Abstract
Background
A receptor tyrosine kinase for ephrin ligands, EPHB2, is expressed in normal colorectal tissues and colorectal cancers (CRCs). The aim of this study was to investigate EPHB2 expression over CRC progression and determine its prognostic significance in CRC.
Methods
To measure EPHB2 mRNA and protein expression, real-time polymerase chain reaction and immunohistochemistry were performed in 32 fresh-frozen and 567 formalin-fixed paraffin-embedded CRC samples, respectively. We further investigated clinicopathological features and overall and recurrence-free survival according to EPHB2 protein expression.
Results
The EPHB2 level was upregulated in CRC samples compared to non-cancerous tissue in most samples and showed a strong positive correlation with AXIN2. Notably, CD44 had a positive association with both mRNA and protein levels of EPHB2. Immunohistochemical analysis revealed no difference in EPHB2 expression between adenoma and carcinoma areas. Although EPHB2 expression was slightly lower in invasive fronts compared to surface area (p < .05), there was no difference between superficial and metastatic areas. EPHB2 positivity was associated with lymphatic (p < .001) and venous (p = .001) invasion, TNM stage (p < .001), and microsatellite instability (p = .036). Kaplan–Meier analysis demonstrated that CRC patients with EPHB2 positivity showed better clinical outcomes in both overall (p = .049) and recurrence-free survival (p = .015). However, multivariate analysis failed to show that EPHB2 is an independent prognostic marker in CRCs (hazard ratio, 0.692; p = .692).
Conclusions
Our results suggest that EPHB2 is overexpressed in a subset of CRCs and is a significant prognostic marker.
Keywords: EPHB2, Colorectal neoplasms, Immunohistochemistry, Prognosis
The receptor tyrosine kinases (RTKs) are transmembrane glycoproteins that are important regulators of cell growth, differentiation, and migration. The 16 vertebrate EPH receptors form the largest subgroup of RTKs and are subdivided into EPHA and EPHB kinases depending on homology and binding specificity to their cell-bound ligands called ephrins [1]. In cooperation with membrane-bound ephrins, EPHs organize tissue patterning, cell positioning, and organ patterning and regulate cell growth during normal and tumor development [2-4]. Studies on EPHs and ephrins initially focused on the patterning of embryonic tissue, but have since also shown important roles in adult tissue and organ maintenance, regeneration, and a variety of other processes, including synaptic plasticity and response to nerve damage, bone remodeling, stem cell location and proliferation, and cancer development [5].
Numerous studies have implicated a role for EPH–ephrin signaling in cancer progression. In recent years, abnormal expression of EPH genes has been reported in a wide range of human malignancies such as neuroblastoma [6] and carcinomas of breast [7], lung [8], gastric [9], prostate [10], ovary [11], colon [12], and skin [13]. Many researchers have suggested a role for EPHs and ephrins in colorectal cancers (CRCs), in particular EPHB2 is the most expressed EPH receptor in the normal intestine and is mainly produced in epithelial cells [14]. Analysis of EPHB2 and EPHB3 in colon tumor mouse models demonstrated that upregulated expression resulted in dysregulated epithelial cell migration in the crypt villus junction, indicating a role for EPH receptors in colon cancer [15]. In this study, we investigated the expression of EPHB2 in human CRC samples and assessed its expression profile during CRC progression. In addition, we examined the prognostic significance of EPHB2 positivity in a large cohort of human CRC patients.
MATERIALS AND METHODS
Subjects
CRCs were collected from 567 patients who underwent surgical resection at Seoul National University Hospital (SNUH) between 2004 and 2006. Clinicopathological data of patient age, sex, tumor location, size, histological subtype, differentiation, presence of lympho-vascular invasion, American Joint Committee on Cancer/International Union against Cancer (AJCC/UICC) cancer stage (seventh edition), time of death, tumor recurrence, and follow-up time were obtained by reviewing the clinical and pathological reports. Fifty-six CRC samples were obtained from patients at Jeju National University Hospital (JNUH); 22 cases were CRCs arising from preexisting adenomas, and 34 cases were conventional CRCs with lymph node metastasis. The histopathologic features were evaluated by two gastrointestinal pathologists (J.M.B and G.H.K) for CRCs from SNUH and by one pathologist (B.G.J) for CRCs from JNUH. In addition, 32 paired, fresh-frozen CRC tissues and matched normal tissues were obtained from the Jeju National University Hospital Biobank, a member of the National Biobank of Korea, for which informed consent was obtained from all subjects. All procedures were in accordance with the Helsinki Declaration of 1964 and later versions, and this study was approved by the Institutional Review Boards of SNUH (IRB No. 1502-029-647) and JNUH (IRB No. 2017-04-001).
Tissue microarray construction
Thirteen tissue microarrays (TMAs) containing 642 CRCs from SNUH were generated as previously described [16]. In brief, through histologic examination, the representative cancer areas containing more than 70% of cells were marked in each case. For each CRC, core tissue biopsies (2 mm in diameter) were obtained from individual formalin-fixed paraffin-embedded samples (donor blocks) and arranged in a new recipient paraffin block (tissue array block) using a trephine apparatus (SuperBioChips Laboratories, Seoul, Korea). With CRCs from JNUH, two TMAs including 22 pairs of adenoma and carcinoma areas and five TMAs including 34 ulcerofungating CRCs with lymph node metastasis were constructed with 4 mm cores. For ulcerofungating CRCs, superficial and invasive portions, and metastatic cancers in the lymph node were included.
Immunohistochemistry
Immunohistochemistry was performed on 4-μm TMA sections using a Ventana BenchMark XT Staining systems (Leica Microsystems, Wetzlar, Germany) according to the manufacturer’s instructions. The primary antibodies used were anti-EPHB2 (1:700, R&D Systems, Minneapolis, MN, USA), anti-CD44 (1:100, Novocastra Laboratories Ltd., Newcastle upon Tyne, UK), and anti-β-catenin (1:800, 17C2, Novocastra Laboratories). The expression of EPHB2 and CD44 was determined by examining the tumor cell membrane. For each tumor, the intensity and percentage of tumor cells expressing EPHB2 (n = 567) or CD44 (n = 87) were evaluated. Histoscores (H-scores) were calculated by multiplying the intensity score (0, negative; 1, weak; 2, moderate; and 3, strong) and percentage of positive tumor cells (range, 0 to 100), ranging from 0 to 300. For statistical analyses for EPHB2, we used a cutoff of 40 on the basis of the distribution of the H-scores (median value, 40). CRCs with H-score of 40 or lower were classified as negative, while cases with H-score higher than 40 were classified as positive. β-Catenin staining was considered positive when more than 10% of tumor cell nuclei were strongly stained.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from 32 paired fresh-frozen CRCs and matched non-cancerous colon tissues using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). RNA (1 μg) was subjected to reverse-transcription with oligo-dT primers and the GoScript reverse transcription system (Promega, Madison, WI, USA). Complementary DNA was used to conduct real-time polymerase chain reaction (PCR) with Premix EX Taq (Takara Bio, Shiga, Japan) following the manufacturer’s instructions. The cycling conditions were as follows: initial denaturation for 30 seconds at 95˚C, followed by 40 cycles of 95˚C for 1 second and 60˚C for 5 seconds in a StepOne Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The TaqMan expression assays were performed for the following genes: Hs00362096_m1 (EPHB2), Hs00610344_m1 (AXIN2), Hs02379687_s1 (CD24), Hs01075684_m1 (CD44), Hs01009250_m1 (PROM1/CD133), Hs00233455_m1 (ALCAM/CD166), and Hs02786624_g1 (GAPDH). GAPDH served as the endogenous control.
BRAF and KRAS mutation and microsatellite instability analysis
Thorough microscopic examination, representative cancer areas in each case were marked, microdissected, and then incubated at 55˚C with lysis buffer and proteinase K for 2 days. KRAS codons 12 and 13 and BRAF codon 600 mutations were determined by PCR-restriction fragment length polymorphism and direct sequencing techniques. Among the 567 CRCs, 29 and five samples were excluded from the KRAS and BRAF mutation analyses, respectively, due to an insufficient DNA amount. The microsatellite instability (MSI) status of each cancer tissue was determined using five National Cancer Institute recommended markers (BAT25, BAT26, D2S123, D5S346, and D17S250). The MSI status of each case was classified into three groups: MSI-high (two or more unstable markers among the five markers), MSI-low (one unstable marker), or microsatellite stable (no unstable marker).
CpG island methylator phenotype analysis
CpG island methylator phenotype (CIMP) status was determined by MethyLight assay. Sodium bisulphite modification of genomic DNA samples was conducted for all 567 CRC tissues, as previously described [17]. The methylation statuses of eight CIMP-specific CpG islands (MLH1, NEUROG1, CRABP1, CACNA1G, CDKN2A [p16], IGF2, SOCS1, and RUNX) were quantified. CIMP-high was defined as having five or more hypermethylated markers, and CIMP-low was defined as having four or fewer hypermethylated markers out of eight. CIMP-negative was defined as having no hypermethylated marker.
Statistical analysis
All data were analyzed using SPPSS statistical software ver. 18.0 (SPSS Inc., Chicago, IL, USA) and Prism ver. 5.0 (Graph-Pad Software, San Diego, CA, USA). The correlations between EPHB2 and cancer stem cell markers or CD44 were evaluated by the Spearman correlation test. Between-group comparisons of the real-time PCR data were performed using Student’s t-test. The correlations between EPHB2 positivity and clinicopathological parameters were tested using Pearson’s chi-square test or Fisher exact test. Survival curves were estimated using the Kaplan-Meier method, and log-rank test was used to compare groups. Using Cox proportional hazards model, multivariate analyses were performed to identify independent prognostic factors. A p-value of < .05 was considered statistically significant.
RESULTS
EPHB2 is upregulated in human CRCs
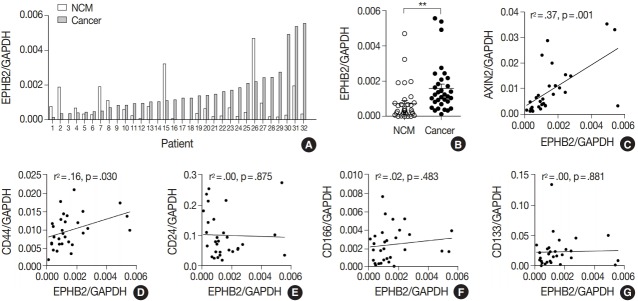
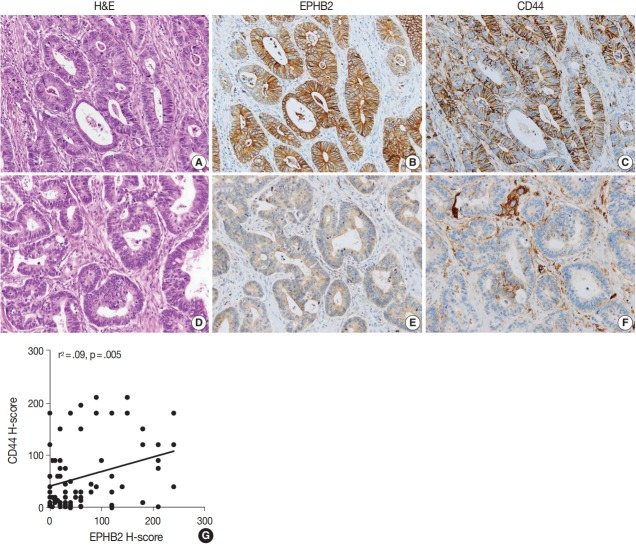
To measure the expression level of EPHB2 in CRCs, we performed real-time PCR with 32 pairs of fresh-frozen human CRC samples and adjacent normal colon tissues. Comparing individual cases, EPHB2 mRNA expression was higher in CRC samples than in non-cancerous tissues in most samples (77%, 25 out of 32 cases) (Fig. 1A), and the mean EPHB2 level was also significantly higher in CRCs than in matched colon tissues (p < .01) (Fig. 1B). EPHB2, a Wnt signaling target gene, showed a strong positive correlation with AXIN2, a direct target of the Wnt pathway (p < .001), confirming enhanced Wnt signaling activity in CRCs (Fig. 1C). As EPHB2-high cancer cells have been suggested as a cancer stem cell population in CRCs [18], we investigated the correlations between EPHB2 expression and expression of other candidate cancer stem cell (CSC) markers CD24, CD44, CD133, and CD166. Notably, we found that only CD44 showed a positive association with EPHB2 (p = .030) (Fig. 1D–G), which was confirmed at the protein level by a positive correlation between EPHB2 and CD44 H-scores in CRCs (n = 87) (Fig. 2).
Fig. 1.
EPHB2 expression in colorectal cancers (CRCs) and matched normal colon tissues. (A, B) Real-time polymerase chain reaction analysis of EPHB2 mRNA level in CRCs and corresponding non-cancerous mucosa (NCM). (C) A correlation between EPHB2 and AXIN2 expression. (D–G) Correlations between EPHB2 and candidate cancer stem cell markers.
Fig. 2.
Representative cases showing high EPHB2 and CD44 expression (A–C) and low EPHB2 and CD44 expression (D–F) in colorectal cancers. (G) Scatter plot with a regression line showing a positive correlation between EPHB2 and CD44 expression. H-scores, Histoscores.
EPHB2 expression during CRC progression
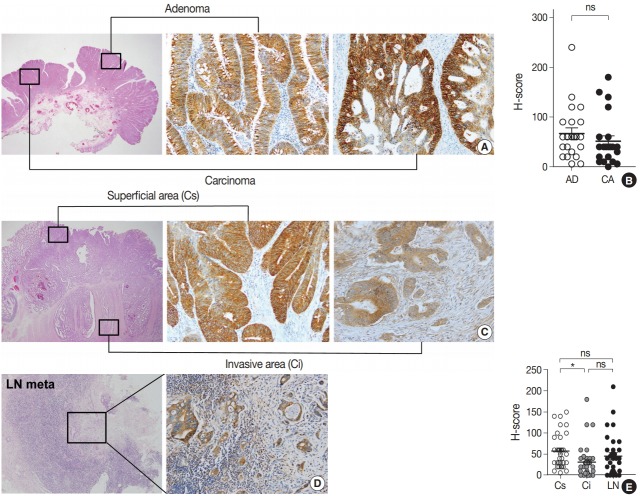
To explore the expression profile of EPHB2 expression during CRC progression, we collected 22 cases of CRCs arising from pre-existing adenomas and measured the H-scores of EPHB2 in adenoma and carcinoma portions (Fig. 3A). However, no difference in H-scores was observed between the adenoma and carcinoma areas (Fig. 3B). Next, we collected 34 ulcerofungating CRCs with lymph node metastasis and examined the H-scores in three spots for each cancer: the superficial fungating area, invasive fronts, and metastatic cancer cells (Fig. 3C, D). EPHB2 expression was slightly lower in invasive fronts compared to surface area (p < .05), but did not differ between superficial and metastatic areas (Fig. 3E).
Fig. 3.
Expression profile of EPHB2 during colorectal cancer (CRC) progression. (A, B) EPHB2 expression in adenoma and carcinoma portions in CRCs arising in pre-existing adenomas. (C–E) EPHB2 expression in three spots of ulcerofungating CRCs; superficial area, invasive fronts, and metastatic cancers. *p<.05. ns, not significant; AD, adenoma; CA, carcinoma; LN, lymph node metastasis; Cs, superficial cancer: Ci, invasive cancer.
Clinicopathological and prognostic significance of EPHB2 expression in CRCs
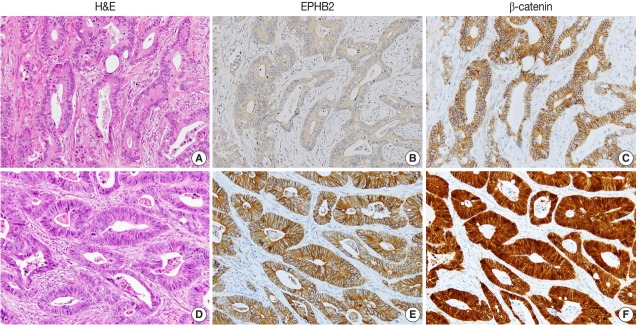
To investigate the prognostic value of EPHB2 in CRCs, immunohistochemical staining was performed on 12 TMAs, and 567 cases were included for the analysis. The clinicopathological relevance of EPHB2 positivity is summarized in Table 1. EPHB2 expression was significantly higher in CRCs from old patients (p = .002). EPHB2 positivity exhibited negative correlations with lymphatic (p < .001) and venous (p = .001) invasion, T category (p < .001), N category (p < .001), M category (p = .013), and tumor stage (p < .001). Nuclear β-catenin staining was observed more frequently in EPHB2-positive CRCs (Table 1, Fig. 4). EPHB2 positivity was higher in CRCs with MSI-high than in those with MSI-negative/low, whereas EPHB2 expression had no correlation with mutation status of BRAF, KRAS, or CIMP (Table 2). Kaplan-Meier analysis demonstrated that CRC patients with EPHB2 positivity showed better clinical outcomes in both overall (p = .048) (Fig. 5A) and recurrence-free survival (p = .015) (Fig. 5B). However, multivariate analysis failed to show EPHB2 as an independent prognostic marker in CRCs (hazard ratio, 0.692; p = .692) (Table 3).
Table 1.
Associations between EPHB2 and clinicopathological characteristics
| Total | EPHB2 |
p-value | ||
|---|---|---|---|---|
| Negative | Positive | |||
| No. of patients | 567 (100) | 259 (46) | 308 (54) | |
| Age (yr) | ||||
| ≥ 60 | 319 (56) | 127 (40) | 192 (60) | .002a |
| < 60 | 248 (44) | 132 (53) | 116 (47) | |
| Sex | ||||
| Male | 343 (61) | 161 (47) | 182 (53) | .491a |
| Female | 224 (39) | 98 (44) | 126 (56) | |
| Location | ||||
| Proximal | 148 (26) | 65 (44) | 83 (56) | .632a |
| Distal | 419 (74) | 194 (46) | 225 (54) | |
| Differentiation | ||||
| WD | 44 (8) | 16 (36) | 28 (64) | .064b |
| MD | 504 (89) | 230 (46) | 274 (54) | |
| PD | 19 (3) | 13 (68) | 6 (62) | |
| Lymphatic invasion | ||||
| Negative | 323 (57) | 110 (34) | 213 (66) | < .001a |
| Positive | 244 (43) | 149 (61) | 95 (39) | |
| Venous invasion | ||||
| Negative | 493 (87) | 212 (43) | 287 (57) | .001a |
| Positive | 74 (13) | 47 (64) | 27 (36) | |
| T category | ||||
| T1 | 20 (3) | 4 (20) | 16 (80) | < .001b |
| T2 | 90 (16) | 25 (28) | 65 (72) | |
| T3 | 408 (72) | 197 (48) | 211 (52) | |
| T4 | 49 (9) | 33 (67) | 16 (33) | |
| N category | ||||
| N0 | 298 (53) | 111 (37) | 187 (63) | < .001b |
| N1 | 151 (27) | 68 (45) | 83 (55) | |
| N2 | 118 (21) | 80 (68) | 38 (32) | |
| M category | ||||
| M0 | 473 (83) | 205 (43) | 268 (57) | .013a |
| M1 | 94 (17) | 54 (57) | 40 (43) | |
| Tumor stagec | ||||
| I | 91 (16) | 24 (26) | 67 (74) | < .001b |
| II | 186 (33) | 76 (41) | 110 (59) | |
| III | 196 (35) | 105 (54) | 91 (46) | |
| IV | 94 (17) | 54 (57) | 40 (43) | |
| β-catenin | ||||
| No nuclear stain | 212 (37) | 110 (52) | 102 (48) | .024a |
| Nuclear stain | 355 (63) | 149 (42) | 206 (58) | |
Values are presented as number (%).
Fisher’s exact test;
Pearson chi-square test;
American Joint Committee on Cancer 7th edition.
Fig. 4.
Representative cases showing negative EPHB2 and nuclear β-catenin expression (A–C) and positive EPHB2 and nuclear β-catenin (D–F) expression in colorectal cancers.
Table 2.
Associations between EPHB2 expression and molecular characteristics
| Total | EPHB2 |
p-value | ||
|---|---|---|---|---|
| Negative | Positive | |||
| No. of patients | 567 (100) | 259 (46) | 308 (54) | |
| CIMP | ||||
| Negative/Low | 528 (93) | 237 (45) | 291 (55) | .163a |
| High | 39 (7) | 22 (56) | 17 (44) | |
| MSI | ||||
| Negative/Low | 519 (91) | 244 (47) | 275 (53) | .036a |
| High | 48 (9) | 15 (31) | 33 (69) | |
| KRAS (n = 538) | ||||
| Wt | 390 (73) | 181 (46) | 209 (54) | .347a |
| Mt | 148 (27) | 62 (42) | 86 (58) | |
| BRAF (n = 562) | ||||
| Wt | 531 (95) | 243 (46) | 288 (54) | .443a |
| Mt | 31(5) | 12 (39) | 19 (61) | |
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; Wt, wild type; Mt, mutation.
Pearson chi-square test.
Fig. 5.
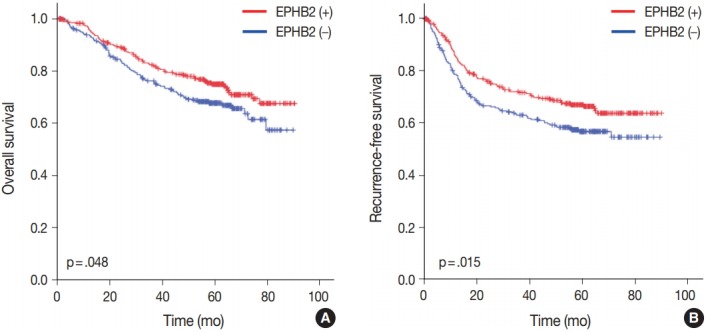
Prognostic significance of EPHB2 expression in overall (A) and recurrence-free (B) survival in colorectal cancers.
Table 3.
The results of multivariate analysis for survival rate in colorectal cancers
| Variable | HR | 95% CI | p-valuea |
|---|---|---|---|
| Age (> 60 yr/< 60 yr) | 1.720 | 1.247–2.374 | .001 |
| Site (distal/proximal) | 0.741 | 0.536–1.025 | .070 |
| Differentiation (poor/moderate/well) | 1.405 | 0.849–2.326 | .186 |
| Lymphatic invasion (positive/negative) | 1.058 | 0.754–1.485 | .742 |
| Venous invasion (positive/negative) | 1.766 | 1.230–2.535 | .742 |
| Tumor stage (IV/III/II/I) | 2.789 | 2.248–3.460 | < .001 |
| EPHB2 (positive/negative) | 0.692 | 0.679–1.293 | .692 |
HR, hazard ratio; CI, confidence interval.
Cox proportional hazard model.
DISCUSSION
A number of studies have reported that a direct correlation is present between loss of EPHB2 and CRC progression [12,19-21], and EPHB2 decline was shown to be attributed to promoter methylation [22]. To verify downregulation of EPHB2 with CRC progression, we attempted to find the difference in EPHB2 expression between adenoma and carcinoma portions and between superficial, invasive fronts, and metastatic cancers in the lymph nodes. However, we only observed a slight reduction in EPHB2 level in invasive fronts compared to superficial tumor cells. On the other hand, EPHB2 expression remained persistent during adenoma-carcinoma transition and lymph node metastasis. Our results suggest that enhanced EPHB2 level in CRCs tends to persist during cancer progression. The discrepancy may be in part explained by the difference in CRC samples and methods, and further studies are required to clarify the alterations in EPHB2 expression during CRC progression and metastasis.
The prognostic significance of EPHB2 expression in CRCs has been controversial. Some reports have shown that EPHB2 overexpression is associated with prolonged survival in CRCs [19,23]. On the contrary, Merlos-Suarez et al. [18] have suggested that EPHB2-high expression identifies CRC stem cells and increased risk of recurrence. With a large cohort of human CRCs in this study, we demonstrated that membranous EPHB2 expression is negatively correlated with lympho-vascular invasion and was a prognostic marker associated with better overall and recurrence-free survival. A tumor suppressor role for EPHB2 has been described in various cancers including prostate, breast cancers, and CRC. In particular, it has been suggested that EPH2 compartmentalizes the expansion of CRC cells through a mechanism dependent on E-cadherinmediated adhesion; in turn, this compartmentalization restricts the spread of EPHB2-expressing CRC cells [24]. Therefore, it is reasonable to hypothesize that EPHB2-mediated signaling prevents cancer progression by suppressing cancer migration and invasion, leading to better clinical outcomes for EPHB2-positive CRC patients.
In the intestinal epithelium, EPHB2 signaling controls the positioning of cell types along the crypt-villus axis [15]. In addition, EPHB2 belongs to the signature genes enriched in the intestinal stem cells, one of the β-catenin and Tcf4 target genes regulated by the Wnt signaling pathway. The most common mutation in CRCs is inactivation of the adenomatous polyposis coli gene, resulting in abnormal Wnt activation. Thus, it is possible to speculate that enhanced Wnt signaling activity might be responsible for elevated EPHB2 expression in CRCs, which was supported by our finding of a strong correlation of EPHB2 mRNA expression with AXIN2, another Wnt pathway target gene. Moreover, we observed that nuclear β-catenin expression, a hallmark of abnormal Wnt activity, was significantly associated with EPHB2 expression, suggesting that the Wnt pathway is likely to be responsible for EPHB2 upregulation in CRCs.
EPHB2-high cells were used to isolate CRC cells with intestinal stem cell-like phenotype positioned at the bottom of tumor structures reminiscent of crypts, which displayed robust tumor-initiating capacity as well as long-term self-renewal potential, indicating a CSC population [18]. As several genes such as CD24, CD44, CD133, and CD166 have also been proposed as CSC markers in CRCs, we investigated whether EPHB2 expression is associated with any of these candidate CSC markers and found that only CD44 has a positive correlation with EPHB2 mRNA expression, and this finding was consistent with the immunohistochemical analysis showing a positive correlation between H-scores of EPHB2 and CD44. It would be worthwhile to explore the interactions between CD44 and EPHB2 in CRCs through double staining or an in vitro study.
In conclusion, upregulated EPHB2 expression persists during adenoma-carcinoma transition and lymph node metastasis, but slightly declines in the invasive fronts of CRCs. EPHB2 positivity is negatively correlated with lymphatic and venous invasion and TNM stage, but positively correlated with MSI-high. In addition, EPHB2 is significantly associated with better clinical outcomes in CRCs. Taken together, our findings suggest that EPHB2 can be used as a prognostic marker for CRC patients.
Acknowledgments
This work was supported by a grant from the Korean Society of Pathologists in 2015.
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Herath NI, Boyd AW. The role of Eph receptors and ephrin ligands in colorectal cancer. Int J Cancer. 2010;126:2003–11. doi: 10.1002/ijc.25147. [DOI] [PubMed] [Google Scholar]
- 2.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janes PW, Adikari S, Lackmann M. Eph/ephrin signalling and function in oncogenesis: lessons from embryonic development. Curr Cancer Drug Targets. 2008;8:473–9. doi: 10.2174/156800908785699315. [DOI] [PubMed] [Google Scholar]
- 4.Lackmann M, Boyd AW. Eph, a protein family coming of age: more confusion, insight, or complexity? Sci Signal. 2008;1:re2. doi: 10.1126/stke.115re2. [DOI] [PubMed] [Google Scholar]
- 5.Boyd AW, Bartlett PF, Lackmann M. Therapeutic targeting of EPH receptors and their ligands. Nat Rev Drug Discov. 2014;13:39–62. doi: 10.1038/nrd4175. [DOI] [PubMed] [Google Scholar]
- 6.Tang XX, Zhao H, Robinson ME, et al. Implications of EPHB6, EFNB2, and EFNB3 expressions in human neuroblastoma. Proc Natl Acad Sci U S A. 2000;97:10936–41. doi: 10.1073/pnas.190123297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox BP, Kandpal RP. Invasiveness of breast carcinoma cells and transcript profile: Eph receptors and ephrin ligands as molecular markers of potential diagnostic and prognostic application. Biochem Biophys Res Commun. 2004;318:882–92. doi: 10.1016/j.bbrc.2004.04.102. [DOI] [PubMed] [Google Scholar]
- 8.Kinch MS, Moore MB, Harpole DH., Jr Predictive value of the EphA2 receptor tyrosine kinase in lung cancer recurrence and survival. Clin Cancer Res. 2003;9:613–8. [PubMed] [Google Scholar]
- 9.Nakamura R, Kataoka H, Sato N, et al. EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci. 2005;96:42–7. doi: 10.1111/j.1349-7006.2005.00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan M, Xu C, Zhang F, Ye C. Aberrant methylation of EphA7 in human prostate cancer and its relation to clinicopathologic features. Int J Cancer. 2009;124:88–94. doi: 10.1002/ijc.23890. [DOI] [PubMed] [Google Scholar]
- 11.Herath NI, Spanevello MD, Sabesan S, et al. Over-expression of Eph and ephrin genes in advanced ovarian cancer: ephrin gene expression correlates with shortened survival. BMC Cancer. 2006;6:144. doi: 10.1186/1471-2407-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batlle E, Bacani J, Begthel H, et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–30. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 13.Hafner C, Becker B, Landthaler M, Vogt T. Expression profile of Eph receptors and ephrin ligands in human skin and downregulation of EphA1 in nonmelanoma skin cancer. Mod Pathol. 2006;19:1369–77. doi: 10.1038/modpathol.3800660. [DOI] [PubMed] [Google Scholar]
- 14.Hafner C, Schmitz G, Meyer S, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50:490–9. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- 15.Batlle E, Henderson JT, Beghtel H, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–63. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Kim KJ, Rhee YY, et al. Expression status of wild-type HSP110 correlates with HSP110 T17 deletion size and patient prognosis in microsatellite-unstable colorectal cancer. Mod Pathol. 2014;27:443–53. doi: 10.1038/modpathol.2013.160. [DOI] [PubMed] [Google Scholar]
- 17.Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109:1004–12. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Guo DL, Zhang J, Yuen ST, et al. Reduced expression of EphB2 that parallels invasion and metastasis in colorectal tumours. Carcinogenesis. 2006;27:454–64. doi: 10.1093/carcin/bgi259. [DOI] [PubMed] [Google Scholar]
- 20.Lugli A, Spichtin H, Maurer R, et al. EphB2 expression across 138 human tumor types in a tissue microarray: high levels of expression in gastrointestinal cancers. Clin Cancer Res. 2005;11:6450–8. doi: 10.1158/1078-0432.CCR-04-2458. [DOI] [PubMed] [Google Scholar]
- 21.Kumar SR, Scehnet JS, Ley EJ, et al. Preferential induction of EphB4 over EphB2 and its implication in colorectal cancer progression. Cancer Res. 2009;69:3736–45. doi: 10.1158/0008-5472.CAN-08-3232. [DOI] [PubMed] [Google Scholar]
- 22.Alazzouzi H, Davalos V, Kokko A, et al. Mechanisms of inactivation of the receptor tyrosine kinase EPHB2 in colorectal tumors. Cancer Res. 2005;65:10170–3. doi: 10.1158/0008-5472.CAN-05-2580. [DOI] [PubMed] [Google Scholar]
- 23.Jubb AM, Zhong F, Bheddah S, et al. EphB2 is a prognostic factor in colorectal cancer. Clin Cancer Res. 2005;11:5181–7. doi: 10.1158/1078-0432.CCR-05-0143. [DOI] [PubMed] [Google Scholar]
- 24.Cortina C, Palomo-Ponce S, Iglesias M, et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat Genet. 2007;39:1376–83. doi: 10.1038/ng.2007.11. [DOI] [PubMed] [Google Scholar]