Abstract
Most metazoan organisms have evolved a mildly acidified and calcium diminished sorting hub in the early secretory pathway commonly referred to as the Endoplasmic Reticulum-Golgi intermediate compartment (ERGIC). These membranous vesicular-tubular clusters are found tightly juxtaposed to ER subdomains that are competent for the production of COPII-coated transport carriers. In contrast to many unicellular systems, metazoan COPII carriers largely transit just a few hundred nanometers to the ERGIC, prior to COPI-dependent transport on to the cis-Golgi. The mechanisms underlying formation and maintenance of ERGIC membranes are poorly defined. However, recent evidence suggests an important role for Trk-fused gene (TFG) in regulating the integrity of the ER/ERGIC interface. Moreover, in the absence of cytoskeletal elements to scaffold tracks on which COPII carriers might move, TFG appears to promote anterograde cargo transport by locally tethering COPII carriers adjacent to ERGIC membranes. This action, regulated in part by the intrinsically disordered domain of TFG, provides sufficient time for COPII coat disassembly prior to heterotypic membrane fusion and cargo delivery to the ERGIC.
Keywords: liquid droplets, membrane trafficking, phase separation, Sar1, Sec12, Sec23, Tango1
1. Introduction
The endoplasmic reticulum (ER) is arguably the most versatile subcellular organelle found in metazoan cells, responsible for the vast majority of phospholipid and membrane protein biosynthesis, calcium storage, and interorganellar communication.[1,2] Homeostasis requires coordination between ongoing deposition of cargoes into the ER lumen, their proper redistribution to other compartments in the cell, and turnover in cases where cargoes lack sufficient quality for function. Dysregulation in any of these pathways can result in activation of an unfolded protein response (UPR), which acutely alters cellular physiology in an attempt to rebalance ER function or promote cell death in cases where long-term ER stress cannot be resolved.[3] The COPII coat machinery is largely responsible for protein export from the ER and has been shown to respond to UPR signaling both under pathological conditions and in situations where cellular physiology demands upregulation of bio-synthetic protein transport.[4,5] The coat is comprised of two distinct layers (the inner Sar1-Sec23-Sec24 lattice and the outer Sec13-Sec31 cage), which function in concert with numerous regulatory factors to select cargoes and simultaneously promote membrane remodeling to accommodate cargoes within transport carriers.[6] With increasingly complex demands on COPII carrier formation, the metazoan early secretory pathway has developed new points of regulation, including an intermediate destination between the ER and Golgi, which enables a second cargo sorting step and provides additional quality control.[7] However, the cost of augmenting this system has been far from negligible, requiring a unique step of transport that allows for plasticity in carrier size, facilitates rapid carrier uncoating, and restricts diffusion away from ERGIC (ER-Golgi intermediate compartment) membranes, all without assistance from canonical motor proteins and cytoskeletal filaments. To achieve this, metazoans appear to have separately evolved two distinct families of proteins, Trk-fused gene (TFG) and Tango1/cTage5, which are both used to enable efficient transport of small and large cargoes between the ER and ERGIC in mammals.[8,9]
1.1. Time to Put Your Coat on!
Membrane subdomains capable of producing COPII-coated transport carriers (sometimes referred to as transitional ER) are found throughout the ER network, although they become more highly dispersed near the periphery of cells (Figure 1). Morphologically, these regions of the ER are devoid of ribosomes and often exhibit negative membrane curvature with respect to flanking portions of the bilayer (Figure 2). This curvature may be driven by the clustering of transmembrane cargo proteins, integral membrane cargo receptors (including members of the p24-, Erv-, ERGIC-, and Tango1/cTage5-receptor families), and local accumulation of the guanine nucleotide exchange factor (GEF) Sec12, which also spans the ER membrane and loads GTP onto Sar1 to drive COPII carrier budding.[10,11] The precise mechanism by which COPII subunit assembly sculpts ER subdomains to create transport carriers remains unknown. However, membrane penetration by the amino-terminal amphipathic helix of Sar1 is critical to induce positive curvature. Specifically, when bound to GTP, Sar1 stably penetrates the lipid bilayer to a depth of ≈5 Å, just below the phospholipid headgroups of the outer leaflet.[12] This type of insertion has been demonstrated to be conducive to outward membrane bending, which is likely stabilized by the sequential recruitment of Sec23-Sec24 heterodimers that bind directly to activated Sar1 and Sec13-Sec31 heterotetramers that associate with the inner coat.[11,13,14] Together, the COPII components form flexible lattices that are sufficient to locally remodel membranes into different shapes, while simultaneously concentrating secretory cargo proteins within the nascent transport carriers via interactions with various cargo receptors.[15]
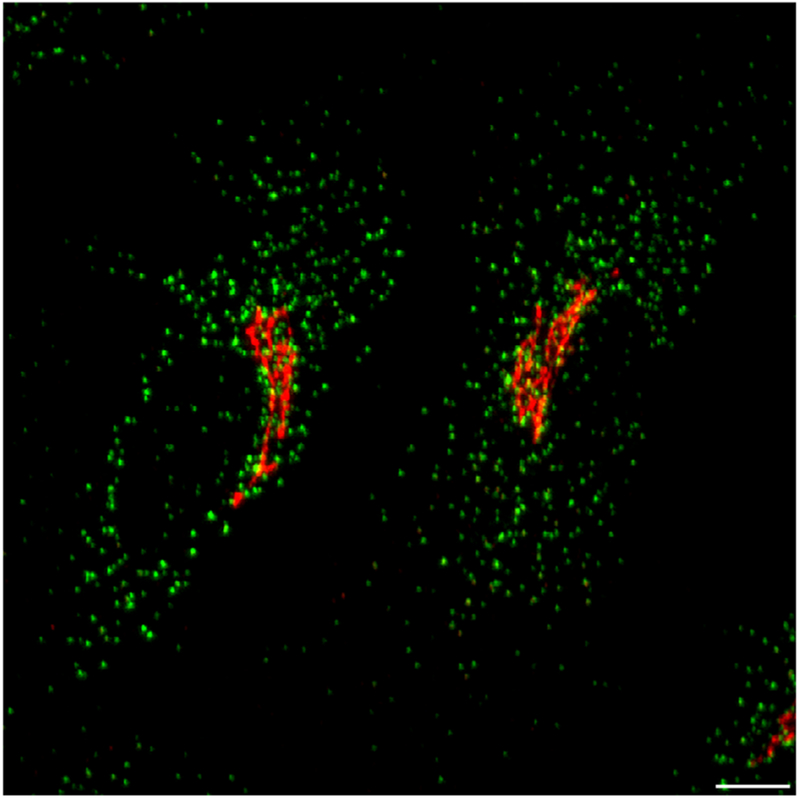
Figure 1.
Organization of the early secretory pathway in cultured human cells. Confocal imaging highlighting the unique distributions of COPII and the cis-Golgi in human cells. Scale bar, 5 μm.
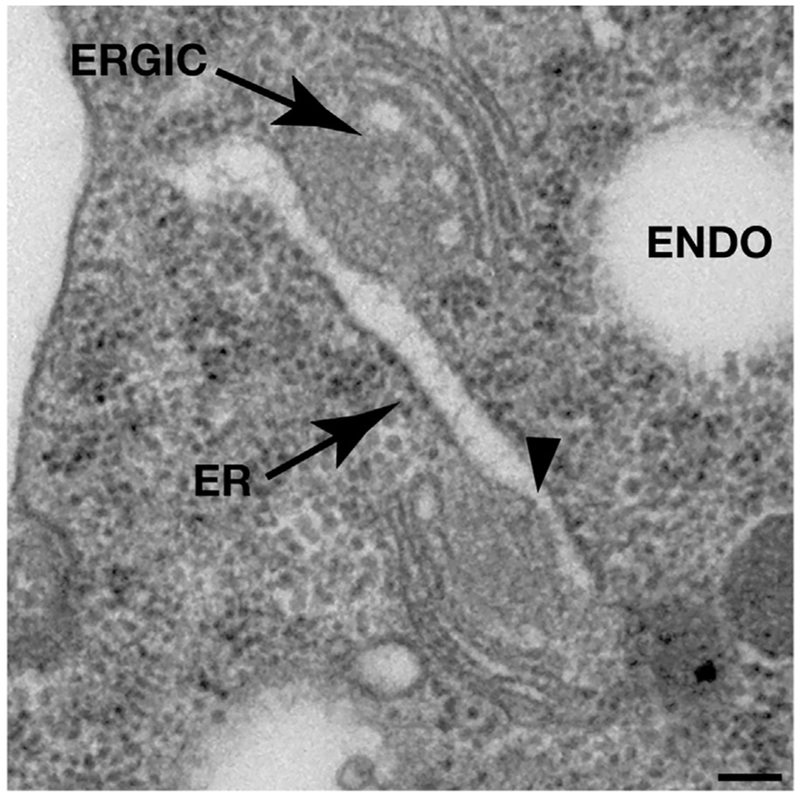
Figure 2.
Morphology of COPII budding sites on the ER in metazoans. Intact C. elegans were high pressure frozen, freeze substituted and embedded in plastic prior to thin section electron microscopy analysis. Arrows highlight the ER and ERGIC membranes in the one-cell stage embryo, and negative membrane curvature associated with COPII budding sites is indicated with an arrowhead. Additional examples of COPII budding sites that are found in mammalian cells have been described previously.[21] ENDO, endosome. Scale bar, 100 nm.
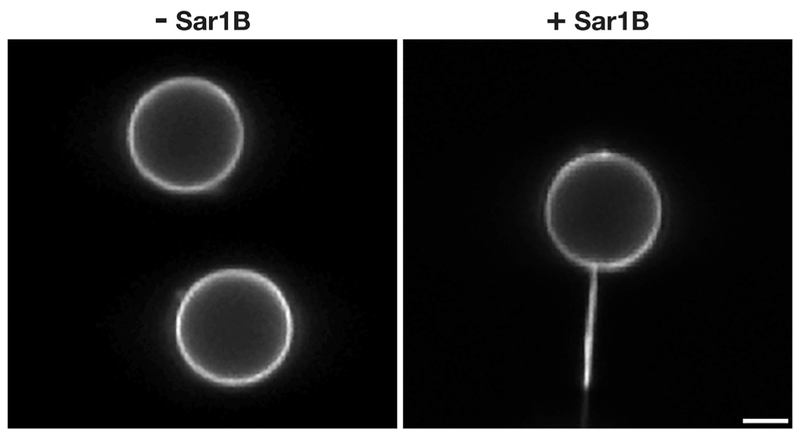
Biochemical, fluorescence, and electron microscopy-based approaches have demonstrated that most inner and outer COPII coat components are enriched on nascent buds emerging from the ER, as well as on large and small transport carriers that have undergone scission.[16–24] However, the distribution of Sar1 remains less clear, with some studies arguing that the protein is restricted to ER subdomains, while others indicate that Sar1 continues to associate with transport carriers after leaving the ER. Biochemical methods aimed at measuring the levels of COPII proteins on purified transport carriers are challenging to interpret, since components of the coat itself stimulate GTPase activity on Sar1, promoting its dissociation during cell fractionation and isolation.[20] Immuno-electron microscopy studies in pancreatic beta cells previously suggested that Sar1 is enriched on vesicular carriers, but detection of gold particles at the trans-Golgi and nucleus raised concerns regarding antibody specificity.[25] Currently, two models exist to describe the function of Sar1 in the biogenesis of COPII transport carriers. The first is based on the premise that Sar1 action is restricted to the ER, where it promotes membrane tubulation and ultimately accumulates at the neck of nascent COPII-coated buds to facilitate bilayer scission upon GTP hydrolysis. This idea is supported by studies demonstrating the ability of recombinant Sar1 to create membrane tubules (Figure 3), as well as more recent work demonstrating that Sar1 is capable of sensing membrane curvature.[12,26,27] Additionally, the intrinsic GTPase activity of Sar1 is stimulated in the presence of elevated curvature, suggesting a mechanism by which it may drive transport carrier scission.[12] In a second model, Sar1 not only acts at the ER, but also on free transport carriers en route to the ERGIC. At this latter location, Sar1-mediated GTP hydrolysis has been speculated to contribute to COPII lattice disassembly, a requisite step prior to fusion with ERGIC membranes.[28] Although these two models are not mutually exclusive, a more detailed description of Sar1 distribution in situ will nonetheless provide much needed clarification.
Figure 3.
Human Sar1B promotes membrane tubulation in vitro. Fluid supported bilayers with excess membrane reservoir (SUPER) templates[59] were imaged using confocal optics in the presence or absence of purified human Sar1B (1 μM) bound to GTP. Scale bar, 2 μm.
Similarly, we continue to await improved, more native views of COPII carrier morphology in cells at high resolution. Although it is clear that purified mammalian COPII components facilitate the formation of ≈60–80 nm coated vesicles in vitro,[29] the existence of spherical transport carriers budding directly from the ER in mammalian cells remains controversial. Instead, most published data suggest the presence of COPII-coated tubules emanating from the ER, often in a dumb-bell shaped configuration, with constrictions spaced ≈60 nm apart from one another.[15,17,21,23,30,31] These data suggest that scission reactions may occur along the entire length of COPII-decorated tubules, potentially generating free carriers only at their tips. With advances in cryo-electron microscopy-based approaches, including those that leverage the use of focused ion beam technology, we should soon have a clearer picture of the transport intermediates generated via the action of COPII in cells.
COPII coat recruitment to ER subdomains is subject to tight regulation, with GTP-bound Sar1 representing only one of several interactions necessary for proper inner and outer lattice assembly. At least two additional families of ER-associated factors are also responsible for coordinating the formation of transport carriers, Sec16 and Tango1/cTage5 complexes. Both localize specifically to ER subdomains, interact with one another and the Sar1 GEF Sec12, but fail to be incorporated into transport carriers as they emerge, likely due to their relatively weak affinities for COPII coat subunits.[32–35] Sec16 and Tango1/cTage5 also bind directly to the inner COPII coat component Sec23, which has been proposed to restrict outer coat assembly until maturation of the transport carrier. Ultimately, however, recruitment Sec13-Sec31 heterotetramers outcompetes Sec16, Tango1, and cTage5, leaving them behind as the COPII-coated carrier undergoes fission from the ER.[36]
The Tango1/cTage5 family of proteins has also been implicated in the formation of “specialized” transport carriers, which can accommodate large cargoes, including collagens that are ≈300 nm in length. Fairly elaborate mechanisms have been proposed to describe how Tango1/cTage5 complexes may facilitate this action, based on the premise that the inner COPII coat exists in two distinct complexes after assembly on ER membranes, bound either to Tango1/cTage5 or Sec13-Sec31.[37] However, direct evidence for this is lacking, and localization studies consistently show the presence of both Tango1/cTage5 and Sec13-Sec31 at all ER subdomains harboring Sec23-Sec24.[9,35] Instead, a simpler explanation may reconcile the role of Tango1/cTage5 complexes in early secretory pathway function. If all COPII transport carriers form as elongated tubules, constraints on cargo incorporation become largely eliminated, and only the constriction/scission reaction must be appropriately controlled to ensure efficient packaging of large secretory proteins. In this scenario, Tango1 functions as a critical receptor for collagen export, via its interaction with the collagen-specific chaperone Hsp47, but does not regulate carrier size. However, a more general role for Tango and cTage5 must also be considered, given their constitutive presence at sites of COPII carrier formation.[38] Recent evidence points to a potential role for Tango1 in the direct regulation of Sec16 distribution and function.[35] Additionally, Tango1/cTage5 complexes recruit the Sar1 GEF Sec12 to sites of COPII carrier formation.[34] These data argue for a broader role for Tango1/cTage5 in ER export, beyond regulation of large cargo transport. Consistent with this idea, elimination of cTage5 in pancreatic beta cells results in impaired insulin trafficking,[39] and depletion of Tango1 delays export of small cargoes from the ER, including VSVG.[35] The question now shifts to what actually regulates constriction and scission of large COPII transport carriers. Importantly, reconstitution of collagen export using ER microsomes has recently been demonstrated, opening the door to defining the minimal components necessary.[24] The addition of one or more cytosolic components appears necessary for the budding reaction, suggesting an origin for the factor(s) that control the timing of carrier scission. In particular, the E3 ubiquitin ligase-substrate adaptor complex, CUL3-KLHL12, which localizes to only a subset of COPII budding sites, potentially plays a key role in this process by modifying the COPII cage.[40] Additional factors, including the calcium-binding co-adaptors PEF1 and ALG2, may also participate to control the timing of COPII carrier scission.[41–43]
1.2. May I Take Your Coat?
Unlike long-range transport processes that require cytoskeletal tracts, the canonical action of the metazoan COPII coat complex is confined to generating transport carriers at the ER that are directionally moved only a few hundred nanometers prior to fusion with ERGIC membranes. Live-cell imaging studies support this idea, demonstrating that COPII components exhibit limited mobility, which mimics that of adjacent ER membranes.[23] Additionally, photobleaching studies have suggested that COPII components undergo rapid turnover at the ER/ERGIC interface, recycling back into a large cytosolic pool that is available for continuous rounds of COPII assembly.[44] Current models depicting the ER/ERGIC interface fail to highlight the spatial and temporal regulation of COPII coat disassembly required for fusion with the ERGIC. For example, a step-wise mechanism based largely on work done in the yeast Saccharomyes cerevisiae, which lack ERGIC membranes, proposes that the changing availability of Sec23 to bind specific components of the cis-Golgi localized TRAPPI complex guides COPII transport carriers after stochastic coat disassembly.[20,45] With many of the key factors in this model sparsely localized to the ERGIC in mammalian cells as compared to the cis-Golgi in yeast, the field lacks a consensus on what drives COPII uncoating at the ER/ERGIC interface. However, recent studies suggest a key role for TFG in this process (Figure 4). Specifically, TFG was shown to bind directly to Sec23 through a shared interface with Sec31, promoting disassembly of the outer coat during the trafficking of COPII carriers within the ER/ERGIC interface.[9] Additionally, post-translation modifications on COPII coat proteins may further facilitate their disassembly, although neither biochemical or structural data are currently available to support this idea.[20,40]
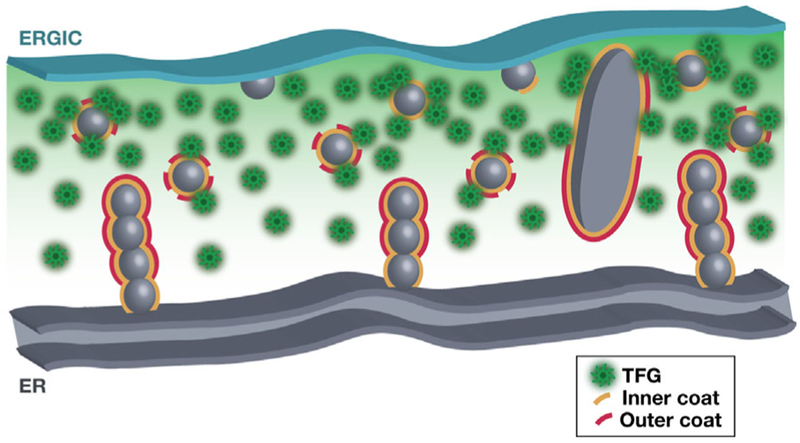
Figure 4.
Model depicting COPII-mediated transport at the ER/ERGIC interface. Cartoon showing the anterograde movement of COPII-coated transport carriers between the ER and juxtaposed ERGIC membranes. The ongoing formation of cargo-laden carriers promotes TFG (green) recruitment and concentration, potentially leading to phase separation and local tethering until dissociation of all COPII components (yellow and red) and exposure of specific SNARE molecules, which mediated fusion with ERGIC membranes.
Strikingly, depletion of TFG not only results in the stabilization of COPII-coated transport carriers, but also their aberrant appearance away from the ER/ERGIC interface, suggesting an additional role for TFG in tethering COPII carriers prior to full uncoating and fusion with ERGIC membranes.[9,23] Consistent with this idea, recombinant TFG is sufficient to cluster inner COPII-coated liposomes in vitro.[9] In particular, the carboxyl-terminal intrinsically disordered domain of TFG is critical for this activity. Previous work suggested that this region of TFG is capable of self-assembly into larger structures (≈200–300 nm in size), raising the possibility that elevated concentrations of TFG can lead to phase separation within the cytoplasm, potentially forming a membrane-less liquid droplet.[9,23] By generating a phase separated liquid droplet at the interface between ER and ERGIC membranes, COPII transport carriers may become transiently trapped, providing sufficient time for the inner coat to disassociate (Figure 4). This process may be aided by GTP hydrolysis on remaining Sar1 present on the carrier and/or post-translational modifications to Sec23-Sec24 heterodimers. Upon release of the inner COPII coat, affinity for the phase separated ER/ERGIC interface is diminished, enabling fusion of the naked transport carriers with tightly juxtaposed ERGIC membranes. In this model, the capture and release of COPII transport carriers parallels synaptic vesicle clustering mediated by synapsin phase separation at pre-synaptic terminals.[46]
Although the ER is replete with non-fusogenic membrane contact sites, multiple protein complexes usually act in trans to physically tether the ER with other organelles. For example, ER-localized extended-synaptotagmins (e-Syts) tether to the plasma membrane, generating a closely apposed membrane contact site (≈10–30 nm) that facilitates glycerolipid metabolism and maintenance of membrane identity through the transfer of specific phosphoinositides.[47,48] In contrast, given the significant distance separating the ER and ERGIC, current data do not support the idea that these organelles are classically tethered. Instead, we propose that ERGIC membranes remain juxtaposed to the ER via continuous efflux of COPII carriers that uncoat and fuse. The constitutive action of COPI at ERGIC membranes restricts the size of this compartment and enables cargoes to move on to the cis-Golgi, using microtubule tracks to traverse the relatively long distances involved. Ablating these tracks with a microtubule severing agent blocks anterograde trafficking from the ERGIC without impeding anterograde COPII transport, resulting in the enlargement of pre-Golgi structures.[49] Nevertheless, impairing transport at any step along the early secretory pathway quickly leads to gridlock, resulting in an accumulation of cargoes within the ER lumen and an accompanying UPR signal that can lead to cell death.
1.3. Do Not Let the Door Hit You on the Way Out!
Beyond the roles for COPII in secretory cargo transport from the ER described thus far, new evidence suggests that COPII assembly can also be initiated at ERGIC membranes during nutrient deprivation and the induction of autophagy. Using a cell-free assay, it was previously shown that the ERGIC provides a key membrane reservoir to promote LC3 lipidation, an early marker of autophagosome formation.[50] Subsequently, COPII was implicated in the budding of LC3 lipidation-active membranes from ERGIC,[51] which are now hypothesized to fuse with other membrane sources, including Atg9-positive vesicles, to generate the phagophore. Ultimately, elongation of the phagophore enables cytoplasmic engulfment of proteins/organelles, which are delivered to lysosomes for breakdown to serve as a nutrient source under conditions of stress. Perhaps somewhat surprisingly, this entire process requires the redistribution of the Sar1 GEF Sec12 to ERGIC membranes, in a manner that requires cTage5, but cTage5 remains tightly associated with the ER under these conditions.[52] The mechanism by which Sec12 is freed from cTage5 and delivered to ERGIC membranes during starvation remains unclear but may involve COPII-mediated transport and phosphatidylinositol 3-kinase signaling, which is also required to generate LC3 lipidation-active membranes from the ERGIC.[51]
For many years, it has been debated whether the ERGIC represents an independent, stable organelle in the early secretory pathway or merely a subdomain of the ER or cis-Golgi. Its absence in yeast cells and plants further fueled speculation that the ERGIC is only a transient intermediate. However, live cell imaging studies argue otherwise, indicating instead that the ERGIC is a long-lived compartment.[53] Additionally, it is now clear that ERGIC membranes play key roles in COPI-dependent sorting of anterograde and retrograde cargo, formation of autophagosomes, and secretory protein quality control. The mildly acidic nature of the compartment facilitates liberation of certain cargoes from their sorting receptors (e.g., procathepsin Z release from ERGIC-53), while it enhances the engagement of other proteins for their cognate receptors (e.g., ER resident proteins binding to KDEL receptors to enable retrieval).[54] Additionally, diminished calcium levels within the ERGIC promotes the release of glycoprotein cargoes, such as blood clotting factors V and VIII, from the calcium-dependent cargo receptor ERGIC-53.[55,56] Overall, the distinct luminal environment of the ERGIC, as compared to the ER or Golgi, set it apart as a unique organelle, with critical importance to the efficiency of secretory efflux from cells.
2. Conclusions
Although we have learned a great deal about the metazoan COPII trafficking pathway over the past decade, there still remains a remarkable amount of information to uncover regarding its regulation during cell proliferation, differentiation, and development. What additional sorting receptors exist, and how do they couple with COPII machinery? How is scission activity on COPII transport carriers regulated? How is COPII lattice/cage construction regulated in vivo? What processes underlie full uncoating of COPII carriers prior to fusion with ERGIC membranes? How do different COPII subunit isoforms contribute to tissue homeostasis? Delineating these mechanisms will be of enormous importance to understanding the wide range of human disease states that arise as a result of deficits in COPII function.[57] It is now clear that COPII-mediated transport has evolved significantly. Although the core components of the system are highly conserved, the emergence of an intermediate compartment between the ER and Golgi in metazoan organisms clearly distinguishes them from most unicellular species. Thus, study of membrane traffic between the ER and ERGIC must occur using metazoan models, including Caenorhabditis elegans, Drosophila, zebrafish, rodents, and human cell lines. Moreover, particular emphasis must be placed upon the role of post-translational modifications such as phosphorylation, ubiquitinylation, and glycosylation, all of which have been identified on COPII subunits, as well as regulators of COPII coat assembly and disassembly.[58] Combining these efforts with structural approaches, both in vitro and in situ, will be critical to gaining a clear understanding of the mechanisms underlying the earliest step in secretory cargo trafficking.
Acknowledgments
This work was supported in part by a grant from the NIH (GM110567 to AA). The authors thank members of the Audhya lab for suggestions and critically reading this manuscript.
Abbreviations
- ER
endoplasmic reticulum
- ERGIC
Endoplasmic Reticulum-Golgi intermediate compartment
- GEF
guanine nucleotide exchange factor
- UPR
unfolded protein response
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Saheki Y, De Camilli P, Annu. Rev. Biochem. 2017, 86, 659. [DOI] [PubMed] [Google Scholar]
- [2].Biwer LA, Isakson BE, Acta Physiol. (Oxf) 2017, 219, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hetz C, Papa FR, Mol. Cell. 2018, 69, 169. [DOI] [PubMed] [Google Scholar]
- [4].Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP, EMBO J. 2008, 27, 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kirk SJ, Cliff JM, Thomas JA, Ward TH, J. Leukoc. Biol. 2010, 87, 245. [DOI] [PubMed] [Google Scholar]
- [6].Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R, Nat. Cell Biol. 2011, 14, 20. [DOI] [PubMed] [Google Scholar]
- [7].Lorente-Rodriguez A, Barlowe C, Cold Spring Harb. Perspect. Biol. 2011, 3, a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rios-Barrera LD, Sigurbjornsdottir S, Baer M, Leptin M, Proc. Natl. Acad. Sci. USA 2017, 114, E10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hanna MG, Block S, Frankel EB, Hou F, Johnson A, Yuan L, Knight G, Moresco JJ, Yates JR, Ashton R, Schekman R, Tong Y, Audhya A, Proc. Natl. Acad. Sci. USA 2017, 114, E7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gomez-Navarro N, Miller E, J. Cell Biol. 2016, 215, 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jarsch IK, Daste F, Gallop JL, J. Cell Biol. 2016, 214, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanna MG, Mela I, Wang L, Henderson RM, Chapman ER, Edwardson JM, Audhya A, J. Biol. Chem. 2016, 291, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Noble AJ, Zhang Q, O’Donnell J, Hariri H, Bhattacharya N, Marshall AG, Stagg SM, Nat. Struct. Mol. Biol. 2013, 20, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, Balch WE, Cell 2008, 134, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zanetti G, Prinz S, Daum S, Meister A, Schekman R, Bacia K, Briggs JA, Elife 2013, 2, e00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Orci L, Ravazzola M, Meda P, Holcomb C, Moore HP, Hicke L, Schekman R, Proc. Natl. Acad. Sci. USA 1991, 88, 8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bannykh SI, Rowe T, Balch WE, J. Cell Biol 1996, 135, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T, Cell 1998, 93, 263. [DOI] [PubMed] [Google Scholar]
- [19].Martinez-Menarguez JA, Geuze HJ, Slot JW, Klumperman J, Cell 1999, 98, 81. [DOI] [PubMed] [Google Scholar]
- [20].Lord C, Bhandari D, Menon S, Ghassemian M, Nycz D, Hay J, Ghosh P, Ferro-Novick S, Nature 2011, 473, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zeuschner D, Geerts WJ, can Donselaar E, Humbel BM, Slot JW, Koster AJ, Klumperman J, Nat. Cell Biol 2006, 8, 377. [DOI] [PubMed] [Google Scholar]
- [22].Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, Schwarze K, Yates JR, Eimer S, Audhya A, Nat. Cell Biol 2011, 13, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johnson A, Bhattacharya N, Hanna M, Pennington JG, Schuh AL, Wang L, Otegui MS, Stagg SM, Audhya A, EMBO J. 2015, 34, 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, Schekman R, J. Cell Biol 2017, 216, 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE, J. Cell Biol 1994, 125, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bacia K, Futai E, Prinz S, Meister A, Daum S, Glatte D, Briggs JA, Schekman R, Sci. Rep 2011, 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hariri H, Bhattacharya N, Johnson K, Noble AJ, Stagg SM, J. Mol. Biol 2014, 426, 3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sato K, Nakano A, FEBS Lett. 2007, 581, 2076. [DOI] [PubMed] [Google Scholar]
- [29].Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R, J. Biol. Chem 2005, 280, 7758. [DOI] [PubMed] [Google Scholar]
- [30].Kirk SJ, Ward TH, Semin. Cell Dev. Biol 2007, 18, 435. [DOI] [PubMed] [Google Scholar]
- [31].Sesso A, de Faria FP, Iwamura ES, Correa H, J. Cell Sci 1994, 107, 517. [PubMed] [Google Scholar]
- [32].Saito K, Yamashiro K, Ichikawa Y, Erlmann P, Kontani K, Malhotra V, Katada T, Mol. Biol. Cell 2011, 22, 2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yorimitsu T, Sato K, Mol. Biol. Cell 2012, 23, 2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Saito K, Yamashiro K, Shimazu N, Tanabe T, Kontani K, Katada T, J. Cell Biol 2014, 206, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Maeda M, Katada T, Saito K, J. Cell. Biol 2017, 216, 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma W, Goldberg J, Proc. Natl. Acad. Sci. USA 2016, 113, 10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Raote I, Ortega Bellido M, Pirozzi M, Zhang C, Melville D, Parashuraman S, Zimmermann T, Malhotra V, J. Cell Biol 2017, 216, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu M, Feng Z, Ke H, Liu Y, Sun T, Dai J, Cui W, Pastor-Pareja JC, J. Cell Biol 2017, 216, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fan J, Wang Y, Liu L, Zhang H, Zhang F, Shi L, Yu M, Gao F, Xu Z, J. Cell Biol 2017, 216, 4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M, Nature 2012, 482, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McGourty CA, Akopian D, Walsh C, Gorur A, Werner A, Schekman R, Bautista D, Rape M, Cell 2016, 167, 525. [DOI] [PubMed] [Google Scholar]
- [42].Takahara T, Inoue K, Arai Y, Kuwata K, Shibata H, Maki M, J. Biol. Chem 2017, 292, 17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Helm JR, Bentley M, Thorsen KD, Wang T, Foltz L, Oorschot V, Klumperman J, Hay JC, J. Biol. Chem 2014, 289, 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Forster R, Weiss M, Zimmermann T, Reynaud EG, Verissimo F, Stephens DJ, Pepperkok R, Curr. Biol 2006, 16, 173. [DOI] [PubMed] [Google Scholar]
- [45].Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S, Nature 2007, 445, 941. [DOI] [PubMed] [Google Scholar]
- [46].Milovanovic D, De Camilli P, Neuron 2017, 8, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fernandez-Busnadiego R, Saheki Y, De Camilli P, Proc. Natl. Acad. Sci. USA 2015, 112, E2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P, Nat. Cell Biol 2016, 18, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J, Nature 1997, 389, 81. [DOI] [PubMed] [Google Scholar]
- [50].Ge L, Melville D, Zhang M, Schekman R, Elife 2013, 2, e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ge L, Zhang M, Schekman R, Elife 2014, 3, e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ge L, Zhang M, Kenny SJ, Liu D, Maeda M, Saito K, Mathur A, Xu K, Schekman R, EMBO Rep. 2017, 18, 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ben-Takaya H, Miura K, Pepperkok R, Hauri HP, J. Cell Sci 2005, 118, 357. [DOI] [PubMed] [Google Scholar]
- [54].Appenzeller C, Andersson H, Kappeler F, Hauri HP, Nat. Cell Biol 1999, 1, 330. [DOI] [PubMed] [Google Scholar]
- [55].Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, Moussalli MJ, Hauri HP, Ciavarella N, Kaufman RJ, Ginsburg D, Cell 1998, 93, 61. [DOI] [PubMed] [Google Scholar]
- [56].Zheng C, Page RC, Das V, Nix JC, Wigren E, Misra S, Zhang B, J. Biol. Chem 2013, 288, 20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Venditti R, Wilson C, De Matteis MA, Trends Cell Biol. 2014, 24, 9. [DOI] [PubMed] [Google Scholar]
- [58].Cox NJ, Unlu G, Bisnett BJ, Meister TR, Condon BM, Luo PM, Smith TJ, Hanna M, Chhetri A, Soderblom EJ, Audhya A, Knapik EW, Boyce M, Biochemistry 2018, 57, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pucadyil TJ, Schmid SL, Biophys. J 2010, 99, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]