Abstract
Posttranslational histone modifications serve critical roles in gene regulation by determining the functional status of chromatin. Histone‐modifying enzymes often work in large multiprotein complexes. A paper in this issue of The EMBO Journal describes a new chromatin‐modifying complex called PEAT that acts via histone deacetylation. The PEAT complex is involved in heterochromatin formation and gene repression but also appears to have a locus‐specific activating role, possibly through promoting histone acetylation.
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Plant Biology; RNA Biology
Eukaryotic genomes are organized into chromatin, which serves a critical role in controlling gene activity. The fundamental unit of chromatin is the nucleosome, where around 150 base pairs of DNA wrap around a core consisting of histone proteins. Histones are subject to posttranslational modifications, which register and determine the functional status of chromatin. These posttranslational modifications include methylation, acetylation, phosphorylation, and addition of other chemical groups that are established by histone modifier enzymes. Existing modifications are detected by reader proteins, which perform various downstream functions.
Certain patterns of histone modifications are commonly found in repressive chromatin regions, also known as heterochromatin. They are established not only on repressed genes, but also on transposons, repetitive sequences, and other genetic elements, which are actively silenced to maintain genome integrity. These repressive chromatin marks are established by several interconnected silencing pathways that trigger the addition of repressive histone modifications and the removal of activating ones. Many of these silencing pathways are partially or fully conserved among eukaryotes, and much progress in discovering and characterizing them has been made in model organisms, including the plant Arabidopsis thaliana (Pikaard & Mittelsten Scheid, 2014).
In this issue of EMBO Journal, Tan et al (2018) describe a new Arabidopsis silencing complex, PEAT. This is a significant discovery because forward genetic screens have been mostly saturated and it is nowadays relatively uncommon to discover a new major silencing factor. The first subunit of PEAT, named Enhancer of Polycomb‐Related 1 (EPCR1), was identified via a large‐scale, highly sensitive reverse genetics approach. A search for its interacting partners identified four protein families that together form the core of the PEAT complex: PWWP, EPCR, ARID, and TRB (Fig 1). The presence of these subunits within PEAT is consistent with a role in gene silencing as all of them have known or predicted roles in binding to chromatin (Wilsker et al, 2002; Doyon & Côté, 2004; Qin & Min, 2014; Zhou et al, 2018). However, two additional classes of proteins associate with PEAT: histone acetyltransferases, HAM1 and HAM2, as well as histone deacetylases, HDA6 and HDA9. There are multiple partially redundant paralogs present for all PEAT subunits, indicating that PEAT is likely not a single complex but a family of closely related complexes with variable subunit compositions and possibly also variable functions.
Figure 1. Composition and function of the PEAT complex.
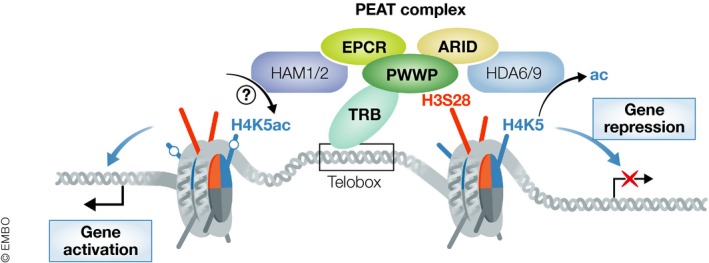
The PEAT complex is composed of PWWP, EPCR, ARID, and TRB proteins. It also associates with HAM1 and HAM2 acetyltransferases as well as HDA6 and HDA9 histone deacetylases. It is recruited to individual loci by sequence‐specific DNA binding of TRB and an interaction of PWWP with unphosphorylated serine 28 of histone H3. PEAT contributes to establishing repressive chromatin marks by deacetylating lysine 5 of histone H4 (H4K5). It also has a locus‐specific activating role possibly related to histone acetyltransferase activity of HAM1 and HAM2.
The composition of PEAT indicates that it binds to specific regions of chromatin and adds or removes acetyl groups from histones. This is consistent with observations that the complex displays two contradictory roles—transcriptional activation and silencing. Tan et al clearly show that the primary role of PEAT on chromatin is repressive and support this with several observations in PEAT subunit mutants. These include a reduction in transposon silencing in heterochromatin, increase in histone H4 lysine 5 acetylation (H4K5ac) on transposons, reduction in heterochromatin compaction, as well as reduction in DNA methylation within genes. However, there are also indications that PEAT can contribute to gene activation in pericentromeric heterochromatin, where repressive features (siRNA production and DNA methylation) are increased in PEAT subunit mutants.
There is no doubt that PEAT has an important biological role as mutants in its subunits have strong morphological phenotypes. Like most chromatin‐modifying complexes, its role likely cannot be attributed to a specific developmental or physiological process. Instead, it is defined by locus‐specific recruitment of PEAT to individual genes and enzymatic reactions performed on chromatin within those genes. The locus‐specificity of PEAT appears to be determined by a combination of sequence‐specificity and recognition of pre‐existing chromatin modifications. Binding of specific DNA sequences is a known feature of PEAT subunit telomere repeat binding (TRB). This multifunctional protein not only binds telomeres but is also associated with telobox motifs within specific gene promoters (Schrumpfová et al, 2016; Zhou et al, 2018). Although not directly tested by Tan et al, it is possible that TRB recruits PEAT to specific genomic regions. In addition to specific DNA sequences, PEAT recruitment may require pre‐existing chromatin modifications. This function may be performed by the PWWP protein, which contains a domain related to well‐known histone modification readers, including tudor and chromodomain (Qin & Min, 2014). Arabidopsis PWWP proteins, also known as PWWP‐Domain Interactor of Polycombs (PWO), specifically recognize histone H3 lacking phosphorylation of serine 28 (H3S28; Hohenstatt et al, 2018). This means that the combination of telobox motifs, absence of H3S28p and other, yet undiscovered factors, may determine which genomic loci will be targeted by PEAT (Fig 1).
Recruitment of PEAT is likely followed by enzymatic reactions performed by proteins associated with the complex (Fig 1). Genetic evidence supports histone deacetylation by HDA6 and HDA9 as the primary enzymatic reaction performed by PEAT. The most likely consequence of this reaction is recruitment of other repressive factors, chromatin compaction, and transcriptional repression. However, the role of HAM1 and HAM2 acetyltransferases in PEAT is less clear. One possibility is that PEAT may facilitate histone acetylation by associating with HAM1 and HAM2. Two opposing activities could coexist or could be mutually exclusive and locus‐specific. This is consistent with genetic data suggesting an activating role of PEAT within pericentromeric heterochromatin. An alternative possibility is that PEAT may sequester HAM1 and HAM2 and prevent their activity within the NuA4 complex (Doyon & Côté, 2004).
A big open question about PEAT is its relationship with the Polycomb Repressive Complex 2 (PRC2), which is supported by the observation that two PEAT subunits also interact with PRC2. One of those subunits is PWWP (PWO), which binds the catalytic subunits of PRC2 and has been implicated in their recruitment to chromatin (Hohenstatt et al, 2018). The second is TRB, which interacts with PRC2 subunits and telobox motifs in DNA (Zhou et al, 2018). These interactions recruit PRC2 and allow the deposition of methylation of lysine 27 of histone H3 (H3K27me3) at specific target genes (Zhou et al, 2018). Because PEAT and PRC2 share two subunits involved in determining their sequence‐specificity, these complexes may work on at least partially overlapping subsets of genes and reinforce each other. PRC2 may establish a repressive histone modification, H3K27me3, and PEAT remove the activating histone modification, H4K5ac. However, the locus‐specificity and recruitment of PEAT remain mysterious and may end up being much more complicated than what can be inferred from currently available data. Gaining deeper insights into these mechanisms remains an important goal for the future.
The EMBO Journal (2018) 37: e100573
See also: https://doi.org/10.15252/embj.201798770 (October 2018)
References
- Doyon Y, Côté J (2004) The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Hohenstatt ML, Mikulski P, Komarynets O, Klose C, Kycia I, Jeltsch A, Farrona S, Schubert D (2018) PWWP‐DOMAIN INTERACTOR OF POLYCOMBS1 interacts with polycomb‐group proteins and histones and regulates arabidopsis flowering and development. Plant Cell 30: 117–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Mittelsten Scheid O (2014) Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 6: a019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Min J (2014) Structure and function of the nucleosome‐binding PWWP domain. Trends Biochem Sci 39: 536–547 [DOI] [PubMed] [Google Scholar]
- Schrumpfová PP, Vychodilová I, Hapala J, Schořová Š, Dvořáček V, Fajkus J (2016) Telomere binding protein TRB1 is associated with promoters of translation machinery genes in vivo. Plant Mol Biol 90: 189–206 [DOI] [PubMed] [Google Scholar]
- Tan L‐M, Zhang C‐J, Hou X‐M, Shao C‐R, Lu Y‐J, Zhou J‐X, Li Y‐Q, Li L, Chen S, He X‐J (2018) The PEAT protein complexes are required for histone deacetylation and heterochromatin silencing. EMBO J 37: e98770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D, Patsialou A, Dallas PB, Moran E (2002) ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13: 95–106 [PubMed] [Google Scholar]
- Zhou Y, Wang Y, Krause K, Yang T, Dongus JA, Zhang Y, Turck F (2018) Telobox motifs recruit CLF/SWN‐PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis . Nat Genet 50: 638–644 [DOI] [PubMed] [Google Scholar]


