Abstract
Neural stem cell (NSC) cultures have been considered technically challenging for time-lapse analysis due to high motility, photosensitivity, and growth at confluent densities. We have tested feasibility of long-term live-cell time-lapse analysis for NSC migration and differentiation studies. Here, we describe a method to study the dynamics of cell cycle, migration, and lineage selection in cultured multipotent mouse or human NSCs using single-cell tracking during a long-term, 7–14 day live-cell time-lapse analysis. We used in-house made PDMS inserts with five microwells on a glass coverslip petri-dish to constrain NSC into the area of acquisition during long-term live-cell imaging. In parallel, we have defined image acquisition settings for single-cell tracking of cell cycle dynamics using Fucci-reporter mouse NSC for 7 days as well as lineage selection and migration using human NSC for 14 days. Overall, we show that adjustments of live-cell analysis settings can extend the time period of single-cell tracking in mouse or human NSC from 24–72 h up to 7–14 days and potentially longer. However, we emphasize that experimental use of repeated fluorescence imaging will require careful consideration of controls during acquisition and analysis.
Keywords: Human neural stem cell, Live-cell imaging, Single-cell tracking, Fate mapping, Cell lineage decision, Cell cycle
1. Introduction
The application of human neural stem cells (hNSC) as cell therapeutics targeted towards central nervous system injuries or degenerative diseases is being heavily investigated. Cell line to cell line variability in human cell manufacturing has created a demand for detailed preliminary in vitro screening methods of newly developed cell lines. Live-cell analysis can provide multi-modal information about cellular function at the single-cell level. NSC proliferation and migration has been extensively studied using time-lapse imaging in rodent tissue slice cultures [11,19,22,10,26]. Several groups have also utilized live-cell time-lapse imaging followed by single-cell tracking for studying in vitro cultured NSC. The majority of these have been performed in rodent primary NSC cultures [17,18,20,2,13,25,12] spanning 4–14 days of time-lapse image acquisition. A subset of these studies have investigated cell cycle by single-cell tracking of Fucci-cell cycle reporter expressing mouse NSC [22,21,3]; however, analyses of Fucci-mouse NSC (Fucci-mNSC) in vitro have focused on short term experiments (24–72 h). In parallel, few studies have investigated single-cell tracking in time-lapse imaging of human NSC (hNSC) [1,7], and these have again been constrained to short term experiments (24–48 h). Here, we describe a method for long-term (7–14 day) live-cell time-lapse imaging and single-cell tracking of cell cycle, migration, and lineage selection in in vitro NSC, using a combination of multipotent mouse Fucci-expressing and human NSC to illustrate these assessments.
2. Material and methods
2.1. Isolation and culture of mouse and human neural stem cells
Multipotent mouse NSC were isolated from the cortices of fluorescent ubiquitination-based cell-cycle indicator expressing (Fucci) mice [22,23] embryos at gestational age E11. Fucci-mNSC were cultured as neurospheres in serum free DMEM (Gibco) basal medium supplemented with bFGF (20 ng/ml, Invitrogen) and EGF (10 ng/ml, Invitrogen) for two passages. After this period, cells were transitioned on poly-L-ornithine (5 μg/mL, Sigma) and laminin (10 μg/mL, Invitrogen) coated flasks in serum free X-VIVO 15 (BioSciences) based growth medium supplemented with 20 ng/ml bFGF, 2 ng/ml EGF and LIF (10 ng/ml, Chemicon).
Multipotent hNSC were isolated from human fetal brain at 16–20 weeks gestational age and cultured as neurospheres in supplemented X-VIVO 15 medium as previously described [24]. After passage number five, the cells were transitioned to a monolayer culture on poly-L-ornithine and laminin coated flasks as described above.
2.2. PDMS inserts with microwells
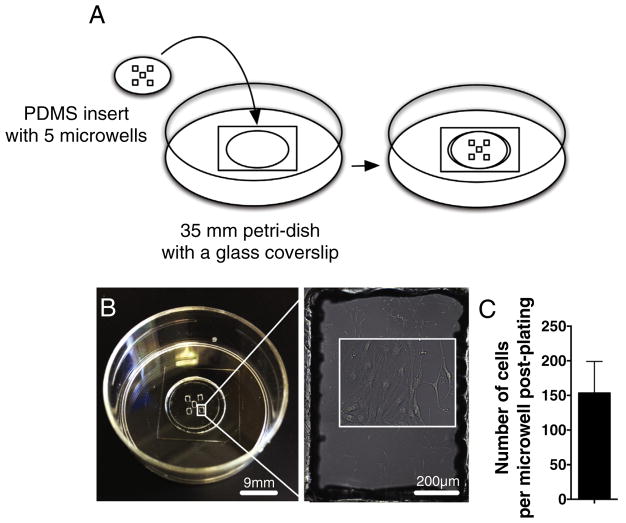
PDMS insert templates with five 1.2 mm × 0.8 mm microwells (Fig. 1A,B) were designed using SolidWorks and AutoCAD (Autodesk, Inc.) design software. They were dimensioned to match a 3 × 3 tiled field of view at 20× magnification on a VivaView FL Incubator Microscope (Olympus America, Inc). For polydimethylsiloxane (PDMS) sheet manufacture, silicon elastomeric base was mixed with curing agent (Sylgard® 184, Dow Corning) in a ratio of 10:1, degassed in a vacuum chamber for 30 min, and added on 60 mm polystyrene petri-dish (4–5 ml/dish), which was rotated in slow motion for even PDMS surface distribution before baking at +65 °C overnight. The following day, PDMS sheets with approximate thickness of 500 μm were covered with a perforated sticky wrap sheet (Plasdent), and the inserts were cut by a VeraLASER 2.3 (Universal Laser Systems, Inc.) using 1 mm as 1 unit for the insert template, 30% laser power intensity, 10% speed and 1000% pulse per inch settings without z-axis cutting. The perforated wrap was removed from the inserts prior to sonication in 70% EtOH at +35 °C, 60 sonics/min, 5 degas pulses/s using Ultrasonic Cleaner FS30 (Fisher Scientific). Sonicated PDMS inserts were sterilized under UV-light for 30 min and stored in 70% EtOH at room temperature. Prior to use, 35 mm petridishes with a 14 mm glass coverslip 1.5 with thickness of 0.16–0.19 mm (MatTek Corporation) were coated with 5 μg/ml poly-L-ornithine (Sigma) at +37 °C overnight, washed with sterile H2O and air-dried the following day. Using sterile tweezers, each PDMS insert was rinsed in sterile H2O, and when still slightly wet adhered onto the poly-L-ornithine coated glass coverslip petridish and air-dried. Next, the microwells of each PDMS insert were coated with 10 μg/ml laminin (Invitrogen) at +37 °C overnight, and air-dried the following day. For live cell imaging, NSC cultures were detached using trypsin-EDTA (Cell-Applications) and plated into the microwells at a density of 100 cells/well (0.5 μl/microwell) of X-vivo based growth medium using extra-long low retention pipette tips (VWR International). 10–15 min post-plating, petri-dishes containing microwell inserts were carefully filled with 2 ml of growth medium, and NSC were cultured for two days at +37 °C and 5% CO2. Prior to time-lapse imaging, NSC were washed, the medium was changed to X-VIVO 15 based differentiation medium supplemented with 10 ng/ml GDNF (PeproTech), 10 ng/ml BDNF (PeproTech) and 1 ng/ml bFGF, and petri-dishes with the inserts were loaded into the VivaView FL Incubator Microscope. The total volume of differentiation medium added into each petri-dish for NSC 7 day experiments was 3 ml. 14 day experiments were plated into 2 ml of differentiation medium and fed by adding 500 μl of prewarmed differentiation medium every fifth day throughout the experiment. A diagram of the PDMS insert manufacturing and use is shown as Supplemental Fig. 1.
Fig. 1.
Microwell application constrains NSC into the area of acquisition and allows simultaneous imaging of larger sample numbers. A) A PDMS insert with five 1.2 mm × 0.8 mm microwells can be attached on a glass coverslip of 35 mm petri-dish to constrain the cells and minimize the area of image acquisition. Inset shows NSC inside the microwell in higher magnification. B) An example of a PDMS microwell plated with NSC. C) An average NSC density on microwells at first day of live-cell imaging suggest that the cell attachment is replicable at plating density of 100 cells per well. n = 4 independent biological experiments, mean ± error bars SEM.
2.3. Live-cell imaging
Time-lapse images were captured using VivaView FL Incubator Microscope LCV110 version 7.7.5 (Olympus America, Inc) at +37 °C and 5% CO2. Images of Fucci-mNSC were captured with 20× objective as 3 × 3 image montages in 20 min intervals for 7 days. To assess feasibility of the long-term fluorescence live-cell imaging in Fucci-mNSC, we tested several image acquisition parameters for laser intensity (25–100%) and exposure (100–800 ms) (Supplemental Table 1). The laser intensities/exposure times used in this study were as follows: 25%/600 ms (GFP), 25%/100 ms (HC red), and 25 ms differential interference contrast (DIC). Time-lapse of Fucci-mNSC DIC only control was acquired as described above using only 25 ms DIC. DIC focus in all experiments was checked daily and adjusted if needed.
In parallel, time-lapse images of hNSC were captured with 20× objective 8 ms DIC as 3 × 3 image montages in 30 min intervals for 14 days during in vitro differentiation. The image capture was paused between the image acquisition cycles for every fifth day and the cell cultures were fed using a pipette from the small window on the inner door of the incubator without opening the door and removing the dish from the incubator. The loading tray of VivaView FL Incubator Microscope has motorized special petridish lids for software controlled open-close function. VivaView FL model LCV110 exhibited no drift in XY coordinates or the areas of image acquisition were observed. DIC focus was checked daily and adjusted if needed.
2.4. Immunocytochemistry post-live-cell imaging
After the last image acquisition timepoint at 14 days in vitro differentiation, hNSC in the microwell were fixed with 4% paraformaldehyde. The inserts were carefully removed, and the cells were permeabilized and immunostained with primary antibodies raised against Map2 (1:50, Sigma), GFAP (1:1000, Dako) or Olig2 (1:100, R&D) followed by secondary antibodies conjugated with 555, 488, or 647 fluorochromes (ThermoFisher Scientific). Hoechst (Sigma) was used as a nuclear counter stain. 3 × 3 multichannel image montages of immunostained cells were captured using an Olympus IX inverted microscope with camera and 20× objective to create a composite image of the post-immunostaining and the last image acquisition timepoint, 14 days at in vitro differentiation.
2.5. Image montages and movie compiling
File names of the acquired time-lapse images were changed to be specified for the x- and y-directions, the channels, and the timepoints using NameChanger (MRR software), and compiled channel by channel into a movie using Imaris image processing and analysis software version 7.5.2 (Bitplane, Inc).
For lineage analysis of hNSC, the composite image of cells post-immunostaining representing the final 14 day timepoint and the preceding timepoints were compiled into Imaris (Bitplane, Inc.) in reverse chronological order.
2.6. Total number of cells and proportional analysis of cell cycle phase
The total number of Fucci-mNSC per timepoint and the relative proportions of the cells in G1 or S, G2 or M phases per timepoint were quantified from the image montages at 0-, 48-, 72-, 96 h, and 7 days during in vitro culture. G1 or S, G2 or M phase were characterized based on the Fucci-nuclear fluorescence reporter expression.
The data is shown as an average of four or three independent biological experimental replicate, respectively. The data of each independent biological experimental replicate was collected from a single microwell on separate petri-dishes, that were either imaged separately (fluorescence time-lapse) or simultaneously (DIC-only time-lapse).
2.7. Single-cell tracking of cell survival, number cell divisions, length of cell cycle and migration
Based on the literature, the number of in vitro cultured NSC analyzed per time-lapse experiment followed by single-cell tracking ranges from approximately 20 to 200 cells. Thus, we chose a total number of 30 randomly selected Fucci-mNSC, referred to as parent NSC (all in G1 within the first frame of the movie corresponding to 0 h timepoint) per each independent biological experimental replicate to be tracked frame-by-frame using the manual tracking feature of Imaris software version 7.5.2 (Bitplane, Inc) for up to 7 days; tracking experiments included the progeny of all selected parent cells. In each frame, a visually determined and manually selected physical center of the cell was used as a reference point for cell tracking. After cell division both daughter cells were tracked until the end of the experiment unless the cell died or was lost to identification.
Cell survival of the tracked Fucci-mNSC is shown as an average percentage of live cells relative total number of tracked cells of four independent biological experimental replicate. The number of dividing parent Fucci-mNSC is shown as a percentage normalized to the total number of parent mNSC of four independent biological experimental replicates. The total number of cell divisions within the parent NSC and their progeny is shown as an average of cell divisions per parent NSC in four independent biological experimental replicate. The time mNSC spent in the G1 and S, G2, or M phases per cell division is shown as an average of three independent biological experimental replicate. Manual tracking of single cells is fundamentally dependent on the accuracy of individuals to reproducibly identify live cells in a DIC image with and without a Fucci fluorescence reporter. We validated the accuracy of our analysis using a standardized image set with independent blinded observers to identify live versus dead cells in DIC/Fucci settings. Accuracy between the individuals analyzing DIC visualized cells in high cell density was less than 95% and accuracy between the DIC analysis and Hoechst based verification of the cell numbers was 98%.
Imaris software calculates the instantaneous speed of an object for each timepoint by taking into account the position of the object at a specific timepoint and the amount of time spent in that position. The average speed of the object is calculated by dividing the track length by the time between first and last spot in the track. Migration track speed of Fucci-mNSC between the timepoint of interest is shown as an average of four independent biological experimental replicates. Track length, track displacement and track straightness of Fucci-mNSC are shown as an average of four independent biological experimental replicate.
2.8. Single-cell analysis of hNSC lineage selection, number of cell divisions, and migration statistics
Three to seven human cells of each neuronal lineage were randomly selected based on the fate marker expression mapped using the composite image post-immunostaining and the final time-lapse image at 14 day. These cells were back-tracked using Imaris software manual tracking and the time-lapse movies compiled in reverse chronological order. If the cells had divided, the sister cells were also tracked and characterized using the post-immunostaining composite image. The total number of cell divisions during in vitro differentiation was compared between the three neuronal lineages. Migration track length, straightness, and speed were compared between the parent hNSC and the progeny of each lineage. Data is shown as an average of 3–4 individual cells per lineage.
3. Results
3.1. Microwells allow long term live-cell imaging analysis in a small subset of NSC
To assess feasibility of long-term live-cell imaging of mouse and human NSC, we tested several incubator/imaging systems. According to our evaluation both Olympus VivaView FL Incubator Microscope and Olympus FluoView FV10i gave the most consistent long-term survival results, where as the stage top heaters/incubators were not suitable for tracking cells past 72 h. NSC are highly motile during in vitro culture. To prevent NSC migration out of the area of acquisition during long-term live-cell imaging, we designed a PDMS insert with five 1.2 mm × 0.8 mm microwells that can be attached into a 35 mm petri-dish with a 14 mm glass coverslip to fence the cells (Fig. 1A,B). We matched the microwell size so that the area within a single microwell could be captured as a 3 × 3 image montage using an Olympus VivaView FL Incubator Microscope with a 20× objective. Ultra-small well size allowed us to focus analysis of NSC population dynamics in a small subset of cells that were constrained to the 3 × 3 “field of view”. Plating density for both human and mouse NSC was established at 100 cells per microwell to enable sufficient cells for analysis while maintaining a final density permissive for cell tracking experiments. Two days post-plating in growth conditions, the average number of attached NSC was 153 ± 46 cells per microwell (Fig. 1C) indicating replicable cell attachment between four independent biological experimental replicates. In parallel, no cells were found to migrate out of the microwells in any of the time-lapse experiments ranging from 7 to 14 days, so long as the original microwell adherence was good. Overall, these data suggest that the microwells are a practical approach for constraining NSC for live-cell imaging experiments.
3.2. Live-cell imaging of NSC dynamics during in vitro culture
3.2.1. Feasibility of single-cell tracking
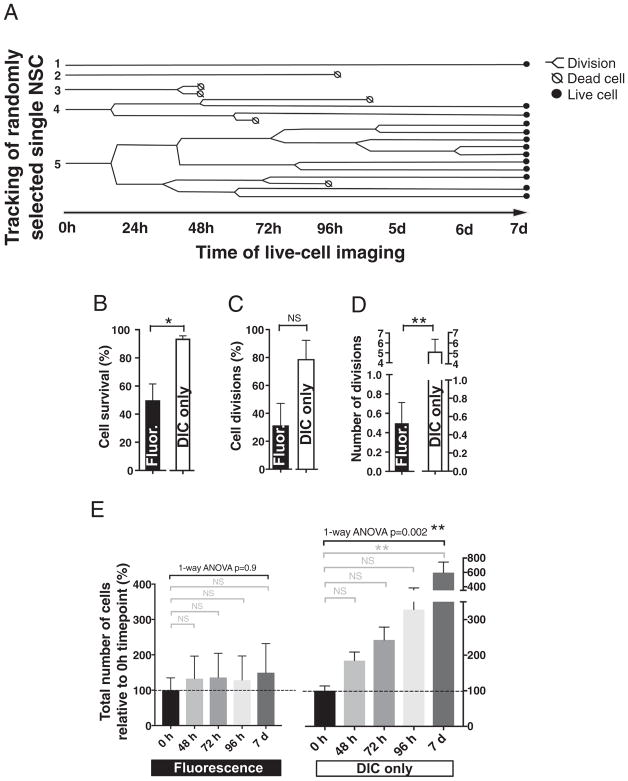
We isolated NSC from cortex of Fucci mice [22,23] at embryonic stage 11. At passage 5, the cells were plated in PDMS microwells and two days post-plating exposed to in vitro differentiation conditions followed by time-lapse image acquisition using Olympus VivaView FL Incubator Microscope for GFP, RFP and DIC with 20× objective for 7 days. The acquired images were compiled into a time-lapse movie and 30 cells per independent biological experiments were randomly selected within the first frame of the movie corresponding to 0 h timepoint. Each cell was tracked frame-by-frame using the manual tracking feature of Imaris software. Based on the single-cell tracking data we generated a pedigree map showing cell divisions or death within the selected Fucci-mNSC population over the course of the fluorescence time-lapse experiment (Fig. 2A).
Fig. 2.
Feasibility of single-cell tracking and the effect of repeated fluorescence excitation on survival and proliferation of Fucci-mNSC in vitro. A) An example pedigree map showing cell divisions or death within the fluorescence imaged, randomly selected Fucci-mNSC population over the course of 7 day experiment. During repeated fluorescence image acquisition, the single-cell tracked Fucci-mNSC exhibit B) decreased cell survival, but C) no significant change in the proportion of parent NSC that divide during the experiment was detected. However, D) the average number of cell divisions per parent Fucci-mNSC was decreased when compared to the control cells imaged with DIC only during a 7 day in vitro experiment. Student’s 2-tailed t-test, *p ≤ .01, **p ≤ .008, n = 4 independent biological experimental replicates, mean ± error bars SEM. D) The number of Fucci-mNSC exposed to repeated fluorescence excitation remained unchanged during a 7 day experiment relative to 0 h timepoint, while the DIC-only control group exhibited increasing number of cells towards the end of the experiment. Black brackets 1-way ANOVA **p ≤ .002, grey brackets Tukey’s t-test **p ≤ .002, n = 4 independent biological experimental replicates, mean ± error bars SEM.
3.2.2. Effect of repeated fluorescence excitation on cell divisions and phases of cell cycle
Tracking of single cell in spatio-temporal motion is challenging and affected by several variables, such as object detection, illumination, background subtraction and unpredictable motion. In addition, fluorescence excitation causes phototoxicity and increased cell death during live-cell imaging [14], which can further restrict the length of time-lapse analysis and the use of current tools for analyzing cell cycle. Although increasing length of the image acquisition interval can decrease phototoxicity, we selected 20min interval to enable accurate tracking of cell position from timepoint to time-point, and detection of transitions between cell cycle phases. To find image acquisition parameters that balanced cell death and fluorescence signal detection we tested several laser intensities and exposure times (Supplemental Table 1). The final settings used in this study for Fucci-mNSC were selected based on qualitative analysis and are described under the Material and Methods. Consistent with the literature, the effects of repeated fluorescence excitation were observed in all conditions evaluated. With the settings used here, the DIC only imaged Fucci-mNSC control group exhibited an average survival rate of 91% ± 2% within the tracked cells during a 7 day experiment, while the fluorescence time-lapse caused a 0.5-fold decrease in the survival of the tracked Fucci-mNSC (Fig. 2B). The proportion of the tracked parent mNSC that divided during the experiment was not significantly changed (Fig. 2C), however the average number of cell divisions per parent mNSC was reduced by 10-fold in comparison to a DIC only imaged control group during a 7 day experiment (Fig. 2D). In parallel, the total number of Fucci-mNSC per microwell remained unchanged during the 7 day fluorescence time-lapse experiment, where as DIC-only imaged control group exhibited a increasing number of cells towards the end of the experiment (Fig. 2E). These data demonstrate that it is possible to establish fluorescence time-lapse image acquisition parameters that enable imaging of Fucci-mNSC through at least 7 days in vitro, however, repeated fluorescence image acquisition affects NSC survival and proliferation. Awareness of these caveats and use of appropriate controls will be important considerations in any experimental design.
3.2.3. Evaluation of cell cycle phase and dynamics
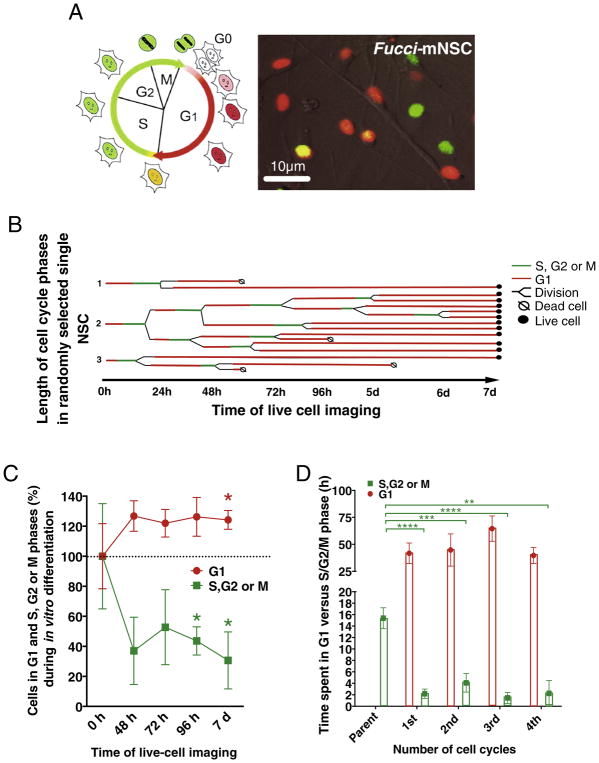
Neural progenitors exit the cell cycle in a developmentally regulated manner to allow for terminal differentiation [9,5]. We and others have shown that in vitro differentiation conditions trigger a decrease in the proportion of BrdU labeled NSC within 2–3 days of monolayer culture [16,8]. However, BrdU labeling and other most common markers for analyzing cell proliferation are not sensitive enough measure cell cycle phase. We evaluated the feasibility of following NSC cell cycle dynamics during a 7 day in vitro differentiation period using fluorescence live-cell imaging and Fucci-mNSC. As previously published, the Fucci-reporter system exploits cell-cycle-dependent proteolysis of ubiquitination oscillators, Cdt1 and Geminin fused with red and green-emitting fluorescent proteins [22]. Nuclei of the Fucci-mNSC in G1 phase appear red, during G1 to S transition the cell nuclei turn yellow, and in S, G2 orMphase nuclei appear green (Fig. 3A–B). Immediately after cytokinesis and after terminal differentiation at G0, nuclei of the Fucci-mNSC appear non-fluorescent (Fig. 3A). The frame-by-frame single-cell tracking of Fucci-mNSC allowed us to generate a pedigree map to visualize the length of G1 and S, G2 orMphases within the selected NSC population over the course of 7 day fluorescence time-lapse in in vitro differentiation conditions (Fig. 3B). Quantification of the total number of Fucci-mNSC in G1 and S, G2 or M phase at 0-, 48-, 72-, 96 h, and 7 day timepoints revealed that the proportion of cells in G1 phase increased while S, G2 or M phase decreased between 72 h and 7 day timepoints relative to the 0 h timepoint (Fig. 3C). In parallel, statistical analysis of the single-cell tracking data revealed that relative to parent cells the progeny of Fucci-mNSC spent less time in S, G2 or M phase (Fig. 3D). These data suggest that the long-term fluorescence live-cell imaging of Fucci-mNSC with continuous time-lapse up to 7 days can be used for studying cell cycle dynamics in cultured NSC. However, as noted above, experiments employing live-cell time-lapse imaging with repeated fluorescence excitation will require additional controls, and the effect of repeated fluorescence excitation should be carefully considered.
Fig. 3.
Fucci-reporter as a live-cell imaging tool for assessing mNSC cell cycle dynamics during in vitro differentiation. A) Fucci-expressing mNSC exhibit red nuclei when in G1 phase, during G1 to S transition cell nuclei turn yellow, and in S, G2 or M phases nuclei appear green. B) An example pedigree map to visualize length of G1 and S, G2 or M phase within the randomly selected Fucci-mNSC population over the course of 7 day fluorescence time-lapse in in vitro differentiation conditions. C) The proportion of Fucci-mNSC in G1 phase increased while S, G2 or M phase decreased between 72 h and 7 day timepoints relative to the 0 h timepoint, 1-sample t-test, *p ≤ .05, n = 3 independent biological experimental replicates, mean ± error bars SEM. In parallel, D) the progeny of Fucci-mNSC spent less time in S, G2 or M phase relative to parent cells. Student’s 2-tailed t-test *p ≤ .05, **p ≤ .01, ***p ≤ .001, ****p ≤ .0001, n = 3 independent biological experimental replicates, mean ± error bars SEM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2.4. Repeated fluorescence excitation and cell migration
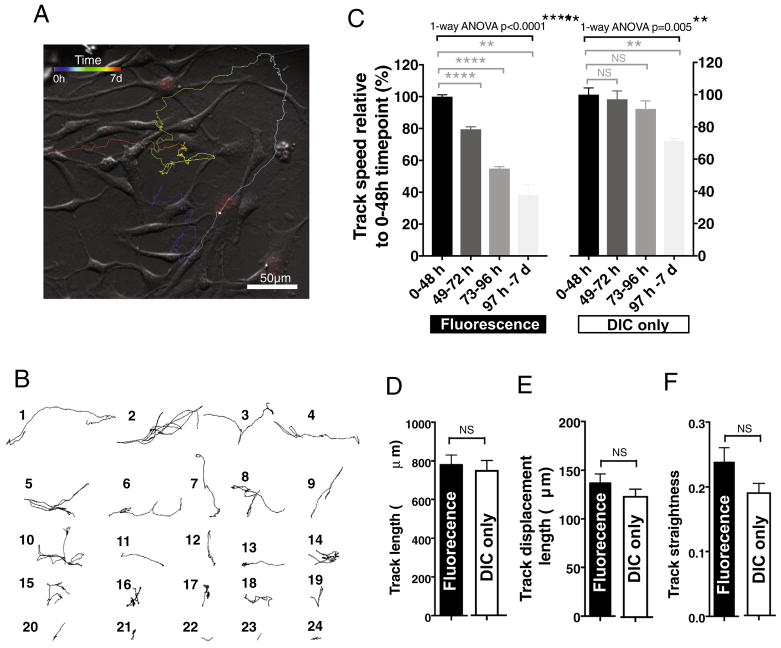
Imaris software generates several cell motility measures, such as track speed, track length, straightness, and displacement. Cell movement can be also visualized using a dragon tail display showing individual migration tracks of each cell over the course of the experiment (Fig. 4A) or as single-cell tracks over the course of experiment (Fig. 4A). We assessed the feasibility of tracking cell migration and the effect of repeated fluorescence excitation on Fucci-mNSC motility measures. mNSC in both fluorescence and DIC time-lapse groups exhibited the highest speed of migration between 0 and 48 h during in vitro differentiation (Fig. 4B), with an average of 0.005 μm/s ± 0.00006 μm/s or 0.006 μm/s ± 0.0003 μm/s, respectively. Speed in both groups decreased significantly over the course of the experiment (Fig. 4B). No difference was detected in distance the cells migrated over the course of the 7 day experiment between the fluorescence and DIC time-lapse groups (Fig. 4C), with an average of 780 μm± 50 μm or 753 μm ± 49 μm, respectively. In parallel, no significant changes were found in track displacement (the distance between the start and end point of the analyzed cell Fig. 4E) or track straightness (track displacement divided by track length; Fig. 4F) between the groups. Track displacement and straightness are both measures that can be used for comparing directionality of the cell movement when the experimental set-up consists of multiple cell lines or study groups. These data suggest that tracking Fucci-mNSC migration in the microwells is feasible, however, while the fluorescence time-lapse imaging settings used in this study did not alter track length, displacement or straightness over time, there may be a small impact on speed of migration.
Fig. 4.
Live-cell fluorescence time-lapse and single-cell tracking of Fucci-mNSC migration and motility in vitro. A) An example of a dragon tail display showing a single Fucci-mNSC migration track in which temporal changes in cell location are indicated as color temperature scale. B) Example migration tracks of 24 individual Fucci-mNSC during a 7 day of live-cell fluorescence time-lapse in in vitro differentiation conditions. C) Fucci-mNSC in both fluorescence and DIC time-lapse groups exhibited highest speed of migration between 0 and 48 h and the cells in both groups exhibited decreased speed over the course of 7 day in vitro experiment. Black brackets 1-ANOVA ****p ≤ .0001, **p ≤ .005, grey brackets Tukey’s t-test ***p ≤ .0002, **p ≤ .004. In parallel, no difference was detected in D) distance the cells migrated, E) track displacement or F) track straightness over the course of 7 day between the groups. Student’s 2-tailed t-test p ≥ .05, n = 4 independent biological experimental replicates, mean ± error bars SEM.
3.2.5. Dynamics of lineage selection and migration
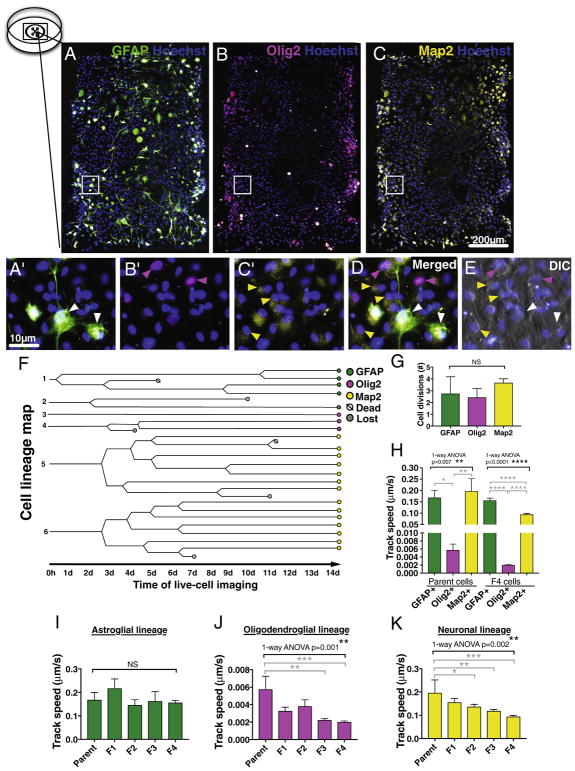
Last, we tested the feasibility of long-term live-cell imaging analysis to assess human NSC differentiation capacity and lineage preference in vitro. Multipotent hNSC were isolated from fetal brain at 16–20 weeks gestational age and cultured as previously described [24]. At passage 5–8, hNSC were plated into microwells, and two days post-plating exposed to in vitro differentiation conditions followed by time-lapse image acquisition as described under Materials and Methods. In comparison to rodent cells, hNSC require a longer time for in vitro differentiation. Our previous data suggest that a small proportion of hNSC remain uncommitted even after a 14 day in vitro differentiation period [15], however, evaluation of GFAP+ cells in combination of stem cell markers Sox2 and Nestin indicates that by this timepoint GFAP expression correlates with astroglial lineage commitment rather than maintenance of neural progenitors [6]. After the last image acquisition at the 14 day timepoint, cells were fixed and triple-immunostained for lineage characterization using astroglial marker GFAP, oligodendroglial maker nuclear Olig2, or neuronal marker Map2 (Fig. 5A–D). Using the composite image of the post-immunostaining and final DIC time-lapse timepoint, we randomly selected three to seven cells in each lineage exhibiting either GFAP, nuclear Olig2, or Map2 expression to be back-tracked from the time-lapse movies compiled in reverse chronological order. Alignment of the layers for the composite image was good, as shown in the composite image of the Hoechst and final DIC time-lapse timepoint in the triple-immunostained microwell (Fig. 5E). Based on the single-cell back-tracking data, we generated a hNSC fate map showing cell divisions, death, and lineage of each analyzed cell and their parent cells during the two week in vitro differentiation period (Fig. 5F). No significant difference was detected between lineages in the total number of cell divisions during the 14 day differentiation (Fig. 5G). In parallel, no significant changes were detected in migration track length or straightness between parent cells and the progeny of each lineage (One-way ANOVA, p ≤ .05). However, parent hNSC of the oligodendroglial nuclear Olig2+ lineage cells exhibited significantly decreased migration track speed when compared to those in the astroglial GFAP+ or neuronal Map2+ lineages (Fig. 5H). When migration track speed was compared between the parent NSC and the progeny in each lineage, no changes were detected in the astroglial GFAP+ lineage (Fig. 5I) and only the cells committed to oligodendroglial Olig2+ lineage (Fig. 5J) or neuronal Map2+ lineage (Fig. 5K) exhibited significantly reduced migration speed suggesting possible cell lineage-specific changes during in vitro differentiation. Overall, these data suggest that differentiation capacity, lineage preference and migration of hNSC can be tracked for assessment live-cell imaging analysis, at least up to 14 days in vitro.
Fig. 5.
Single cell tracking can be used for mapping hNSC differentiation capacity and lineage preference in vitro. An example of hNSC immunostained against lineage specific markers A) GFAP, B) Olig2, and C) Map2 combined with nuclear stain Hoechst in a microwell after 14 day DIC time-lapse in in vitro differentiation conditions. Pictures are pseudo-colored, insets shown as higher magnification in A′–C′, and as merged layers in D. E) Composite image of the Hoechst and final DIC time-lapse timepoint in the triple-immunostained microwell shows good alignment of the layers. Arrow heads point to cells positive for astroglial GFAP+ cells (green), oligodendroglial lineage cells expressing nuclear Olig2 (pink) and neuronal Map2+ cells (yellow). F) An example of hNSC cell lineage map showing cell divisions and fate of each analyzed cell during 14 day DIC time-lapse in in vitro differentiation conditions. G) No significant difference was found in total number of cell divisions between each neuronal lineage during 14 day DIC time-lapse. 1-way ANOVA p = .36, n = 3–7 individual cells/lineage. H) In oligodendroglial nuclear Olig2+ lineage, both parent cells and the progeny exhibited significantly decreased migration speed when compared to those in astroglial GFAP+ or neuronal Map2+ lineage cells. Black bracket 1-way ANOVA **p = .007 or ****p ≤ .0001, grey brackets Tukey’s post hoc *p = .02, **p = .009, or ****p ≤ .0001. In parallel, when migration track speed was compared between the parent hNSC and the progeny in each lineage, I) no changes was detected in astroglial GFAP+ lineage cells, however the cells in both J) oligodendroglial Olig2+ lineage and K) neuronal Map2+ lineage cells were decelerating during 14 day DIC time-lapse in in vitro differentiation conditions. Black brackets 1-way ANOVA **p ≤ .002, grey brackets Dunnett’s post hoc test *p = .04, **p ≤ .005, ***p ≤ .0006, n = 3–4 individual cells/lineage, Mean ± error bars SEM. F indicates the progeny after each cell division. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion and conclusions
Long-term live-cell analysis of NSC cultures has been considered technically challenging due to high migratory capacity and photosensitivity. Additionally, high density NSC culture makes single-cell tracking difficult. Here, we describe the technical feasibility of a long-term (7–14 day) live-cell analysis model in which cell cycle dynamics, migration, and lineage selection can be studied in multipotent NSC in vitro.
Several commercial and non-profit cell quantification and tracking software programs are currently available. In this study, we used a commercial tracking program (Imaris), which offers both automated continuous tracking and manual tracking features. Imaris software for automated tracking was not able to differentiate NSC from background with the contrast settings used for our time-lapse. Additionally, errors in automated continuous tracking increased dramatically with increasing NSC density over time. However, we found the manual single-cell tracking feature feasible for assessing NSC survival, proliferation, lineage selection or migration in vitro.
Repeated fluorescence excitation induced phototoxicity and the manifestation of physiological changes in live cells are well recognized. Our data suggest that adjustments in image acquisition interval, laser power, and time of exposure can permit time-lapse imaging up to 7 days in culture in Fucci-mNSC. Consistent with the literature, repeated fluorescence excitation decreased Fucci-mNSC survival and proliferation compared to DIC only controls, however, effects on Fucci-mNSC migration measures were minimal with the image acquisition settings used in this study. Although the timing of cell cycle arrest in Fucci-mNSC during the fluorescence live-cell imaging was similar to that detected with BrdU labeling during in vitro differentiation [16,8], repeated fluorescence excitation did alter cell cycle dynamics in this experimental setting. Critically, baseline changes in live cells post-fluorescence time-lapse imaging are rarely reported in the literature, which may lead to a misconception that photodamage does not occur during short-term experiments. Furthermore, it is important to recognize that repeated fluorescence excitation on live cells must be considered and controlled for when interpreting experimental data. However, when combined with appropriate controls, these methods can provide insight into specific questions about NSC cell cycle and migration in vitro.
Previous live-cell time-lapse-, single-cell tracking studies performed in hNSC have not only been conducted in short-term experiments (24–48 h) utilizing image acquisition intervals as short as 3–5 min. Our data indicates that a 30 min DIC acquisition interval is adequate for single-cell tracking of hNSC migration and lineage selection for 14 days or longer. High cell density remains as a particular issue during hNSC time-lapse imaging. Regardless of the low plating density, hNSC cultures became very dense towards the end of experiment, which made single-cell tracking laborious and resulted a 5–14% loss within the tracked cells. Use of genetically modified NSC expressing a less phototoxic long-wavelength nuclear fluorescence reporter in combination with selective sample illumination approaches that limit illumination and fluorophore excitation to the focal plane may be able to alleviate the issues of single-cell tracking in high density cultures observed using DIC.
Although, several techniques to constrain cells within a 2D imaging region exist, the materials used for live-cell culture have specific requirements. Although cell type dependent variation may exist, PDMS is generally considered to be a bio-compatible and non-cytotoxic cell culture material based on the use of medical grade PDMS implants [4]. In this study we used in-house manufactured PDMS microwells and found these practical for the human and mouse NSC long-term live-cell imaging. The custom size of microwells reduced both number of cells plated and number of tiled images per timepoint, and easy detachment of the inserts post-fixation enabled secondary immunocytochemical analysis of the time-lapse captured cells. Based on our experience, manufacturing PDMS inserts with microwells is a simple procedure. Inhouse PDMS sheet fabrication in not required, since prefabricated sheets with a thickness of 0.024–0.04″ are commercially available (SSP or Interstate Specialty Products), however, prefabricated sheets would require a laser system for cutting. An alternative commercial approach for those that consider testing microwell applications could be nearly similar detachable 1.5 m m × 2 mm and Ø 0.4 mm silicone microinsert applications with 4 microwells (Ibidi). We have not performed a side-by-side comparison between our microwell application and any of the commercial products. However, it should be noted that the wall structures in some of the Ibidi products are sloped, which may permit some migration of cultured NSC. Additionally, use of in-house manufactured products can significantly reduce cost. For example, the cost of one PDMS insert with 5 microwells together with a commercial glass coverslip petri-dish is approximately 10% of that when compared to a commercial ready-to-use microinsert with 4 microwells. In parallel, use of custom made inserts each with five microwells that fit into a 35 mm petri-dish with glass cover slip can save time and reagent costs by increasing the number of simultaneously imaged samples. For example, incorporation of a PDMS insert with a sample per each five microwells with an live-cell imaging system, such as the VivaView Incubator Microscope employed here, which has a loading tray with built in spacing for eight 35 mm petri-dishes, could increase the number of acquired samples in a single experiment from 8 to 40.
Overall, these proof-of-concept data suggest that long-term live-cell analysis followed by single-cell tracking can be used for studying dynamics of cell cycle, migration, and lineage selection in multipotent NSC. Most importantly, the feasibility of single-cell tracking and fate mapping human NSC in vitro enables further studies addressing some of the fundamental questions on stem cell maintenance and lineage-specific differentiation.
Supplementary Material
Acknowledgments
We want to thank Joseph Requejo, Bryant Le, Dan Haus, Harvey Perez, and Scott Strayer for technical assistance. We also want to thank Mark Eastman, David Jackson, and James Gonya at Olympus America, Inc. for excellent customer service and troubleshooting. This study was supported in part by Christopher Reeve Foundation funding (AAC-2005) to A.J.A; a CIRM Early Translational Award (TR2-01767) to B.J.C; by a CIRM Postdoctoral Training Grant (TG2-01152) to K.M.P; and by CIRM Stem Cell Training Awards (TI-01182) to K.C., A.M.P., C.L.W., and K.S.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ymeth.2017.10.003.
References
- 1.Blong CC, Jeon CJ, Yeo JY, Ye EA, Oh J, Callahan JM, Law WD, Mallapragada SK, Sakaguchi DS. Differentiation and behavior of human neural progenitors on micropatterned substrates and in the developing retina. J Neurosci Res. 2010;88:1445–1456. doi: 10.1002/jnr.22324. [DOI] [PubMed] [Google Scholar]
- 2.Costa MR, Ortega F, Brill MS, Beckervordersandforth R, Petrone C, Schroeder T, Gotz M, Berninger B. Continuous live imaging of adult neural stem cell division and lineage progression in vitro. Development. 2011;138:1057–1068. doi: 10.1242/dev.061663. [DOI] [PubMed] [Google Scholar]
- 3.Daynac M, Morizur L, Kortulewski T, Gauthier LR, Ruat M, Mouthon MA, Boussin FD. Cell sorting of neural stem and progenitor cells from the adult mouse subventricular zone and live-imaging of their cell cycle dynamics. J Vis Exp. 2015 doi: 10.3791/53247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halldorsson S, Lucumi E, Gomez-Sjoberg R, Fleming RM. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 5.Hindley C, Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem J. 2012;444:375–382. doi: 10.1042/BJ20112040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooshmand MJ, Nguyen HX, Piltti KM, Benavente F, Hong S, Flanagan LA, Uchida N, Cummings BJ, Anderson AJ. Neutrophils induce astroglial differentiation and migration of human neural stem cells via C1q and C3a synthesis. J Immunol. 2017;199:1069–1085. doi: 10.4049/jimmunol.1600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keenan TM, Nelson AD, Grinager JR, Thelen JC, Svendsen CN. Real time imaging of human progenitor neurogenesis. PLoS One. 2010;5:e13187. doi: 10.1371/journal.pone.0013187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerosuo L, Piltti K, Fox H, Angers-Loustau A, Hayry V, Eilers M, Sariola H, Wartiovaara K. Myc increases self-renewal in neural progenitor cells through Miz-1. J Cell Sci. 2008;121:3941–3950. doi: 10.1242/jcs.024802. [DOI] [PubMed] [Google Scholar]
- 9.Lu B, Jan L, Jan YN. Control of cell divisions in the nervous system: symmetry and asymmetry. Annu Rev Neurosci. 2000;23:531–556. doi: 10.1146/annurev.neuro.23.1.531. [DOI] [PubMed] [Google Scholar]
- 10.Namba T, Mochizuki H, Suzuki R, Onodera M, Yamaguchi M, Namiki H, Shioda S, Seki T. Time-lapse imaging reveals symmetric neurogenic cell division of GFAP-expressing progenitors for expansion of postnatal dentate granule neurons. PLoS One. 2011;6:e25303. doi: 10.1371/journal.pone.0025303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 12.Ortega F, Costa MR. Live imaging of adult neural stem cells in rodents. Front Neurosci. 2016;10:78. doi: 10.3389/fnins.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortega F, Costa MR, Simon-Ebert T, Schroeder T, Gotz M, Berninger B. Using an adherent cell culture of the mouse subependymal zone to study the behavior of adult neural stem cells on a single-cell level. Nat Protoc. 2011;6:1847–1859. doi: 10.1038/nprot.2011.404. [DOI] [PubMed] [Google Scholar]
- 14.Pattison DI, Davies MJ. Actions of ultraviolet light on cellular structures. EXS. 2006:131–157. doi: 10.1007/3-7643-7378-4_6. [DOI] [PubMed] [Google Scholar]
- 15.Piltti K, Haus D, Do E, Perez H, Anderson A, Cummings B. Computer-aided 2D and 3D quantification of human stem cell fate from in vitro samples using Volocity high performance image analysis software. Stem Cell Res. 2011;7:256–263. doi: 10.1016/j.scr.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Piltti K, Kerosuo L, Hakanen J, Eriksson M, Angers-Loustau A, Leppa S, Salminen M, Sariola H, Wartiovaara K. E6/E7 oncogenes increase and tumor suppressors decrease the proportion of self-renewing neural progenitor cells. Oncogene. 2006;25:4880–4889. doi: 10.1038/sj.onc.1209492. [DOI] [PubMed] [Google Scholar]
- 17.Qian X, Goderie SK, Shen Q, Stern JH, Temple S. Intrinsic programs of patterned cell lineages in isolated vertebrate CNS ventricular zone cells. Development. 1998;125:3143–3152. doi: 10.1242/dev.125.16.3143. [DOI] [PubMed] [Google Scholar]
- 18.Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 19.Raineteau O, Rietschin L, Gradwohl G, Guillemot F, Gahwiler BH. Neurogenesis in hippocampal slice cultures. Mol Cell Neurosci. 2004;26:241–250. doi: 10.1016/j.mcn.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Ravin R, Hoeppner DJ, Munno DM, Carmel L, Sullivan J, Levitt DL, Miller JL, Athaide C, Panchision DM, McKay RD. Potency and fate specification in CNS stem cell populations in vitro. Cell Stem Cell. 2008;3:670–680. doi: 10.1016/j.stem.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Roccio M, Schmitter D, Knobloch M, Okawa Y, Sage D, Lutolf MP. Predicting stem cell fate changes by differential cell cycle progression patterns. Development. 2013;140:459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- 22.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 23.Sakaue-Sawano A, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progressions with fucci technology. Cold Spring Harb Protoc. 2014;5(2014) doi: 10.1101/pdb.prot080408. [DOI] [PubMed] [Google Scholar]
- 24.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97(26):14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter MR, Liu M, Monteleone D, Melunis J, Hershberg U, Goderie SK, Temple S, Cohen AR. Computational image analysis reveals intrinsic multigenerational differences between anterior and posterior cerebral cortex neural progenitor cells. Stem Cell Rep. 2015;5:609–620. doi: 10.1016/j.stemcr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokose J, Ishizuka T, Yoshida T, Aoki J, Koyanagi Y, Yawo H. Lineage analysis of newly generated neurons in organotypic culture of rat hippocampus. Neurosci Res. 2011;69:223–233. doi: 10.1016/j.neures.2010.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.