Abstract
Chronic obstructive pulmonary disease (COPD) is a syndrome caused by damage to the lungs that results in decreased pulmonary function and reduced structural integrity. Pulmonary function testing (PFT) is used to diagnose and stratify COPD into severity groups, and computed tomography (CT) imaging of the chest is often used to assess structural changes in the lungs. We hypothesized that the combination of PFT and CT phenotypes would provide a more powerful tool for assessing underlying morphologic differences associated with pulmonary function in COPD than does PFT alone. We used factor analysis of 26 variables to classify 8,157 participants recruited into the COPDGene cohort between January 2008 and June 2011 from 21 clinical centers across the United States. These factors were used as predictors of all-cause mortality using Cox proportional hazards modeling. Five factors explained 80% of the covariance and represented the following domains: factor 1, increased emphysema and decreased pulmonary function; factor 2, airway disease and decreased pulmonary function; factor 3, gas trapping; factor 4, CT variability; and factor 5, hyperinflation. After more than 46,079 person-years of follow-up, factors 1 through 4 were associated with mortality and there was a significant synergistic interaction between factors 1 and 2 on death. Considering CT measures along with PFT in the assessment of COPD can identify patients at particularly high risk for death.
Keywords: chronic obstructive pulmonary disease, Cox proportional hazards, factor analysis, mortality
Chronic obstructive pulmonary disease (COPD) is defined by reduced pulmonary function and is associated with reduced quality of life, more hospitalizations, and higher risk for death (1, 2). Cigarette smoking is the major environmental risk factor for development of COPD (3). Although COPD is defined by a ratio of less than 0.70 of forced expiratory volume at 1 second (FEV1) to forced vital capacity (FVC), there is substantial heterogeneity in the clinical and pathological manifestations of the disease (4, 5). Identifying the pathophysiologic processes reflecting underlying disease heterogeneity could lead to more targeted therapies for prevention and treatment.
Chest computed tomography (CT) can aid visualization and quantification of anatomic features of the lung; however, some key features of interest are correlated. This presents challenges in the use of these features as covariates in multivariable statistical models but also presents an opportunity to better define multidimensional pathologic processes in COPD. These CT features may represent primary structural abnormalities in the lung that lead to reduced pulmonary function and, ultimately, COPD diagnosis.
By taking advantage of the correlation structure of chest CT and pulmonary function data, factor analysis can reduce the number of variables to a small and manageable set of uncorrelated factors (6). We hypothesized that this smaller set of continuous vectors may represent disease axes that can be used to identify the underlying pathophysiologic heterogeneity within COPD. We hypothesized that this method, applied to a large heterogeneous dataset of COPD, would result in novel insights into disease phenotypes and prediction of mortality.
METHODS
The Genetic Epidemiology of COPD Study
The Genetic Epidemiology of COPD (COPDGene) Study is a multicenter (21 clinical sites in the United States), observational study designed so genetic factors associated with COPD can be identified and COPD-related phenotypes characterized (7). Into this study were recruited 10,192 adult current and former smokers who were non-Hispanic whites (two-thirds of cohort) or who were black (one-third of cohort), ages 44–81 years, with at least a 10 pack-year history of smoking. Participants with known COPD were recruited from outpatient pulmonary clinics and other smokers were recruited through personal contact with friends and relatives of clinic patients, advertisements, and outreach to community groups and other organizations. Study centers were instructed to target recruitment of participants without COPD from community sources rather than pulmonary clinics serving other lung diseases. Nonclinic-based recruitment identified participants with and without COPD and identified undetected COPD in many participants. Exclusion criteria were as follows: women who were pregnant, a history of lung disease other than asthma, surgical removal of at least 1 lung lobe, active cancer treatment or suspected lung cancer, chest radiation therapy, metal objects in the chest, recent COPD exacerbation treated with antibiotics or steroids (these patients were invited to participate at a later date), recent eye surgery, past myocardial infarction or other cardiac hospitalization, recent chest or abdominal surgery, inability to use albuterol, a first- or second-degree relative participating in the study, or multiple racial categories. All participants provided written informed consent, and the overall study was approved by the institutional review boards at all of the participating centers.
All participants were assessed for pulmonary function using spirometry and lung morphology using high-dose inspiration and low-dose expiration chest CT imaging. Although COPDGene is enriched for COPD cases, participants included people with and without COPD and represented a full range of pulmonary function (Web Figure 1, available at https://academic.oup.com/aje). Thus, COPDGene provides a broad spectrum of pulmonary phenotypes, based on findings of CT imaging, after specific exclusion criteria were applied (7), allowing unique insights into this population of current and former smokers.
Table 1 details characteristics of the COPDGene participants included in the present analysis compared with the full cohort. We excluded subjects without complete data on the phenotypes of interest or death follow-up. There were a total of 5,612 non-Hispanic white and 2,545 black participants with complete data.
Table 1.
Characteristics of Individuals Included in This Analysis With Complete Factor Data Compared With the Entire Cohort, Genetic Epidemiology of Chronic Obstructive Pulmonary Disease Study, United States, 2008–2011
| Characteristic | Analysis Cohort (n = 8,157) | COPDGene Cohort (n = 10,192) | P Value | ||||
|---|---|---|---|---|---|---|---|
| Mean (SD) | No. | % | Mean (SD) | No. | % | ||
| Age, years | 59.7 (9.0) | 59.6 (9.0) | 0.45 | ||||
| Female sex | 3,775 | 46.3 | 4,742 | 46.5 | 0.74 | ||
| Black race | 2,545 | 31.2 | 3,408 | 33.4 | 0.001 | ||
| Current smoker | 4,276 | 52.4 | 5,414 | 53.1 | 0.35 | ||
| Pack-years of smoking | 44.4 (24.9) | 44.2 (25.0) | 0.59 | ||||
| COPD (yes) | 3,604 | 44.2 | 4,484 | 44.0 | 0.07 | ||
| FEV1% predicteda | 76.8 (25.3) | 76.4 (25.6) | 0.29 | ||||
| FEV1/FVC | 0.67 (0.16) | 0.67 (0.16) | 1.00 | ||||
| COPD classificationb | 0.88 | ||||||
| PRISM | 992 | 12.2 | 1,257 | 12.4 | |||
| Gold 0 | 3,561 | 43.7 | 4,388 | 43.3 | |||
| Gold 1 | 651 | 8.0 | 794 | 7.8 | |||
| Gold 2 | 1,574 | 19.3 | 1,922 | 19.0 | |||
| Gold 3 | 918 | 11.2 | 1,162 | 11.5 | |||
| Gold 4 | 461 | 5.6 | 606 | 6.0 | |||
Abbreviations: FEV1%, percentage of forced expiratory volume at 1 second; COPDGene, the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease Study; GOLD, Global Initiative for Chronic Obstructive Lung Disease; PRISM, preserved ratio impaired spirometry; SD, standard deviation.
a Based on Third National Health and Nutrition Examination Survey reference values (9).
b GOLD stage is missing for 63 patients in the COPDGene cohort because spirometry could not be completed or spirometry data were missing. Percentages are calculated on the basis of a total of 10,129 patients.
Pulmonary function variables
Pulmonary function testing was performed following American Thoracic Society guidelines (8) using the Easy-One spirometer (ndd Medical Technologies, Andover, Massachusetts). Spirometry was performed at baseline and repeated after the administration of 180 μg of inhaled albuterol. FEV1, FVC, peak expiratory flow, and forced expiratory flow at 25%–75% of the FVC were obtained for all participants, FEV1 percent (FEV1%) predicted and FVC percent (FCV%) predicted values were calculated using the Third National Health and Nutrition Examination Survey reference values (9). The FEV1/FVC ratio was calculated using the absolute measures in liters. FEV1% predicted, FVC% predicted, peak expiratory flow, forced expiratory flow at 25%–75% of the FVC, and FEV1/FVC measure unique aspects of pulmonary function and each was included in the factor analysis.
CT variables
CT images of the chest were acquired at full inspiration and relaxed exhalation, as described previously (10, 11). Multiple CT-based metrics of the lung were obtained, including densitometric assessments of the lung parenchyma to provide objective assessments of emphysema-like tissue (CT threshold of −950 Hounsfield units (HU) and −910 HU expressed as a percentage of total lung parenchyma) and gas trapping (i.e., expiration-to-inspiration attenuation ratio, which is thought to reflect small airways disease). The percentage of lung tissue below a threshold of −856 HU was used to represent a quantitative metric of gas trapping on the expiratory CT scan.
Additional measures of central airway morphology were also used to provide objective assessments of airway wall thickening. One such measure is the wall area percentage. This is calculated as the 100 multiplied by the ratio of the airway wall area divided by the total bronchial cross-sectional area (wall plus lumen). Multiple investigations have demonstrated that increases in these measures (e.g., increased wall area percentage) reflect airway wall thickening and spirometric and clinical impairment. These measures are commonly obtained at select sites in the tracheobronchial tree, such as the third-generation (i.e., segmental) airways. Measures of airway wall thickness and the square root of the wall area of derived airways with lumen circumference of 10 mm and 15 mm were calculated as described previously (12, 13). Total lung capacity was measured in liters using volumetric CT imaging obtained during a breath hold at full inspiration with the subject supine. Functional residual capacity was measured in liters using volumetric CT scans obtained at the end of relaxed exhalation while supine. CT variables included in the factor analysis were designed to represent the broad range of CT phenotypes related to COPD.
Other covariates
Height was measured in centimeters using a stadiometer, weight was measured in kilograms,, and body mass index (BMI) was calculated by dividing the weight by height squared (using weight in kilograms and height in meters) (14). Current smoking status and pack-years of smoking were determined by questionnaire. Self-reported physician diagnoses of comorbid conditions were also determined by questionnaire.
Statistical methods for factor analysis
Before conducting the factor analysis, the distributions of variables were assessed for normality and Box-Cox transformations were considered for each nonnormally distributed variable. See Web Table 1 for transformations that were performed. Note that for all transformations, the scale direction was preserved to facilitate interpretation of the loading scores in clinically relevant terms.
Before performing factor analyses in the full cohort, we stratified the cohort by sex and race (non-Hispanic white or black) and assessed the dimensionality of the variables, each centered at 0 and scaled to have variance of 1, using principal components analysis based on the number of eigenvalues that were greater than 1. In addition, factor analysis was performed in the 4 strata we compared for factor similarities and differences. Horn’s parallel analysis was also conducted based on factor analysis fit to minimize the sum of squares of off-diagonal residuals of the resulting correlation matrix (15). Factor scores were computed using the Varimax rotation. Analyses were all conducted in R, version 3.1.1 (R Foundation for Statistical Computing, Vienna, Austria) using the psych package.
All-cause mortality
Assessment of death in COPDGene was conducted using multiple approaches. A longitudinal follow-up data collection effort was conducted using automated telephony and web-based survey instruments every 6 months for all available participants (16). Participant contact through this system resulted in identification of deceased participants and subsequent follow-up request for confirmation of death. Searches based on the Social Security Death Index (SSDI) are also conducted at regular intervals in COPDGene. Individual study-center institutional review board restrictions allowed an SSDI search to be conducted for 8,675 subjects in October 2016 by a central study search and by 9 sites performing their own searches. Results were aggregated centrally. Assessment of vital status (i.e., alive vs. dead) was backdated 3 months to account for the expected lag time between death and its appearance in the SSDI dataset. We included 333 participants who were unable to be searched through SSDI but were active participants in the longitudinal follow-up (participant returned a longitudinal follow-up survey within 7 months of the search). Participant follow-up time was the time between their baseline study visit and SSDI identified death, report of death from institutional review board–restricted study centers, or most recent, active, longitudinal follow-up participation. A total of 1,454 participants have been lost to follow-up (i.e., they have no SSDI identifier and no study contact has been made after the baseline study visit).
The Cox proportional hazard model, based on time to death, was used to model prediction of death in the sample. Continuous factor scores were tested for interactions as well as nonlinear associations with death.
RESULTS
The study population for the current analysis with complete data is similar to the overall cohort with respect to age (P = 0.5), sex (P = 0.7), smoking status (P = 0.4), and history of pack-years of smoking (P = 0.6), but differed with respect to race, with fewer black participants included (P = 0.001) (Table 1). There were no differences in COPD case status (P = 0.07), pulmonary function (P for FEV1 = 0.3 and P for FEV1/FVC = 1.0), or Global Initiative for Chronic Obstructive Lung Disease stage (a measure of COPD severity) (P = 0.9) between the study population with complete data and the overall cohort.
Principal component analysis was performed separately in the subgroups (male non-Hispanic whites, female non-Hispanic whites, male blacks, female blacks) to assess the dimensionality of the underlying factor model and all models yielded 5 to 6 eigenvalues greater than 1. The first 6 principal components explained between 82% and 85% of the variability for all groups. Horn’s parallel analysis indicated no more than 7 factors existed, and that no more than 6 principal components explained variability beyond background noise.
Beginning with the white male group (n = 2,973), factor analysis was conducted starting with 7 factors with factors subsequently removed until all factors had absolute factor loadings greater than 0.7. This yielded a 5-factor model. Likewise, in each of the other subgroups, a 5-factor model was supported by the data. Correlating these factor loadings among all subgroups revealed 5 consistent factors. Using the factor model from the white male subgroup, factor scores were derived for each of the other subgroups. These were then correlated with the scores derived from their respective subgroup analyses. The correlations were all quite high: 0.84 for factor 2 in female black participants and greater than 0.96 for all other subgroups and all 5 factors (Web Table 2). These correlations support the same underlying factor model for all subgroups. Given evidence for the same underlying factors explaining correlation among the variables, a single-factor model was fit on the basis of the combined set of data, using the same approach.
From the factor analysis with varimax rotation, 5 factors were identified (Table 2). These 5 factors explained 80% of the total variance of the 26 variables included in the final analysis, with the first factor accounting for 37% of the variance of these measures and the remaining factors accounting for progressively less of the total variance—17%, 10%, 9%, and 7% of the total variance, respectively, for factors 2 through 5. These factors accounted for a majority of the individual-measure variances of pulmonary function (72%–98%), inspiration CT density measures (52%–99%), expiration CT measures (74%–99%), but substantially less of the specific airway disease measurements (33%–36%) (Table 2).
Table 2.
Factor Loadings for a 5-Factor Model Based on the Combined Data Set (n = 8,157), Genetic Epidemiology of Chronic Obstructive Pulmonary Disease Study, United States, 2008–2011
| Variable | Emphysema Disease Axis | Airway Disease Axis | Gas Trapping | CT Intensity Variability | TLC and FRC | Proportion of Variance Explaineda |
|---|---|---|---|---|---|---|
| Pulmonary function | ||||||
| FEV1/FVC | −0.63b | −0.61b | −0.23 | 0.12 | 0.05 | 0.83 |
| FEV1% predictedc | −0.41 | −0.89b | −0.1 | 0.03 | −0.08 | 0.98 |
| FVC% predictedc | −0.06 | −0.82b | 0.06 | −0.05 | −0.19 | 0.72 |
| Peak expiratory flow | −0.4 | −0.7b | −0.19 | 0.08 | 0.16 | 0.72 |
| Forced expiratory flow 25%–75% | −0.51b | −0.7b | −0.22 | 0.07 | 0.11 | 0.81 |
| TLC (predicted-race adjusted) | −0.04 | −0.08 | −0.03 | 0.01 | 0.95b | 0.9 |
| FRC (predicted-race adjusted) | 0.05 | −0.02 | 0.04 | 0.03 | 0.94b | 0.88 |
| Inspiratory CT | ||||||
| Less than −856 HU | 0.86b | −0.18 | 0.03 | −0.44 | 0.01 | 0.96 |
| Less than −910 HU | 0.96b | −0.05 | 0.11 | −0.2 | 0 | 0.98 |
| Less than −950 HU | 0.96b | 0.08 | 0.18 | 0.1 | 0 | 0.96 |
| Inspiration histogram, 15th percentile | −0.94b | −0.01 | −0.12 | 0.19 | 0.01 | 0.93 |
| Emphysema, lower one-third, % | 0.92b | 0.04 | 0.12 | 0.02 | 0.05 | 0.87 |
| Emphysema, upper one-third, % | 0.91b | 0.09 | 0.19 | 0.15 | −0.04 | 0.89 |
| Inspiration intensity, mean | −0.88b | 0.06 | −0.06 | 0.41 | −0.01 | 0.94 |
| Inspiration intensity, SD | −0.03 | 0.26 | 0.12 | 0.66b | −0.01 | 0.52 |
| Exp/insp attenuation ratio | 0.31 | 0.41 | 0.81b | −0.24 | −0.01 | 0.99 |
| Expiratory CT | ||||||
| Less than −910 HU | 0.75b | 0.33 | 0.54b | 0.15 | 0.03 | 0.98 |
| Less than −950 HU | 0.73b | 0.33 | 0.48 | 0.3 | 0.04 | 0.97 |
| Gas trapping, % | 0.68b | 0.23 | 0.65b | −0.05 | 0.03 | 0.95 |
| Expiration histogram, 15th percentile | −0.66b | −0.25 | −0.58b | 0.1 | −0.01 | 0.84 |
| Expiration intensity, mean | −0.62b | −0.27 | −0.64b | 0.36 | 0 | 0.99 |
| Expiration intensity, SD | 0.1 | −0.1 | −0.19 | 0.82b | 0.05 | 0.74 |
| Airway measurements | ||||||
| Wall area, % segmental | −0.07 | 0.57b | 0.13 | 0.07 | −0.08 | 0.36 |
| Pi 10 | −0.16 | 0.51b | 0.13 | 0.16 | −0.1 | 0.34 |
| Pi 15 | −0.21 | 0.51b | 0.1 | 0.14 | −0.02 | 0.33 |
| BMI | −0.26 | 0.07 | −0.13 | 0.50b | 0.01 | 0.33 |
| Proportion of variance explained | 0.37 | 0.17 | 0.1 | 0.09 | 0.07 | |
| Cumulative variance explained | 0.37 | 0.53 | 0.64 | 0.72 | 0.8 |
Abbreviations; BMI, body mass index; CT, computed tomography; Exp, expiration; FEV1/FVC, ratio of forced expiratory volume at 1 second to forced vital capacity; FEV1%, percentage of forced expiratory volume at 1 second; FRC, functional residual capacity; FVC%, percentage of forced vital capacity; HU, Hounsfield unit; Insp, inspiration; Pi 10, airway wall thickness at an internal perimeter of 10 mm; Pi 15, airway wall thickness at an internal perimeter of 15 mm ; SD, standard deviation; TLC, total lung capacity.
a Proportion of variance in the row variable explained by the 5 factors.
b Factors loading ≥│0.5│.
c Based on Third National Health and Nutrition Examination Survey reference values (9).
CT measures of quantitative measures of emphysema on CT scan loaded strongly on factor 1, with the highest factor-loading scores being inspiratory CT volume less than −910 HU and less than −950 HU (factor loading = 0.96 for both) (Table 2). Measures of emphysema distribution also loaded highest on factor 1, as did the analogous measures of density on the expiration CT scans. Pulmonary function measures FEV1/FVC, FEV1% predicted, and forced expiratory flow 25%–75% loaded negatively on factor 1 (−0.63, −0.41, and −0.51, respectively). The airway measurements did not load strongly on factor 1. Based on the factor-loading scores, high CT measures of lung density with concomitant low pulmonary function, we interpret factor 1 to represent a multidimensional (i.e., low attenuation areas and lower pulmonary function) emphysema disease axis.
Factor 2 was represented by strong factor loadings for the physiologic pulmonary function measures, with factor-loading scores range from −0.61 to −0.89, except for total lung capacity and functional residual capacity (Table 2). Airway measurements of CT morphology loaded highest on factor 2: 0.57 for segmental wall area percentage and 0.51 for square root of the wall area of derived airways with lumen circumference of 10 mm and 15 mm. The measures of CT density, particularly from the inspiratory CT scans, did not load strongly on factor 2. The morphologic measures of the airways, combined with low pulmonary function loading on factor 2, indicated to us that factor 2 represents a multidimensional airway disease axis.
Physiologic measures of pulmonary function did not load strongly on factors 3 or 4. CT measures of low attenuation on expiratory CT scans, which are not present on inspiration CT images, indicate gas trapping and represent a gas-trapping disease axis (factor-loading score for expiration-to-inspiration attenuation ratio, 0.81). Measures of CT density variability measured by the standard deviation of the CT histogram and BMI loaded on factor 4 and therefore represent a complex axis capturing risk associated with BMI (low BMI possibly suggesting cachexia, high BMI suggesting obesity), as well as CT “noise” potentially capturing risk associated with both high and low attenuation present in individuals (e.g., low attenuation attributable to emphysema combined with high attenuation attributable to fibrotic lung diseases). Total lung capacity and functional residual capacity are the only variables that loaded strongly on factor 5 (Table 2). The coefficients for the derivation of factor scores of this model are provided in Web Table 3.
Relationships between disease axes and all-cause mortality
A total of 950 deaths occurred over a mean follow-up time of 6.3 years, representing 46,079 person-years of follow-up. Older age (P < 0.0001), male sex (P = 0.018), being a current versus former smoker (P = 0.048), and pack-years of smoking (P = 0.0003) were all positively associated with death (Table 3). Lower BMI was associated with higher risk for death (P < 0.0001) in this population of current and former smokers enriched for more severe COPD. The emphysema disease axis was associated with higher risk for death (P = 0.045). The airway disease axis was associated with the greatest risk for death (P < 0.0001) and, in addition to the linear term, a squared term for the airway disease axis was also significantly associated with greater risk for death (P = 0.027).
Table 3.
Cox Proportional Hazard Model of Death, Survival Follow-up December 2016, the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease Study Cohort, United States, 2008–2011
| Variable | β Estimate | Standard Error | P Value | Hazard Ratio | 95% CI |
|---|---|---|---|---|---|
| Age | 0.03058 | 0.00556 | <0.0001 | 1.031 | 1.02, 1.04 |
| Male sex | 0.00334 | 0.00141 | 0.0178 | 1.003 | 1.00, 1.01 |
| Current smoker | 0.19166 | 0.09702 | 0.0482 | 1.211 | 1.002, 1.465 |
| Pack-years of smoking | 0.30179 | 0.08248 | 0.0003 | 1.352 | 1.15, 1.59 |
| BMI (1 unit) | −0.0569 | 0.00867 | <0.0001 | 0.945 | 0.929, 0.961 |
| High blood pressure | 0.17596 | 0.08129 | 0.0304 | 1.192 | 1.017, 1.398 |
| Emphysema disease axisa | 0.11584 | 0.05784 | 0.0452 | 1.123 | 1.002, 1.258 |
| Airway disease axisb | 0.64179 | 0.05853 | <0.0001 | 1.900 | 1.694, 2.131 |
| Interaction between the emphysema and airway disease axes | 0.16892 | 0.0474 | 0.0004 | ||
| Airway disease axis squared | 0.07635 | 0.0345 | 0.0269 | 1.079 | 1.009, 1.155 |
| Gas trapping | 0.03526 | 0.05231 | 0.5004 | 1.036 | 0.935, 1.148 |
| CT intensity variability/noise | 0.30924 | 0.04911 | <0.0001 | 1.362 | 1.237, 1.500 |
| TLC and FRC | −0.0398 | 0.04004 | 0.3205 | 0.961 | 0.888, 1.039 |
Abbreviations: BMI, body mass index; CI, confidence interval; CT, computed tomography; FRC, functional residual capacity; TLC, total lung capacity.
a Hazard ratio for factor 1 is presented for factor 2 = 0.
b Hazard ratio for factor 2 is presented for factor 1 = 0.
We explored both the airway and emphysema axes by assessing their relationship with death, using deciles of each axis. The nonlinear risk observed across deciles of both axes prompted us to test for a statistical relationship (Web Figure 2). Furthermore, there was a statistically significant synergistic interaction between the emphysema disease axis and the airway disease axis (P = 0.001) on death risk. Neither the gas-trapping factor nor the total lung capacity and functional residual capacity factor were related to all-cause mortality (P = 0.5 and P = 0.3, respectively). The CT intensity variability factor that included BMI was positively associated with a higher risk of death (P < 0.0001).
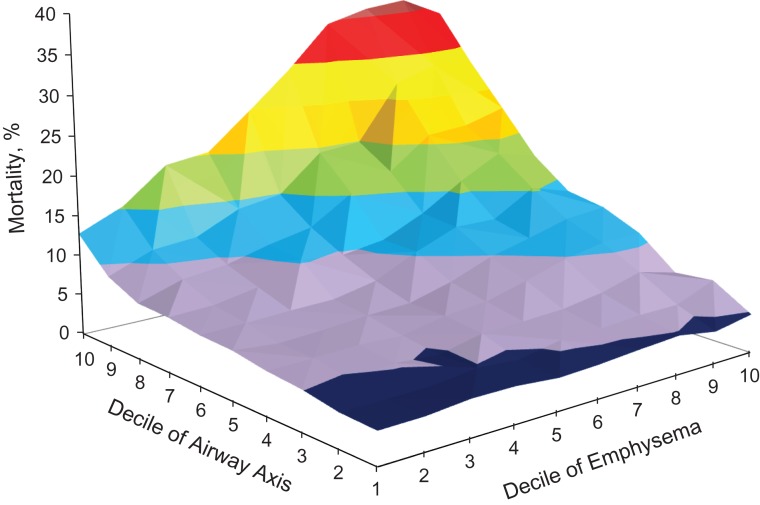
The complex relationship among the emphysema and airway disease axes with death is summarized in Figure 1. The z-axis represents the probability of all-cause mortality ranging from less than 5% to 40% for each decile of loading score for factors 1 and 2 in a Cox proportional hazards model adjusted for age, sex, current smoking, pack-years of smoking, BMI, high blood pressure, each of the 5 factors, the interaction between factors 1 and 2, and a quadratic term for factor 2. As can be seen in Figure 1 in dark blue (the decile of emphysema axis ranging from 1, small loading score, to 10, large loading score), there was not a significant increase in death for low levels of the emphysema disease axis, and even at high levels of the emphysema disease axis, the death rate was not elevated at the lower end of the distribution of the airway disease axis. The airway disease axis was strongly associated with death at all levels of the emphysema axis (decile of airway axis in Figure 1 ranging from 1, small loading score, to 10, large loading score) and with the increase in the death rate being more than a simple linear function. The synergistic interaction between the emphysema and airway disease axes can be seen as the greater mortality rate associated with the higher levels of both (i.e., the progression dark blue to the upper, rear quadrant of the surface plot shown in red in Figure 1.
Figure 1.
The relationship among the emphysema and airway disease axes with death, Genetic Epidemiology of Chronic Obstructive Pulmonary Disease Study, United States, 2008–2011. The z-axis represents the probability of all-cause mortality, ranging from 4% (dark blue), 5%–10% (purple), 10%–15% (blue), 15%–20% (green), 20%–25% (orange), 25%–30% (yellow), 30%–35% (red), to greater than 35% (dark red) for each decile of loading score for factors 1 (emphysema axis) and 2 (airway axis) in a Cox proportional hazards model adjusted for age, sex, being a current smoker, pack-years of smoking, body mass index (calculated by dividing the weight in kilograms by height in meters squared), high blood pressure, each of the 5 factors, the interaction between factors 1 and 2, and a quadratic term for factor 2. The x- and y-axes represent deciles of each axis, ranging from 1, representing a small loading score, to 10, representing a large loading score.
DISCUSSION
COPD has long been recognized as a heterogeneous disease (4). In recent uses of multidimensional analyses, researchers have characterized this heterogeneity by clustering individuals into discrete phenotypic categories (5). Our approach attempts to identify multidimensional vectors on the basis of combined spirometric and CT data, with each person contributing to each vector depending upon their values for all variables within this multivariable analysis. This provides a continuous distribution for each vector representing an underlying physiologic process, which can be interpreted based on the factor loadings of each individual variable.
Conducting factor analysis of subjects in the COPDGene Study revealed 5 unique, multidimensional factors from the correlation structure of pulmonary function and morphologic measures obtained from chest CT imaging. Spirometric measures contributed to 2 of the factors, which were defined on the basis of morphologic measures from the CT images: the emphysema disease axis, characterized by low attenuation areas from inspiration CT, and the airway disease axis, characterized by measures of airway wall thickness. These 2 disease axes were associated with death. Furthermore, a synergistic interaction became apparent such that high levels of both factors were associated with the greatest risk for death.
Morphologic measures from chest CT imaging and measures of pulmonary function were included in this analysis. Vector labels from chest CT variables indicate the observed morphologic differences. For example, the emphysema axis represents strong factor loadings of low attenuation area on inspiration CT scans (i.e., less than −950 HU and less than −910HU), and the airway axis represents strong factor loadings of airway thickness (i.e., segmental wall area percentage, square root of the wall area of a derived airway with lumen circumferences of 10 mm and 15 mm). These were labeled as disease axes on the basis of their strong inverse factor loadings of pulmonary function (i.e., FEV1/FVC for the emphysema and airway disease axes and FEV1% predicted for the airway disease axis).
Low pulmonary function is a well-established risk factor for death (2), and the results of our study are consistent with that observation. In addition, these analyses partition pulmonary function variables into the proportion associated with an emphysema disease axis and the proportion associated with an airway disease axis. To illustrate, reading Table 2 from left to right, the loading scores reported for the row labeled FEV1 show a negative relationship with the emphysema disease axis in column 1 (loading score = −0.41), a stronger negative relationship with the emphysema disease in column 2 (loading score = −0.89), and a weaker negative relationship with the gas-trapping axis in column 3. To assess the impact of these disease axes on death independent of pulmonary function, FEV1% predicted and the FEV1/FVC ratio were removed from the disease axes and were included directly as independent variables in the Cox proportional hazard model along with the disease axes. The airway disease axis remained independently associated with death rate, whereas the emphysema disease axis was only marginally associated with overall death rate after adjustment for pulmonary function (data not shown). These analyses indicate an important role for pulmonary function on death associated with emphysema as well as an independent role associated with the airway disease axis.
Limitations of this study include the use of several measures of CT intensity on inspiration and expiration that have not been directly shown to have clinical relevance. These measures fell into predictable factors, however, which suggests the values chosen (e.g., inspiration percentage of the lung less than −856, less than −910, and less than −950) were correlated measures of a similar disease process with clinical relevance. COPDGene is a large study of current and former smokers with a history of smoking cigarettes for more than 10 pack-years. This likely makes the generalizability of the results questionable when applied to a population with shorter history of cigarette smoking. The COPDGene Study has experienced loss to follow-up over this time, and this can induce bias. Table 1 indicates that all characteristics except race were not statistically different between the main cohort and the cohort used for the factor analysis.
Disease axes, compared with disease clusters, may be difficult to interpret in a clinical setting, making the direct applicability of this approach complex for physicians. This approach does avoid the potential misinterpretation of assessing individual variables in the presence of highly correlated data. However, future work should include assessment of inflection points in the risk for all-cause mortality in each of the factors that define high-risk subgroups on the basis of the continuous disease axes. Although clustering approaches have not achieved strong separation for COPD subtypes, inflection points in risk may suggest reasonable clinical cutpoints for individuals. Disease axes may provide important insights into the pathophysiologic processes leading to COPD and death associated with COPD and provide potential targets for intervention for prevention or treatment of COPD.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Epidemiology, Colorado School of Public Health, University of Colorado Anschutz Medical Campus, Aurora, Colorado (Gregory L. Kinney, Kendra A. Young, John E. Hokanson); Department of Mathematical and Statistical Sciences, University of Colorado Denver, Denver, Colorado (Stephanie A. Santorico); Human Medical Genetics and Genomics Program, University of Colorado School of Medicine, Aurora, Colorado (Stephanie A. Santorico); Division of Biostatistics and Bioinformatics, Office of Academic Affairs, National Jewish Health, Denver, Colorado (Stephanie A. Santorico, Douglas C. Everett); Channing Division of Network Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Michael H. Cho, Peter J. Castaldi, Raul San José Estépar, James C. Ross, Edwin K. Silverman, George R. Washko); Department of Electrical & Computer Engineering, Northeastern University, Boston, Massachusetts (Jennifer G. Dy); Department of Medicine, National Jewish Health, Denver, Colorado (Barry J. Make, Elizabeth A. Regan, James D. Crapo); Department of Radiology, National Jewish Health, Denver, Colorado (David A. Lynch); and Department of Biostatistics and Informatics, University of Colorado Anschutz Medical Campus, Aurora, Colorado (Sharon M. Lutz).
This project was supported by grants R01 HL089897, R01 HL089856, and K08 HL097029 (to S.M.L.) from the National Heart, Lung, and Blood Institute. The Genetic Epidemiology of COPD Study is also supported by the COPD Foundation through contributions made to an industry advisory board composed of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Conflict of interest: E.K.S. has received honoraria and consulting fees from Merck, grant support and consulting fees from GlaxoSmithKline, and honoraria from Novartis. These funding sources played no role in the design of the study or the decision to submit the manuscript for publication. The other authors report no conflicts.
Abbreviations
- BMI
body mass index
- COPD
chronic obstructive pulmonary disease
- COPDGene
Genetic Epidemiology of Chronic Obstructive Pulmonary Disease
- CT
computed tomography
- FEV1
forced expiratory volume at 1 second
- FEV1%
percentage of forced expiratory volume at 1 second
- FVC
forced vital capacity
- FVC%
percentage of forced vital capacity
- HU
Hounsfield unit
- SSDI
Social Security Death Index
REFERENCES
- 1. Friedlander AL, Lynch D, Dyar LA, et al. Phenotypes of chronic obstructive pulmonary disease. COPD. 2007;4(4):355–384. [DOI] [PubMed] [Google Scholar]
- 2. Silverman EK. Exacerbations in chronic obstructive pulmonary disease: do they contribute to disease progression? Proc Am Thorac Soc. 2007;4(8):586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Løkke A, Lange P, Scharling H, et al. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61(11):935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanders KJC, Ash SY, Washko GR, et al. Imaging approaches to understand disease complexity: chronic obstructive pulmonary disease as a clinical model. J Appl Physiol (1985). 2018;124(2):512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castaldi PJ, Benet M, Petersen H, et al. Do COPD subtypes really exist? COPD heterogeneity and clustering in 10 independent cohorts. Thorax. 2017;72(11):998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Stat. 2010;2(4):433–459. [Google Scholar]
- 7. Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J. 2005; 26(2):319–338. [DOI] [PubMed] [Google Scholar]
- 9. Hankinson JL, Odencrantz JR, Fedan KB, et al. Spirometric reference values from a sample of the general US population. Am J Respir Crit Care Med. 1999;159(1):179–187. [DOI] [PubMed] [Google Scholar]
- 10. Han MK, Kazerooni EA, Lynch DA, et al. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261(1):274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diaz AA, Come CE, Ross JC, et al. Association between airway caliber changes with lung inflation and emphysema assessed by volumetric CT scan in subjects with COPD. Chest. 2012;141(3):736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim WJ, Silverman EK, Hoffman E, et al. CT metrics of airway disease and emphysema in severe COPD. Chest. 2009;136(2):396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakano Y, Wong JC, de Jong PA, et al. The prediction of small airway dimensions using computed tomography. Am J Respir Crit Care Med. 2005;171(2):142–146. [DOI] [PubMed] [Google Scholar]
- 14. Xu J, Murphy SL, Kochanek KD, et al. Deaths: final data for 2013. Natl Vital Stat Rep. 2016;64(2):1–119. [PubMed] [Google Scholar]
- 15. Horn JL. A rationale and test for the number of factors in factor analysis. Psychometrika. 1965;30(2):179–185. [DOI] [PubMed] [Google Scholar]
- 16. Stewart JI, Moyle S, Criner GJ, et al. Automated telecommunication to obtain longitudinal follow-up in a multicenter cross-sectional COPD study. COPD. 2012;9(5):466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.