Abstract
Ammonium, stemming from renal ammoniagenesis, is a major urinary proton buffer and is excreted along the collecting duct. This process depends on the concomitant secretion of ammonia (NH3) by the NH3 channel RhCG and of protons (H+) by the vacuolar-type H+-ATPase pump. Thus, urinary ammonium content and urinary acidification are tightly linked. However, mice lacking Rhcg excrete more alkaline urine despite lower urinary ammonium, suggesting an unexpected role of Rhcg in urinary acidification.
RhCG and the B1 and B2 H+-ATPase subunits could be coimmunoprecipitated from kidney. In ex vivo microperfused cortical collecting ducts (CCD) H+-ATPase activity was drastically reduced in the absence of Rhcg. Conversely, overexpression of RhCG in HEK293 cells resulted in higher H+ secretion rates and increased B1 H+-ATPase mRNA expression. However, in kidneys from Rhcg−/− mice the expression of only B1 and B2 subunits was altered. Immunolocalization of H+-ATPase subunits together with immuno-gold detection of the A H+-ATPase subunit showed similar localization and density of staining in kidneys from Rhcg+/+ and Rhcg−/− mice. In order to test also for a reciprocal effect of intercalated cell H+-ATPases on Rhcg activity, we assessed Rhcg and H+ATPase activities in mCCD from ATP6v1B1−/− mice and showed reduced H+-ATPase activity without altering Rhcg activity.
Thus, RhCG and H+-ATPase are located within the same cellular protein complex. RhCG may modulate H+-ATPase function and urinary acidification, whereas H+-ATPase activity does not affect RhCG function. This mechanism may help to coordinate NH and H+secretion beyond the physico-chemical driving forces.
INTRODUCTION
The renal collecting duct excretes acid into urine by parallel secretion of protons (H+) and ammonia (NH3) by vacuolar H+-ATPases (H+-ATPase) and the ammonia transporter RhCG 1–3. Renal acid excretion is critical for maintaining normal acid-base homeostasis and is increased during acidosis or following a systemic acid-load 2.
Highly conserved through evolution from bacteria to humans, RhCG belongs to the Rh (Rhesus) glycoprotein family and has been characterized by in vitro and ex vivo studies as a channel selectively permeable to NH3 but not to ammonium (NH +4) 4–6. In the kidney, RhCG expression specifically spreads from the late distal convoluted tubule (DCT) to the inner medullary collecting duct (IMCD), on both basolateral and apical poles of most epithelial cells (with the exception of selective apical expression in non-type A intercalated cells) 7–11. We recently showed that RhCG is responsible for a major part of NH3 transported through apical and basolateral membranes of the collecting duct resulting in a drastic reduction of transepithelial NH transport in collecting ducts from Rhcg−/− mice 11–12. As a consequence of this defect, chronically acid-loaded Rhcg−/− mice cannot eliminate NH +4 into urine and develop a severe incomplete distal renal tubular acidosis (dRTA) characterized by low blood pH and HCO − concentration 11–12. Similarly, mice with selective deletion of Rhcg from only the collecting duct or intercalated cells show reduced urinary ammonium excretion 13–14.
While RhCG is responsible for luminal NH3 secretion, the renal H+ -ATPase is critical to actively secrete H+ into urine 15–16. H+-ATPases are ubiquitous multi-subunits proteins composed of a cytosolic V1 catalytic domain, responsible for ATP hydrolysis and a membrane-associated V0 domain mediating H+ translocation 16–17. In the collecting duct, H+-ATPases are localized at the luminal side of type-A intercalated cells and basolateral side of non-type A intercalated cells. The V1 domain contains 8 subunits A-H including the tissue-specific subunit isoforms B1 and B2. The B1 isoform is highly enriched in intercalated cells of kidney and its genetic mutation or deletion cause a form of distal renal tubular acidosis 18–20. The V0 domain contains 6 different subunits, a, d, c, c’, c”, and e including a1, a2, a3 and a4. Also, the a4 isoform is highly expressed in kidney 21–22 and mutations in human and its deletion in mice are associated with renal tubular acidosis 23–25.
Urinary acidification and ammonium content are closely linked26. Secretion of NH3 and H+ by RhCG and H+-ATPases may be coordinated since protonation of NH3 and subsequent trapping of NH +4 are critical for the efficient excretion of NH4 + 26–29. The partial overlap of both proteins in type-A intercalated cells may allow even for a direct functional coupling or interaction30. We have recently shown in two differently constructed mouse models lacking Rhcg that urinary pH is more alkaline during stimulation of urinary acidification and NH + 4excretion 11–12. Since Rhcg−/− mice have highly reduced NH3 secretion, the lack of NH3 buffering capacity would be expected to result in a rather more acidic urinary pH, as shown in another mouse model with intact Rhcg-mediated NH secretion but reduced medullary NH4+ accumulation31. Thus, we hypothesized that the absence of Rhcg may reduce proton secretion by H+- ATPases.
RESULTS
Rhcg and B1 and B2 subunits of H+-ATPase are part of one protein complex in native kidney
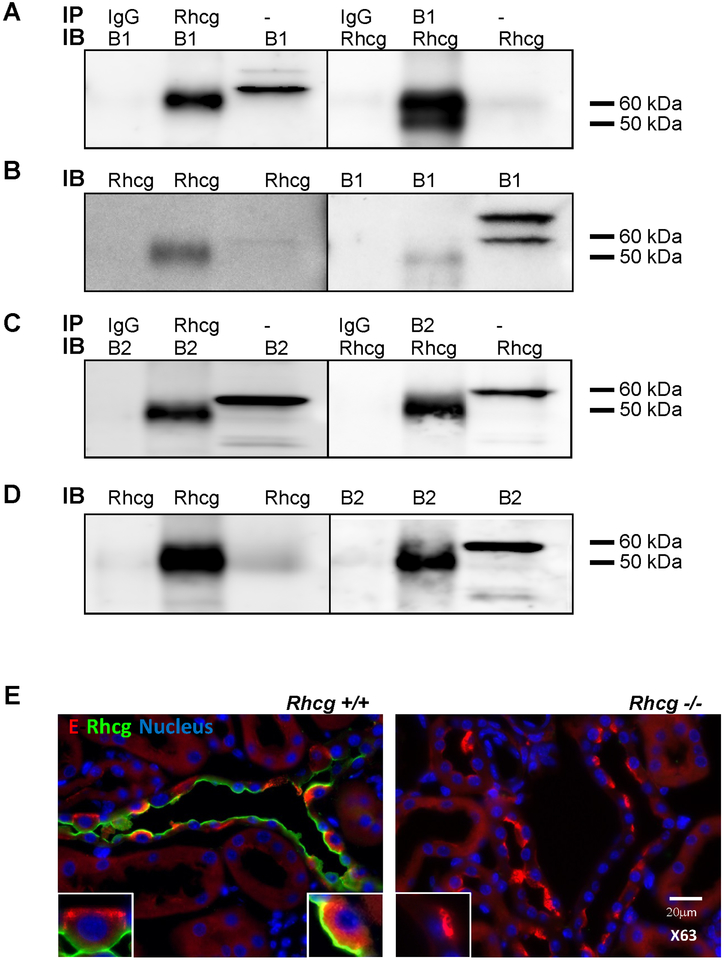
To assess whether RhCG and H+-ATPase were located within the same cellular protein complex in intercalated cells we performed coimmunoprecipitation of RhCG and B1 and B2 H+-ATPase subunits from native rat kidney medulla (Figure 1). Immunoprecipitation with antibodies against RhCG or B1 (panel A and B) or RhCG or B2 (panel C and D) after chemical crosslinking yielded protein complexes that contained both proteins whereas immunoprecipitation with control IgG did not detect RhCG or the B subunits. Thus, RhCG, B1 and B2 containing H+-ATPases are part of the same protein complex in kidney.
FIGURE 1. The B1 and B2 H+-ATPase subunits and Rhcg coexist in the same protein complex.
Co-Immunoprecipitation was performed on rat kidney medulla using crosslinking. Panel A: Co-IP was performed with anti-Rhcg (lane 2) and anti-B1 (lane 5) polyclonal antibodies or with an isotypic IgG antibody (lanes 1 and 4). Lanes 3 und 6 were loaded with 50 mg rat kidney medulla protein. Immunoblotting (IB) was performed against the B1 H+-ATPase subunit (left side) or against RhCG (right side). Panel B: the Co-IP membrane was stripped and reblotted with antibodies against Rhcg and B1 H+-ATPase antibodies as control for the IP. The upper band for B1 (approx 70 kDa) is considered unspecific. Panel C: Co-IP was performed with anti-Rhcg (lane 2) and anti-B2 (lane 5) polyclonal antibodies or with an isotypic IgG antibody (lanes 1 and 4). Lanes 3 und 6 were loaded with 50 mg rat kidney medulla protein. Immunoblotting (IB) was performed against the B2 H+-ATPase subunit (left side) or against RhCG (right side). Panel D: the Co-IP membrane was stripped and reblotted with antibodies against Rhcg and B2 H+-ATPase antibodies as control for the IP. Panel E: Coimmunostaining of Rhcg and the E H+-ATPase subunit in kidney tissue from Rhcg+/+ and Rhcg−/− mice. Immunostaining of E H+-ATPase subunit (in red) and Rhcg (green) was performed on kidney sections from Rhcg+/+ and Rhcg−/− mice. Nucleus were labelled with DAPI staining. Inserts show higher magnification of single intercalated cells.
We further confirmed partial colocalization of Rhcg and B1 subunits at the apical pole of intercalated cells in mouse kidney by immunohistochemistry (Figure 1, panel E).
Rhcg−/− mice show a reduced capacity to excrete H+ from kidney CCD cells
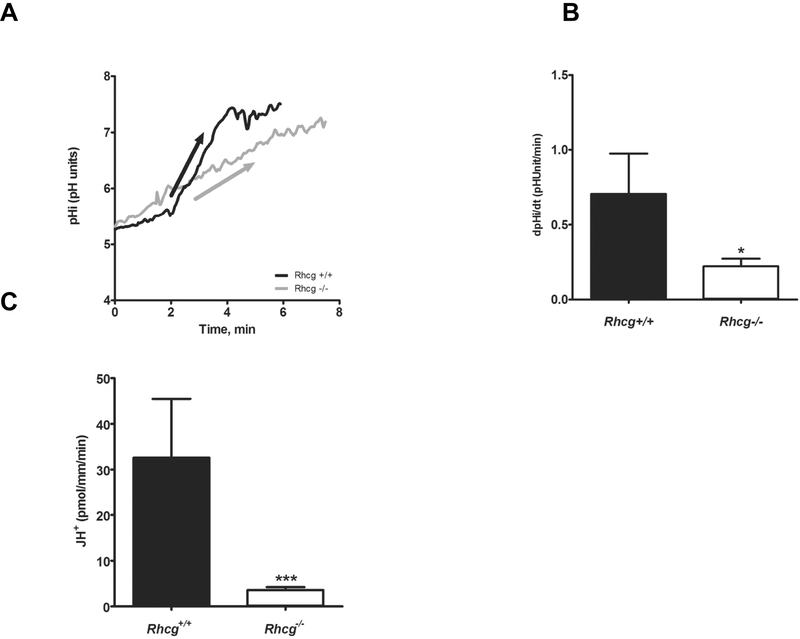
We recently showed in ex vivo microperfused collecting ducts from Rhcg−/− mice a reduced NH3 transepithelial flux due to decrease in apical and basolateral NH3 permeabilities 11–12. Here, using the same approach, we assessed H+ transport activity in microperfused CCDs from Rhcg+/+ and Rhcg−/− mice challenged for 2 days with an HCl load in food to maximize transport activities. Luminal NH4Cl prepulses were performed and H+ transport activity was assessed from the steep pHi recovery rates after maximal intracellular acidification when NH4Cl was removed from the lumen 32–33. The initial steep rate of alkalinisation reflects H+ extrusion, mostly by H+-ATPases 33 (Figures 2A and2B). We also observed in WT mice an early, almost immediate but slow pHi recovery which was not observed in KO mice suggesting that it is related to the function of Rhcg. As the initial pHi values were slightly different in CCDs from Rhcg−/− and Rhcg+/+ mice, we also measured intracellular buffering power and calculated H+ fluxes (JH+) across the membrane to directly compare transport rates. Similar to pHi recovery rates, JH+ was greatly reduced in CCDs from Rhcg−/− by 89% (Rhcg+/+: 32.6 ± 12.8 pmol/mm/min vs. Rhcg−/−: 3.6 ± 0.63 pmol/mm/min, p ≤ 0.0001) (Figure 2C). Thus, the more alkaline urine observed in Rhcg−/− during acid-loading results from impaired proton secretion along the collecting duct.
FIGURE 2. H+ secretion is reduced in cortical collecting duct cells from Rhcg−/− mice.
Cortical collecting ducts were isolated from kidneys of Rhcg+/+ and Rhcg−/− mice after 2 days of HCl-loading, microperfused in vitro, and intracellular pH (pHi) monitored. NH4Cl (20 mM) was applied from the luminal perfusate. (A) Original pHi tracing from a CCD exposed to a luminal NH4Cl pulse. The arrow indicates the rate of pHi change measured and calculated. Exposure to an NH4Cl pulse caused a large intracellular acidification (not shown) followed by an alkalinization phase leading to pHi recovery. The initial slope (ΔpHi/Δt) of the alkalinization phase was measured . (B) Bar graph summarizing ΔpHi/Δt after removal of the luminal NH4Cl pulse (n = 8–12 tubules/genotype). (C) Bar graph summarizing H+-fluxes calculated based on intracellular buffering power and pHi recovery rates.
The absence of Rhcg alters B1 and B2-H+-ATPase subunits expression
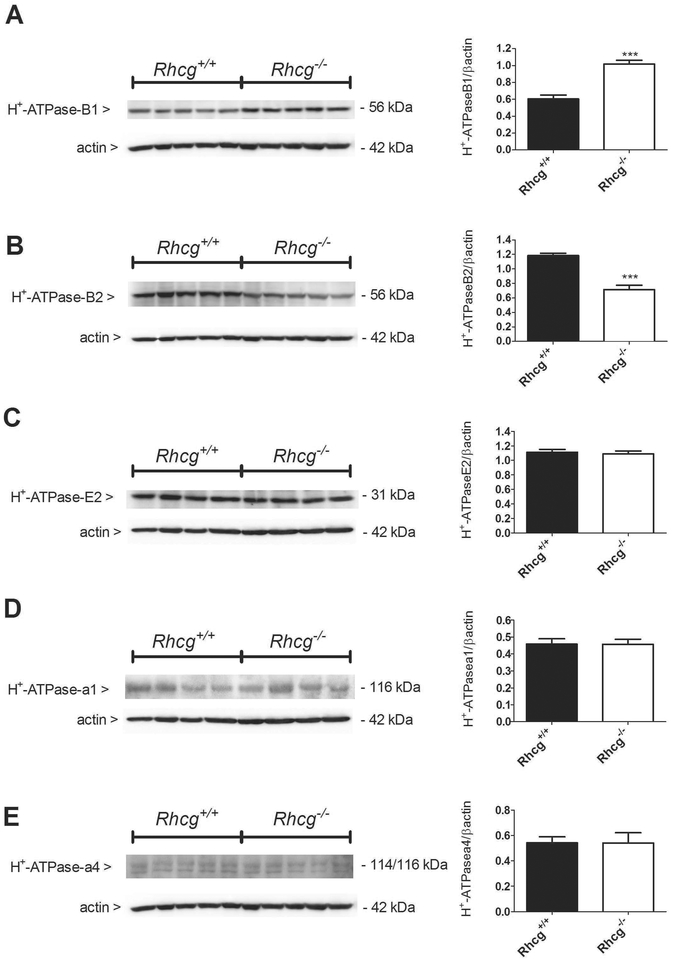
Next, we investigated whether the reduced H+ flux observed in microperfused CCDs from Rhcg−/− mice could be linked to altered expression of H+-ATPase subunits. At the mRNA level, we could not detect any variation of the intercalated cell-enriched B1 H+-ATPase isoform (suppl. figure 1A), and the more ubiquitous B2 (suppl. Figure 1B) and a4 (suppl. Figure 1C) H+-ATPase isoforms between Rhcg+/+ and −/− mice. In membrane fractions from medullary kidney tissue to enrich the proportion of H+- ATPases originating from intercalated cells, the amount of the intercalated cell specific B1 isoform (Figure 3A) was increased whereas the B2 isoform (Figure 3B) was decreased in kidney tissue from Rhcg−/− mice (p ≤ 0.01). However, the protein abundance of the ubiquitous E2 (Figure 3C) and a1 isoforms (Figure 3D) as well as the kidney-enriched a4 (Figure 3E) subunits of the H+-ATPase was not different between the 2 genotypes.
FIGURE 3. Increased B1 and decreased B2 H+-ATPase subunit abundance in Rhcg−/− mice.
Protein expression of the B1 (A), B2 (B), E2 (C), a1 (D) and a4 (E) subunits of the H+-ATPase was assessed by Western blotting in medullary kidney tissue. In Rhcg−/− mice, B1-H+-ATPase protein abundance was upregulated (A), while B2 H+-ATPase was downregulated (B). Protein expression of the a1, a4, and E subunits remained unchanged (C-E). Values are mean ± SEM (n= 5 mice), ***p ≤ 0.001.
Rhcg does not regulate H+-ATPase localization
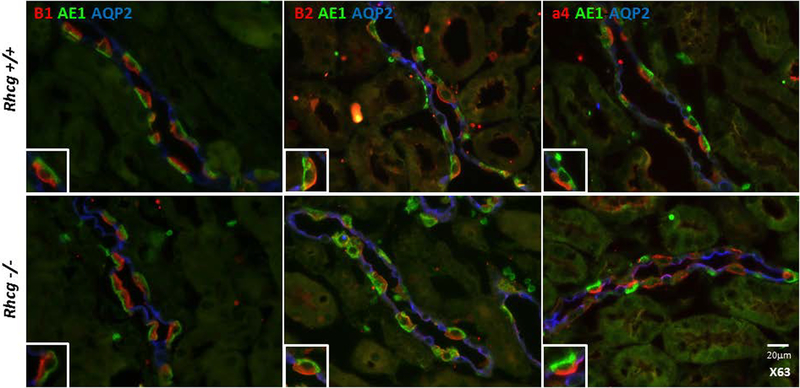
Reduced H+-ATPase activity may result from altered subcellular localization of H+-ATPases. Immunostaining for the B1, B2, E and a4 H+-ATPase subunits showed no apparent difference in subcellular localization of the 3 proteins in type-A intercalated cells in Rhcg+/+ and Rhcg−/− kidneys (Figure 4 and suppl. Figure 2). Electron microscopy with immunogold staining for the ubiquitious A subunit similarly detected no difference in the subcellular localization of this H+-ATPase subunit (Figure 5). The absence of Rhcg did not affect the immunoreactivity for the A subunit in type-A intercalated cells of Rhcg−/− mice. H+-ATPase staining was observed mostly in intercalated cells and was associated with the luminal membrane as well as with intracellular vesicles as described previously 16. Thus, the absence of Rhcg is not associated with a redistribution of H+-ATPases in type-A intercalated cells.
FIGURE 4. Unchanged H+-ATPase subunit localization in Rhcg−/− type A intercalated cells.
Immunostaining of B1, B2 and a4 subunits (in red) of three different H+-ATPase subunits (B1, B2, and a4) was performed on kidney sections from Rhcg+/+ and Rhcg−/− mice. Type A intercalated cells were identified by the presence of the anion exchanger AE1 (green) and principal cells using the apical water channel AQP2 (in blue). Inserts show higher magnification of single intercalated cells.
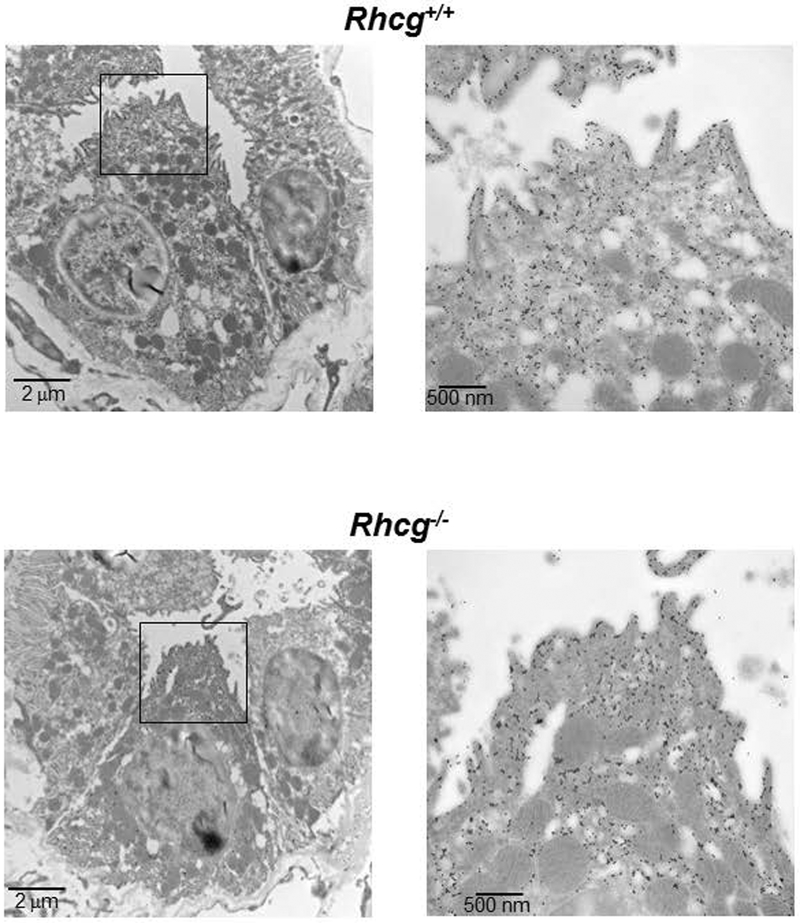
FIGURE 5. Subcellular localization of H+-ATPases in type A intercalated cells is similar in Rhcg+/+ and Rhcg−/− mice.
Immunogold staining for the A subunit of the H+-ATPase revealed a similar density of H+-ATPases at the apical membrane and in apically localized vesicles of type A intercalated cells in Rhcg+/+ (upper panels) and Rhcg−/− (lower panels) mice. Magnification of the boxed areas of the overview electron micrograph are shown on the right.
The overexpression of RhCG in HEK293 cells increases H+ secretion rates
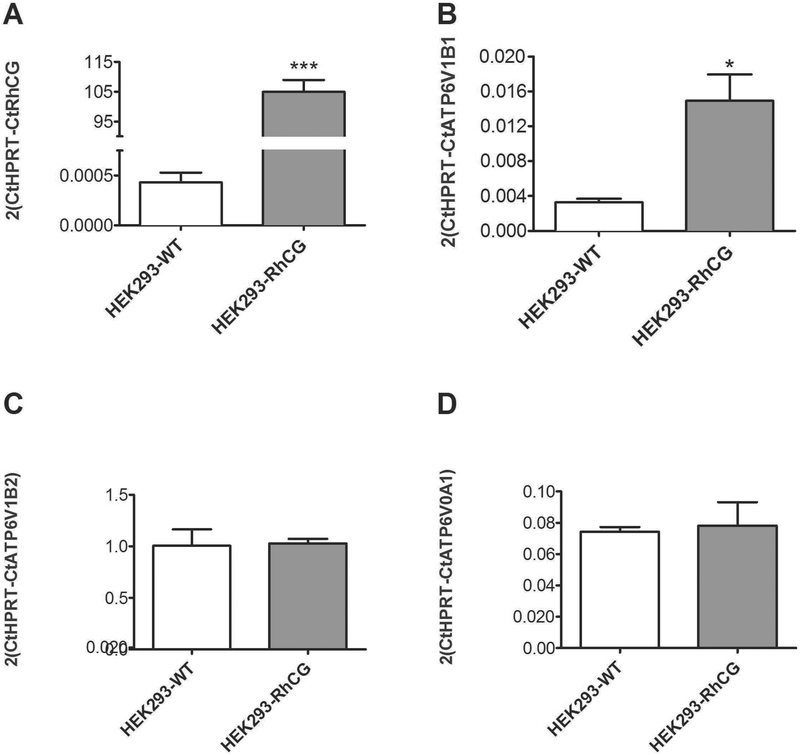
To test if the expression of RhCG was associated with stimulation of H+-ATPase activity, we used HEK293 cells, which express H+-ATPases and exhibit H+-ATPase-mediated H+ secretion across the plasma membrane 34. Non-transfected HEK293 cells and HEK293 cells stably transfected with human RhCG were used to examine the effect of RhCG expression on H+-ATPase expression and activity. qRTPCR confirmed high expression levels of RhCG in RhCG-transfected HEK293 cells as reported previously (Figure 6A) 5. Next, we compared the expression levels of transcripts encoding different subunits of H+-ATPase, ATP6V1B1, ATP6V1B2 and ATP6V0A1, and found an exclusive increase in mRNA expression of ATP6V1B1 in HEK-RhCG compared to HEK-WT cells (Figure 6B–D).
FIGURE 6. Overexpression of RhCG in HEK293 cells stimulates ATP6V1B1 expression.
.(A) HEK293 cells were left untransfected (HEK293-WT) or transfected to stably express RhCG (HEK293-RhCG) and RhCG expression was confirmed using RT-qPCR. mRNA expression of ATP6V1B1 (B), ATP6V1B2 (C) and ATP6V0A1 (D) were measured by RT-qPCR in HEK293-WT and RhCG overexpressing cells HEK293-RhCG. ATP6V1B1 mRNA was higher in HEK293-RhCG cells versus HEK293-WT. The mRNA abundance of the other H+-ATPase subunits, ATP6V1B2 and ATP6V0A1, were unchanged. Values are mean ± SEM (n= 5 dishes), *p ≤ 0.05, *** ≤ 0.001
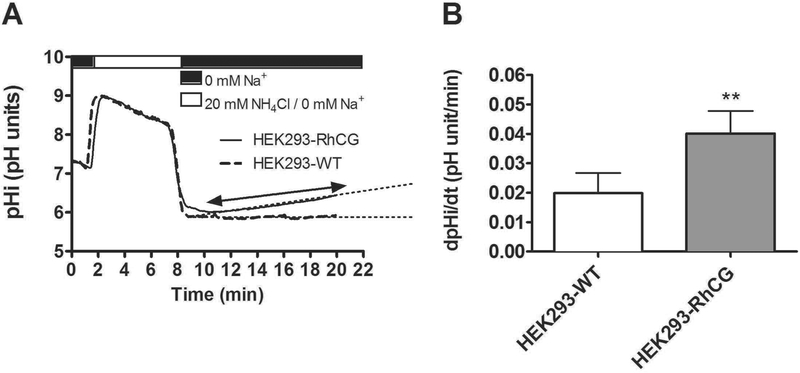
Following NH4Cl prepulse acidification in a HCO3− and Na+ free solution, we measured intracellullar pH recovery rates, similar to experiments in microperfused CCDs (Figure 7A). We have previously demonstrated that this pHi recovery is mediated solely by H+-ATPases and sensitive to the H+-ATPase blocker bafilomycin34. The initial pHi of both groups of cells measured was in the same range (pH 7.16 ± 0.001 for non-transfected HEK293 cells and pH 7.18 ± 0.002 for RhCG expressing RhCG cells, Figure 7A) which allowed a direct comparison of ΔpHi/Δt. In HEK-RhCG cells, the rate of pHi-recovery after NH4Cl removal was 52 % higher than the one observed in non-transfected HEK (HEK-RhCG: (0.666 ± 0.001)*10−3 pH units/min vs. non-transfected HEK: (0.322 ± 0.006)*10−3 pH units/min, p≤ 0.01) (Figure 7B). This result suggests that the presence of RhCG can stimulate H+ secretion and is in agreement with our data showing that H+ secretion is impaired in collecting ducts from Rhcg−/− mice.
FIGURE 7. RhCG overexpression in renal HEK293 cells increases H+ excretion.
H+-ATPase activity was measured in non-transfected HEK293 cells (HEK293-WT) and HEK293 cells stably transfected with human RhCG (HEK293-RhCG) using the NH4Cl (20 mM) prepulse after loading cells with BCECF. (A) Original pHi tracings from WT) and RhCG overexpressing HEK293 cells. Dashed lines indicate the slope measured to calculate pHi changes over time.(B) Bar graph summarizes the H+ extrusion rate of HEK-WT compared to HEK-RhCG after NH4Cl pulse (n = 5 cells/plate, 3 dishes/condition). Data are mean ± SEM, **p ≤ 0.01,
Lack of the Atp6v1b1 H+-ATPase subunit reduces H+-ATPase activity without effect on NH3 permeability
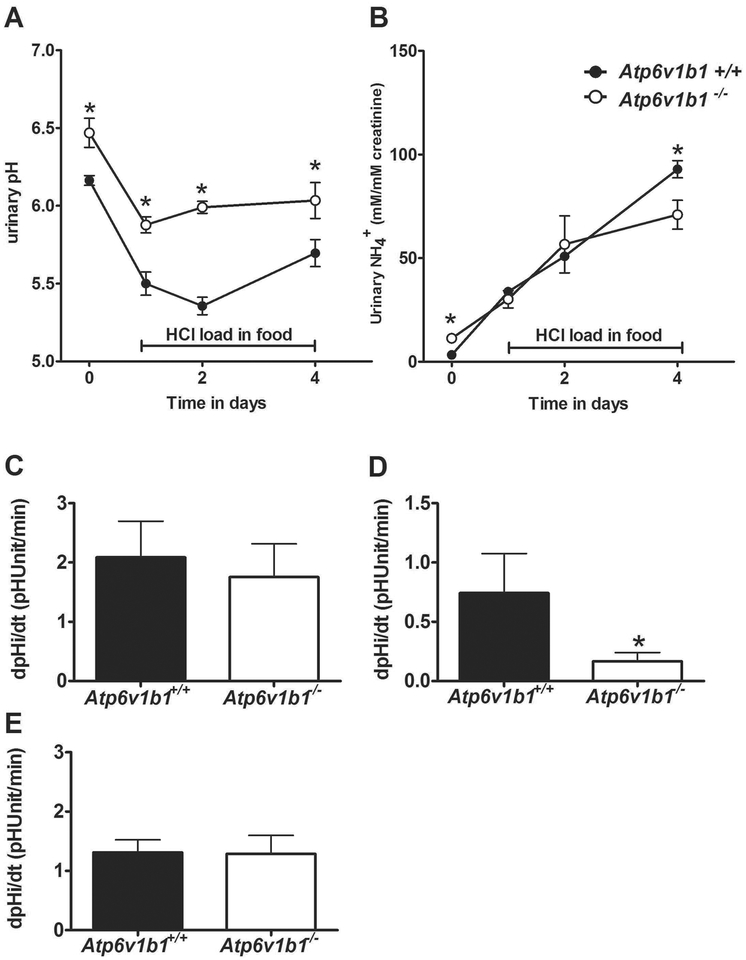
In order to assess whether H+-ATPase activity could affect Rhcg activity, we used a genetic animal model with reduced H+-ATPase activity, mice lacking the Atp6v1b1 (B1) subunit 19, 35–36. Atp6v1b11−/− mice challenged with a HCl containing diet showed a more alkaline urinary pH compared to Atp6v1b1+/+ mice (Figure 8A and Supplementary table 1) as demonstrated previously19. However, urinary NH +4 excretion was similar between both genotypes before and during acute HCl treatment. But, after 4 days of acid inducing diet, Atp6v1b1−/− presented with a significant 23% reduction of NH +4 excretion (Figure 8B). To assess whether collecting ducts from Atp6v1b11−/− mice had altered NH3 permeability, we investigated NH3 membrane permeabilities and H+ secretion in ex vivo microperfused isolated CCDs from Atp6v1b1+/+ and −/− mice receiving HCl diet for 2 days. We confirmed the expected reduction in H+ secretion in Atp6v1b1−/− mice (Figure 8C) but did not detect any differences in NH3 membrane permeability across the apical (Figure 8D) or basolateral (Figure 89E) membranes. Thus, in this genetic model, reduced H+- ATPase activity is not paralleled by reduced NH3 permeability.
FIGURE 8. Atp6v1b1−/− mice lacking the B1 H+-ATPase subunit have decreased H+-ATPase activity but preserved apical and basolateral NH3 permeabilities.
Widltype (Atp6v1b1+/+) and Atp6v1b1−/− mice were observed in metabolic cages and urinary pH (A) and urinary ammonium excretion normalized to creatinine (B) monitored during baseline and an acid-load (HCl) for 4 days (n = 7/genotype). Cortical collecting ducts from mice acid-loaded for 2 days were microperfused in vitro and H+ secretion (C) and apical NH3 permeability (D) measured after a luminal NH4Cl prepulse (20 mM) (n = 10 tubules/genotype). (E) Similarly, basolateral NH3 permeability assessed after an NH4Cl prepulse from the bath (6 mM) was not different between genotypes (n = 6 tubules/genotype). *p ≤ 0.05
DISCUSSION
The present study provides a new and additional explanation for the well-known association between urinary acidification and ammonium excretion 26. We demonstrate that the ammonia channel RhCG is required for maximal urinary acidification and that RhCG stimulates H+-ATPase activity.
Secretion of protons acidifies urine which provides a major driving force for NH3 secretion and trapping of NH +4 after protonation. Thus, in this physico-chemical model, H+-secretion is the primary process which then affects the chemical gradient for NH3 secretion.
Urinary acidification depends on H+ excretion along the collecting duct mediated mostly by H+-ATPases and to a lesser extent by H+-K+-ATPases. In intercalated cells, H+-ATPases are the primary pump at the apical membrane mediating type A intercalated cell H+-excretion after an intracellular acid-load 33, 37. The B1 H+-ATPase subunit is specifically represented and colocalizes with RhCG at the luminal side of renal type A intercalated cells 10. Based on previous observations that Rhcg null mice had more alkaline urine despite reduced NH secretion rates 11–12, we hypothesized that Rhcg was required for maximal urinary acidification. Here, we observed a reduced apical H+ secretion rate in acid-loaded Rhcg−/− mice suggesting a form of regulation of H+-ATPases by RhCG or its activity.
In Rhcg−/− mice, reduced H+-ATPase activity was not caused by down-regulation of B1 or other H+-ATPase subunits at mRNA or protein level. On the contrary, B1 was upregulated which may represent a compensatory process. The B2 H+-ATPase subunit was downregulated and as previously been shown to exhibit counterregulation in B1 deficient mice 38. These data suggest that the reduction in apical H+ flux observed in Rhcg−/− mice CDs is not the consequence of a lower amount of H+-ATPases formed but rather explained by H+-ATPases which are not fully functional due to either an inadequate assembling of the subunits or improper insertion of the pumps at the plasma membrane. Immunohistochemistry and immunogold staining for several subunits of the V1 and V0 domains of the H+-ATPase at the apical pole of intercalated cells did not find any evidence for altered subcellular localization and the presence of both V0 and V1 domain subunits suggests also intact assembly of pumps.
In vitro studies in HEK293 cells revealed that overexpression of RhCG stimulates H+ extrusion paralleled by higher mRNA expression of the B1 H+-ATPase subunit. HEK293 cells express H+-ATPases and exhibit H+ extrusion only after intracellular acidification in a slow, time dependent manner that is independent from trafficking of H+-ATPases to the plasma membrane 34. Thus, H+-ATPase activity is increased in the presence of RhCG in vivo and in vitro and this effect may involve also increased abundance of some subunits, at least in vitro.
We next tested in Atp6v1b1−/− mice whether H+ATPases would have a reciprocal regulatory effect on Rhcg activity. As previously reported, we observed higher urinary pH and highly reduced apical H+ transport in acute and chronic HCl acid-loaded Atp6v1b1−/− mice compared to wildtype littermates 19, 35–36. However, the absence of Atp6v1b1 and reduced H+-ATPase activity did not affect the NH3 transport measured on both basolateral and apical cellular membranes. Accordingly, urinary NH +4 excretion was not different in 2 days acid-loaded Atp6v1b1−/− compared to wildtype. However, only after a more chronic acid-load, urinary NH +4 excretion was reduced in Atp6v1b1−/− mice compared to Atp6v1b1+/+ animals but to a much smalleri extent than in Rhcg−/− mice under the same conditions 11. The modest decrease in urinary NH +4 excretion could be due to the more alkaline urinary pH reducing the driving force for NH +4 secretion along the collecting system. Thus, RhCG stimulates H+-ATPase activity whereas reduced H+-ATPase activity does not directly impact on RhCG activity.
H+-ATPase activity is regulated by assembly/disassembly of the pump, trafficking to and from the membrane, and by phosphorylation at different sites which may occur in response to changes in acid-base status and/or endocrine factors 3, 15, 39–51. RhCG may modulate H+-ATPase activity through at least two different mechanisms. NH3 stimulates H+/K+-ATPase possibly involving increased insertion of the H+/K+-ATPase via a SNARE-dependent process 52. RhCG participates in basolateral NH3 uptake 11 and loss of RhCG activity may reduce intracellular NH3 levels and NH3 dependent regulation of luminal H+-extrusion. Alternatively, RhCG may stimulate H+-ATPase activity through (in)direct protein-protein interactions. The B1 H+-ATPase subunit possesses a PDZ domain to interact with other proteins 46. Whether RhCG interacts with other protein has not been reported to date. However, the related RhBG protein, expressed at the basolateral side of renal CCD cells 53, has been shown to be anchored to the membrane via the actin cytoskeleton-binding protein ankyrin-G 54 and to be part of a structural complex where the anion exchanger 1 (AE1) responsible for the secretion of HCO −3 into blood by the intercalated cells can bind to both RhBG and ankyrin-G 55. This complex is essential for the activity of RhBG and AE1..Likewise, H+-ATPases exhibit interactions with the actin cytoskeleton, via the C subunit 56 and most likely via binding sites for F-actin that are present in the amino-terminal domains of both B1 and B2 isoforms 57 Thus, RhCG and H+-ATPases may be held together in one protein complex through interactions via the actin cytoskeleton. There is emerging evidence for large protein complexes involved in the regulation of H+-ATPases as well as coordinating the development of intercalated cells. These processes are critical for the kidneys ability to maintain and adapt acid excretion. On the one hand, rare mutations may exist in some patients and their discovery will help to diagnose hitherto unexplained cases of inborn dRTA. On the other hand, dysregulation of these networks may contribute to reduce acid excretion observed in patients with more common forms of CKD. Nevertheless, a better understanding of the components of these networks is required.
In summary, we show that RhCG and H+ATPases are located within the same cellular protein complex in the kidney and this interaction is required for maximal urinary acidification by H+-ATPases, a prerequisite for efficient NH3 secretion and urine excretion of NH +4. Thus, this novel mechanism provides a molecular explanation for the tight association between urinary acidification and ammonium excretion where RhCG mediates NH3 secretion and maximizes H+-secretion.
MATERIALS AND METHODS
Co-Immunoprecipitation
Co-immunoprecipitation was performed using the Pierce Cross-link Magnetic beads kit (Pierce, USA) according to the manufacturers’ instructions. Fifty mg of medulla from rat kidney were homogenized using an electric homogenizer in 3 ml Pierce IP Lysis/Wash Buffer (pH 7.4, 0.025M Tris, 0.15M NaCl, 0.001M EDTA, 1% NP40, 5% glycerol). Rabbit anti-B1 H+-ATPase 33 and rabbit anti-Rhcg (Proteintech, USA) polyclonal antibodies were bound to Pierce Protein A/G Magnetic Beads, and crosslinked using DSS reagent. Finally, crosslinked complexes were eluated with the pH 2.0 Pierce Elution Buffer, and neutralized with the Pierce Neutralization Buffer at pH 8.5.
Animals - Rhcg+/+ and −/− mice were generated and bred as described before 11. Also, Atp6v1b1+/+ and −/− mice were bred as described before 19, 35. Mice were genotyped by PCR directly on a 3 μl 25 mM NaOH biopsy digestion product.
Genomic DNA was amplified using primer pairs specific for Rhcg exon 1: forward (AGACCCCACAATGGAAAGCTATAA) and wildtype reverse (CAACCAGAACTCCCCAGTGTCAGA) and knock-out reverse (ATGGGCTGACCGCTTCCTCGTGCTTTAC) and Atp6v1b1 primer pairs specific for the Neo cassette: forward (CACGACGAGATCCTCGCCGTC) and reverse (GCGCAGCTAGTGCTCGACGTTG) and an Atp6v1b1 3′ flanking sequence not included within the targeting vector: forward (CTGGCACTGACCACGGCTGAG) and reverse (CCAGCCTGTGACTGAGCCCTG). The products were separated by electrophoresis on 1% agarose gels (Rhcg mutant product: 522 bp, wildtype product: 376 bp, Atp6v1b1 mutant product: 300 bp, wildtype product: 200bp).
All animal experiments were performed in acid-loading conditions for which mice were given 0.2 M HCl added to powdered standard food for 2 days. All experiments were performed according to Swiss Animal Welfare laws and were approved by the local veterinary authority (Veterinäramt Zürich).
In vivo experiments
All experiments were performed using age- and sex-matched Atp6v1b1 wildtype (Atp6v1b1+/+) and Atp6v1b1 knockout (Atp6v1b1−/−) littermate mice (3–4 month-old), housed in metabolic cages (Techniplast, Switzerland). Mice were given deionized water ad libitum and were fed with a standard powdered laboratory chow (Kliba, Augst, Switzerland). Mice were allowed to adapt to metabolic cages for 3 days and a first retro-orbital blood sample was taken for blood gas analysis at baseline. Then, two 24 hrs urine samples were collected under light mineral oil in the urine collector to determine daily urinary parameters. Mice were then allowed to recover for 2 weeks before given a HCl-containing diet (0.2 M HCl added to powdered standard food) in normal cages. Food, water intake, and urine excretion was monitored following the same procedures as under baseline conditions. Urine collections were performed on the first, second and fourth day of acid-loading. Retro-orbital blood samples were taken on the second and fourth day of the HCl diet under light anesthesia with isoflurane. All animal experiments were conducted according to Swiss Laws of Animal Welfare and approved by the local Zurich Veterinary Authority (Kantonales Veterinäramt Zürich).
Analytic procedures
Blood pH, pCO2, and electrolytes were measured with a pH/blood-gas analyzer (ABL77 Radiometer). Urinary pH was measured with a pHmeter (Metrohm AG, Canada) and creatinine by a modified kinetic Jaffé colorimetric method 58. Urinary NH + was measured with the Berthelot protocol 59. Urine inorganic phosphate (Pi) concentration was determined by the phosphomolybdate method 60.
Electron microscopy and immunogold labeling
Mice were anesthetized with 2.5 % Isoflurane/pressurized air and perfused through the left ventricle with PBS (0.9% NaCl in 10 mM phosphate buffer, pH 7.4) followed by paraformaldehyde-lysine-periodate fixative (PLP: 4% paraformaldehyde, 75 mM lysine HCl, 10 mM sodium periodate, and 0.15 M sucrose in 37.5 mM sodium phosphate)61. After 5 minutes the kidneys were removed and postfixed for an additional hour on ice in the same fixative solution. Following fixation, the kidneys were stored at 4°C in PBS.
For immunogold staining, small pieces of Rhcg+/+ and −/− kidneys were dehydrated through a graded series of ethanol to 100% ethanol and then embedded in LR White resin (Electron Microscopy Sciences, Fort Washington, PA, USA). Thin sections were incubated on drops of primary anti-H+-ATPase (A-subunit, 1:200) antibody for 2 h. After rinsing with PBS, the grids were incubated on drops of goat anti-rabbit IgG coupled to 15nm gold particles (Ted Pella, Redding, CA, USA) for 1 h. Following several rinses with distilled water, the grids were stained with 2% uranyl acetate for 10 min, rinsed in distilled water, and dried. Sections were examined in a JEM-1011 transmission electron microscope (JEOL Ltd, Tokyo, Japan).
Immunohistochemistry
Rhcg+/+ and −/− mice were anesthetized with ketamine/xylazine and perfused through the left ventricle with PBS followed by paraformaldehyde-lysine-periodate (PLP) fixative 61. Kidneys were removed and fixed overnight at 4°C by immersion in PLP. Kidneys were washed three times with PBS, and 3 μm cryosections were cut after cryoprotection with 0.9 M sucrose in PBS for at least 1 h. Immunostaining was carried out as described 62. The following primary antibodies were used: guinea pig anti-AE1 1:500 63, rabbit anti-a4 H+-ATPase 1:500 33, rabbit anti-B1 H+-ATPase 33, rabbit anti-B2 H+-ATPase 1:500 64, mouse monoclonal ant E H+-ATPase 1:200 (gift from S. Gluck), rabbit anti Rhcg 65 1:1000 and goat anti AQP-2 (Santa Cruz Biotechnology) 1:1000. The secondary antibodies were donkey anti-rabbit 594, donkey anti-goat 488, donkey anti-rabbit 488, donkey anti-mouse 594 and donkey anti-guinea pig 647 (Molecular Probes) at 1:1000.
Immunoblotting
Crude total membranes were obtained from Rhcg+/+ and −/− mouse renal medulla homogenized in 250 mM sucrose, 10 mM Tris-HCl, pH 7.5, and in the presence of protease inhibitors (Roche Protease Inhibitor Cocktail) 62. Fourty micrograms of crude membrane proteins were solubilized in Laemmli loading buffer containing 10% DTT and separated on 8 to 10% polyacrylamide gels. For immunoblotting, proteins were transferred electrophoretically to polyvinylidene fluoride membranes (Immobilon-P, Millipore Corp., Bedford, MA, USA). After blocking with 5 % milk powder in Tris-buffered saline/0.1% Tween-20 for 60 min, blots were incubated with primary antibodies: rabbit anti-mouse ATP6V1B1 (B1)(1:5000) 33, rabbit anti-ATP6V1B2 (B2) (1:5,000) 64, rabbit anti-ATP6V0a4 33, 1:5000, rabbit anti-ATP6V0a1 (1:5000), a kind gift of Dr Xie, Dallas, TX, USA) 66, and mouse anti-ATP6V1E2 (1:5000, a kind gift of Dr. S. L Gluck, University of California, San Francisco, CA, USA) 67, and mouse monoclonal anti-β-actin antibody (Sigma, St. Louis, MO; 1:10000) overnight at 4°C. After washing and blocking with 5 % milk powder for 60 min, membranes were incubated for 2 h at room temperature with secondary goat anti-rabbit or donkey anti-mouse antibodies 1:5000 linked to alkaline phosphatase (Promega, Madison, WI, USA). The protein signal was detected with the appropriate substrate (Millipore Corp, Bedford, MA, USA) using the las-4000 image analyzer system (Fujifilm Life Science USA). All images were analyzed using Advanced Image Data Analyzer AIDA (Raytest, Straubenhardt, Germany) to calculate the protein of interest/β-actin ratio.
Cell culture
Non-transfected HEK293 (human embryonic kidney 293) or stably transfected with human RhCG (HEK-RhCG) cell lines were generated as previously described 6 and grown at 37°C, 5% CO2 in DMEM (Dulbecco’s modified Eagle’s medium)/Glutamax I (Invitrogen) supplemented with 10% (v/v) fetal bovine serum. Selection of HEK-RhCG cells was performed in culture medium supplemented with 0.3 mg/ml hygromycin (Invitrogen). For all pH experiments, cells were passaged, disseminated onto coverslips and grown to sub-confluency for 48 hours. The final medium exchange was performed 24 hours before the experiments were started. Medium pH was maintained at pH 7.4.
RNA extraction and reverse transcription
Snap-frozen kidneys (five kidneys for each condition) or non-transfected HEK and HEK-RhCG cells (3 petri dishes of 5×106 cells for each condition) were homogenized in RLT-Buffer (Qiagen, Basel, Switzerland) supplemented with β-mercaptoethanol to a final concentration of 1%. Total RNA was extracted from 200 μl aliquots of each homogenized sample using the RNeasy Mini Kit (Qiagen,Basel, Switzerland) according to the manufacturer’s instructions. Quality and concentration of the isolated RNA preparations were analyzed on the ND-1000 spectrophotometer (Nano-Drop Technologies). Total RNA samples were stored at –80°C. Each RNA sample was diluted to 100 ng/μl and 3 μl used as a template for reverse transcription using the TaqMan Reverse Transcription Kit (Applied Biosystems, Forster City, CA, USA). For reverse transcription, 300 ng of RNA template were diluted in a 40 μl reaction mix that contained (final concentrations) RT buffer (1 x), MgCl2 (5.5 mM), random hexamers (2.5 μM), RNase inhibitor (0.4U/μl), the multiscribe reverse transcriptase enzyme (1.25U/μl), dNTP mix (500 μM each), and RNase-free water.
Reverse Transcription and semiquantitative real-time PCR
Semiquantitative real-time RT-qPCR was performed on the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Forster City, CA). Primers and probes were chosen using BLAST tool: Ensemble (http://www.ensembl.org/index.html) to result in amplicons no longer than 150 bp spanning intron-exon boundaries to exclude genomic DNA contamination. The specificity of all primers was first tested on mRNA derived from kidney and always resulted in a single product of the expected size (data not shown). Probes were labeled with the reporter dye FAM at the 5’ end and the quencher dye TAMRA at the 3’ end (Microsynth, Balgach, Switzerland). Primers and probes sequences are summarized in supplementary table 3.
Real-Time PCR reactions were performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Forster City, CA). Briefly, 3 μl cDNA, 0.8 μl of each primer (25 μM), 0.4 μl labeled probe (5 μM), 5 μl RNAse-free water, and 10 μl TaqMan Universal PCR Master Mix reached 20 μl of final reaction volume. Reaction conditions were denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15s and annealing/elongation at 60°C for 60s with auto ramp time. All reactions were run in triplicate. For analyzing the data, the threshold was set to 0.2 as this value had been determined to be in the linear range of the amplification curves for all mRNAs in all experimental runs. The expression of the genes of interest was calculated in relation to hypoxanthine guanine phosphoribosyl transferase (mouse HPRT or human HPRT). Relative expression ratios were calculated as R = 2(Ct(HPRT) − Ct(test gene), where Ct represents the cycle number at the threshold 0.02.
Intracellular pH (pHi) measurements on HEK293 cells
For pHi measurements, individual slides seeded with non-transfected HEK or HEK-RhCG cells were transferred into a perfusion chamber (~ 3–5 ml/min flow rate) and mounted on an inverted microscope (Zeiss Axiovert 200, Carl Zeiss, Feldbach, Switzerland) equipped with a video imaging system (Visitron, Munich, Germany) for the duration of the experiment. The temperature of the chamber was maintained at 37°C by an electronic feedback circuit. The control bath solution was initially a HEPES-buffered Ringer solution (125 mM NaCl/ 5 mM KCl/ 1 mM CaCl2/ 1.2 mM MgSO4/ 2 mM NaH2PO4/ 32.2 mM HEPES/ 5 mM Glucose). Cells were loaded with the acetoxymethyl ester of the pH sensitive dye BCECF (10 μM, 2’,7’-bis(2-carboxyl)-5-(and-6)-carboxyfluorescein, Invitrogen, Switzerland) for 15 min and washed to remove all non-deesterified dye. pHi was measured by microfluorometry by exciting the BCECF dye with a spot of light alternately at 490 and 440 nm while monitoring the emission at 532 nm with a video-imaging system 68. H+-ATPase activity was measured after induction of a sharp intracellular acidification using the NH4Cl (20 mM) prepulse technique 32, 68. Bicarbonate-free solutions were used and Na+ was replaced by N-methyl-D-glucamine (NMDG). Each experiment was calibrated for pHi using the nigericin/high K+ method 69 and the obtained ratios were converted to pHi. Na+-independent pHi recovery rates in response to an acid load were calculated in non-transfected HEK293 and HEK293 stably transfected cells with RhCG. Data are shown as changes in pHi (ΔpHi) per minute.
Microperfusion of isolated cortical collecting ducts
Male Rhcg+/+, Rhcg−/−, Atp6v1b1+/+, and Atp6v1b1−/− mice were given a HCl-containing diet (a 50:75 mixture (w/V) of normal powdered chow, Kliba Nafag and 0.33 M HCl) in normal cages for 2 days. Mice were anesthetized with Xylazin/Ketamin i.p. Both kidneys were cooled in situ with control bath solution (see below) for 1 min and then removed and cut into thin coronal slices for tubule dissection. Cortical CDs (CCDs) were dissected from the cortex at 10°C in the control solution under a stereo-microscope.
Intracellular pHi measurement in microperfused cortical collecting ducts
The isolated tubules were transferred to the bath chamber on the stage of an inverted microscope (IX81, Olympus, Japan) in the control solution containing (in mM) 138 NaCl, 1.5 CaCl2, 1.2 MgSO4, 2 K2HPO4, 10 HEPES, 5.5 glucose, 5 alanine, pH 7.40) and then mounted on concentric pipettes and perfused in vitro with Na+- free, ammonium-free solution where N-methyl-D-Glutamine+ (NMDG+) replaced Na+. The average tubule length exposed to bath fluid was limited to 300 – 350 μm in order to prevent motion of the tubule. CCDs were loaded with 5 μM of the fluorescent probe BCECF for ~20 min at 37°C in the control bath solution. Intracellular pH (pHi) recordings were made over regions of interest comprising of several cells, however, the signal came mostly from intercalated cells that had taken up more BCECF-AM than principal cells. The loading solution was then washed out by initiation of bath flow and the tubule was equilibrated with dye-free control bath solution for 5 min. Bath solution was delivered at a rate of 20 ml/min and warmed to 37°C by water jacket immediately upstream to the chamber. After this temperature equilibration in control solution, tubules were first transiently acidified by peritubular Na+ removal (Na-free, ammonium-free solution) (10 min duration), replaced by NMDG+ to avoid exit of NH + 4by basolateral Na+-coupled transport. This maneuver was performed in the luminal absence of Na+. During the fluorescence recording, perfusion solution was delivered to the perfusion pipette via a chamber under an inert gas (N2) pressure (around 1 bar) connected through a manual 6-way valve. With this system, opening of the valve instantaneously activates flow of solutions. The majority of the fluid delivery to the pipette exits the rear of the pipette system through a drain port at 4 ml/min. This method results in a smooth and complete exchange of the luminal or the peritubular solution in less than 3 to 4 s 70. After the fluorescence signal stabilization, luminal medium was instantly (at the rate of 4 ml/min in the draining) replaced by a Na+-free solution containing 20 mM NH4Cl (and 118 mM NMDG-Cl) that elicited a rapid intracellular alkalinization, followed by a sharp acidification. The rate of intracellular alkalinization has been associated with the entry of NH3 whereas the subsequent phase of intracellular acidification in the continued presence of extracellular NH4Cl reflects mostly NH + entry 12, 32. After the acidification reached a plateau, NH4Cl was removed from the lumen to further acidify cells and initiate direct H+-excretion with a maximum rate. All measurements were done at maximum rate for the determination of H+-ATPase activity. The measured light intensities were digitized with the CellM&CellR Imaging hardware system (Olympus, Japan) for further analysis as previously described 11–12, 53. For peritubular ammonium pulses, the peritubular solution was changed to a 6 mM NH4Cl solution, pH 7.40 in the presence of 1 mM furosemide and 2.5 mM ouabain in the bath.
Intrinsic buffering capacity determination
The intrinsic buffering capacity (ßi) of CCD cells was determined for the Rhcg strain, as previously reported 11, 71, using a 40 mM NH4Cl basolateral pulse to acidify cells. To exclude HCO −3/CO as a buffering component and block Na+- dependent pHi regulatory mechanisms, Na+-free, HEPES-buffered solutions were used in perfusate and bath containing 1 mM amiloride and bath and perfusate also contained 100 μM Sch28080 and 200 nM Concanamycin A. Addition of 40 mM NH4Cl to the bath induced an increase followed by a decrease in pHi. Cellular buffering power (βi) was measured in CCDs from Rhcg+/+ and Rhcg−/− mice.
Statistics
Statistical comparisons were tested by unpaired t-test using Graphpad Prism (GraphPad Software). P values < 0.05 were considered as statistically significant.
Supplementary Material
Acknowledgements
This study was supported by grants from the Swiss National Science Foundation to C.A. Wagner (31003A_138143 and 31003A_155959). The use of the Zurich Integrative Rodent Physiology (ZIRP) Core Facility is gratefully acknowledged. We thank Dr. Dominique Goossens (INTS, Paris) for the generous gift of the anti-Rhcg polyclonal antibody
Footnotes
DISCLOSURE
All the authors declared no competing interests.
REFERENCES
- 1.Weiner ID, Hamm LL. Molecular mechanisms of renal ammonia transport. Annu Rev Physiol 2007; 69: 317–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner CA, Devuyst O, Bourgeois S, et al. Regulated acid-base transport in the collecting duct. Pflugers Arch 2009; 458: 137–156. [DOI] [PubMed] [Google Scholar]
- 3.Wagner CA, Devuyst O, Belge H, et al. The rhesus protein RhCG: a new perspective in ammonium transport and distal urinary acidification. Kidney Int 2011; 79: 154–161. [DOI] [PubMed] [Google Scholar]
- 4.Gruswitz F, Chaudhary S, Ho JD, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A 2010; 107: 9638–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mouro-Chanteloup I, Cochet S, Chami M, et al. Functional reconstitution into liposomes of purified human RhCG ammonia channel. PLoS One 2010; 5: e8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zidi-Yahiaoui NCI, Genetet S, Le Van Kim C,, Cartron JPCY, Ripoche P, Mouro-Chanteloup I. Functional analysis of human RhCG: comparison with E. coli ammonium transporter reveals similarities in the pore and differences in the vestibule. Am J Physiol Cell Physiol 2009; 297: 537–547. [DOI] [PubMed] [Google Scholar]
- 7.Han KH, Croker BP, Clapp WL, et al. Expression of the ammonia transporter, rh C glycoprotein, in normal and neoplastic human kidney. J Am Soc Nephrol 2006; 17: 2670–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HY, Verlander JW, Bishop JM, et al. Basolateral Expression of the ammonia transporter family member, Rh C Glycoprotein, in the Mouse Kidney. Am J Physiol Renal Physiol 2009; 296: F543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verlander JW, Miller RT, Frank AE, et al. Localization of the ammonium transporter proteins RhBG and RhCG in mouse kidney. Am J Physiol Renal Physiol 2003; 284: F323–337. [DOI] [PubMed] [Google Scholar]
- 10.Eladari D, Cheval L, Quentin F, et al. Expression of RhCG, a new putative NH3 /NH4 + transporter, along the rat nephron. J Am Soc Nephrol 2002; 13: 1999–2008. [DOI] [PubMed] [Google Scholar]
- 11.Bourgeois S, Bounoure L, Christensen EI, et al. Haploinsufficiency of the ammonia transporter Rhcg predisposes to chronic acidosis: Rhcg is critical for apical and basolateral ammonia transport in the mouse collecting duct. J Biol Chem 2013; 288: 5518–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biver S, Belge H, Bourgeois S, et al. A role for Rhesus factor Rhcg in renal ammonium excretion and male fertility. Nature 2008; 456: 339–343. [DOI] [PubMed] [Google Scholar]
- 13.Lee HW, Verlander JW, Bishop JM, et al. Effect of intercalated cell-specific Rh C glycoprotein deletion on basal and metabolic acidosis-stimulated renal ammonia excretion. Am J Physiol Renal Physiol 2010; 299: F369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HW, Verlander JW, Handlogten ME, et al. Effect of collecting duct-specific deletion of both Rh B Glycoprotein (Rhbg) and Rh C Glycoprotein (Rhcg) on renal response to metabolic acidosis. Am J Physiol Renal Physiol 2014; 306: F389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar H+-ATPase. Physiol Rev 2004; 84: 1263–1314. [DOI] [PubMed] [Google Scholar]
- 16.Breton S, Brown D. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 2013; 28: 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forgac M Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 2007; 8: 917–929. [DOI] [PubMed] [Google Scholar]
- 18.Karet FE, Finberg KE, Nelson RD, et al. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 1999; 21: 84–90. [DOI] [PubMed] [Google Scholar]
- 19.Finberg KE, Wagner CA, Bailey MA, Paunescu TG, Breton S, Brown D, Giebisch G, Geibel JP, Lifton RP. The B1 subunit of the H+ATPase is required for maximal urinary acidification. Proc Nat Acad Sci USA 2005; 102: 13616–13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhayat NA, Schaller A, Albano G, et al. The Vacuolar H+-ATPase B1 Subunit Polymorphism p.E161K Associates with Impaired Urinary Acidification in Recurrent Stone Formers. J Am Soc Nephrol 2016; 27: 1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz N, Dave MH, Stehberger PA, et al. Differential localization of vacuolar H+-ATPases containing a1, a2, a3, or a4 (ATP6V0A1–4) subunit isoforms along the nephron. Cell Physiol Biochem 2007; 20: 109–120. [DOI] [PubMed] [Google Scholar]
- 22.Stehberger P, Schulz N, Finberg KE, Karet FE, Giebisch G, Lifton RP, Geibel JP, Wagner CA Localization and regulation of the ATP6V0A4 (a4) vacuolar H+-ATPase subunit defective in an inherited form of distal renal tubular acidosis. J Am Soc Nephrol 2003; 14: 3027–3038. [DOI] [PubMed] [Google Scholar]
- 23.Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, Scherer SW, Karet FE. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet 2000; 26: 71–75. [DOI] [PubMed] [Google Scholar]
- 24.Hennings JC, Picard N, Huebner AK, et al. A mouse model for distal renal tubular acidosis reveals a previously unrecognized role of the V-ATPase a4 subunit in the proximal tubule. EMBO Mol Med 2012; 4: 1057–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norgett EE, Golder ZJ, Lorente-Canovas B, et al. Atp6v0a4 knockout mouse is a model of distal renal tubular acidosis with hearing loss, with additional extrarenal phenotype. Proc Natl Acad Sci U S A 2012; 109: 13775–13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrong O, Davies HE. The excretion of acid in renal disease. Q J Med 1959; 28: 259–313. [PubMed] [Google Scholar]
- 27.Weiner ID, Verlander JW. Ammonia Transporters and Their Role in Acid-Base Balance. Physiol Rev 2017; 97: 465–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitts RF. The Renal Regulation of Acid Base Balance with Special Reference to the Mechanism for Acidifying the Urine. II. Science 1945; 102: 81–85. [DOI] [PubMed] [Google Scholar]
- 29.Pitts RF. The Renal Regulation of Acid Base Balance with Special Reference to the Mechanism for Acidifying the Urine. Science 1945; 102: 49–54. [DOI] [PubMed] [Google Scholar]
- 30.Quentin F, Eladari D, Cheval L, et al. RhBG and RhCG, the putative ammonia transporters, are expressed in the same cells in the distal nephron. J Am Soc Nephrol 2003; 14: 545–554. [DOI] [PubMed] [Google Scholar]
- 31.Stettner P, Bourgeois S, Marsching C, et al. Sulfatides are required for renal adaptation to chronic metabolic acidosis. Proc Natl Acad Sci U S A 2013; 110: 9998–10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roos A, Boron WF. Intracellular pH. Physiol Rev 1981; 61: 296–434. [DOI] [PubMed] [Google Scholar]
- 33.Wagner CA, Lukewille U, Valles P, Breton S, Brown D, Giebisch GH, Geibel JP. A rapid enzymatic method for the isolation of defined kidney tubule fragments from mouse. Pflugers Arch 2003; 446: 623–632. [DOI] [PubMed] [Google Scholar]
- 34.Lang K, Wagner CA, Haddad G, Burnekova O, Geibel J. Intracellular pH activates membrane-bound Na+/H+ exchanger and vacuolar H+-ATPase in human embryonic kidney (HEK) cells. Cell Physiol Biochem 2003; 13: 257–262. [DOI] [PubMed] [Google Scholar]
- 35.Rothenberger F, Velic A, Stehberger PA, et al. Angiotensin II stimulates vacuolar H+-ATPase activity in renal acid-secretory intercalated cells from the outer medullary collecting duct. J Am Soc Nephrol 2007; 18: 2085–2093. [DOI] [PubMed] [Google Scholar]
- 36.Paunescu TG, Russo LM, Da Silva N, et al. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am J Physiol Renal Physiol 2007; 293: F1915–1926. [DOI] [PubMed] [Google Scholar]
- 37.Milton AE, Weiner ID. Intracellular pH regulation in the rabbit cortical collecting duct A-type intercalated cell. Am J Physiol 1997; 273: F340–347. [DOI] [PubMed] [Google Scholar]
- 38.Paunescu TG, Da Silva N, Marshansky V, et al. Expression of the 56-kDa B2 subunit isoform of the vacuolar H+-ATPase in proton-secreting cells of the kidney and epididymis. Am J Physiol Cell Physiol 2004; 287: C149–162. [DOI] [PubMed] [Google Scholar]
- 39.Alzamora R, Thali RF, Gong F, et al. PKA regulates vacuolar H+-ATPase localization and activity via direct phosphorylation of the a subunit in kidney cells. J Biol Chem 2010; 285: 24676–24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaulieu V, Da Silva N, Pastor-Soler N, et al. Modulation of the actin cytoskeleton via gelsolin regulates vacuolar H+-ATPase recycling. J Biol Chem 2005; 280: 8452–8463. [DOI] [PubMed] [Google Scholar]
- 41.Gong F, Alzamora R, Smolak C, et al. Vacuolar H+-ATPase apical accumulation in kidney intercalated cells is regulated by PKA and AMP-activated protein kinase. Am J Physiol Renal Physiol 2010; 298: F1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastor-Soler N, Beaulieu V, Litvin TN, et al. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem 2003; 278: 49523–49529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander EA, Brown D, Shih T, et al. Effect of acidification on the location of H+-ATPase in cultured inner medullary collecting duct cells. Am J Physiol Cell Physiol 1999; 276: C758–763. [DOI] [PubMed] [Google Scholar]
- 44.Belleannee C, Da Silva N, Shum WW, et al. Role of purinergic signaling pathways in V-ATPase recruitment to apical membrane of acidifying epididymal clear cells. Am J Physiol Cell Physiol 2010; 298: C817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breton S, Brown D. Cold-induced microtubule disruption and relocalization of membrane proteins in kidney epithelial cells. J Am Soc Nephrol 1998; 9: 155–166. [DOI] [PubMed] [Google Scholar]
- 46.Breton S, Nsumu NN, Galli T, et al. Tetanus toxin-mediated cleavage of cellubrevin inhibits proton secretion in the male reproductive tract. Am J Physiol Renal Physiol 2000; 278: F717–725. [DOI] [PubMed] [Google Scholar]
- 47.Brown D, Paunescu TG, Breton S, et al. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol 2009; 212: 1762–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merkulova M, Paunescu TG, Azroyan A, et al. Mapping the H+(V)-ATPase interactome: identification of proteins involved in trafficking, folding, assembly and phosphorylation. Sci Rep 2015; 5: 14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Păunescu TG, Ljubojevic M,, Russo LM, Winter C, McLaughlin MM, Wagner CA,, Breton S, Brown D, cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells. Am J Physiol Renal Physiol 2010; 298: F643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner CA, Friedrich B, Setiawan I, et al. The use of Xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell Physiol Biochem 2000; 10: 1–12. [DOI] [PubMed] [Google Scholar]
- 51.Winter C, Kampik NB, Vedovelli L, et al. Aldosterone stimulates vacuolar H+-ATPase activity in renal acid-secretory intercalated cells mainly via a protein kinase C-dependent pathway. Am J Physiol Cell Physiol 2011; 301: C1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank AE, Wingo CS, Andrews PM, Ageloff S, Knepper MA, Weiner ID Mechanisms through which ammonia regulates cortical collecting duct net proton secretion. Am J Physiol Renal Physiol 2002; 282: F1120–1128. [DOI] [PubMed] [Google Scholar]
- 53.Chambrey R, Goossens D, Bourgeois S, et al. Genetic ablation of Rhbg in the mouse does not impair renal ammonium excretion. Am J Physiol Renal Physiol 2005; 289: F1281–1290. [DOI] [PubMed] [Google Scholar]
- 54.Sohet F, Colin Y, Genetet S, Ripoche P, Métral S, Le Van Kim C, Lopez C Phosphorylation and ankyrin-G binding of the C-terminal domain regulate targeting and function of the ammonium transporter RhBG. J Biol Chem 2008; 26: 26557–26567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Genetet S, Ripoche P, Le Van Kim C, et al. Evidence of a structural and functional ammonium transporter RhBG.anion exchanger 1.ankyrin-G complex in kidney epithelial cells. J Biol Chem 2015; 290: 6925–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vitavska O, Wieczorek H, Merzendorfer H, A novel role for subunit C in mediating binding of the H -V-ATPase to the actin cytoskeleton. J Biol Chem 2003; 278: 18499–18505. [DOI] [PubMed] [Google Scholar]
- 57.Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, Gluck SL The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem 2000; 275: 32331–32337. [DOI] [PubMed] [Google Scholar]
- 58.Seaton B, Ali A. Simplified manual high performance clinical chemistry methods for developing countries. Med Lab Sci 1984; 41: 327–336. [PubMed] [Google Scholar]
- 59.Berthelot M Violet d’aniline. Rep Chim App 1859; 1: 284. [Google Scholar]
- 60.Fiske CH, SubbaRow Y. The colorometric determination of phosphorus. J Biol Chem 1925; 66: 375–400. [Google Scholar]
- 61.McLean IW, Nakane PK Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem 1974; 22: 1077–1083. [DOI] [PubMed] [Google Scholar]
- 62.Wagner CA, Finberg KE, Stehberger PA, et al. Regulation of the expression of the Cl−/anion exchanger pendrin in mouse kidney by acid-base status. Kidney Int 2002; 62: 2109–2117. [DOI] [PubMed] [Google Scholar]
- 63.Stehberger PA, Shmukler BE, Stuart-Tilley AK, et al. Distal renal tubular acidosis in mice lacking the AE1 (Band3) Cl−/HCO −3 exchanger (Slc4a1). J Am Soc Nephrol 2007; 18: 1408–1418. [DOI] [PubMed] [Google Scholar]
- 64.Mohebbi N, Kovacikova J, Nowik M, et al. Thyroid hormone deficiency alters expression of acid-base transporters in rat kidney. Am J Physiol Renal Physiol 2007; 293: F416–427. [DOI] [PubMed] [Google Scholar]
- 65.Eladari D, Cheval L, Quentin F, Bertrand O, Mouro I, Cherif-Zahar B, Cartron JP, Paillard M, Doucet A, Chambrey R Expression of RhCG, a New Putative NH3/NH +4 Transporter, along the Rat Nephron. J Am Soc Nephrol 2002; 13: 1999–2008. [DOI] [PubMed] [Google Scholar]
- 66.Peng SB, Li X, Crider BP, Zhou Z, Andersen P, Tsai SJ, Xie XS, Stone DK. Identification and reconstitution of an isoform of the 116-kDa subunit of the vacuolar proton translocating ATPase. J Biol Chem 1999; 274: 2549–2555. [DOI] [PubMed] [Google Scholar]
- 67.Bastani B, Purcell H, Hemken P, et al. Expression and distribution of renal vacuolar proton-translocating adenosine triphosphatase in response to chronic acid and alkali loads in the rat. J Clin Invest 1991; 88: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohebbi N, Benabbas C, Vidal S, et al. The proton-activated G protein coupled receptor OGR1 acutely regulates the activity of epithelial proton transport proteins. Cell Physiol Biochem 2012; 29: 313–324. [DOI] [PubMed] [Google Scholar]
- 69.Thomas JA, Buchsbaum RN, Zimniak A, Racker E. Intracellular pH measurements in Ehrlich ascites tumor cells utilizing spectroscopic probes generated in situ. Biochemistry 1979; 18: 2210–2218. [DOI] [PubMed] [Google Scholar]
- 70.Watts BA 3rd, Good DW. Apical membrane Na+/H+ exchange in rat medullary thick ascending limb. pH-dependence and inhibition by hyperosmolality. J Biol Chem 1994; 269: 20250–20255. [PubMed] [Google Scholar]
- 71.Milton AE, Weiner ID. Regulation of B-type intercalated cell apical anion exchange activity by CO2/HCO3. Am J Physiol 1998; 274: 1086–1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.