T, Bordych C, Grobe K, 2011. Sonic hedgehog shedding results in functional activation of the solubilized protein. Developmental cell 20, 764–774. doi: 10.1016/j.devcel.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Ohlig S, Pickhinke U, Sirko S, Bandari S, Hoffmann D, Dreier R, Farshi P, Götz M, Grobe K, 2012. An emerging role of Sonic hedgehog shedding as a modulator of heparan sulfate interactions. The Journal of biological chemistry 287, 43708–43719. doi: 10.1074/jbc.M112.356667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew CA, Asp E, Emerson CP, 2014. A new role for Hedgehogs in juxtacrine signaling. Mechanisms of development 131, 137–149. doi: 10.1016/j.mod.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park WJ, Kessler,von DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA, 1996. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34. [DOI] [PubMed] [Google Scholar]
- Qi X, Schmiege P, Coutavas E, Wang J, Li X, 2018. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 15, 3059. doi: 10.1038/s41586-018-0308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, Casillas C, Alfaro AC, Jägers C, Roelink H, 2016. Patched1 and Patched2 inhibit Smoothened non-cell autonomously. Elife 5, e17634. doi: 10.7554/eLife.17634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelink H, 2018. Sonic Hedgehog Is a Member of the Hh/DD-Peptidase Family That Spans the Eukaryotic and Bacterial Domains of Life. Journal of Developmental Biology 6, 12. doi: 10.3390/jdb6020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, Beachy PA, Jessell TM, 1995. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455. [DOI] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Vélez JI, Solomon BD, Pineda-Alvarez DE, Lacbawan F, Zhou N, Ouspenskaia M, Paulussen A, Smeets HJ, Hehr U, Bendavid C, Bale S, Odent S, David V, Muenke M, 2009. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum. Mutat 30, E921–35. doi: 10.1002/humu.21090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP, 2007. Patched1 regulates hedgehog signaling at the primary cilium. Science (New York, N.Y 317, 372–376. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, 1998. Combinatorial Gli gene function in floor plate and neuronal inductions by Sonic hedgehog. Development (Cambridge, England) 125, 2203–2212. [DOI] [PubMed] [Google Scholar]
- Sagai T, Amano T, Tamura M, Mizushina Y, Sumiyama K, Shiroishi T, 2009. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development (Cambridge, England) 136, 1665–1674. doi: 10.1242/dev.032714 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H, 1997. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development (Cambridge, England) 124, 1313–1322. [DOI] [PubMed] [Google Scholar]
- Sharpe HJ, Wang W, Hannoush RN, de Sauvage FJ, 2015. Regulation of the oncoprotein Smoothened by small molecules. Nature chemical biology 11, 246–255. doi: 10.1038/nchembio.1776 [DOI] [PubMed] [Google Scholar]
- Shaw A, Gipp J, Bushman W, 2009. The Sonic Hedgehog pathway stimulates prostate tumor growth by paracrine signaling and recapitulates embryonic gene expression in tumor myofibroblasts. Oncogene 28, 4480–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Tokhunts R, Baubet V, Goetz JA, Huang ZJ, Schilling NS, Black KE, MacKenzie TA, Dahmane N, Robbins DJ, 2009. Sonic hedgehog mutations identified in holoprosencephaly patients can act in a dominant negative manner. Hum. Genet 125, 95–103. doi: 10.1007/s00439-008-0599-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J, 2005. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes & development 19, 626– 641. doi: 10.1101/gad.325905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA, 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897. [DOI] [PubMed] [Google Scholar]
- Tokhunts R, Singh S, Chu T, D’Angelo G, Baubet V, Goetz JA, Huang Z, Yuan Z, Ascano M, Zavros Y, Thérond PP, Kunes S, Dahmane N, Robbins DJ, 2010. The full-length unprocessed hedgehog protein is an active signaling molecule. The Journal of biological chemistry 285, 2562–2568. doi: 10.1074/jbc.M109.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traiffort E, Dubourg C, Faure H, Rognan D, Odent S, Durou M-R, David V, Ruat M, 2004. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J. Biol. Chem 279, 42889–42897. doi: 10.1074/jbc.M405161200 [DOI] [PubMed] [Google Scholar]
- Tukachinsky H, Petrov K, Watanabe M, Salic A, 2016. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proceedings of the National Academy of Sciences of the United States of America 113, E5866–E5875. doi: 10.1073/pnas.1606719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore DR, Wong SL, Roche RS, 1994. The efficiency of processing and secretion of the thermolysin-like neutral protease from Bacillus cereus does not require the whole prosequence, but does depend on the nature of the amino acid sequence in the region of the cleavage site. Mol. Microbiol 12, 747–759. [DOI] [PubMed] [Google Scholar]
- Xiao X, Tang J-J, Peng C, Wang Y, Fu L, Qiu Z-P, Xiong Y, Yang L-F, Cui H-W, He X-L, Yin L, Qi W, Wong CCL, Zhao Y, Li B-L, Qiu W-W, Song B-L, 2017. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Molecular cell 66, 154–162.e10. doi: 10.1016/j.molcel.2017.02.015 [DOI] [PubMed] [Google Scholar]
- Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ, 2008. A paracrine requirement for hedgehog signalling in cancer. Nature 455, 406–410. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenthal A, de Sauvage FJ, Shivdasani RA, 2001. Downregulation of Hedgehog signaling is required for organogenesis of the small intestine in Xenopus. Developmental biology 229, 188–202. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP, 2001. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell 105, 781–792. [PubMed] [Google Scholar]