Abstract
Background
MicroRNAs (miRNAs) are short, non-coding RNAs that mediate post-transcriptional gene regulation. They are commonly deregulated in human malignancies, including non-small cell lung cancer (NSCLC). The aim of this study is to investigate miRNA expression in T790M-mutated NSCLC resistant to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors.
Methods
Six cases of resected NSCLC harboring the T790M mutation were examined. We performed miRNA time polymerase chain reaction (PCR) array profiling using EGFR T790M-mutated NSCLC and L858R-mutated NSCLC. Once identified, miRNAs that were differentially expressed between the two groups were validated by quantitative real-time polymerase chain reaction (qRT-PCR).
Results
miRNA PCR array profiling revealed three up-regulated miRNAs whose expression levels were altered 4.0-fold or more in the EGFR T790M mutation group than in the L858R group: miR-1 (fold change, 4.384), miR-196a (fold change, 4.138), and miR-124 (fold change, 4.132). The three differentially expressed miRNAs were validated by qRT-PCR, and they were found to be overexpressed in the T790M group relative to L858R group. In particular, expression levels of miR-1 and miR-124 were significantly higher in the T790M group (p-value of miR-1 = .004, miR-124 = .007, miR-196a = .096).
Conclusions
MiR-1, miR-124, and miR-196a are overexpressed in EGFR T790M mutated NSCLC.
Keywords: MicroRNAs, Lung neoplasms, EGFR T790M mutation
Lung cancer is the most common cause of cancer death worldwide, and advanced-stage non-small cell lung cancer (NSCLC) is considered an incurable disease [1]. Recently, the identification of genetic alterations present in lung cancer has resulted in the development of new treatment options for NSCLC and improved survival of NSCLC patients [2]. Among these genetic alterations, the epidermal growth factor receptor (EGFR) activating mutation is the most widely used target for therapy. However, most patients who initially benefit from the EGFR tyrosine kinase inhibitor (TKI) experience subsequent disease progression, and in more than 50% of cases, the mechanism of resistance is the T790M point mutation in the EGFR gene [3-5]. As treatment with EGFR-TKIs has become routine for advanced lung cancer, the need to better understand the EGFR T790M mutation has increased.
MicroRNAs (miRNAs) are a class of short, non-coding RNAs that mediate post-transcriptional gene regulation. They are involved in nearly all biologic processes, and deregulation of miRNA is correlated with many diseases, including cancer [6]. Multiple previous studies have observed distinct patterns of miRNA expression across tumor types and revealed that up- or down-regulation of miRNA expression is indicative of a specific cancer [7,8]. Moreover, a growing number of evidence indicates that certain miRNA profiles distinguish poor-prognosis cancers, and specific miRNA signatures can predict the clinical outcomes of tumors [9-11]. Recent research has suggested that miRNAs have therapeutic capacities and can be used in cancer treatment [12-14]. All things considered, miRNAs are clinically useful biomarkers and potential therapeutic agents.
The aim of this study is to investigate the expression of miRNAs in EGFR-TKI resistant T790M mutation-positive lung cancer. For this purpose, we performed miRNA array profiling and compared miRNA expression between (1) NSCLC with the EGFR-TKI sensitive mutation (L858R) and (2) NSCLC with the EGFR-TKI resistant mutation coexisting with the EGFR-TKI sensitive mutation (T790M/L858R). Using this method, we identified three differentially expressed miRNAs between the two groups. In this paper, we report on these differentially expressed miRNAs with a prediction of common target genes and discuss the possible role of each miRNA in the biology of lung cancer containing the EGFR T790M mutation.
MATERIALS AND METHODS
Sample collection
Pathology files from three institutions (Pusan National University Hospital, Pusan National University Yangsan Hospital, and Inje University Haeundae Paik Hospital) and from the time period between January 2011 and June 2016 were reviewed to identify NSCLC harboring the EGFR T790M mutation. After the exclusion of biopsy samples due to insufficient tumor material, six out of 1,445 lung cancer patients who had undergone surgical resection were enrolled (Pusan National University Hospital, 4 cases; Pusan National University Yangsan Hospital, 1 case; and Inje University Haeundae Paik Hospital, 1 case). All the included cases were adenocarcinomas harboring preexisting EGFR T790M mutations before exposure to EGFR-TKI. All patients had coexisting TKI sensitive EGFR L858R point mutations, and no patients were known to have coexisting exon 19 deletion mutations. All patients underwent curative resection as their first treatment and did not have a history of EGFR-targeted therapy, neo-adjuvant chemotherapy, or radiation therapy. EGFR mutation testing was conducted as follows: Direct sequencing of the EGFR gene was performed in three patients (Pusan National University Hospital, 1 case; Pusan National University Yangsan Hospital, 1 case; Inje University Haeundae Paik Hospital, 1 case). At the Pusan National University Hospital, pyrosequencing was used in the case of one patient, and the peptide nucleic acid–mediated polymerase chain reaction (PCR) clamping method was used to detect the mutation in two patients. All six patients had an EGFR-TKI resistant T790M mutation and a coexisting TKI sensitive L858R mutation. For the control group, EGFR L858R mutant adenocarcinoma tissues from eight patients who underwent lung resection surgery were randomly selected. Additionally, four cases of EGFR wild-type adenocarcinoma and three non-neoplastic lung tissues were randomly selected and used as control in the miRNA array profiling. This study was approved by the institutional review board of Pusan National University Hospital (C1608-003-001), and informed consent from patients was waived.
miRNA extraction and cDNA synthesis
Hematoxylin and eosin (H&E) stained slides were prepared from routinely processed tissue sections using 10% buffered formalin and then reviewed to confirm the diagnosis. Five 10-μm sections were cut from a representative paraffin block of each tumor and mounted on glass slides. The tumor area intended for tissue dissection was marked on the unstained slides using the matched H&E stained slide; therefore, only the tumor portion was subjected to miRNA analysis. Manually dissected tissue samples were placed in 1.5 mL microcentrifuge tubes and deparaffinized in xylene. After washing with ethanol, the total miRNA of the tissue was extracted and purified using a miRNeasy formalin-fixed paraffin-embedded (FFPE) kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The concentration of each isolated miRNA was measured by a Nano-Drop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the samples that showed optimal purity and quantities were subjected to further analysis. Then, miRNA was processed using an miScript II reverse transcription kit (Qiagen) and cDNA was synthesized.
miRNA array profiling
The synthesized cDNA was used as a template for subsequent real-time PCR. The reaction mix was prepared using a miScript SYBR Green PCR Kit (Qiagen). The miRNA PCR array method was employed for comprehensive miRNA expression profiling. We used the miScript miRNA PCR Array Human Cancer Pathway Finder (MIHS-102Z, Qiagen), which is composed of a 96-well plate containing 84 human cancer specific miRNA primers and 12 controls. The selected miRNAs are known to be associated with the diagnosis, staging, progression, or prognosis of various cancers. The primers used for miRNA array profiling are described in Supplementary Table S1. Using the RT2 PCR array loading reservoir (Qiagen), equal amounts of the reaction mix were added to each well of the array plate. Then, real-time PCR was performed in the ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA), and 84 miRNAs were simultaneously quantified in each sample. For miRNA PCR array profiling, we used four cases of EGFR T790M- and L858R-mutated adenocarcinoma as test samples and five cases of EGFR L858R-mutated adenocarcinoma as control samples. Additionally, four cases of EGFR wild-type adenocarcinoma and three non-neoplastic lung tissues were also used as controls in this array profiling.
Data analysis
The miRNA profiling data were analyzed by web-based miRNA PCR Array Data Analysis software (http://pcrdata-analysis.sabiosciences.com/mirna/arrayanalysis.php) to identify differentially expressed miRNAs between the groups. This software automatically performs fold-change calculations from the user’s uploaded Ct values obtained in miRNA PCR.
Validation of differentially expressed miRNAs
We validated the differentially expressed miRNAs selected from miRNA array profiling results with a larger sample group that included six cases of T790M/L858R-mutated adenocarcinoma and eight cases of L858R-mutated adenocarcinoma. In the T790M-positive group, four out of six cases were the same cases included in the microarray profiling. In the control group without the T790M mutation, all eight cases were different from those of microarray profiling. miRNA was processed using the miRNeasy FFPE (Qiagen) and TaqMan MicroRNA reverse transcription (Applied Biosystems) kits. For the targeted quantification of up- or down-regulated miRNAs, preformulated primers included in the TaqMan MicroRNA Assays (Applied Biosystems) were used. The PCR reaction mix was prepared with TaqMan Universal Master Mix II (Applied Biosystems) and the ABI 7500 Real-Time PCR system (Applied Biosystems) was used for quantitative real-time PCR (qRT-PCR) analysis. The miRNA expression level was normalized to endogenous control gene RNU48. Following the calculation of the ΔCt values, miRNA expressions of T790M/L858R-mutated and L858R-mutated samples were compared by t-test using the statistical program R. A p < .05 was considered statistically significant.
Target gene prediction and gene set enrichment analysis
We evaluated common target genes of differentially expressed miRNAs using MicroRNA.org (http://www.microrna.org) [15]. Gene set enrichment analysis was done with the top 10%-ranked genes in the common target gene list using MSigDB v6.0 [16].
RESULTS
Clinicopathologic factors of the study population
Six cases of EGFR T790M-positive and eight cases of T790M-negative pulmonary adenocarcinoma were included in the study. The clinicopathologic information of the two patient groups are described in Supplementary Tables S2 and S3. In the T790M-positive group, all patients were female (age range, 47 to 72; mean, 63). Tumor size ranged from 2.0 to 3.6 cm (mean, 2.78). Five adenocarcinomas exhibited an acinar pattern, while 1 was an adenocarcinoma with a papillary pattern. In the control group without the EGFR T790M mutation, six patients were female and two were male. The patients’ ages ranged from 44 to 72 years (mean, 61). Mean tumor size was 2.29 cm (range, 1.4 to 4.0 cm). The predominant histologic patterns were acinar (5 cases), lepidic (2 cases), and papillary (1 case). A representative microphotograph of T790M mutated adenocarcinoma (case 5) is shown in Fig. 1.
Fig. 1.
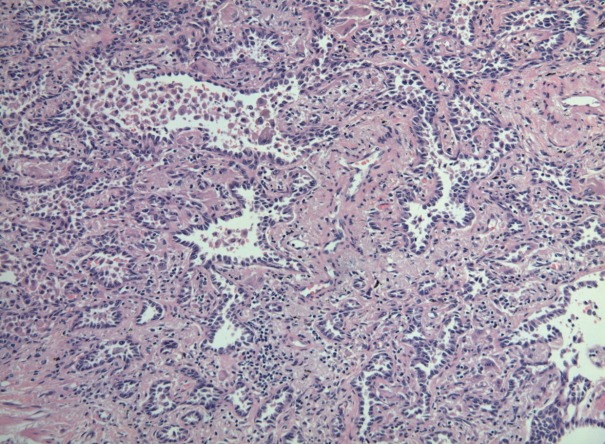
Representative microphotograph of EGFR T790M mutated adenocarcinoma (predominantly acinar pattern of case 5). EGFR, epidermal growth factor receptor.
miRNA array profiling analysis
We identified three up-regulated miRNAs whose expression levels were altered by 4.0-fold or more in EGFR T790M/L858R mutated lung cancer tissues compared to L858R mutated tissues (Table 1). These were miR-1 (fold change, 4.3841), miR-196a (fold change, 4.1380), and miR-124 (fold change, 4.1321). There were no down-regulated miRNAs in the T790M-positive lung cancer group. Compared to EGFR wildtype lung cancer, T790M/L858R mutated lung cancer was found to have 20 up-regulated miRNAs, including miR-1 and miR-196a (Table 2). No significantly differentially expressed miRNAs were identified between the L858R and EGFR wildtype groups. When non-neoplastic lung tissue was used as a control sample, there were seven overexpressed miRNAs in the T790M/L858R group, one of which was miR-196a (Table 3).
Table 1.
miRNAs overexpressed in EGFR T790M/L858R mutated lung cancer (vs L858R mutation)
| miRNA | Fold change |
|---|---|
| miR-196a | 4.1380 |
| miR-124 | 4.1321 |
| miR-1 | 4.3841 |
EGFR, epidermal growth factor receptor.
Table 2.
miRNAs overexpressed in EGFR T790M/L858R mutated lung cancer (vs EGFR wild type)
| miRNA | Fold change |
|---|---|
| miR-196a | 5.9922 |
| miR-148b | 5.0038 |
| miR-146b | 5.0653 |
| miR-183 | 7.0982 |
| miR-148a | 5.7195 |
| miR-218 | 6.0127 |
| miR-29b | 4.2675 |
| miR-135b | 8.4407 |
| miR-21 | 7.1416 |
| miR-10b | 4.9795 |
| miR-155 | 4.2678 |
| miR-1 | 7.1204 |
| miR-27b | 5.0205 |
| miR-7 | 6.0372 |
| miR-9 | 7.0287 |
| let-7f | 5.9577 |
| miR-10a | 5.8620 |
| miR-16 | 4.2105 |
| miR-98 | 5.0532 |
| miR-126 | 5.0032 |
EGFR, epidermal growth factor receptor.
Table 3.
miRNAs overexpressed in EGFR T790M/L858R mutated lung cancer (vs non-neoplastic lung tissue)
| miRNA | Fold change |
|---|---|
| miR-196a | 17.5784 |
| miR-96 | 4.4977 |
| miR-183 | 7.3777 |
| miR-135b | 9.3675 |
| miR-21 | 6.1286 |
| miR-9 | 9.7106 |
| miR-210 | 4.2718 |
EGFR, epidermal growth factor receptor.
Comparison of miRNA expression between the T790M/L858R and L858R groups (miR-1, miR-196a, miR-124)
The expression of miR-1, miR-196a, and miR-124, which were found to be differentially expressed in array profiling, was validated by qRT-PCR. The three miRNAs were overexpressed in the T790M/L858R group when compared to the L858R group. In particular, the expression of miR-1 and miR-124 was significantly higher in the T790M group (p-value of miR-1 = .004, miR-124 = .007) (Fig. 2A, B). Expression of miR-196a was higher in the T790M group than in the L858R group, but not significantly so (p = .096) (Fig. 2C).
Fig. 2.
Expression of miRNAs (A, miR-1; B, miR-124; and C, miR-196a) in EGFR T790M and L858R groups. EGFR, epidermal growth factor receptor.
Prediction of common target genes of miR-1, miR-124, and miR-196a
We investigated putative common target genes of these three miRNAs using microRNA.org, and found 2,010 candidate genes. Then, the MSigDB web tool was used to conduct gene set enrichment analysis of the top 10%-ranked genes in the common target gene list. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that 18 gene sets were significantly enriched (false discovery rate < 0.05), including the sets of several cancer-related and signaling pathways (Supplementary Table S4).
DISCUSSION
Lung cancer is the leading cause of cancer mortality worldwide. Despite extensive efforts to improve the survival rate, the 5-year survival for NSCLC is only 21% [1]. Moreover, 57% of lung cancers are diagnosed at an advanced stage, with a 5-year survival rate of 4% [1]. Over the last few decades, the development of targeted agents has provided marked survival benefits to lung cancer patients whose tumors harbor specific genomic alterations [17]. In particular, agents targeting EGFR-activating mutations are the most widely used targeted therapy, and EGFR mutation analysis of NSCLC is now routine in standard clinical practice. It is well known that patients carrying a point mutation in exon 21 (L858R) or a deletion in exon 19, which account for approximately 90% of EGFR-activating mutations, have significant survival benefit when treated with EGFR-TKI [18]. However, the majority of patients who initially respond to EGFR-TKIs acquire resistance to the drug, with the most important mechanism of resistance being a T790M gatekeeper mutation in the EGFR gene [4]. This mutation leads to decreased drug binding through steric hindrance and increased binding affinity with ATP [19,20]. The EGFR T790M mutation is a current focus of study for many investigators attempting to overcome EGFR-TKI resistance [21,22]. Osimertinib, which was recently developed to treat patients with EGFR T790M mutation-positive NSCLC, is being tested in clinical trials [23]. However, the complex biology of EGFR-TKI resistance is not thoroughly understood, and new approaches are being explored [24]. We speculated that altered miRNA expression of T790M-mutated NSCLC might provide us with new insights into EGFR-TKI resistance.
miR-1 is known to be a muscle-specific miRNA that is preferentially involved in muscle development. However, many studies have also demonstrated that miR-1 functions as a tumor suppressor, regulating several oncogenic pathways in human cancers [25,26]. According to previous research, the expression of this miRNA was significantly down-regulated in several cancers, and restoration of miR-1 in cancer cells resulted in inhibition of proliferation, migration, and invasion, indicating its therapeutic potential [27]. miR-124 has also been reported to act as a tumor suppressor in human cancers. Several studies have demonstrated that this miRNA is frequently silenced by DNA methylation, and thus, plays a pivotal role in carcinogenesis. Moreover, down-regulation of miR-124 was correlated with tumor progression and its reinstatement was suggested to be a novel therapeutic strategy [28-30]. In the present study, these tumor suppressive miRNAs (miR-1, miR-124) were overexpressed in the T790M/L858R mutation group relative to the L858R mutation group. This result corresponds with previous studies indicating that the natural clinical behavior of the T790M mutated tumor is more indolent except for the poor response to EGFR-TKI therapy [31]. Some studies revealed that patients who acquired the EGFR T790M mutation after TKI therapy had longer survival, suggesting that the T790M mutation may be related to indolent biology and slow tumor growth [32,33]. In the EURTAC (EURopean TArceva vs Chemotherapy) study, the survival of patients with a pre-existing T790M mutation was superior to that of patients without it [34]. Fujita et al. [35] found that presence of the T790M mutation correlated with better outcomes even in patients treated with TKI. Taken together, these data suggest that T790M mutant tumors may have indolent clinical course and the tumor suppressive miRNAs identified in the present study may contribute to this.
Most studies of miR-196a have reported that this miRNA is overexpressed in a variety of human malignancies, especially in pancreatic cancer [36,37]. It has a dominant effect on the inhibition of tumor suppressor genes, finally acting as oncogenes [38]. In this study, we found that miR-196a was up-regulated in T790M/L858R-positive tumors when compared to tumors that were only L858R-positive. We also observed that miR-196a was highly overexpressed in T790M-positive tumor tissue compared to non-neoplastic lung tissue, a finding that is consistent with the previous reports suggesting that miR-196a has an oncogenic function.
In our investigation, we also analyzed the common target genes of 3 miRNAs (miR-1, miR-124, and miR-196a) by gene set enrichment analysis. KEGG pathway enrichment analysis showed that the predicted targets were mainly enriched in cancerrelated gene sets that included renal cell carcinoma, chronic myeloid leukemia, endometrial cancer, and non-small cell lung cancer, as well as in signaling pathway gene sets that included JAK-STAT, ErbB, and RIG-I-like receptor signaling pathways. Four genes (SOS1, PIK3CG, NRAS, and CRKL) were common to most of these gene sets. A review of previous studies revealed that these pathways or genes were reported to be associated with EGFR-TKI resistance. In a study using T790M-mutated NSCLC cell lines, the authors demonstrated that activation of the IL-6R/JAK1/STAT3 pathway induced de novo resistance to irreversible EGFR-TKIs [39]. In another study, oncogenic CRKL was shown to activate the SOS1-RAS-RAF-ERK and SRC-C3G-RAP1 pathways, and overexpression of CRKL in EGFR-mutant cells induced resistance to EGFR-TKI [40]. Wu et al. [41] reported that targeted regulation existed between miR-124 and STAT3, and up-regulation of miR-124 suppresses STAT3. It is also noteworthy that miR-124 was observed to directly target the 3'-untranslated regions of SOS1 mRNA and regulate the behavior of tumor cells [42]. These findings indicate an association between resistance to EGFR-TKI and the up-regulated miRNAs examined in this study, and we conjecture that miR-124 regulates the expression of STAT3 or SOS1 and ultimately targets the EGFR pathway. However, miRNAs target numerous genes, and their mechanism of action is extremely complicated. Therefore, further study is needed to validate the function of these miRNAs.
In this study, we used cases in which the T790M mutation pre-existing, rather than acquired. Acquired EGFR T790M mutations occur in 50%–60% of patients who initially benefited from EGFR-TKIs and are the primary mechanism of resistance to therapy. In some patients, however, EGFR T790M mutations may exist before EGFR-TKI exposure, and these pre-existing mutations are associated with de novo resistance to the drugs [43]. We used samples with pre-existing T790M mutations because repeat biopsies were rarely performed for lung cancer patients whose disease progressed after treatment with EGFR-TKIs. In cases where the biopsy was repeated, only small tissue fragments or cytology specimens were included, so even after confirming the presence of an acquired T790M mutation, the biopsy tissue was inadequate for further miRNA analysis. Therefore, all the available tissues for this study were of pre-existing T790M mutants before EGFR-TKI therapy. We assumed that the fundamental action of the EGFR T790M mutation does not differ between cases of acquired and pre-existing mutations. According to one previous study, a minor clone with a pre-existing T790M mutation is present at a low frequency in NSCLC, and selective pressure from EGFR-TKI therapy causes expansion of these mutant cells [44]. This selection results in the emergence of a resistant phenotype in the entire tumor cell population. Therefore, the acquired T790M mutation can be considered an enriched pre-existing T790M mutation. In a certain point of view, an investigation with a de novo T790M mutation sample might be better than with the samples of acquired T790M mutation, especially when the researcher wants to know the innate nature of EGFR mutation types, since the acquired T790M mutation may be accompanied by additionally accumulated genetic alterations during EGFR-TKI therapy.
We performed miRNA PCR array profiling using NSCLC tissue, and the results showed that 3 miRNAs (miR-1, miR-124, and miR-196a) were up-regulated in the T790M mutation group compared to the L858R mutation group. The miRNA expression levels obtained with qRT-PCR were in accordance with the array profiling results. The results from this study may yield insights into the mechanisms of EGFR-TKI resistance.
Acknowledgments
This work was supported by a 2-year research grant of Pusan National University.
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Electronic Supplementary Material
Supplementary materials are available at Journal of Pathology and Translational Medicine (http://jpatholtm.org).
REFERENCES
- 1.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–89. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 3.Denis MG, Vallée A, Théoleyre S. EGFR T790M resistance mutation in non small-cell lung carcinoma. Clin Chim Acta. 2015;444:81–5. doi: 10.1016/j.cca.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in nonsmall cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 5.Remon J, Morán T, Majem M, et al. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: a new era begins. Cancer Treat Rev. 2014;40:93–101. doi: 10.1016/j.ctrv.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 8.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 10.Pignot G, Cizeron-Clairac G, Vacher S, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479–91. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- 11.Xu F, Zhang H, Su Y, Kong J, Yu H, Qian B. Up-regulation of microRNA-183-3p is a potent prognostic marker for lung adenocarcinoma of female non-smokers. Clin Transl Oncol. 2014;16:980–5. doi: 10.1007/s12094-014-1183-9. [DOI] [PubMed] [Google Scholar]
- 12.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsamiR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446:179–86. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 14.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genomewide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 18.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 19.Sos ML, Rode HB, Heynck S, et al. Chemogenomic profiling provides insights into the limited activity of irreversible EGFR inhibitors in tumor cells expressing the T790M EGFR resistance mutation. Cancer Res. 2010;70:868–74. doi: 10.1158/0008-5472.CAN-09-3106. [DOI] [PubMed] [Google Scholar]
- 20.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 23.Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017;35:1288–96. doi: 10.1200/JCO.2016.70.3223. [DOI] [PubMed] [Google Scholar]
- 24.Sin TK, Wang F, Meng F, et al. Implications of microRNAs in the treatment of gefitinib-resistant non-small cell lung cancer. Int J Mol Sci. 2016;17:237. doi: 10.3390/ijms17020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao H, Zeng J, Li H, et al. MiR-1 downregulation correlates with poor survival in clear cell renal cell carcinoma where it interferes with cell cycle regulation and metastasis. Oncotarget. 2015;6:13201–15. doi: 10.18632/oncotarget.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han C, Yu Z, Duan Z, Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res Int. 2014;2014:428371. doi: 10.1155/2014/428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Liu S, Tian L, et al. miR-124 and miR-506 inhibit colorectal cancer progression by targeting DNMT3B and DNMT1. Oncotarget. 2015;6:38139–50. doi: 10.18632/oncotarget.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 30.Dong LL, Chen LM, Wang WM, Zhang LM. Decreased expression of microRNA-124 is an independent unfavorable prognostic factor for patients with breast cancer. Diagn Pathol. 2015;10:45. doi: 10.1186/s13000-015-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Lee GK, Hwang JA, Yun T, Kim HT, Lee JS. Clinical likelihood of sporadic primary EGFR T790M mutation in EGFR-mutant lung cancer. Clin Lung Cancer. 2015;16:46–50. doi: 10.1016/j.cllc.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–22. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hata A, Katakami N, Yoshioka H, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119:4325–32. doi: 10.1002/cncr.28364. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Molina-Vila MA, Taron M, et al. EGFR compound mutants and survival on erlotinib in non-small cell lung cancer (NSCLC) patients (p) in the EURTAC study. J Clin Oncol. 2012;30(15 Suppl):7522. [Google Scholar]
- 35.Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol. 2012;7:1640–4. doi: 10.1097/JTO.0b013e3182653d7f. [DOI] [PubMed] [Google Scholar]
- 36.Panarelli NC, Chen YT, Zhou XK, Kitabayashi N, Yantiss RK. MicroRNA expression aids the preoperative diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2012;41:685–90. doi: 10.1097/MPA.0b013e318243a905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas. 2011;40:1165–71. doi: 10.1097/MPA.0b013e3182218ffb. [DOI] [PubMed] [Google Scholar]
- 38.Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15:14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SM, Kwon OJ, Hong YK, et al. Activation of IL-6R/JAK1/STAT3 signaling induces de novo resistance to irreversible EGFR inhibitors in non-small cell lung cancer with T790M resistance mutation. Mol Cancer Ther. 2012;11:2254–64. doi: 10.1158/1535-7163.MCT-12-0311. [DOI] [PubMed] [Google Scholar]
- 40.Cheung HW, Du J, Boehm JS, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–25. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Huang W, Chen B, Bai PD, Wang XG, Xing JC. Up-regulation of miR-124 inhibits invasion and proliferation of prostate cancer cells through mediating JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:2338–45. [PubMed] [Google Scholar]
- 42.Lv Z, Yang L. MiR-124 inhibits the growth of glioblastoma through the downregulation of SOS1. Mol Med Rep. 2013;8:345–9. doi: 10.3892/mmr.2013.1561. [DOI] [PubMed] [Google Scholar]
- 43.Chen LY, Molina-Vila MA, Ruan SY, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: a systematic review and metaanalysis. Lung Cancer. 2016;94:46–53. doi: 10.1016/j.lungcan.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–8. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



