Background
The phenomenon by which a given stimulus in the maternal environment has a permanent effect on the structure, physiology and metabolism of a fetus is called ‘fetal programming’, and the long‐term effects on adult health are thought to be consequences of these events. Animal models have been extensively used to study the basic physiological principles underlying fetal programming, and the results served as a guide to elucidate this phenomenon in humans. Currently, the consequences of maternal stimuli on the fetus, such as nutrient supply, chemical use, psychological fluctuations, immunological history, among others, are well known, and the consequences for fetal metabolism have been raised for different types of uterine environmental stimulus from different non‐human primates. Numerous animal and human studies have shown elevated prenatal maternal physiological stress in response to external adversities such as predational, climatic, social or nutritional stress causing various effects on offspring phenotype (Coe & Lubach, 2008). For example, stress experienced by the pregnant Macaca assamensis has programming effects that accelerate offspring growth accompanied by decelerated motor skill acquisition and reduced immune function of the offspring (Berganhel et al. 2016).
Intriguingly, the mechanisms whereby these maternal stimuli influence the risk of future metabolic disease in the offspring are poorly understood. The in utero environment can substantially modify how the fetal genome is expressed and may stimulate or inhibit mechanisms responsible for various fetal metabolic pathways. In addition, a large number of studies are now focusing on the epigenetic contributions to disease manifestation arising from developmental programming. However, the functional significance of these epigenetic regulators and their dynamic and complex interactions regulating gene expression are just beginning to be explored, and, over recent years, there has been an increasing focus on developing models of maternal obesity. In this context, a recent article published in The Journal of Physiology by Puppala et al. (2018) has advanced this research line with non‐human primates, in this case baboons (Papio hamadryas). In their study, the authors examined the molecular mechanisms (genetic and epigenetic changes) by which maternal obesity impacts the developing baboon fetal liver resulting in dysregulation of key metabolic pathways that impact lipid metabolism.
Presentation of data
The aim of Puppala et al. 2018 was to examine the molecular mechanisms by which a high‐fat maternal diet affects the developing fetal liver. To that end, a group of 32 healthy female baboons of similar age, body dimensions and weight were fed with diets differentiated for the supply of fats and sugars for at least 9 months before pregnancy. Of these, 16 randomly assigned females received a regular diet and became part of the control group (CON), while 16 females received a high‐fat, high‐fructose diet for weight gain before and during pregnancy, forming the maternal obesity (MO) group. The MO females exhibited greater waist and hip circumference, and elevated LDL‐cholesterol and triglycerides. The fetuses’ livers were extracted and part of the tissue was removed for histological, genomic, epigenomic and transcriptomic analysis.
The authors used state of the art genomic and epigenomic methods to determine which genes increased or decreased in activity and identified differential expression of miRNAs that regulate these genes in the fetal liver following exposure to maternal obesity. A bioinformatic approach was taken to identify the cellular signalling pathways, important for liver metabolic function, that were altered by these genomic and epigenetic alterations. Furthermore, microscopical studies were conducted to quantify the amount of stored fat and sugar present in the fetal liver cells, as well as to assess the shape of the cells, which is an indicator of liver cell health.
As hypothesized, they found evidence that MO affects the fetal liver through changes in pathways central to metabolism and liver metabolic function. Among these changes were dysregulation of the TCA cycle, proteasome, oxidative phosphorylation, glycolysis and Wnt/β‐catenin signalling pathways, determined using gene expression quantification, miRNA expression quantification and histological comparisons between the fetal liver of obese and non‐obese mothers (Fig. 1). The authors found 933 differentially expressed genes between fetuses of obese and non‐obese mothers and some of these differentially expressed genes are at the end of the aforementioned five important pathways. Of the 67 differentially expressed miRNAs, inversely expressed with target genes, found between CON and MO fetuses, 11 targeted 13 genes in these key pathways, recognizing the importance of these markers for the proper functioning of these pathways, which would be compromised in fetuses of obese mothers. In addition, they identified in the MO fetuses short‐term consequences, such as an increase in the amount of the hormone insulin in the blood, a threefold accumulation of lipid content in MO fetal livers and a marginal decrease in hepatic glycogen content in MO fetal livers compared with CON fetal livers. Approximately half of the MO fetal livers presented severe steatosis. Other features of liver injury, including lobular and portal inflammation, hepatocyte ballooning and fibrosis, were absent in both groups.
Figure 1.
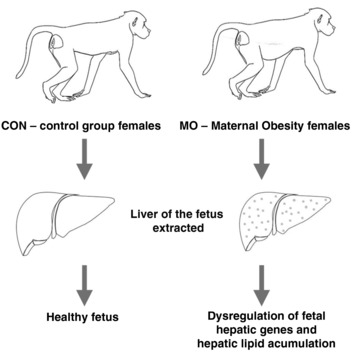
Influence of maternal obesity on baboon fetal liver lipid content
Significance of the article
miRNAs are small non‐coding endogenous RNAs that are the major regulators of gene expression in a versatile way, being key players in control of various metabolic processes and diseases involved in cell differentiation and proliferation, organ development, cell cycle regulation and energy metabolism (Neri, 2016). Puppala et al. (2018) is the first study in which unbiased transcriptome analysis has been used to identify potential miRNA epigenetic regulators of metabolic disruption in livers of non‐human primate fetuses with maternal obesity.
Overall, the data indicate that molecular approaches such as characterization of epigenetic modifications and miRNA expression play a key role in identifying the fundamentals of fetal programming related to maternal obesity. These epigenetic modifications involve persistent and hereditary modification of the genome without changes in the DNA sequence itself, increasing the risk of developing cardiovascular disorders, neurological diseases and lifelong obesity (Neri, 2016). In fact, molecular techniques such as transcriptomics, based on RNA sequencing (RNA‐Seq), are powerful strategies to relate the genotype to the phenotype of a cell and provide significant insight into the underlying mechanisms of fetal programming.
Although the study of Puppala et al. (2018) has limitations in its ability to show significant changes during early stages of fetal development, it was shown that during the critical time of fetal development, the epigenetic changes could influence gene expression and modulate the offspring's phenotype later, with the liver being one of the main targets for metabolic programming during this development. This result encourages future studies in MO postnatal offspring to determine the extent of this dysregulation on fetal hepatic metabolism. Puppala et al. (2018) provide further substantial evidence that maternal diet can have a profound impact on the epigenome and determine gene expression patterns and health conditions of the offspring throughout the life‐course. From now on, studies of altered epigenetic marking will be of profound importance for the mechanistic understanding of the role of nutrition in health but especially for studies of the developmental origins of health (Mathers & McKay, 2009).
This study provides further evidence that maternal health affects the physiological development of the fetus, increasing the risk of illness and disease throughout life. Moreover, epigenetic prenatal maternal stress effects on the offspring's phenotype are evidenced by the authors, ensuring a fertile soil for discussion of the evolutionary, adaptive or maladaptive effects of the programming events, in addition to providing new information about this phenomenon that may serve for the investigation of similar effects of high‐fat maternal diet in humans. Considering the results of Puppala et al. (2018) and the significant difference between baboon and human diets, especially in relation to the consumption of highly processed and industrialized foods by humans, questions emerge concerning the possible consequences of prolonged ingestion of ultra‐processed nourishment (added with unhealthy ingredients) for the development of fetuses and for long‐term health effects in humans.
Additional information
Competing interests
The authors declare no conflicts of interest.
Author contributions
All authors have approved the final version of the manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
No funding was received for this work.
Acknowledgements
We would like to acknowledge Dr Henrique Marques‐Souza for his guidance and critical insight towards the preparation of this Journal Club submission, and for the teachings obtained during the discipline ‘NH024 – Critical Study of Scientific Articles’ (Unicamp). We would also like to thank Dr Marcelo Alves da Silva Mori for assistance in the preparation of this article, and Bianca Laura da Silva Ramos and Giovane Vedovatti for the drawings.
Edited by: Kim Barrett & Laura Bennet
Linked articles This Journal Club article highlights an article by Puppala et al. To read this article, visit http://doi.org/10.1113/JP275422.
References
- Berghänel A, Heistermann M, Schülke O & Ostner J (2016). Prenatal stress effects in a wild, long‐lived primate: predictive adaptive responses in an unpredictable environment Proc Biol Sci 283, 20161304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe CL & Lubach GR (2008). Fetal programming: Prenatal origins of health and illness. Curr Dir Psychol Sci 17, 36–41. [Google Scholar]
- Mathers JC & McKay JA (2009). Epigenetics – Potential contribution to fetal programming In Early Nutrition Programming and Health Outcomes in Later Life, ed. Koletzko B, Decsi T, Molnár D. & de la Hunty A, pp 119–123. Springer, Dordrecht. [DOI] [PubMed] [Google Scholar]
- Neri C & Edlow A G (2016). Effects of maternal obesity on fetal programming: Molecular approaches. Cold Spring Harb Perspect Med 6, a026591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppala S, Li C, Glenn JP, Saxena R, Gawrieh S, Quinn A, Palarczyk J, Dick EJ, Nathanielsz PW & Cox LA (2018). Primate fetal hepatic responses to maternal obesity: epigenetic signalling pathways and lipid accumulation. J Physiol, 10.1113/JP275422. [DOI] [PMC free article] [PubMed] [Google Scholar]


