Abstract
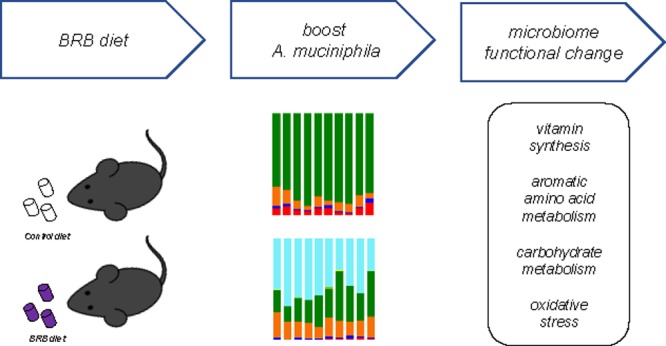
Gut microbiome plays an essential role in host health through host–gut microbiota metabolic interactions. Desirable modulation of beneficial gut bacteria, such as Akkermansia muciniphila, can confer health benefits by altering microbiome-related metabolic profiles. The purpose of this study is to examine the effects of a black raspberry-rich diet to reshape the gut microbiome by selectively boosting A. muciniphila population in C57BL/6J mice. Remarkable changes of the mouse gut microbiome were revealed at both compositional and functional levels with an expected increase of A. muciniphila in concert with a profound impact on multiple gut microbiome-related functions, including vitamin biosynthesis, aromatic amino acid metabolism, carbohydrate metabolism, and oxidative stress. These functional alterations in the gut microbiome by an easily accessed freeze-dried black raspberry-supplemented diet may provide novel insights on the improvement of human health via gut microbiome modulation.
Introduction
Our gut microbiota, which consists of up to 100 trillion microorganisms, encodes 100 times more genes than our own genome.1 The human body interacts with the gut bacteria in numerous ways. For instance, microbial fermentation of dietary fiber produces short-chain fatty acids, such as butyrate, acetate, and propionate, which are not only energy substrates but also involved in diverse signaling pathways.2 However, growing evidence shows that the gut microbiome and its metabolic functions can be readily perturbed by exposure to environmental toxic agents, such as heavy metals or pesticides.3−5 Disrupted gut microbiome (dysbiosis) is frequently associated with adverse health outcomes, such as obesity or a compromised immune system,6,7 and we define such perturbed gut microbiome as an unhealthy one. An unhealthy gut microbiome caused by environmental insults would lead to numerous adverse health effects, and its compositional and functional characteristics can be partially responsible. For example, the Firmicutes/Bacteroidetes ratio in gut microbial composition is associated with obesity and the gut microbiome in obese individuals has enhanced microbial functions for energy extraction than that present in lean individuals.8 Bäckhed et al. described the healthy microbiome as a microbial community with structure and function that presumably confer health benefits for the host.9 Therefore, it is of significance to be able to modulate the gut microbiome toward a bacterial community profile that promotes “healthy” outcomes. Currently, a simple, effective, and selective approach with the potential for broad applicability has not been adequately explored yet. In recent years, a series of patterns for gut microbiome alterations associated with various diseases have been documented but the functional impact of those changes still remains elusive.9 A number of gut bacteria have proven health-associated functions and clinical efficacy. For instance, probiotic strains present in Lactobacillus and Bifidobacterium genera are able to confer a health benefit in human when treating gastrointestinal disorders.9 Thus, encouraging beneficial and inhibiting detrimental bacteria would be a reasonable approach to transform the gut microbiome into one that confers health benefits, and the investigation on the consequent functional changes resulting from compositional alterations is also warranted.
A number of recent studies have demonstrated that Akkermansia muciniphila, commonly found in human and mouse gut microbiome, plays a key role in regulating glucose homeostasis, inflammation, gut barrier function, adipose tissue metabolism, fat mass storage, and host metabolic functions in diverse animal models and human.10−20 It has been shown that the development of obesity and associated metabolic disorders could be attenuated by A. muciniphila treatment.18 In this context, methods to selectively increase A. muciniphila are highly desired. We propose a simple approach for the modulation of the gut microbiome by boosting A. muciniphila with a black raspberry (Rubus occidentalis, BRB)-rich diet. We hypothesize that BRBs could lead to a notable increase of A. muciniphila, thus modulating the gut microbiome and its metabolic functions to provide health benefits to the host. We used whole, ripe, freeze-dried BRBs in this study with the intent to increase A. muciniphila, hence reshaping the mouse gut microbiome. The rationale for this choice is that it has been reported that the gut microbiota was shifted in rats fed with a diet supplemented with freeze-dried berries.21,22 In particular, oligofructose and polyphenols are both abundant in berry fruits and administration of oligofructose and dietary polyphenols can increase the A. muciniphila population in mice.23,24 Given the pivotal role played by the gut microbiome in human health as well as the ability of BRBs to modify the gut microbiota, the BRBs have the potential to modulate mouse gut microbiome by increasing the beneficial gut bacteria, such as A. muciniphila, hence promoting host health and decreasing disease risk. Furthermore, it is also relevant and applicable to elucidate metabolic function changes associated with the gut microbiome modulation by BRBs.
In the present study, a BRB-rich diet was employed to directionally modulate the mouse gut microbiome. We combined 16S rRNA gene sequencing and shotgun metagenomics sequencing to investigate the interplay between the gut microbiome and the BRB-supplemented diet. The gut microbial profiles after 7 week BRB diet examined by 16S rRNA sequencing and metagenomics sequencing showed a significantly altered gut microbial composition in C57BL/6 mice with a 157-fold A. muciniphila in concert with a remarkable impact on the microbiome-related functions, including vitamin biosynthesis, aromatic amino acid metabolism, carbohydrate metabolism, and oxidative stress, that can potentially influence host health.
Results
The American Institute of Nutrition (AIN)-76A diet and BRB diet (formulated on AIN-76A with 10% BRB powder) used in the present study were also used in previous studies for the potential chemopreventive agents in BRBs, for instance, simple and complex phenols or sterols.25 It has been reported that the BRB diet is able to ameliorate ulcerative colitis and esophageal tumorigenesis in animal models.26,27 However, the functional effects of the BRB diet on the gut microbiome have not been examined. With an increasingly strong link between the gut microbiome and human health, characterization of the functional changes driven by the BRB diet in the gut microbiome may provide mechanistic insights regarding the effects by black raspberries on host health and disease. In addition, oligofructose and polyphenols are both important components in berry fruits. Administration of oligofructose and dietary polyphenols can boost the A. muciniphila population in mice,23,24 it is expected to see boosted A. muciniphila after the BRB diet treatment.
Composition and Diversity of Mouse Gut Microbial Communities Were Strikingly Impacted Following Consumption of a BRB-Rich Diet
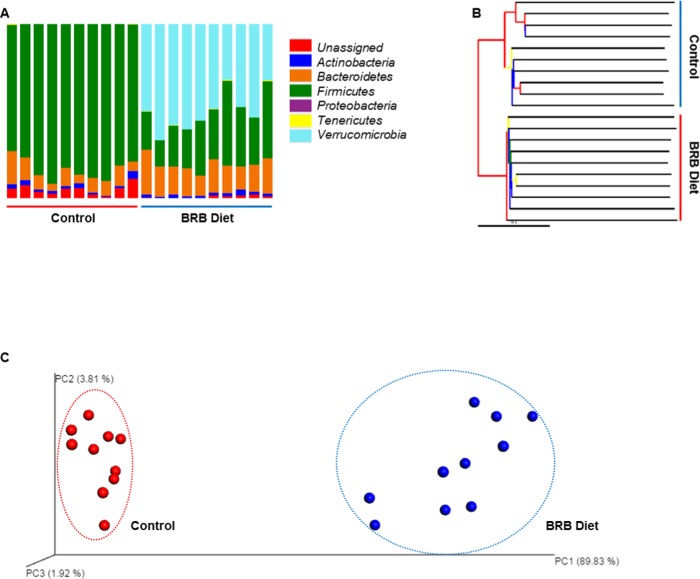
Taxonomic summary bar plots (Figures 1A and S1) show the identified gut bacteria assigned at the phylum level from 16S rRNA sequencing reads, with each color representing a bacterial phylum. The abundance of several bacterial phyla and the general composition of mouse gut microbiota were distinctly different between the control and BRB diet groups. In the control group, Firmicutes (83.41 ± 5.97%) and Bacteroidetes (9.49 ± 4.24%) covered the majority of the mouse gut microbes, followed by Actinobacteria (2.00 ± 1.09%), Tenericutes (0.12 ± 0.13%), Verrucomicrobia (<0.01%), and Proteobacteria (<0.01%). In contrast, in the BRB group, the abundance of the Firmicutes was reduced to 29.68 ± 9.92%, whereas the Bacteroidetes was increased to 17.18 ± 3.79%. This notable ratio change of Firmicutes to Bacteroidetes is consistent with the previous study that lower Firmicutes and higher Bacteroidetes were observed in lean subjects.28 Other bacterial phyla found in the gut microbiota of BRB-fed mice were Actinobacteria (1.68 ± 0.69%), Tenericutes (0.18 ± 0.17%), and Proteobacteria (<0.01%). In particular, the abundance of Verrucomicrobia (50.50 ± 10.65%) was remarkably higher compared to that in the control group. The jackknifed beta diversity and hierarchical clustering analysis via the unweighted pair group method with arithmetic mean (UPGMA) (Figure 1B) indicated that the control and BRB diet groups were typically clustered into their groups. Consistently, three-dimensional principal coordinate analysis (PCoA) plot (Figure 1C) shows that the structure of gut microbial community is distinct from each other. The control and BRB groups were well separated and clustered together in their distinct groups; 89.83, 3.81, and 1.92% variation were explained by principle component (PC)1, PC2, and PC3, respectively. In addition, Figure S2 shows the gut bacterial genera that were altered significantly after BRB diet treatment compared to controls (p < 0.05). There were 17 altered genera in total, with 7 increased and 10 decreased genera. Interestingly, all 10 decreased bacterial genera were from phylum Firmicutes, whereas 2 genera from phylum Bacteroidetes significantly increased, which is consistent with a decreased Firmicutes/Bacteroidetes ratio.
Figure 1.
(A) Gut microbial composition at phylum level in the BRB diet and control groups, with each color representing one phylum. (B) Hierarchical clustering analysis by UPGMA with the UPGMA distance tree constructed at a distance of 0.1. (C) Three-dimensional PCoA plot, based on the UniFrac distance metric and beta diversity, shows that the BRB diet and control group are well separated.
Highly Enriched A. muciniphila Population and Functional Metagenome Comparison
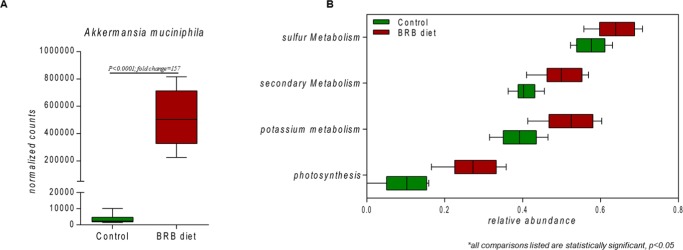
As hypothesized, the normalized gene counts show that the A. muciniphila population in the mouse gut microbiome increased by approximately 157-fold in the BRB group compared to that in control (Figure 2A). Furthermore, the gut microbiome-related functions have been profoundly altered by BRBs supported by the gut bacterial metagenome comparisons. Figure 2B shows that several metabolic subsystems at level 1 of the SEED subsystem were significantly changed in the mouse gut microbiome, including photosynthesis, potassium metabolism, secondary metabolism, and sulfur metabolism. In particular, bacterial functions related to the biosynthesis of vitamins and aromatic amino acids exhibited significant enrichment. Carbohydrate metabolism, especially sugar alcohol and di- and oligosaccharide utilization, was also significantly changed. In addition, several bacterial genes encoding antioxidative enzymes, as well as programmed cell death (PCD) proteins, were shifted between the control and BRB diet groups.
Figure 2.
(A) Comparison of A. muciniphila population between control and BRB diet groups. (B) Comparative metagenomic analysis at the level 1 SEED subsystem with photosynthesis, potassium metabolism, secondary metabolism, and sulfur metabolism being significantly altered by BRB treatment (p < 0.05).
Enrichment of Genes Involved in Vitamin Synthesis
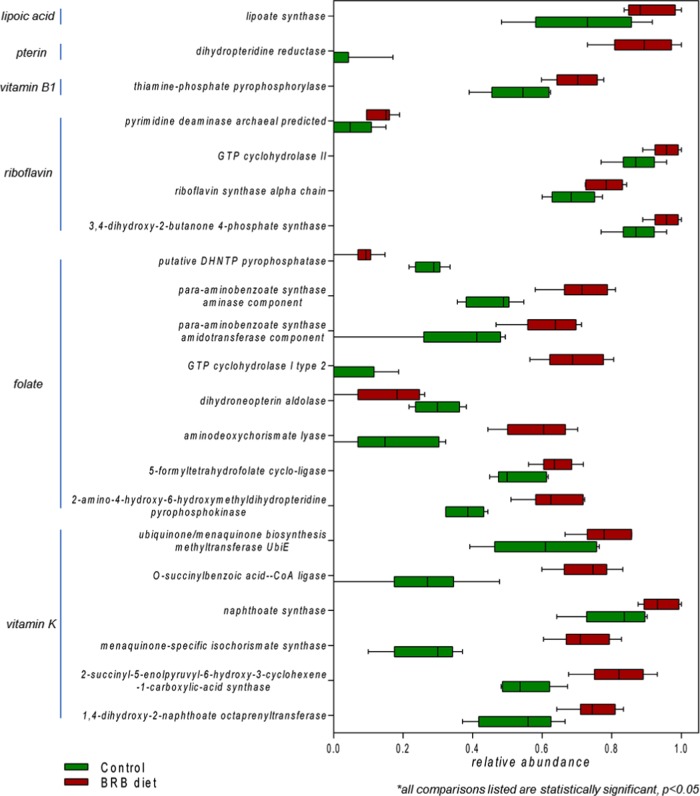
The gut microbiome is an important source of vitamins (especially vitamins B and K) that are essential to human health.29 In this study, we discovered a significant enrichment of bacteria genes encoding key enzymes involved in the pathways of vitamin synthesis. Figure 3 displays the abundance distribution of genes involved in the biosynthetic pathways of riboflavin, folate, and vitamin K. The relative abundances of GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase, which are rate-limiting enzymes in riboflavin synthesis,30 significantly increased in the BRB diet group compared to those in controls. Likewise, a series of genes encoding enzymes involved in the biosynthesis of folate and vitamin K (phylloquinone and menaquinone) were also significantly enriched (p < 0.05). Besides these, the relative abundances of bacterial genes of the biosynthesis of other vitamins or cofactors, such as vitamin B1 (thiamine-phosphate pyrophosphorylase), pterin (dihydropteridine reductase), and lipoic acid (lipoate synthase), were all significantly enriched in the BRB diet group compared to those in controls (Figure 3).
Figure 3.
Enrichment of gut bacterial genes involved in vitamin biosynthesis in the gut microbiome of mice fed on the BRB diet. (All comparisons listed are statistically significant, p < 0.05.)
Enrichment of Genes Involved in Aromatic Amino Acid Synthesis
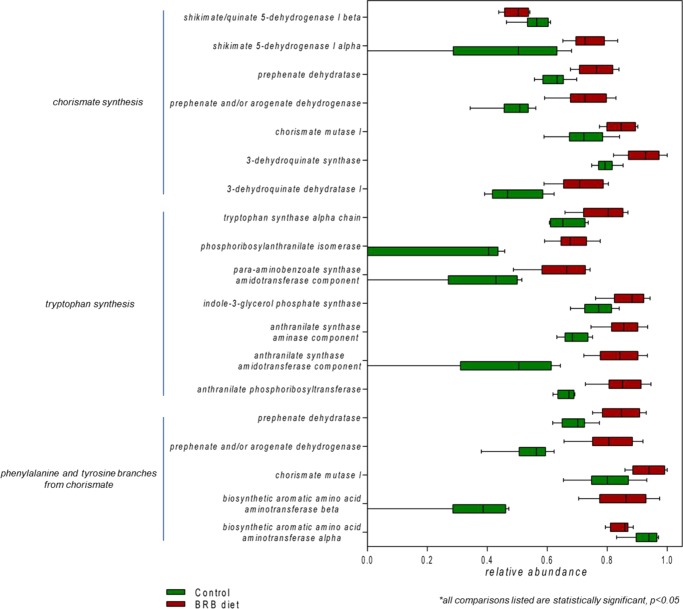
We next looked into the gut bacterial synthesis of amino acids. The metabolism of aromatic amino acid was significantly enriched with BRB diet treatment. The synthesis of chorismate, an important precursor for aromatic amino acids, was elevated with multiple genes encoding its biosynthetic enzymes being enriched (Figure 4). Likewise, the relative abundances of bacterial genes encoding enzymes involved in the biosynthesis of phenylalanine and tyrosine significantly increased. In addition, the biosynthesis of tryptophan, which is an essential amino acid, was also enriched in the BRB diet group. Specifically, the bacterial gene encoding the β subunit of tryptophan synthase that catalyzes the last two steps in tryptophan biosynthesis was enhanced.31 In addition, the relative abundances of genes encoding tryptophan-biosynthetic enzymes, such as anthranilate phosphoribosyltransferase, indole-3-glycerol-phosphate synthase, anthranilate synthase, and phosphoribosylanthranilate isomerase, were all significantly elevated.
Figure 4.
Enrichment of gut bacterial genes involved in aromatic amino acid metabolism in the gut microbiome of mice fed on the BRB diet. (All comparisons listed are statistically significant, p < 0.05.)
BRBs Decreased Bacterial Genes Involved in Sugar Alcohol and Di- and Oligosaccharide Utilization
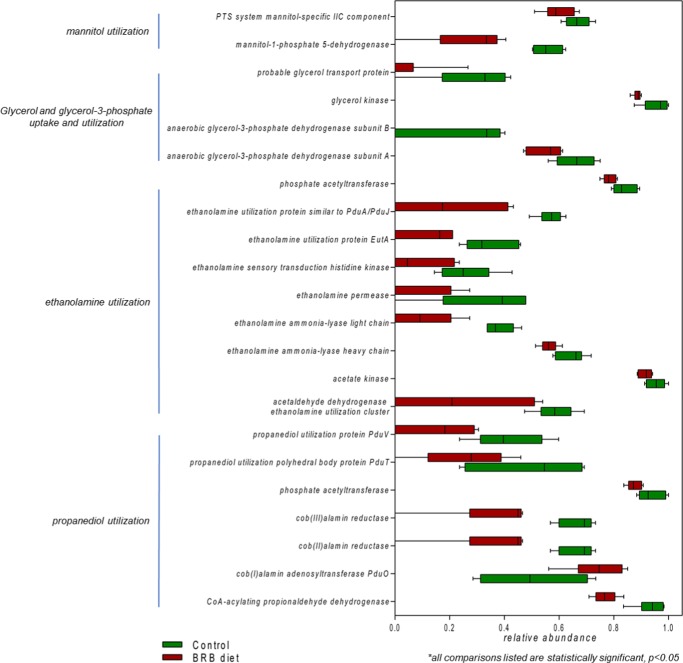
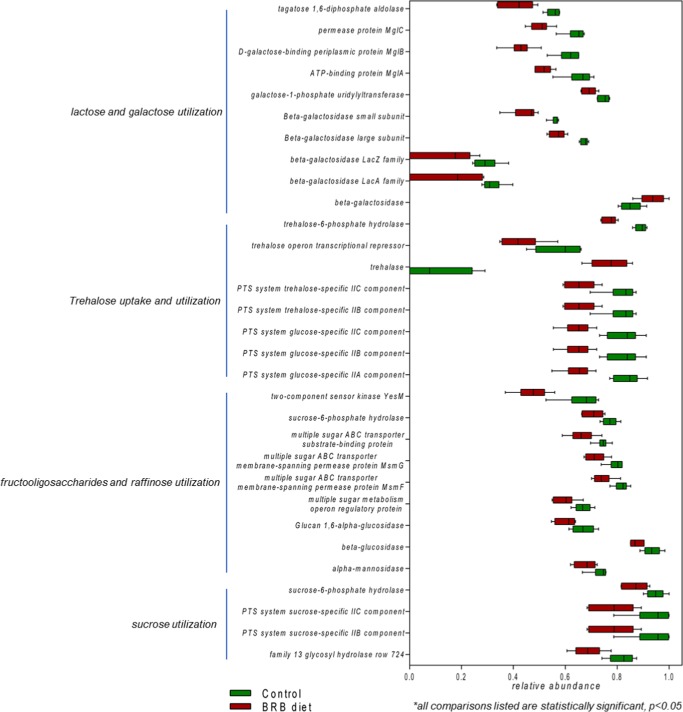
A significant decrease in the abundance of genes involved in sugar alcohol and di- and oligosaccharide utilization was found in mouse gut microbiome of the BRB diet group compared to that in the control. Specifically, for sugar alcohols, the utilization of propanediol, ethanolamine, glycerol and glycerol-3-phosphate, and mannitol decreased (Figure 5). Meanwhile, for di- and oligosaccharides, the utilization of trehalose, fructooligosaccharides and raffinose, sucrose, lactose, and galactose also significantly decreased (Figure 6). This obvious reduction in sugar alcohol and di- and oligosaccharide utilization indicated significant downregulation in an extensive repertoire of gut bacterial genes involved in the utilization of a variety of sugars and might influence the carbohydrate metabolism and energy extraction efficiency in gut microbiota.
Figure 5.
Reduction of gut bacterial genes involved in sugar alcohol utilization in the gut microbiome of mice fed on the BRB diet. (All comparisons listed are statistically significant, p < 0.05.)
Figure 6.
Reduction of gut bacterial genes involved in di- and oligosaccharide utilization in the gut microbiome of mice fed on the BRB diet. (All comparisons listed are statistically significant, p < 0.05.)
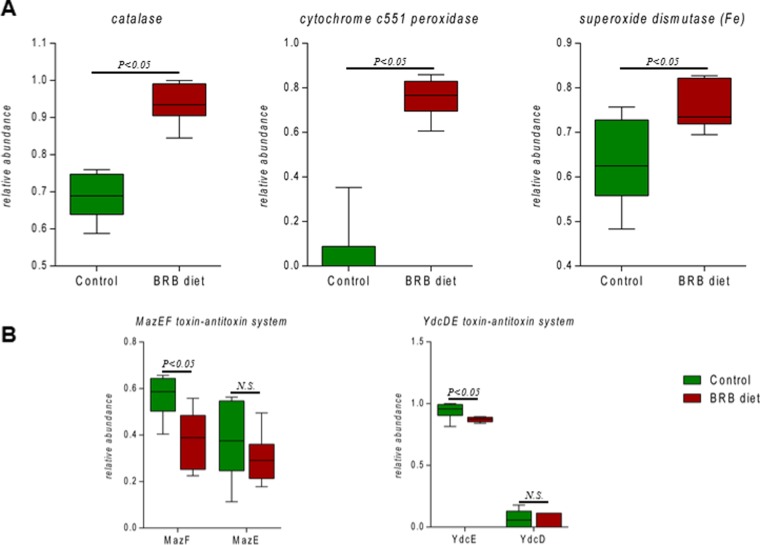
Enrichment of Genes Encoding Antioxidative Enzymes and Reduced Programmed Cell Death (PCD)
We compared several genes encoding antioxidative enzymes in gut bacteria between the BRB diet and control groups. As shown in Figure 7A, the relative abundances of catalase, cytochrome c551 peroxidase, and superoxide dismutase (Fe) were significantly upregulated in the BRB group compared to those in the control. In addition, the relative abundances of PCD system toxin genes mazF and ydcE decreased in the BRB diet group, whereas antitoxin genes mazE and ydcD did not show significant difference (Figure 7B). Toxin–antitoxin systems are the most studied forms of PCD in bacteria. For example, the mazEF module found in Escherichia coli is a typical toxin–antitoxin system that triggers PCD.32 The mazEF system comprises two adjacent genes mazE and mazF, encoding an antitoxin and a toxin, respectively.33 The mediation of the mazEF system could trigger PCD under conditions, such as oxidative stress.34 Meanwhile, the ydcDE system found in Bacillus subtilis is very similar to the mazEF system with ydcD and ydcE encoding an antitoxin and a toxin, respectively.35 Together, these data indicate that the antioxidative capacity of the gut bacteria was reinforced and the progress of PCD was inhibited.
Figure 7.
Distribution of gut bacterial genes encoding antioxidative enzymes ((A) all comparisons listed are statistically significant, p < 0.05) and genes involved in programmed cell death (B).
Discussion
Mounting evidence shows that host health and susceptibility to diseases can be greatly influenced by the developmental trajectory of the gut bacteria, including alterations of the microorganisms and their collective genomes.36 To improve the host fitness through effective modulation of the gut microbiome is of importance for the protection from metabolic disorders associated with dysbiosis. Therefore, it is imperative to develop an effective approach by which we can directionally modify the gut microbiota, particularly its associated functions. In this study, we used high-throughput 16S rRNA gene sequencing and shotgun metagenomics gene sequencing to investigate the effects of a simple approach for the modulation of gut microbiome with a BRB-rich diet. The results clearly demonstrated that the structure of the mouse gut microbial community changed by the BRB diet treatment with enriched A. muciniphila population, more importantly, the gut microbial metagenome experienced significant changes that could potentially confer benefits for the host by altering bacterial metabolic functions. These findings supported our hypothesis that BRB is able to modulate mouse gut microbiome probably by inducing A. muciniphila population and may provide novel insights regarding the gut microbiome as a therapeutic target to develop new and effective approaches to cope with gut microbiome-related adverse outcomes.
An easily accessed freeze-dried black raspberry-supplemented diet was used to selectively increase A. muciniphila, hence modulating the mouse gut microbiome. This selection is grounded in the fact that BRB is associated with many beneficial health effects, and more importantly, the abundance of A. muciniphila can be potentially boosted by BRBs. Specifically, after 7 week BRB diet treatment, we identified a notable increase of A. muciniphila by 157-fold compared to that in controls. In addition, the functional metagenome profiles were also significantly changed after the BRB diet treatment, with metabolic alterations in bacterial pathways involved in vitamin synthesis, aromatic amino acid synthesis, carbohydrate utilization, and oxidative stress, indicating that the BRB diet not only induced alterations in the gut microbiota at the abundance level, but also had a profound impact on its metabolic functions.
It is previously reported that obesity is associated with the ratio of Firmicutes to Bacteroidetes. In particular, the abundance of Bacteroidetes is lower, whereas the abundance of Firmicutes is higher in obese than that in lean subjects.28 The compositional pattern of gut microbiota in mice fed with BRB diet is opposite to that observed in obesity. The ratio of Firmicutes to Bacteroidetes is lower in the BRB group compared to that in controls. Likewise, it is previously reported that the A. muciniphila population decreased in obese children and increased after weight loss and was negatively correlated with BMI.37−39 In the present study, the A. muciniphila population was highly induced by BRB. Along with the progress of obesity, besides a characteristic bacterial compositional pattern, a series of metabolic disorders, such as diabetes and metabolic syndrome, would ensue,40 suggesting an association between the changing trajectory of gut bacteria and the deterioration of metabolic disorders. Thus, it is likely that the development of metabolic disorders could be reversed by the gut microbiome modulation and its functional reprogramming. That the abundance of Firmicutes decreased, whereas Bacteroidetes increased in mice fed with BRB diet in the present study may imply less susceptibility to metabolic disorders. In addition, it is suggested recently that the gut microbiome plays an important role in the development of obesity through its capacity for energy harvest from the diet.28 Carbohydrate utilization is considered to be involved in the energy metabolism of the gut microbiota, hence affecting its efficiency of energy extraction. After the BRB diet treatment, we discovered a major decrease in carbohydrate utilization especially sugar alcohols and di- and oligosaccharides in mouse gut microbiome, which indicated alterations in bacterial energy extraction efficiency and carbohydrate utilization strategy. Therefore, BRB may have the potential to protect the host from metabolic deterioration by steering the developmental trajectory of gut microbiota in a direction opposite to that in obesity.
Gut microbiome modulation by BRB can benefit the host health through enhanced bacterial biosynthesis of important biomolecules, such as vitamins and essential amino acids. These biomolecules play an indispensable role in maintaining cellular functions of the host; however, most of them cannot be synthesized by human body.41 Therefore, the deficiency or even suboptimal levels would inevitably result in health problems.42 In this study, we discovered that the abundance of multiple bacterial genes encoding key enzymes involved in biosynthesis of vitamin K, riboflavin, and folate were significantly upregulated in the BRB diet group. Modulation of the gut microbiome could be a novel approach to optimize the body level of vitamins. For example, sufficient body levels of folate would prevent neural tube defects42 and folate can be obtained via the gut bacterial biosynthesis. Likewise, a recent study suggested that besides vitamin D, vitamin K can also improve health by conferring protective effects on cardiovascular and skeletal systems, hence decreasing osteoporosis risk.43 The gut microbiome exerts impact not only within the gastrointestinal tract but also on bone health, which manifested the idea that a series of host–microbe metabolic axes connect the gut microbiome and distant organs, such as liver, bone, and brain, via metabolic interactions.36 In addition, bacterial genes involved in tryptophan synthesis also showed significant enrichment. As an essential amino acid, tryptophan is linked to the regulation of food intake, mood disorders, and immune responses.44 Clearly, improved bacterial production of vital biomolecules would confer health effects; thus, these findings may provide new insights regarding the micronutrient supplement via gut microbiome modulation.
As expected, highly enriched A. muciniphila population (157-fold compared to controls) was induced by BRBs in the mouse gut microbiota, far more than its usual constitution in mouse cecal microbial community.45A. muciniphila was first described as a new strain with mucin-degrading ability in 2004 and has been linked to human health ever since.46 For example, increased expression of genes involved in immune response was shown in mice with colonization of A. muciniphila.45 Likewise, the abundance of A. muciniphila is negatively correlated with body weight and type 1 diabetes in human and mice.10 In the present study, we achieved a remarkable A. muciniphila increase in mice fed with the BRB diet. As a typical mucin-degrading bacterium, A. muciniphila is capable of using mucin as carbon, nitrogen, and energy sources.46 And in the epithelial tissue of intestine, goblet cells are responsible for the major mucin production.47 Previous studies showed that the number of goblet cells and the thickness of mucosal tissue increased in rats fed with oligofructose.48 Therefore, oligofructose, commonly found in berry fruits,49 may be the key factor for the increase of A. muciniphila for its capability to induce mucin production. The hypothesis is supported by the study in which administration of oligofructose increased the A. muciniphila population ∼100 times, which is comparable to the present study.23 Another study enumerated mycolytic bacteria in inflammatory bowel disease patients and discovered that A. muciniphila decreased, whereas another two mucolytic bacteria Ruminococcus gnavus and Ruminococcus torques increased.50 In the present study, the BRB diet group showed an increase of A. muciniphila and decrease of Ruminococcus (Figure S2). In addition, lower levels of A. muciniphila population was recognized in patients with metabolic disorders.19 Increased A. muciniphila is also associated with decreased inflammation in mice.51 Given the antiobesity and antidiabetes functions of A. muciniphila, modulation of gut bacteria by increasing A. muciniphila abundance may attenuate or even eliminate metabolic disorders.
As demonstrated above, the mouse gut microbiome was substantially changed by BRBs at both compositional and functional levels, with a range of potential beneficial effects resulting from bacterial metabolic alterations. Many studies focused on the perturbation and dysfunction of the gut microbiome caused by environmental stress or imbalanced diets. However, a better comprehension of the host–gut microbiome interactions would lead us to develop therapeutic strategies via desirable modulation of gut bacterial metabolism. The BRB diet used in this study to modulate the mouse gut microbiome is only a pilot example that the gut bacteria can be shaped and optimized for human health. More importantly, we demonstrated that it is of promise to improve host health through the interplay between the gut microbiome and host via effective interventions, although answers to the underlying mechanisms of the gut microbiome change as well as the realization of precise manipulation of gut bacterial functions await future studies.
Materials and Methods
Animals and BRB Diet Preparation
A total of 20 specific-pathogen free C57BL/6 mice (∼8 week old), purchased from Jackson Laboratories, were housed in the animal facility of the University of Georgia for 1 week to acclimate with standard pelleted rodent diet and tap water ad libitum provided (protocol number: A2013 06-033-Y3-A3). The BRB diet was prepared essentially as described in ref (52). Briefly, whole ripe BRBs of the Jewel variety were freeze-dried and ground into powder. BRB powder was stored at −20 °C until being incorporated into custom purified American Institute of Nutrition (AIN)-76A animal diet pellets (protein, 20.8 kcal %; carbohydrate, 67.7 kcal %; and fat, 11.5 kcal %)53 by 10% w/w concentration at the expense of corn starch. The diets were stored at 4 °C until being fed to animals. In the beginning of the diet treatment, mice were randomly assigned to either control (AIN-76A diet) or treatment group (BRB diet). They were housed under the environmental conditions of 22 °C, 40–70% humidity, and a 12:12 h light/dark cycle and were provided water ad libitum throughout the experiment period. Regular monitoring for health conditions was twice a week. After 7 weeks, fecal samples from individual mouse were collected and kept at −80 °C immediately for further analysis. The animal protocol was approved by the University of Georgia Institutional Animal Care and Use Committee.
16S rRNA Gene Sequencing
16S rRNA gene sequencing was performed as described in ref (3). DNA was isolated from fecal pellets of individual mouse using PowerSoil DNA isolation kit according to the manufacturer’s instruction. Then, the DNA was amplified using 515F and 806R primers54 targeting the V4 regions of 16S rRNA of bacteria, followed by normalization, barcoding procedure, and finally was pooled to construct the sequencing library. The resultant DNA was quantified using Qubit 2.0 fluorometer and then sequenced using Illumina MiSeq (500 cycles v2 kit) in the Georgia Genomics Facility of University of Georgia. Paired-reads were assembled using the Geneious software (Biomatters, Auckland, New Zealand), and operational taxonomic unit picking and diversity analysis was conducted using the Quantitative Insights into Microbial Ecology (QIIME) software.
Shotgun Metagenomics Sequencing
Shotgun metagenomics sequencing was performed as described in ref (4). DNA (10 ng/μL) of individual mouse was fragmented using a Bioruptor UCD-300 sonication device and then the library was constructed using the Kapa Hyper Prep Kit according to the manufacturer’s instruction. The resultant DNA was quantified using a Qubit 2.0 fluorometer and then sequenced using an Illumina NextSeq high-output flow cell in the Georgia Genomics Facility of University of Georgia. Raw sequencing data were uploaded into the Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST, version 3.6) for automated taxonomic and functional profiling with RefSeq and subsystems databases, respectively.55 A p value <0.05 was considered indicative of a significant difference between two groups.
Statistical Analysis of Data
Principle coordinate analysis (PCoA) was applied to differentiate the gut microbiome profiles between control and treatment samples, which examines the difference of beta diversity based on the UniFrac distance metric.56 Also, we used the jackknifed beta diversity and hierarchical clustering analysis via unweighted pair group method with arithmetic mean (UPGMA) to compare the gut microbiome profiles between the control and treatment group. Moreover, the difference of gut microbial composition was assessed by a nonparametric test via the Metastats software (http://metastats.cbcb.umd.edu/), as described previously.57 The metagenomic sequence count data for taxonomic analysis were processed using DESeq2 for statistics analysis.58
Glossary
Abbreviations
- PCoA
principle coordinate analysis
- BRB
black raspberry
- AIN-76A diet
American Institute of Nutrition-76A diet
- PCD
programmed cell death
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00064.
Gut microbial comparisons at phylum level in the BRB diet and control groups (Figure S1); the fold changes of significantly altered gut bacterial genera in the BRB diet group compared to those in the control group (Figure S2) (PDF)
This work was partially supported by the University of Georgia, University of North Carolina at Chapel Hill and the National Institute of Health/National Institute of Environmental Health Sciences (grant number R01ES024950).
Ethics approval and consent to participate: The animal experiment was approved by the University of Georgia Institutional Animal Care and Use Committee.
The authors declare no competing financial interest.
Supplementary Material
References
- Bäckhed F.; Ley R. E.; Sonnenburg J. L.; Peterson D. A.; Gordon J. I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Tremaroli V.; Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Lu K.; Abo R. P.; Schlieper K. A.; Graffam M. E.; Levine S.; Wishnok J. S.; Swenberg J. A.; Tannenbaum S. R.; Fox J. G. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 2014, 122, 284. 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B.; Bian X.; Mahbub R.; Lu K. Sex-specific effects of organophosphate diazinon on the gut microbiome and its metabolic functions. Environ. Health Perspect. 2017, 125, 198. 10.1289/EHP202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B.; Bian X.; Chi L.; Tu P.; Ru H.; Lu K. Editor’s Highlight: Organophosphate Diazinon Altered Quorum Sensing, Cell Motility, Stress Response, and Carbohydrate Metabolism of Gut Microbiome. Toxicol. Sci. 2017, 157, 354–364. 10.1093/toxsci/kfx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H.; Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 2011, 121, 2126–2132. 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau A. L.; Ahern P. P.; Griffin N. W.; Goodman A. L.; Gordon J. I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Bäckhed F.; Fulton L.; Gordon J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F.; Fraser C. M.; Ringel Y.; Sanders M. E.; Sartor R. B.; Sherman P. M.; Versalovic J.; Young V.; Finlay B. B. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 2012, 12, 611–622. 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Everard A.; Cani P. D. Diabetes, obesity and gut microbiota. Best Pract. Res., Clin. Gastroenterol. 2013, 27, 73–83. 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Liu W.; Wang J.; Shi J.; Sun Y.; Wang W.; Ning G.; Liu R.; Hong J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J. Mol. Endocrinol. 2017, 58, 1–14. 10.1530/JME-16-0054. [DOI] [PubMed] [Google Scholar]
- Wang L.; Christophersen C. T.; Sorich M. J.; Gerber J. P.; Angley M. T.; Conlon M. A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.; Shen M.; Zhao X.; Zhu H.; Yang Y.; Lu S.; Tan Y.; Li G.; Li M.; Wang J.; Hu F.; Le S. Anti-obesity Effect of Capsaicin in Mice Fed with High-Fat Diet Is Associated with an Increase in Population of the Gut Bacterium Akkermansia muciniphila. Front. Microbiol. 2017, 8, 272 10.3389/fmicb.2017.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger M.; Everard A.; Gomez-Valades A. G.; Matamoros S.; Ramirez S.; Delzenne N. M.; Gomis R.; Claret M.; Cani P. D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Lin S.; Vanhoutte P. M.; Woo C. W.; Xu A. Akkermansia muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe-/- Mice. Circulation 2016, 133, 2434–2446. 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- Greer R. L.; Dong X.; Moraes A. C.; Zielke R. A.; Fernandes G. R.; Peremyslova E.; Vasquez-Perez S.; Schoenborn A. A.; Gomes E. P.; Pereira A. C.; Ferreira S. R.; Yao M.; Fuss I. J.; Strober W.; Sikora A. E.; Taylor G. A.; Gulati A. S.; Morgun A.; Shulzhenko N. Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat. Commun. 2016, 7, 13329 10.1038/ncomms13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gallego C.; Pohl S.; Salminen S.; De Vos W. M.; Kneifel W. Akkermansia muciniphila: a novel functional microbe with probiotic properties. Benefic. Microbes 2016, 7, 571–584. 10.3920/BM2016.0009. [DOI] [PubMed] [Google Scholar]
- Everard A.; Belzer C.; Geurts L.; Ouwerkerk J. P.; Druart C.; Bindels L. B.; Guiot Y.; Derrien M.; Muccioli G. G.; Delzenne N. M.; de Vos W. M.; Cani P. D. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 9066–9071. 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M.; Belzer C.; de Vos W. M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017, 106, 171–181. 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Dao M. C.; Everard A.; Aron-Wisnewsky J.; Sokolovska N.; Prifti E.; Verger E. O.; Kayser B. D.; Levenez F.; Chilloux J.; Hoyles L.; Consortium M. I.-O.; Dumas M. E.; Rizkalla S. W.; Dore J.; Cani P. D.; Clement K. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir G.; Blanco N.; Xu J.; Ahrné S.; Molin G.; Sterner O.; Nyman M. Formation of short-chain fatty acids, excretion of anthocyanins, and microbial diversity in rats fed blackcurrants, blackberries, and raspberries. J. Nutr. Metab. 2013, 2013, 202534 10.1155/2013/202534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P.; Lam V.; Salzman N.; Huang Y.-W.; Yu J.; Zhang J.; Wang L.-S. Black Raspberries and Their Anthocyanin and Fiber Fractions Alter the Composition and Diversity of Gut Microbiota in F-344 Rats. Nutr. Cancer 2017, 69, 943–951. 10.1080/01635581.2017.1340491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A.; Lazarevic V.; Derrien M.; Girard M.; Muccioli G. G.; Neyrinck A. M.; Possemiers S.; Van Holle A.; François P.; de Vos W. M.; et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011, 60, 2775–2786. 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopchand D. E.; Carmody R. N.; Kuhn P.; Moskal K.; Rojas-Silva P.; Turnbaugh P. J.; Raskin I. Dietary Polyphenols Promote Growth of the Gut Bacterium Akkermansia muciniphila and Attenuate High-Fat Diet–Induced Metabolic Syndrome. Diabetes 2015, 64, 2847–2858. 10.2337/db14-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner G. D. Foodstuffs for preventing cancer: the preclinical and clinical development of berries. Cancer Prev. Res. 2009, 2, 187–194. 10.1158/1940-6207.CAPR-08-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresty L. A.; Morse M. A.; Morgan C.; Carlton P. S.; Lu J.; Gupta A.; Blackwood M.; Stoner G. D. Chemoprevention of esophageal tumorigenesis by dietary administration of lyophilized black raspberries. Cancer Res. 2001, 61, 6112–6119. [PubMed] [Google Scholar]
- Montrose D. C.; Horelik N. A.; Madigan J. P.; Stoner G. D.; Wang L.-S.; Bruno R. S.; Park H. J.; Giardina C.; Rosenberg D. W. Anti-inflammatory effects of freeze-dried black raspberry powder in ulcerative colitis. Carcinogenesis 2011, 32, 343–350. 10.1093/carcin/bgq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Ley R. E.; Mahowald M. A.; Magrini V.; Mardis E. R.; Gordon J. I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1131. 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- LeBlanc J. G.; Milani C.; de Giori G. S.; Sesma F.; Van Sinderen D.; Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Hümbelin M.; Griesser V.; Keller T.; Schurter W.; Haiker M.; Hohmann H.; Ritz H.; Richter G.; Bacher A.; Van Loon A. GTP cyclohydrolase II and 3, 4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J. Ind. Microbiol. Biotechnol. 1999, 22, 1–7. 10.1038/sj.jim.2900590. [DOI] [Google Scholar]
- Raboni S.; Bettati S.; Mozzarelli A. Tryptophan synthase: a mine for enzymologists. Cell. Mol. Life Sci. 2009, 66, 2391–2403. 10.1007/s00018-009-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H.; Amitai S.; Kolodkin-Gal I.; Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006, 2, e135 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizenman E.; Engelberg-Kulka H.; Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 6059–6063. 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R.; Sat B.; Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004, 186, 3663–3669. 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini O.; Mathy N.; Gogos A.; Shapiro L.; Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 2005, 56, 1139–1148. 10.1111/j.1365-2958.2005.04606.x. [DOI] [PubMed] [Google Scholar]
- Nicholson J. K.; Holmes E.; Kinross J.; Burcelin R.; Gibson G.; Jia W.; Pettersson S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Karlsson C. L.; Önnerfält J.; Xu J.; Molin G.; Ahrné S.; Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- Remely M.; Tesar I.; Hippe B.; Gnauer S.; Rust P.; Haslberger A. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benefic. Microbes 2015, 6, 431–439. 10.3920/BM2014.0104. [DOI] [PubMed] [Google Scholar]
- Escobar J. S.; Klotz B.; Valdes B. E.; Agudelo G. M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014, 14, 311. 10.1186/s12866-014-0311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman M. K.; Flier J. S. The adipocyte as an active participant in energy balance and metabolism. Gastroenterology 2007, 132, 2103–2115. 10.1053/j.gastro.2007.03.058. [DOI] [PubMed] [Google Scholar]
- Drouin G.; Godin J.-R.; Pagé B. The genetics of vitamin C loss in vertebrates. Curr. Genomics 2011, 12, 371–378. 10.2174/138920211796429736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said H. M. Intestinal absorption of water-soluble vitamins in health and disease. Biochem. J. 2011, 437, 357–372. 10.1042/BJ20110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe J. H.; Bergman N.; Carrera-Bastos P.; Fontes-Villalba M.; DiNicolantonio J. J.; Cordain L. Nutritional strategies for skeletal and cardiovascular health: hard bones, soft arteries, rather than vice versa. Open Heart 2016, 3, e000325 10.1136/openhrt-2015-000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floc’h N.; Otten W.; Merlot E. Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids 2011, 41, 1195–1205. 10.1007/s00726-010-0752-7. [DOI] [PubMed] [Google Scholar]
- Derrien M.; Van Baarlen P.; Hooiveld G.; Norin E.; Muller M.; de Vos W. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011, 2, 166 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M.; Vaughan E. E.; Plugge C. M.; de Vos W. M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Linden S. K.; Sutton P.; Karlsson N.; Korolik V.; McGuckin M. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleessen B.; Hartmann L.; Blaut M. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br. J. Nutr. 2003, 89, 597–606. 10.1079/BJN2002827. [DOI] [PubMed] [Google Scholar]
- Muir J. G.; Shepherd S. J.; Rosella O.; Rose R.; Barrett J. S.; Gibson P. R. Fructan and free fructose content of common Australian vegetables and fruit. J. Agric. Food Chem. 2007, 55, 6619–6627. 10.1021/jf070623x. [DOI] [PubMed] [Google Scholar]
- Png C. W.; Lindén S. K.; Gilshenan K. S.; Zoetendal E. G.; McSweeney C. S.; Sly L. I.; McGuckin M. A.; Florin T. H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Anhê F. F.; Roy D.; Pilon G.; Dudonné S.; Matamoros S.; Varin T. V.; Garofalo C.; Moine Q.; Desjardins Y.; Levy E.; Marette A. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015, 64, 872–883. 10.1136/gutjnl-2014-307142. [DOI] [PubMed] [Google Scholar]
- Oghumu S.; Casto B. C.; Ahn-Jarvis J.; Weghorst L. C.; Maloney J.; Geuy P.; Horvath K. Z.; Bollinger C. E.; Warner B. M.; Summersgill K. F.; Weghorst C. M.; Knobloch T. J. Inhibition of Pro-inflammatory and Anti-apoptotic Biomarkers during Experimental Oral Cancer Chemoprevention by Dietary Black Raspberries. Front. Immunol. 2017, 8, 1325 10.3389/fimmu.2017.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri J. G. Second report of the ad hoc committee on standards for nutritional studies. J. Nutr. 1980, 110, 1726. 10.1093/jn/110.8.1726. [DOI] [PubMed] [Google Scholar]
- Caporaso J. G.; Lauber C. L.; Walters W. A.; Berg-Lyons D.; Huntley J.; Fierer N.; Owens S. M.; Betley J.; Fraser L.; Bauer M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F.; Paarmann D.; D’Souza M.; Olson R.; Glass E. M.; Kubal M.; Paczian T.; Rodriguez A.; Stevens R.; Wilke A.; et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008, 9, 386 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.; Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. R.; Nagarajan N.; Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.