Abstract
Current methods for the diagnosis of sepsis have insufficient precision, causing regular misdiagnoses. Microbiological tests can help diagnose sepsis but are usually too slow to have an impact on timely clinical-decision making. Neutrophils have high sensitivity to infections, yet measurements of neutrophil surface markers, genomic changes, and phenotype alterations have had only a marginal effect on sepsis diagnosis. Here, we report a microfluidic assay that measures the spontaneous motility of neutrophils in the context of plasma, in one droplet of blood. We measured the performance of the assay in two independent cohorts of critically ill patients suspected of sepsis. In the first cohort, we developed a machine-learning-based scoring system (sepsis score) that segregated patients with sepsis from those without sepsis. In the second cohort, we validated the sepsis score in a double-blinded, prospective case-control study. For the 42 patients across the two cohorts, the assay identified sepsis patients with 97% sensitivity and 98% specificity. The neutrophil assay could potentially be used to accurately diagnose and monitor sepsis in larger populations of at-risk patients.
Introduction
The latest epidemiological studies of sepsis recommend the use of classifications of end-organ-injury, such as the sequential organ failure assessment (SOFA) or quick SOFA (qSOFA) score, to diagnose sepsis1,2. However, the condition remains misdiagnosed in approximately 30% of patients. Poor specificity of current diagnosis capabilities leads to the unnecessary prescription of antibiotics, placing a considerable financial burden on both hospitals and patients, promoting propagation of antibiotic-resistant strains, and directly affecting prognosis3. Poor sensitivity often delays diagnosis and treatment, resulting in worse outcomes for patients4. A multitude of biomarkers has been proposed to aid the diagnosis of sepsis (reviewed in detail by others5,6). However, none of these is currently in clinical use. C-reactive protein (CRP), procalcitonin (PCT), and IL-6 have undergone extensive clinical testing, but display high levels of heterogeneity and an inverse correlation between sample size and diagnostic value. Changes in neutrophil biomarkers, such as an increased expression of the CD64 surface receptor7, have also been explored and found to be sensitive not only to infections but also to major inflammatory states, such as the systemic inflammatory response syndrome (SIRS). A recent study measured CD64 expression on neutrophils from 450 patient samples, using a point-of-care microfluidic device. This approach was found useful for predicting patient prognosis but provided limited utility in sepsis diagnosis8. Similarly, many other markers fail to differentiate between sepsis and systemic inflammatory responses9,10 and thus so far have had a limited practical impact. Microbiological cultures help to diagnose sepsis, but require extended periods of time (2–3 days) to grow out the bacteria, and are not available to inform early diagnosis and treatment.
Neutrophil dysfunction is a hallmark of sepsis, contributing to weak immune responses to the causative infections, as well as additional off-target organ damage11–13. Neutrophils from septic patients lose the ability to respond appropriately to chemotactic signals14,15 and have altered antimicrobial activity16. Pharmaceutically correcting neutrophil behavior during sepsis has been shown to improve outcomes in animal models17, suggesting that neutrophil dysregulation contributes to the severity of sepsis. Our previous studies identified a sepsis-specific spontaneous motility signature displayed by isolated ex vivo neutrophils in straight microfluidic channels, which allowed prediction of sepsis in patients with major burns with 80% sensitivity and 77% specificity15.
Neutrophils are sensitive to a diverse range of circulating factors and integrate these signals to modulate their activation state and behavior accordingly. Thus, their activation during sepsis is likely the cumulative effect of inflammation- and infection-related factors present in the circulation, including many of the putative sepsis biomarkers discussed above. Measuring neutrophil behavior in our previous study revealed changes even after the neutrophils have been isolated from blood15. Therefore, we hypothesized that measuring neutrophil motility using whole blood samples might amplify the behavioral changes that we previously observed in isolated neutrophils.
Here, we engineered a microfluidic device for measuring the functionality of neutrophils in the context of sepsis. We measured neutrophil spontaneous motility through mazes of channels and identified key features of this phenotype that are relevant to sepsis. We show that interactions between neutrophils and plasma are critical for the spontaneous motility phenotype. We find that, when performed in the context of whole blood, neutrophil testing enabled, in a case-control study on patients in intensive care units, a precision of sepsis diagnosis that was better than 98%. Overall, our study brings the neutrophil phenotype into a new focus for research on the pathology of sepsis, which could potentially transform the diagnosis and monitoring of sepsis.
Results
A microfluidic device to assay spontaneous neutrophil motility using a droplet of diluted whole blood
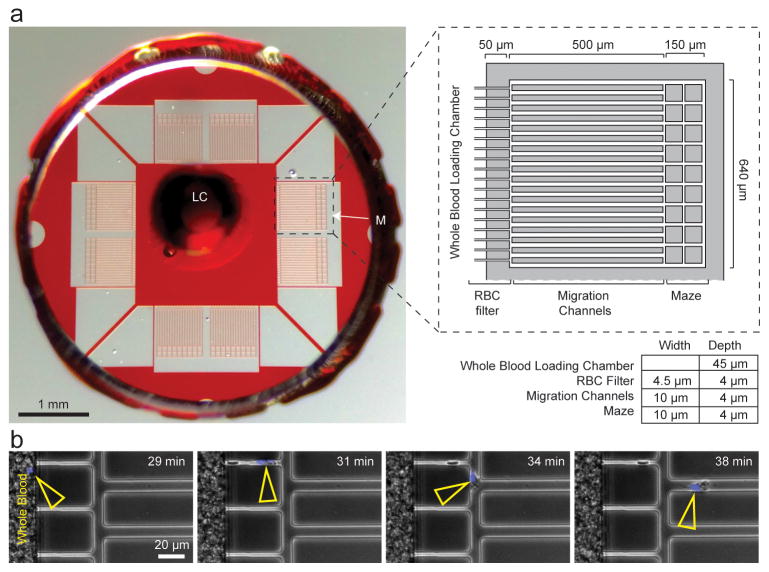
We designed a microfluidic device with channels and mazes (Fig. 1) to measure complex neutrophil motility patterns using whole blood samples taken from critically ill patients. After loading, the diluted blood sample is static and neutrophils motility is autonomous, in the absence of chemoattractant gradients. Several design elements of the microfluidic device enable measurements to be taken using diluted whole blood and with high precision. A single loading chamber contains a small (~1 μL) sample volume (Fig. 1A) and a red blood cell (RBC) filter18 at the periphery of the chamber facilitate the confinement of sample to the center of the device. The RBCs filter employs 4 × 4 μm cross-section channels and mechanically counteracts the RBCs passive movement in the lateral direction during blood sedimentation, preventing entrance or RBCs into the assay field. At the same time, the channels of the RBC filter are large enough to allow the passage of motile neutrophils to the assay field (Fig. 1A, B). The RBC filter also prevents the entrance of other leukocytes that are larger and less deformable compared to human neutrophils. We validated the efficacy of the RBC filter by counting the number of RBCs advancing (1.2 ± 1.9 RBC advancing per channel, N = 1520 channels scored, N = 10 devices). We validated the selectivity for neutrophils by monitoring the shape of the nucleus for leukocytes advancing through the filter. We observed the neutrophil-characteristic poly-lobated nucleus in more than 96% of cells entering the assay field (N = 131/133 cells scored for non-sepsis samples, 336/347 cells scored for sepsis samples. N = 6 devices monitored in detail per condition). Platelets were observed to enter migration channels but did not appear to obstruct neutrophil migration. In combination with the migration channels, the RBC filter allowed us to measure neutrophil motility velocity and directional persistence with high precision and to avoid the mechanical interference of RBCs with moving neutrophils. The design of migration channels as geometrical mazes enabled the measurement of directional choices and allowed neutrophils to reverse the direction of migration relative to the location of the blood droplet (Fig. 1A, Fig. 2A).
Figure 1. A microfluidic device to assay spontaneous neutrophil motility from a droplet of diluted blood.
A) Left panel shows macroscopic image of the microfluidic device indicating blood loading chamber (LC) and one of the 8 assay mazes (M) with scale. Magnified view (right panel, dashed box) shows a detailed diagram of the neutrophil migration maze. The red blood cell (RBC) filter, spontaneous migration channels, and simple mazes are identified.
B) Still images extracted from a time-lapse movie demonstrate neutrophil migration through the filter region while RBCs are blocked.
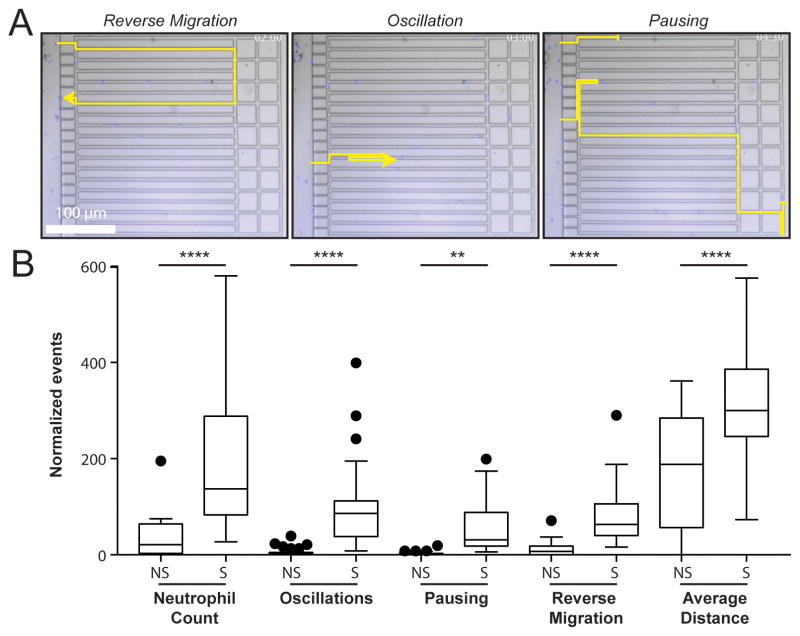
Figure 2. Neutrophils spontaneous migration patterns specific to sepsis.
A) Still images extracted from a time-lapse movie (Movie S1) of spontaneous migration of neutrophils from a septic patient sample, showing examples of behavior identification from neutrophil tracks. Timestamp: hour:min.
B) Neutrophil behaviors per device for non-septic and septic patient samples, normalized by the neutrophil count in the central chamber. A significant increase in scores for the five parameters shown was observed in septic versus non-septic patient samples. (N = 70 samples, N = 23 patients). Box and whisker plots: Tukey’s boxplot (see Methods). Statistics: unpaired, two-tailed t-test. **p ≤ 0.01, ****p ≤ 0.001.
Identification of sepsis-specific neutrophil motility patterns
We hypothesized that specific features of spontaneous neutrophil motility would differentiate septic patients from non-septic patients. To test this hypothesis, we characterized neutrophil motility patterns inside the channels (Movie S1) using a comprehensive set of 13 motility parameters (Table S1). The blood samples for neutrophil testing were obtained from a “derivation” cohort of patients in the intensive care unit at the Massachusetts General Hospital, which included septic and non-septic patients (half of the non-septic patients exhibited SIRS) (Tables S2 and S3).
To identify the motility characteristics that segregated the patients based on their condition, in an unbiased manner, we applied a machine-learning approach (Fig. S1, detailed in the methods section). Initially, unsupervised self-organizing feature mapping (SOFM) approaches were attempted, but could not be applied efficiently because of the small number of samples. A supervised machine learning approach (SVM) was adopted instead. We trained the machine using septic and non-septic patient samples (Fig. S1A, B). We used N = 72 samples obtained from 23 patients at multiple time points during their hospital stay. For machine training, all samples from one patient were in the same group. We assumed that these samples were independent.
From an initial set of 13 variables, this machine learning approach identified a smaller set of five parameters that conferred maximum prediction accuracy (Table S1 - bolded, Fig. 2, Fig. S1C). These included the Neutrophil count (N), the number of Oscillations (O) they exhibited within the migration channels, the time spent Pausing (P) during spontaneous motility, Reverse migration (R) of cells out of the device, and the Average Distance (AD) migrated by the cells. Maximum Distance traveled inside the channels was also found to confer accuracy. However, it was excluded from the score because it was already represented by the Average Distance parameter. The Maximum Distance was also more susceptible to skewing by outliers. Although some individual parameters such as Oscillations already conferred excellent prediction accuracy in the derivation cohort (Fig. S1C), we reasoned that a score combining multiple parameters might provide a more widely applicable diagnostic. The value of each parameter was further verified using a “minus-one” approach, where the contribution of each parameter was tested by removing it from the combinatory score. The final formulation of the Sepsis Score was defined based on a nonlinear formula that incorporated the five-selected neutrophil spontaneous motility parameters. This equation represents an evolution of our previous Neutrophil Activation Score15 and multiplies the number of spontaneously migrating neutrophils by the sum of sepsis-associated behaviors (Sepsis Score = N*(O+P+R+AD)/103). We tested the value of the scoring system by generating receiver operator characteristic (ROC) curves and calculating the area under the receiver operator characteristic curve (AUC). We determined that a threshold Sepsis Score of 30 is optimal for discriminating samples from septic vs. non-septic patients. Overall, the scoring system generated an AUC of 0.98 for comparison of non-sepsis and sepsis patients in this cohort, with 96.8% sensitivity and 97.6% specificity.
Assay robustness
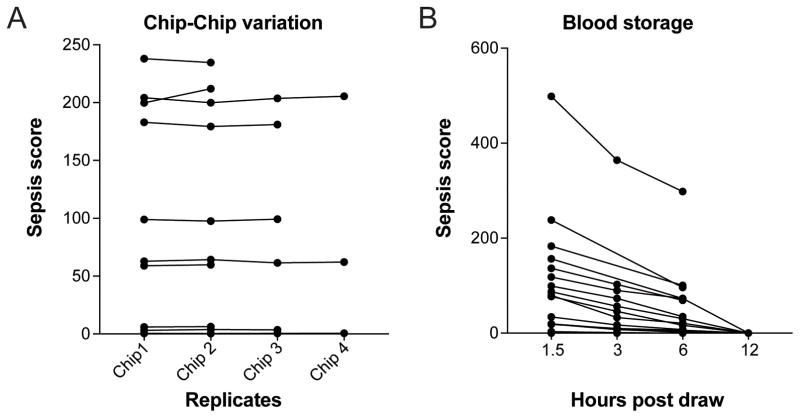
The assay requires minimal handling. A droplet of diluted blood is pipetted directly into the central chamber of the device. Spontaneous neutrophil motility occurs in the absence of neutrophil chemoattractants, further reducing the complexity of assay preparation. The assay requires 4 hours of time-lapse microscopy and 2.5 hours of image processing and analysis to generate a Sepsis Score. Integration of real-time tracking and analysis into the imaging protocol using a dedicated imaging platform might foreseeably reduce this time. Assessment of chip-to-chip reproducibility showed no significant variation between measurements of the same blood sample on distinct chips (average standard deviation was 4.7%, Fig. 3A). Testing the blood is optimal within the first 3 hours following blood collection. It appears that neutrophil activity declines as the blood ages (Fig. 3B), with a global 30–47% drop in all parameters observed from 1.5 to 3 hours after the blood draw. These results suggest that reducing the time between the blood collection and the assay will improve diagnostic performance.
Figure 3. Whole blood quality controls.
A) Chip to Chip variability. We compared Sepsis Scores from assays run in parallel on multiple devices. We observed no significant differences. (N = 10 blood samples)
B) Comparison of Sepsis Scores from assays repeated after various times post-blood draw, with blood stored at room temperature. Sepsis Scores decline as the blood ages. (N = 17 blood samples).
Comparison of spontaneous motility patterns between neutrophils in diluted whole blood and neutrophils purified by immunomagnetic negative selection
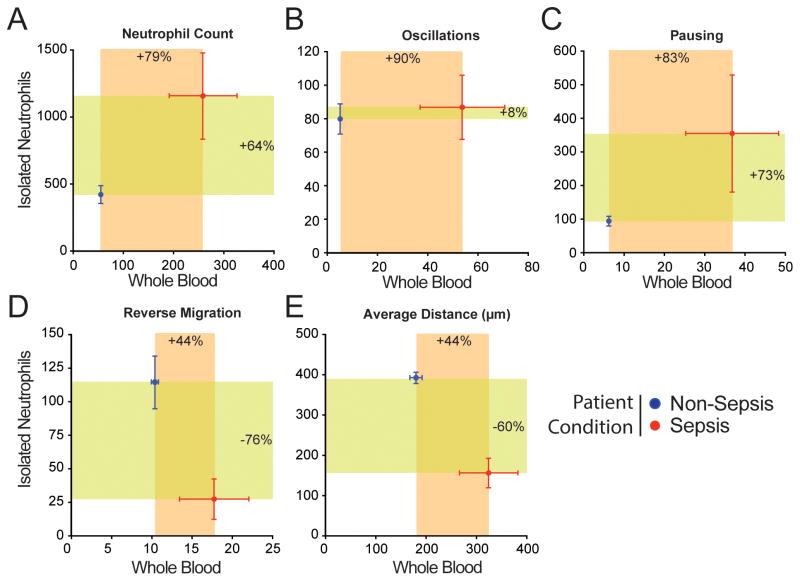
We verified that the performance of the assay is due to the use of whole blood by comparing the motility parameters of neutrophils from the same blood samples, using either diluted whole blood or isolated neutrophils in the assay. The concentration of the isolated neutrophil sample was adjusted to match the density of neutrophils in the blood. This comparison showed that some parameters, such as Neutrophil count and Pausing, were consistently increased in sepsis patients when measured either in blood or isolated neutrophil samples (Fig. 4A, C). Interestingly, Oscillation was better able to separate septic patients from non-septic in the blood assay than in the isolated neutrophil assay (Fig. 4B). The changes in parameter values between septic and non-septic samples were of larger magnitude when analyzed in the context of diluted whole blood compared to isolated neutrophils for all parameters except the Reverse migration. However, Reverse migration and Average Distance migrated were increased for sepsis samples in the blood assay but decreased with sepsis in the isolated neutrophil assay (Fig. 4D, E). Overall, analysis of neutrophil motility patterns in the whole blood assay magnifies the behavioral differences and enables more accurate discrimination between septic and non-septic blood samples.
Figure 4. Comparison of neutrophil behavior in diluted whole blood and isolated neutrophil assays.
Comparison of results from neutrophils in diluted whole blood and isolated neutrophils assays run in parallel for non-septic and septic patient samples. All 5 parameters were significantly increased in whole blood assay. In the isolated neutrophil assay, the Reverse Migration and Average Distance were negatively correlated with sepsis. (N = 70 samples, N = 23 patients). Error bars: Mean ± SEM.
Septic neutrophil spontaneous motility signatures are driven by cell-autonomous pathways and extracellular stimuli
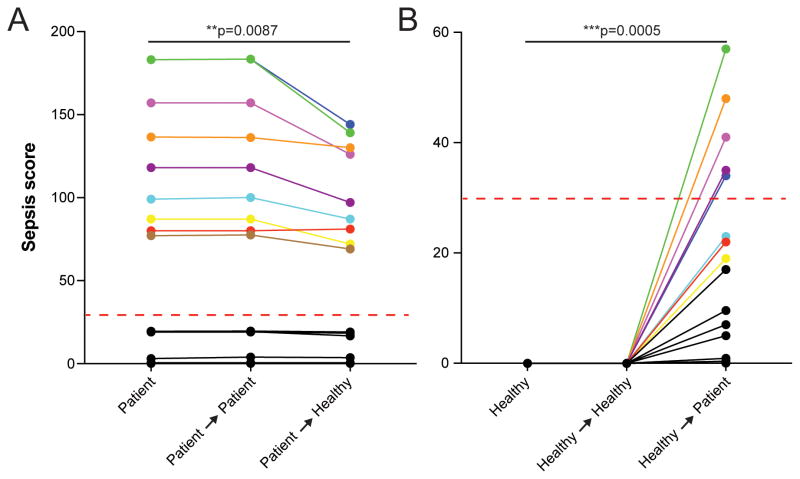
We decoupled the contribution to the neutrophil-autonomous and extracellular, plasma factors to the Sepsis Score by exchanging plasma and neutrophils between blood samples from healthy and septic donors. We found that neutrophils from septic blood continued to exhibit spontaneous motility in the presence of healthy plasma, confirming that cell-autonomous factors in septic neutrophils have the dominant contribution to the Sepsis Score. This observation also suggests that the spontaneous motility phenotype is maintained in the presence of healthy plasma (Fig. 5A). Interestingly, neutrophils from healthy blood displayed spontaneous motility patterns following incubation in septic plasma. This suggests that extracellular factors present in the septic blood also contribute to the Sepsis Score (Fig. 5B). Spontaneous motility was not stimulated by the presence of non-autologous plasma or by the experimental procedure. In control experiments, exchanging plasma between healthy blood samples did not result in stimulation of spontaneous neutrophil motility. Attempts to recapitulate a sepsis-like neutrophil phenotype by spiking whole blood with various individual immune-modulators previously reported to be elevated in septic blood were unsuccessful at inducing spontaneous motility patterns (Table S6). These observations suggest that neutrophils integrate multiple signals over time when exposed to the altered blood environment present during sepsis, which ultimately induces cell-intrinsic pathways that drive the spontaneous motility patterns we observed.
Figure 5. Whole blood assays provide higher accuracy by retaining plasma factors.
A) Plasma factors drive spontaneous neutrophil migration. Transfer of patient blood cells into the plasma from healthy donors reduced the Sepsis Scores. The scores for blood cells from septic patients transferred in the original plasma from the same patients (colored lines) remain within the septic range. N = 10 healthy donors, N = 9 septic patients.
B) Transfer of healthy blood cells into the septic patient plasma significantly increased Sepsis Scores. Five out of eight transferred-neutrophil samples achieved scores within the septic range (>30, dashed red line). N = 10 healthy donors, N = 9 septic patients. Statistics: paired, two-tailed t-test.
Validation of the Sepsis Score in a second, independent patient cohort
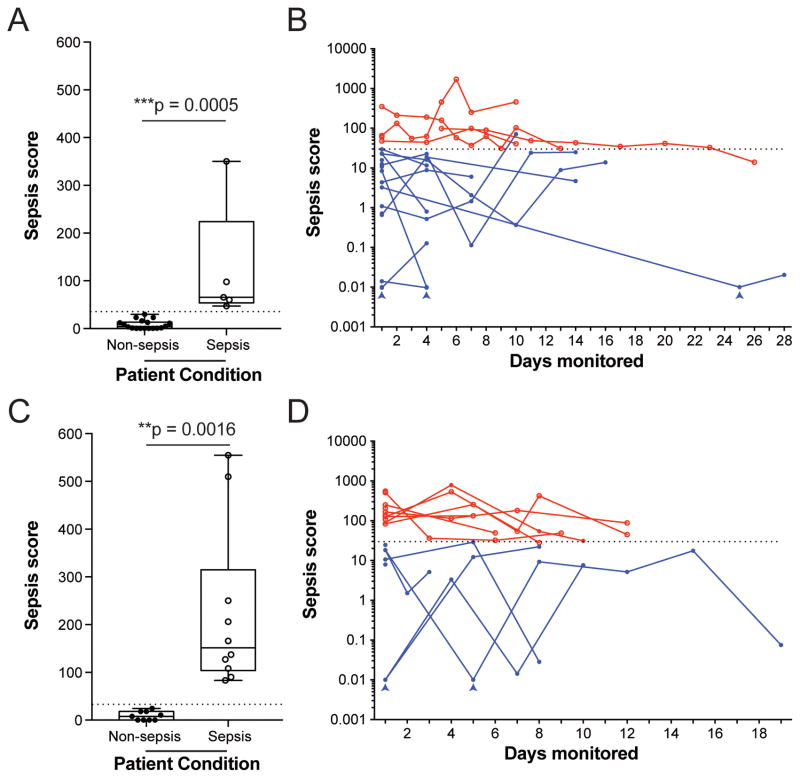
The Sepsis Score performed well at separating sepsis from non-sepsis blood samples from patients in the primary “derivation” cohort. The Sepsis Score for blood samples collected at day 1 discriminated with perfect accuracy the sepsis and non-sepsis patients in the “derivation” cohort (Fig. 6A). The Sepsis score also exhibits a high degree of internal consistency. Continued monitoring of patients during their hospital stay demonstrated that their Sepsis Score appeared to remain elevated for up to 3 weeks (Fig 6B), generally dropping before discharge. To further validate the Sepsis Score, we employed a second, independent patient cohort, managed by a different clinical team than the “derivation” cohort.
Figure 6. Segregation of clinical conditions using the Sepsis Score in the “derivation” and “validation” cohorts.
A) The Sepsis Score in Day-1 blood samples was significantly higher for initial samples from septic (empty dots) versus non-septic patients (full dots) in both the primary “Derivation” cohort (N = 23 patients). Box and whisker plots: Tukey’s boxplot (see Methods). Statistics: unpaired, two-tailed t-test.
B) Patients were followed longitudinally and their Sepsis Score monitored over weeks. Most Sepsis Scores appeared to be dropping toward Non-Septic ranges prior to discharge. Arrowheads indicate scores that were adjusted from 0 to 0.01 for presentation using a logarithmic scale (N=70 samples, N=23 patients, Sepsis scores for sepsis patients are shown in red, Sepsis scores for non-sepsis patients are shown in blue).
C) The Sepsis Score measured in day-1 blood samples was significantly higher for initial samples in the independent, double-blinded, “Validation” cohort (N = 19 patients). Box and whisker plots: Tukey’s boxplot (see Methods). Statistics: unpaired, two-tailed t-test.
D) Patients from the Validation cohort were also followed longitudinally and their Sepsis Score monitored over weeks. Again, most Sepsis Scores appeared to be dropping toward Non-Septic ranges before discharge, but no overlap was observed between Non-Sepsis and Sepsis groups in this cohort. Arrowheads indicate scores that were adjusted from 0 to 0.01 for presentation using a logarithmic scale (N = 50 samples, N = 19 patients, Sepsis scores for sepsis patients are shown in red, Sepsis scores for non-sepsis patients are shown in blue).
We further verified the diagnostic accuracy of the Sepsis Score in a double-blinded, prospective case-control study design. We calculated that the “validation” cohort should include at least 19 patients (10 septic and 9 non-septic) to demonstrate the improved performance of the assay compared that of current sepsis diagnostic standard. These statistical comparisons are based on the 96.8% sensitivity and 97.6% specificity of the assay inferred from the performance in the “derivation” cohort, the known sensitivity and specificity of the current sepsis diagnostic standard using sepsis-3 criteria (64% sensitivity and 65% specificity)19, and a 0.05 probability of a type-I error and 90% power20. Thus, the validation cohort was comprised of 19 patients in intensive care with complex co-morbidities, which were selected as either septic or control cases. Of these, 10 patients were septic and 9 were non-septic (6/9 non-septic patients exhibited SIRS, Tables S4 and S5, Fig. S2). Multiple blood samples were collected from each patient at 1–3 days interval. The clinical and research teams did not know the results of the assay or the status of the patient, until un-blinding at the completion of the study.
Blinded scoring of neutrophil behavior from the first blood samples (day 1) collected from each patient demonstrated that the Sepsis Score provided accurate segregation of non-septic and septic samples (Fig. 6C). Moreover, all 23/23 samples collected at 1–3 days interval from non-septic patients all exhibited Sepsis Scores < 30, while 26/27 septic samples scored > 30, suggesting that the Sepsis Score may provide a valuable tool for monitoring non-septic and septic patients over time (Fig. 6D). In the validation cohort, using the neutrophil assay, sepsis was diagnosed with 96.3% sensitivity and 100% specificity. This result substantiates the applicability of the Sepsis Score to sepsis diagnosis for a broad spectrum of at-risk patients. The analysis of the assay performance in the two cohorts combined (N = 42 patients) shows that the Sepsis Score achieved an overall AUC of 0.99, with 97% sensitivity and 98% specificity.
Discussion
The assay we describe measures the patterns of neutrophil spontaneous motility and enables accurate diagnosis of sepsis in patients. The assay performs the measurements directly from a droplet of diluted blood, preserving the physiological and biochemical environment of the neutrophils. The ability to use whole blood distinguishes this assay from previous ones, which could only probe the motility of patient neutrophils after isolation from blood15. Moreover, we show that retaining plasma in the assay is critical for differentiating neutrophil motility patterns between sepsis and non-sepsis patients. Removal of plasma factors during neutrophil isolation results in changes in neutrophil phenotype and compromises discrimination between sepsis and non-sepsis conditions.
The design of the assay takes advantage of the higher precision measurements inherent to neutrophil motility through narrow microfluidic channels14,15,21. Despite limiting the richness of observable behavioral traits, the confinement of motile neutrophils to channels and the restriction to simple directional decisions results in measurements that are less sensitive to the noise from the small, continuous directional changes that plague traditional cell migration assays. From the set of 13 motility parameters that we defined from these measurements, we used a machine learning approach to identify those that corresponded to retrospective sepsis diagnosis. Three more neutrophil spontaneous motility parameters (Pausing, Reverse migration, and Average Distance) emerged as essential for the Sepsis Score, in addition to the Oscillation phenotype and the number of neutrophils entering the channels, which were previously identified as markers of sepsis using a purified-neutrophil assay15. Two of these parameters were enabled by the design of the microfluidic channels. The mazes at the end of channels allowed more neutrophils to reverse their direction of migration and travel back to the location of the blood droplet. The mazes also allowed the neutrophils to travel larger average distances in the absence of directional cues. Both parameters increased in sepsis vs. non-sepsis when analyzed in blood and decreased when analyzed in isolated neutrophils. Moreover, the fraction of cells to slow down their migration significantly and stop (Pausing) emerged as an important parameter for the sepsis score in this geometrical context.
Neutrophils have long been suspected of playing a role in septic responses22. They are an essential cell type for combating infections, boasting a wide range of potent antimicrobial mechanisms. Their functional deficiencies during sepsis may explain the poor control of infections. Also, deregulated neutrophil entrance into tissues can result in the off-target deployment of these defensive mechanisms, which can cause tissue damage to bystander organs11. Treatment with molecules that correct aspects of neutrophil dysfunction in sepsis17, have been shown to have a positive impact on outcome in animal sepsis models. These studies suggest that neutrophils play a direct role in promoting the cytokine storm and subsequent organ failure that leads to mortality in sepsis. For patients that were followed longitudinally, two out of eleven sepsis patients exhibited Sepsis Scores in the healthy range by the time they were discharged and nine out of eleven showed a decrease (by 76% on average) in Sepsis Score before discharge. This decrease suggests a return of neutrophils towards homeostasis and may correlate with successful treatment. The results also indicate that sepsis may exert an extended temporal impact upon neutrophil phenotype. Further studies will clarify the importance of these correlations and may guide the development of new treatment regimens to correct neutrophil activity and eventually ameliorate sepsis.
The biology underpinning the activation of neutrophils during sepsis remains mostly unexplored. Our study hints at the presence of extracellular factors in septic plasma that drive self-perpetuating alterations of neutrophil functionality. Factors such as activation of CRP23 may impact neutrophils, while also contributing to other phenotypes, e.g. rouleaux formation24. Platelet activation has also been widely reported in sepsis25–27 and has been suggested as a candidate rapid-response element during disease28. A previous study by some of the authors of this work demonstrated that activated platelets could induce spontaneous neutrophil motility and oscillatory motility patterns29. While no single factor has yet been identified that can accurately stratify sepsis from non-sepsis, in combination, these responses likely contribute to the specific neutrophil spontaneous motility signatures that we capture using the whole blood assay presented here. Our preliminary efforts to identify such factors, by the spiking experiments, reveal a complex situation where neutrophil incubation with individual pro-inflammatory cytokines fails to induce the spontaneous motility phenotype. Future identification of inflammatory cocktails and temporal sequences of stimulation able to recapitulate sepsis-like spontaneous motility patterns from healthy blood will provide a useful tool to screen for compounds suppressing this phenotype, which is likely key to improving clinical outcomes. Additionally, pinpointing specific molecules driving the neutrophil responses that we observe may allow alternative diagnostic strategies, where the combination of factors might be rapidly measured directly from the plasma.
The present approach to sepsis diagnosis relies on neutrophil motility changes measured in the presence of plasma. The assay is logistically simple because it circumvents the need for neutrophil isolation procedures. In a clinical setting, the assay will require only training of an operator who will prepare and load the blood sample in the device. Automated imaging and analysis of cell trajectories will then generate a Sepsis Score readout. The formula for the Sepsis Score and threshold values will be further refined and validated in subsequent research studies. The striking performance of the assay in the case-control study brings a new focus on the fundamental role that neutrophils play during sepsis. In addition to the potential for better understanding sepsis pathology, the assay for neutrophil behavior may also become an essential tool for sepsis diagnostic and monitoring. Future validation of the assay in larger and more diverse cohorts of patients at risk for sepsis will eventually lead to valuable new diagnostic tools for the physicians treating septic patients.
Methods
Device design and fabrication
Devices were designed using AutoCAD. Chrome masks for photolithography were printed by FrontRange Imaging. Silicon wafers were fabricated using standard techniques. Briefly, clean silicon wafers were spin-coated with two layers of negative photoresist (SU-8, Microchem, Newton, MA), the first layer 5 μm thick and the second 50 μm. The wafer was then patterned by sequential UV exposure through two photolithography masks, and processed per manufacturer’s instructions. The patterned wafer was then used as a mold for PDMS (Polydimethylsiloxane, Fisher Scientific, Fair Lawn, NJ) soft-lithography to produce the final PDMS devices. Central inlets were punched using a 1.2-mm punch (Harris Uni-coreTM). The whole device was punched out using a 5-mm punch. Devices were then irreversibly bonded to glass-bottom well plates, as previously described30.
Patient Diagnostic Criteria and Study design
This observational study complied with all current human research ethical regulations and was approved by the Massachusetts General Hospital Institutional Review Board. Subjects were consenting adults > 18 years and < 80 years, in two patient cohorts. A “derivation” cohort of 23 patients included trauma and postoperative surgical patients admitted to the surgical intensive care unit (SICU) for trauma or surgical management (injury severity scores greater than 15, critically ill, or postoperative patients with indwelling lines). Data collected from the derivation cohort of patients was used to optimize the analysis and machine learning. Subsequently, a “validation” cohort of 19 patients admitted to Cardiac, Medical, and Surgical Intensive Care Units was enrolled and used for the double-blinded validation of the assay. Patients were evaluated using the recent sepsis-3 guidelines published in 2016, which defined sepsis as end-organ dysfunction caused by the host response to infection1,2. This dysfunction is quantified by the sequential organ failure assessment (SOFA) score, which considers the different organ systems and the degree of dysfunction in each one. Following these recent guidelines, septic shock was defined as hypotension requiring vasopressors to maintain a mean arterial pressure (MAP) greater than 65 mm Hg, and a serum lactic acid above 2 mmol/dL despite adequate fluid resuscitation.
In addition, patients in the first, “derivation” cohort were evaluated based on the older definition of systemic inflammatory response syndrome (SIRS). The SIRS criteria were previously used to define a global inflammatory response, which, in the setting of an infection, comprised the definition of sepsis. In the setting of the SIRS criteria, septic shock included a lactic acidosis and hypotension despite adequate fluid resuscitation. The SIRS criteria included; temperature dysregulation - hyperthermia (> 38°C) or hypothermia (< 36°C) - heart rate > 90 beats/minute, respiratory rate > 20 breaths/minute or PaCO2 < 32 mm Hg, and WBC > 12,000/mm3 or < 4,000/mm3 or > 10% bands.
All blood samples were drawn with consent from patients with existing venous lines expected to remain in place for more than 48 hours. The first sample was drawn within first 7 days of admission to intensive care. Thereafter, a blood sample was drawn every three days for up to two weeks or until the patient was discharged, relocated to another unit, or developed sepsis. During periods of sepsis, samples were drawn daily. Removal of indwelling line automatically excluded (removed) a patient from the study.
Sample preparation
Devices were primed with Iscove’s modified Dubecco’s medium (IMDM) containing 20% fetal bovine serum (FBS) and 1% Fibronectin (from human plasma, Sigma) by pipetting 50 μl in and around each device. A dome of liquid formed on top and the outer edge of the device in contact with the coverslip was also surrounded by liquid. The devices were placed under vacuum for 10 mins, then removed and allowed to equilibrate for at least 15 mins until all channels had filled. IMDM containing 20% FBS was then added to the well containing the device until the device was completely covered. Devices for isolated neutrophils were primed as previously described14,15.
Peripheral blood samples from patients were drawn in 10 mL Heparin-coated vacuum tubes (Vacutainer, Becton Dickinson) from indwelling lines. Some blood samples were split for comparison between the two assays using diluted whole blood (1 mL) and isolated neutrophil (9 mL). Whole blood samples were diluted 1:1 in IMDM with 20% FBS and stained with Hoechst 33342 dye at 32 μM for 15 mins prior to loading. 0.5 μl of stained blood was then pipetted into the center of the device using a gel-loading tip, taking care to draw the tip out of the device while dispensing.
Neutrophils were isolated from whole blood by density separation and negative selection (Neutrophil Enrichment Kit, STEMCELL Technologies, Vancouver, Canada) protocols as previously described14,15. To examine the shape of the nucleus, nucleated cells in blood were stained with 32 μM Hoechst 33342 dye for 10 mins, washed, and re-suspended in IMDM + 20% FBS at 2.0 × 10^7 cells/mL prior to loading. Devices for isolated neutrophils were loaded by pipette using a gel-loading tip until cells were observed to exit the device outlet.
For plasma-swapping experiments, 1 mL of whole blood was centrifuged at 200 g for 10 mins to pellet cells. The platelet-rich plasma supernatants were then drawn from the tubes and transferred into fresh tubes. Platelets were then pelleted at 1900 g for 10 mins and the plasma fraction drawn into new tubes. Cell pellets were then re-suspended in the appropriate exchanged plasma, incubated at room temperature for 30 mins, and an aliquot loaded into the device.
For spiking experiments, peripheral blood from healthy volunteers aged 18 years or older was also collected in Heparin-coated vacuum tubes and delivered at room temperature to the lab within 1 to 6 hours after collection (Research Blood Components). Immune-modulators were added to the media at 2× the target dose before mixing 1:1 with whole blood. After mixing, spiked blood samples were incubated at 37°C for 30 min before loading.
Imaging
Cell motility was imaged at 10× magnification using a fully automated fluorescent Nikon TiE inverted wide field microscope with a bio-chamber heated to 37°C with 5% CO2. Each microfluidic device provided 8 fields of view, each containing one migration maze. Each field was imaged every 2 minutes to allow accurate tracking of cell motility, for 4 hours. Datasets compromised by microscope failure were excluded from the data analysis.
Data analysis
Files were converted to standard AVI format using Nikon Elements or ImageJ. Initial processing was performed to allow bright-field tracking and included removing the background. Cell tracking was performed automatically from bright-field images for most of the time-lapse sequences. The size, velocity, and directionality of moving neutrophils was quantified. These tracks were written to individual CSV files for each imaging field. The specific variables for each track included: track number, video frame, cell diameter, x position, y position, distance, and velocity. Automated cell tracking was performed using ImageCV, TrackPy, and SciKit-Learn packages in Python. Cell motility pattern identification and definitions are detailed in Table S1. All custom tracking and analysis algorithms are available for download at http://dx.doi.org/10.6084/m9.figshare.5572687. Figures were prepared using Illustrator CS5 Version 15.0.0 (Adobe Systems).
Machine learning
We performed supervised learning on neutrophils and patient data from the “derivation” cohort to identify and validate the group of parameters characteristic to sepsis. Support vector machines (SVM) were then used to differentiate septic from non-septic patients in the “derivation” cohort31–34. Briefly, the data was split 1:2:1 (training data: testing data: held-out set) by patient. The algorithm was trained on the training data set. Then, variables were changed and significant variables were determined on the testing data set. These variables included the number of cells, spontaneous motility distance, oscillations, reverse migration, and pausing. With these variables, final graphs and results were produced with the held-out set. To further optimize the sepsis classification parameters, regularized linear discriminant analysis was applied via hold-out analysis with cross-validation and multiple resampling due to the few samples and increasing number of parameters35. With these parameters, error estimation was applied to all possible subsets to find the parameters that resulted in the highest accuracy sepsis prediction36. t-distributed stochastic neighbor embedding (tSNE) graphs37 were produced to visually confirm the split of data by groups (sepsis and non-sepsis). Finally, the test-train split was changed and the analysis is run 500 – 1000 times and a histogram of the AUC values from the held-out data is graphed.
Statistical analysis
The number of subjects needed to test the hypothesis that the level of performance for the proposed Sepsis Score is significantly higher than current diagnostic standards was calculated using asymptotic variance equations from Pepe20. For the calculations, we chose α = 0.05 as the desired minimum type 1 error rate, β = 0.1 the desired minimum type 2 error rate (90% study power). The sensitivity (64%) and specificity (65%) for sepsis of 2016 Sepsis-3 diagnostic criteria were employed as a reference. The desirable sensitivity and specificity levels for the proposed Sepsis Score were estimated from the first part of the study as 96.8% and 97.6%, respectively.
For comparison of patient samples, values are presented as Tukey boxplots. The bottom and top of the box represent the first and third quartiles respectively, and the central line represents the median. Bottom and top whiskers represent the lowest and highest datum within 1.5 interquartile range respectively. Outliers are represented by single points. For statistical analysis, comparison of parameters for sepsis and non-sepsis groups was performed using unpaired, two-tailed t-tests, while comparison of matched samples (Fig. 4) used a paired, two-tailed t-test. Graphing and statistical tests were performed using Prism 7.0a software (GraphPad Software Inc.).
Data availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. Numerical values for the graphs in Fig. 2 to 6 and custom computer codes use to classify the cell migration tracks are available at http://dx.doi.org/10.6084/m9.figshare.5572687. Raw imaging data is available upon request.
Supplementary Material
Summary.
A microfluidic assays measures spontaneous neutrophil motility signatures from a drop of blood and helps diagnose sepsis.
Acknowledgments
We thank Mrs. Carrie Holland, R.N and Kelsey Brait for assistance in recruiting patients. We thank Drs. Mehmet Toner, H Shaw Warren, and Michael Filbin for thoughtful discussions and advice. This project was supported by funding from the National Institutes of Health, National Institute of General Medical Sciences (grant GM092804) and National Institute of Allergy and Infectious Diseases (grant AI113937), and Shriners Hospital for Children. Microfluidic devices were manufactured at the BioMEMS Resource Center at Massachusetts General Hospital, supported by a grant from the National Institute of Biomedical Imaging and Bioengineering (grant EB002503). We also thank for biostatistics assistance from Dr. Douglas Hayden and the National Center for Research Resources and the National Center for Advancing Translational Sciences (grant UL1 TR001102). Dr. Felix Ellett was supported by a fellowship from Shriners Hospital for Children.
Footnotes
AUTHOR CONTRIBUTIONS
FE designed the study, designed and fabricated the microfluidic device, performed experiments, analyzed the data, and wrote the manuscript. JJ performed experiments and cell tracking, analyzed the data, and applied machine-learning approaches to derive the Sepsis Score. JJ and AM performed blood-spiking experiments. MM, YML, and KB coordinated collection of patient samples for the first, “derivation” cohort and provided advice on the study design and manuscript. VS and JL coordinated collection of samples for the “validation” cohort and provided advice on the study and manuscript. DI conceived, designed, and supervised the study and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Vincent JL, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive care medicine. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) Jama. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma S, Kumar A. Antimicrobial management of sepsis and septic shock. Clinics in chest medicine. 2008;29:677–687. ix. doi: 10.1016/j.ccm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Critical care medicine. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 5.Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Critical care. 2010;14:R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Biomarkers for diagnosis of sepsis in patients with systemic inflammatory response syndrome: a systematic review and meta-analysis. SpringerPlus. 2016;5:2091. doi: 10.1186/s40064-016-3591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu KH, Chan MC, Wang JM, Lin LY, Wu CL. Comparison of Fcgamma receptor expression on neutrophils with procalcitonin for the diagnosis of sepsis in critically ill patients. Respirology. 2011;16:152–160. doi: 10.1111/j.1440-1843.2010.01876.x. [DOI] [PubMed] [Google Scholar]
- 8.Hassan U, et al. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nature communications. 2017;8:15949. doi: 10.1038/ncomms15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubljar D, Skvarc M. Effective Strategies for Diagnosis of Systemic Inflammatory Response Syndrome (SIRS) due to Bacterial Infection in Surgical Patients. Infectious disorders drug targets. 2015;15:53–56. doi: 10.2174/1871526515666150320161804. [DOI] [PubMed] [Google Scholar]
- 10.Bone RC, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 11.Brown KA, et al. Neutrophils in development of multiple organ failure in sepsis. Lancet. 2006;368:157–169. doi: 10.1016/S0140-6736(06)69005-3. [DOI] [PubMed] [Google Scholar]
- 12.Fazal N, et al. CINC blockade prevents neutrophil Ca(2+) signaling upregulation and gut bacterial translocation in thermal injury. Biochimica et biophysica acta. 2000;1535:50–59. doi: 10.1016/s0925-4439(00)00082-x. [DOI] [PubMed] [Google Scholar]
- 13.Weiss SJ. Tissue destruction by neutrophils. The New England journal of medicine. 1989;320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 14.Butler KL, et al. Burn injury reduces neutrophil directional migration speed in microfluidic devices. PloS one. 2010;5:e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CN, et al. Spontaneous Neutrophil Migration Patterns during Sepsis after Major Burns. PloS one. 2014;9:e114509. doi: 10.1371/journal.pone.0114509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomkin JS. Neutrophil disorders in burn injury: complement, cytokines, and organ injury. The Journal of trauma. 1990;30:S80–85. doi: 10.1097/00005373-199012001-00019. [DOI] [PubMed] [Google Scholar]
- 17.Kurihara T, et al. Resolvin D2 restores neutrophil directionality and improves survival after burns. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:2270–2281. doi: 10.1096/fj.12-219519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoang AN, et al. Measuring neutrophil speed and directionality during chemotaxis, directly from a droplet of whole blood. Technology. 2013;1:49. doi: 10.1142/S2339547813500040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson SQ. Diagnosing sepsis: a step forward, and possibly a step back. Ann Transl Med. 2017;5:55. doi: 10.21037/atm.2017.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepe MC. The Statistical Evaluation of Medical Tests for Classification and Prediction Oxford Statistical Science Series. Oxford University Press; 2004. pp. 218–228. [Google Scholar]
- 21.Ambravaneswaran V, Wong IY, Aranyosi AJ, Toner M, Irimia D. Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integrative biology : quantitative biosciences from nano to macro. 2010;2:639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhundov AG. On the problem of toxic granulation of the neutrophils in acute suppurative diseases and sepsis. Klinicheskaia meditsina. 1961;39:84–89. [PubMed] [Google Scholar]
- 23.Povoa P. C-reactive protein: a valuable marker of sepsis. Intensive care medicine. 2002;28:235–243. doi: 10.1007/s00134-002-1209-6. [DOI] [PubMed] [Google Scholar]
- 24.Weng X, Cloutier G, Beaulieu R, Roederer GO. Influence of acute-phase proteins on erythrocyte aggregation. The American journal of physiology. 1996;271:H2346–2352. doi: 10.1152/ajpheart.1996.271.6.H2346. [DOI] [PubMed] [Google Scholar]
- 25.Gawaz M, Fateh-Moghadam S, Pilz G, Gurland HJ, Werdan K. Platelet activation and interaction with leucocytes in patients with sepsis or multiple organ failure. European journal of clinical investigation. 1995;25:843–851. doi: 10.1111/j.1365-2362.1995.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 26.Mavrommatis AC, et al. Coagulation system and platelets are fully activated in uncomplicated sepsis. Critical care medicine. 2000;28:451–457. doi: 10.1097/00003246-200002000-00027. [DOI] [PubMed] [Google Scholar]
- 27.Ogura H, et al. Activated platelets enhance microparticle formation and platelet-leukocyte interaction in severe trauma and sepsis. The Journal of trauma. 2001;50:801–809. doi: 10.1097/00005373-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 28.Engelmann B, Massberg S. Thrombosis as an intravascular effector of innate immunity. Nature reviews. Immunology. 2013;13:34–45. doi: 10.1038/nri3345. [DOI] [PubMed] [Google Scholar]
- 29.Frydman GH, et al. Technical Advance: Changes in neutrophil migration patterns upon contact with platelets in a microfluidic assay. J Leukoc Biol. 2016 doi: 10.1189/jlb.1TA1115-517RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones CN, et al. Microfluidic platform for measuring neutrophil chemotaxis from unprocessed whole blood. J Vis Exp. 2014 doi: 10.3791/51215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furey TS, et al. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 32.Suykens JA, Vandewalle J. Least squares support vector machine classifiers. Neural processing letters. 1999;9:293–300. [Google Scholar]
- 33.Vapnik VN. Wiley; New York: 1998. [Google Scholar]
- 34.Vapnik V. Nonlinear Modeling. Springer; 1998. pp. 55–85. [Google Scholar]
- 35.Fan J, Guo S, Hao N. Variance estimation using refitted cross-validation in ultrahigh dimensional regression. Journal of the Royal Statistical Society. Series B, Statistical methodology. 2012;74:37–65. doi: 10.1111/j.1467-9868.2011.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maaten Lvd, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. Numerical values for the graphs in Fig. 2 to 6 and custom computer codes use to classify the cell migration tracks are available at http://dx.doi.org/10.6084/m9.figshare.5572687. Raw imaging data is available upon request.