Abstract
Quantification of fat and muscle on clinically acquired CT scans is critical for determination of body composition, a key component of health. Manual tracing has been regarded as the gold standard method of body segmentation; however, manual tracing is time-consuming. Many semi-automated/automated algorithms have been proposed to avoid the manual efforts. Previous efforts largely focused on segmenting 2D cross-sectional images (e.g., at L3/T4 vertebra locations) rather than on the whole-body volume. In this paper, we propose a fully automated 3D body composition estimation framework for segmenting the muscle and fat from abdominal CT scans. The 3D whole body segmentations were reconstructed from a slice-wise multi-atlas label fusion (MALF) based framework. First, we used a low-dimensional atlas representation to estimate each class for each axial slice. Second, the abdominal wall and psoas muscle were segmented by combining MALF with active shape models and deformable models. Third, skeletal muscle, visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) were measured to assess the areas of muscle and fat tissue. The proposed method was compared to manual segmentation and demonstrated high accuracy. Then, we evaluated the approach on 40 CT scans comparing the new method to a prior atlas-based segmentation method and achieved 0.854, 0.740, 0.887 and 0.933 on Dice similarity index for the skeletal muscle, psoas muscle, VAT and SAT, respectively. Compared with the baseline, our method showed significantly (p < 0.001) higher accuracy on skeletal muscle, VAT and SAT estimation.
Keywords: Skeletal Muscle, Psoas Muscle, Visceral Fat, Subcutaneous Fat, Multi-atlas
1 Introduction
Body composition of fat and muscle mass is an important biomarker in cancer treatment. The quantitative measurement of body composition is related to the efficacy and toxicity of chemotherapy, post therapy functional status, surgical complication rates, length of hospital stay and overall survival [1]. In addition, body composition estimation will lead to a more reliable replacement of basic measures of healthy weight, such as Body Mass Index (BMI). Manual delineation on computed tomography (CT) images has been regarded as the gold standard in body composition estimation [2]. However, manual tracing on muscle and fat regions are time-consuming and cannot be easily applied to large cohorts. Therefore, many automated or semi-automated methods have been proposed to perform the segmentation. The previous studies estimated the body composition of muscle and fat based on the 2D axial slice at the L3 lumbar vertebra position. However, single slice based estimation is a rough approximation of the whole-body composition, which is sensitive to slice selection.
For fat segmentation, the Hounsfield unit (HU) intensity is typically used to distinguish muscle and fat on CT images when performing the manual segmentation (e.g., [−29, 150] for muscle tissue and [−190, −30] for fat tissue [3]). Importantly, the compartment in which adipose tissue resides relates to the clinical significance of that fat. For example, it is found that nonagenarian individuals with and without frailty syndrome presented marked differences in the pericardial and visceral adipose tissue [4]. Therefore, to generate a meaningful measurement, efforts are required to separate fat into visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT). The VAT is the adipose tissue included in intra-abdominal cavity, while SAT is the adipose tissue bounded by the inner abdominal wall musculature and the skin surface [4]. Muscle segmentation presents a greater challenge as the HU for muscle overlaps with other abdominal organs and tissues. Moreover, the variable shape and location of muscle make the segmentation even more difficult.
Previous efforts were typically focused on segmenting either muscle or fat. For fat segmentation, Yao et al. [5] separated the subcutaneous and visceral fat by a single surface at the abdominal wall driven by active contour models (ACM). For muscle segmentation, shape models were typically used. For instance, Tsutomu et al. [6] incorporated a shape prior represented as logistic curves in higher-order graph cut models to segment psoas from CT images. Chung et al. [7] presented a muscle segmentation method in which a thresholded binary image was warped to a mean shape prior by a Free Deformation model. Popuri et al. [3] proposed a FEM-based registration model to perform template-based segmentation of skeletal muscle. Although these methods achieved high accuracy, they were based on 2D cross-sectional images taken at the 3rd lumbar vertebra (L3) or the 4th thoracic vertebra (T4) locations rather than on whole 3D volumes. Zhang et al. [8] presented an atlas-based approach to segment the musculature on CT volumes using five pre-defined muscle atlas models, and then refined by ACM. Xu et al.[9] proposed a slice-wise method called augmented ASM (AASM) that integrated multi-atlas label fusion (MALF) and level set into the active shape model (ASM) framework. To overcome the large intra-slice variations in the abdominal wall along the cranial-caudal direction, this method pre-classifies slice-wise images to five exclusive classes using landmarks. However, for clinical data with large variations on fields of view (FOV), such landmarks are not typically available (Fig. 1).
Fig. 1.
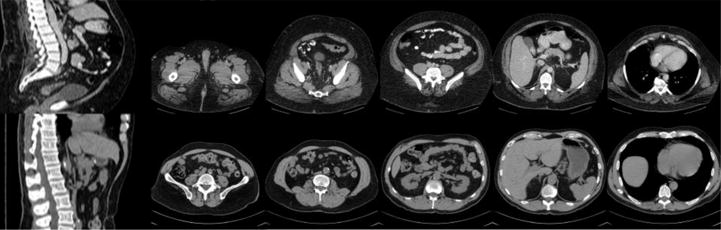
Illustration of challenges in muscle and fat segmentation. The sagittally reconstructed images show differing fields of view while the axial images demonstrated the large intra-slice and inter-slice variations in muscle and fat cross-sectional area on CT images.
In this study, we propose a fully automated framework to segment the skeletal muscle, psoas muscle, VAT and SAT from clinically acquired CT scans. Briefly, we first used a PCA-based low-dimensional representation to estimate the atlas class for the target image. Second, the abdominal wall was segmented using AASM under a slice-wise MALF framework. Here, the abdominal wall is characterized as an enclosed region bounded by the inner surface and outer surface. Then, the psoas muscle was segmented using combination of MALF and a deformable model. Finally, the skeletal muscle, VAT and SAT were extracted using the generated abdominal wall mask and pre-defined HU ranges. The main novelty of our work lies in the application of MALF on the challenging problem of muscle and fat quantification. Considering different anatomies of target regions, an active shape model and a deformable model are used to refine and regularize the initial results of MALF. The whole pipeline is fully automated, without the need of user interaction or parameter adjustment, which gives it potential to be applied in clinic.
2 Methods
The proposed pipeline is shown in Fig. 2. It integrates a slice-wise multi-atlas label fusion framework with an active shape model and a deformable model.
Fig. 2.
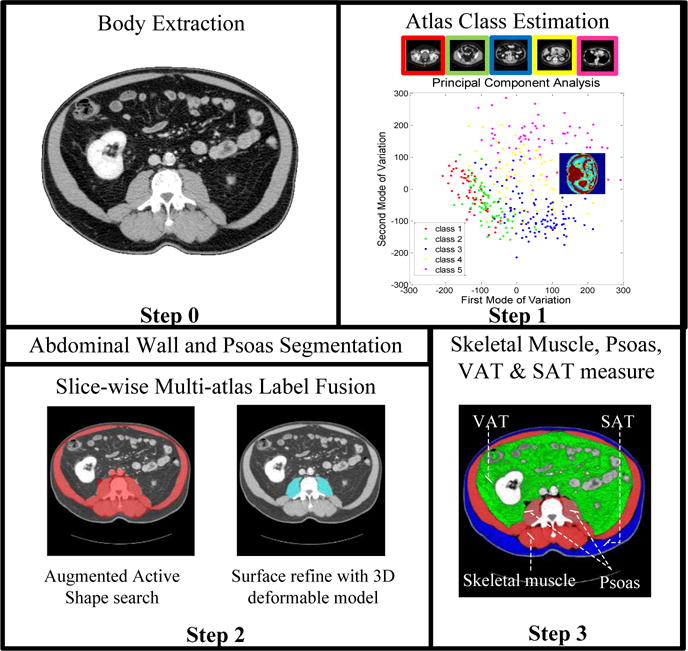
Flowchart of the proposed segmentation framework
2.1 Atlas class estimation via PCA
We use a PCA-based low-dimensional representation to decide the atlas class for each testing image. Following [9], three biomarkers, i.e., xiphoid process (XP), pubic symphysis (PS) and umbilicus (UB) are manually labeled on each training volume. According to the location of these biomarkers, training axial slices are separated into five exclusive classes. For training image j, we assume that we are given the image aj ∈ ℝD and its atlas class cj, where D is the total number of pixels and cj ∈ {1,2,3,4,5}. Given a set of training images, the goal is to assign a target slice image x into the most similar atlas class. Firstly, we align all training images to the same space. Specifically, one image is randomly selected and others are registered to it using an affine transform. Then, an average image is generated from all registered images and all images are registered to the average image again. We denote the aligned training data set as matrix A = (a11, …, a1n1, …, a51, …, a5n5) ∈ ℝD×N, where is the total number of training images. Secondly, by using PCA dimension reduction, the high-dimensional datapoints A are transformed into a low-dimensional representation , where . The PCA is performed by minimizing the cost function , where represents Euclidean distance between the high-dimensional points and . Finally, the target image x is projected to the low-dimensional space and the k -nearest neighbors in Euclidean distance are selected to estimate its class using majority voting.
2.2 Abdominal wall and psoas segmentation
Prior probability map learned from MALF
For each axial slice of the test CT volume, all the training images in the estimated class are considered as atlases to perform slice-wise multi-atlas label fusion with respect to regions of interest, i.e., the abdominal wall and psoas muscle. Each atlas is non-rigidly registered to the target image with Nifty Reg package [10]. Atlas labels are then warped to the target image and combined using the joint label fusion algorithm [11], yielding prior probability maps for the abdominal wall and psoas muscle.
Abdominal wall segmentation
In abdominal wall segmentation, we use AASM [9] to search the optimal shape iteratively. In the training stage, an active shape model and a local appearance model are trained from each atlas class. In testing, the trained ASM, local appearance model and the probability map generated from MALF guide landmarks along current contour move to new positions. Specifically, firstly level set evolution is applied on the probability map to move the current contour. Then, the zero-crossing points along the normal direction of the zero level set are collected as landmarks. These landmarks are updated with the active shape search and used as the initialization of LS evolution for the next iteration. The constrained ASM parameters guarantee the regularization of the shape.
Psoas muscle segmentation
For psoas muscle, we adopt a 3D deformable model that integrates intensity statistical information, a prior probability map, and a gradient map to refine the initial surface. Given a volume image defined on , we denote the indicator function of psoas as , which is obtained by minimizing the energy functional:
| (1) |
The first data term formulates the intensity statistics inside and outside the region, i.e., , where pin and pout are intensity distributions of foreground and background pixels defined by the initial psoas muscle region given by MALF. The second prior term presented as is to provide spatial constraints, where L(x) is the psoas muscle probability map learned from MALF. The last weighted total-variation term acts as regularization term, where . The energy function (1) is globally optimized in a surface evolution way with the continuous max-flow algorithm [12, 13].
2.3 Skeletal muscle and adipose tissue measurement
Using the segmented abdominal wall as a mask, skeletal muscle and fat tissue in each axial slice are extracted based on pre-defined HU ranges according to Ref. [3]. The region that locates inside the inner wall surface with HU [−190,−30] is extracted as VAT, and the region resides outside inner wall surface within the body mask with HU [−190,−30] is extract as SAT. The skeletal muscle tissue is segmented inside abdominal wall mask with HU [−29,−150].
2.4 Baseline method
We take the method of Zhang et al. [8] as baseline, which also focused on whole CT volume segmentation and used slice-wise algorithms. Following [8], we used fuzzy c-means and an active contour model (ACM) to segment VAT and SAT, and then used an atlas-based method to segment skeletal muscle. To avoid the effect of atlas model selection on muscle segmentation, the ACM was applied to refine the results from our MALF as re-implemented.
All experiments in this paper were run on a machine with 8 cores of Intel Xeon W3520 processors and 12 GB RAM available running Ubuntu 14.04.1 LTS (64 bit). Algorithms were implemented in MATLAB 2014b environment. The proposed segmentation method has been made available online in open-source1.
3 Experiments and Results
3.1 Data
Abdominal CT data on 60 patients from two clinical datasets were randomly retrieved in de-identified form under IRB approval (40 patients from PHC and 20 from GIONC). Specifically, 20 scans from PHC were used for training, and the remaining 20 PHC scans and 20 GIONC scans were for testing. The FOVs of PHC scans ranged from 335×335×390 mm3 to 500×500×708 mm3, with resolutions ranging from 0.65×0.65×2.50 mm3 to 0.98×0.98×5.00 mm3. The FOVs of GIONC scans were from 346×346×165 mm3 to 412×412×505 mm3, with resolutions from 0.65×0.65×1.50 mm3 to 0.85×0.85×5.00 mm3. All 60 scans were manually labeled by an experienced rater to generate ground truth of skeletal muscle, psoas, VAT and SAT. Specifically, for the 40 PHC scans, essential biomarkers, i.e., xiphoid process, pubic symphysis, and umbilicus were identified, and the abdominal wall and psoas muscle were delineated on axial slices spaced every 5 cm, generating 177 and 184 axial slices for training and testing, respectively. Besides, 40 axial slices at the L3 position of the 40 testing scans were extracted and labeled. Axial slices from the same patient for training were not used for testing.
3.2 Experimental setting
For all training and testing slices, a body mask was obtained by thresholding the image to remove the background, then selecting the largest 3D connected component to ensure that the CT table was excluded before image analysis. All the images were centered after body extraction. A leave-one-out approach that excluded slices from the same patient was used to optimize the parameters in PCA dimension reduction on the labeled slices from PHC dataset. As a result, 9 modes of variation in the low-dimensional space and neighboring images are chosen to estimate the target image class.
In the AASM-based abdominal wall segmentation, 177 labeled axial slices from 20 patients were used as training set (also atlases) to build the active shape model with default parameters as in [9]. The parameters in energy function (1) of psoas refinement were empirically set as λ2=0.1, .
3.3 Results
Automated results were validated against the manual labels on 184 axial slices taken at different positions in 20 scans (Fig. 3). The Spearman’s rank correlation coefficient between estimated cross-sectional tissue area (cm2) and the truth was 0.91, 0.75, 0.99, and 0.99 for skeletal muscle, psoas, VAT and SAT, respectively. Fig. 4 shows qualitative segmentation results at different position slices from one CT. Although there are large shape variations in axial slices taken at different positions, the automated segmentations of muscle, VAT and SAT region match well with the manual label. Failures of the psoas segmentation occur on the slices taken at the bottom position, where the psoas muscle region is very small and difficult to detect (see Fig. 4, row 1, column 3).
Fig. 3.
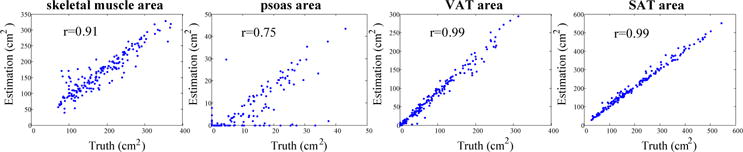
Scatter plots showing the correlation between estimated tissue area by the proposed method and the ground truth on 184 axial slices.
Fig. 4.
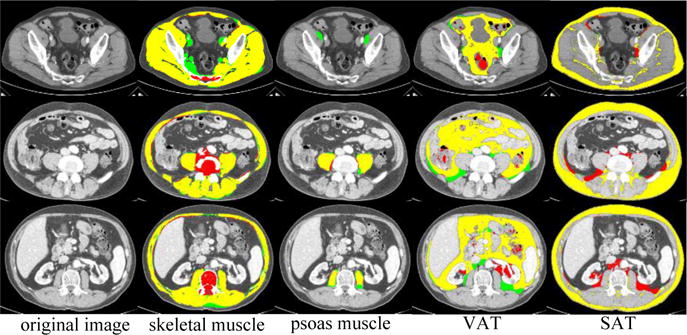
Qualitative results on different axial slices from one subject. Manual label (green), automated label (red) and overlap (yellow).
In Fig. 5, we used the Spearman rank correlation to compare the estimated tissue area by the proposed method with the ground truth on axial slices taken at L3 position from 40 testing scans. Average tissue areas of three and five axial slices centered at L3 are also measured.
Fig. 5.
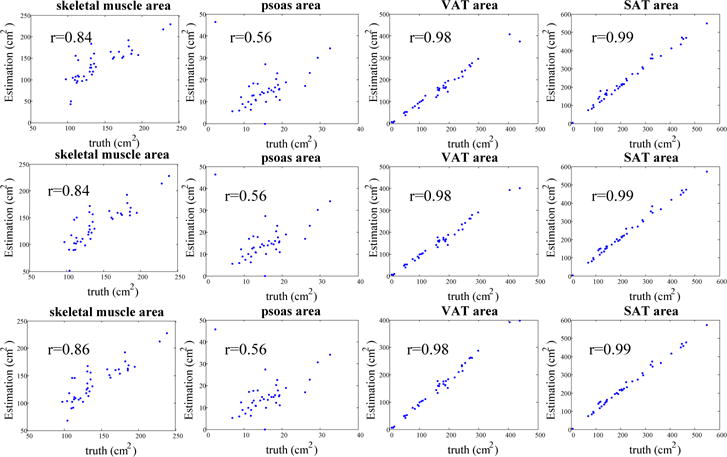
Scatter plots showing the correlation between estimated tissue area by the proposed method and the ground truth. The first row shows tissue area estimated on one slice at middle L3 position. The second and third row show the average tissue area estimated at three and five slices at the L3 position, respectively.
In Table 1, we compared our method with the baseline method [8] on 40 L3 axial slices from testing scans. Both the proposed method and baseline focused on whole CT scan segmentation and used slice-wise algorithms. As shown in Table 1, our method achieved significantly (p <0.001) higher Dice similarity index (Dice) values for the skeletal muscle, VAT and SAT using Wilcoxon signed rank test.
Table 1.
Comparison of tissue area estimation by the proposed method and Zhang et al. [8] on 40 L3 axial slices.
| Tissue | Skeletal Muscle |
Psoas Muscle |
VAT | SAT |
|---|---|---|---|---|
| Manual area (cm2) | 142.7±35.0 | 15.6±5.9 | 168.4±97.5 | 224.6±126.5 |
| Proposed Dice | 0.854±0.110 | 0.740±0.259 | 0.887±0.075 | 0.933±0.046 |
| Proposed area error (cm2) | 17.1±14.9 | 4.5±7.1 | 12.0±14.0 | 15.5±14.4 |
| Zhang [8] Dice | 0.758±0.128 | – | 0.828±0.054 | 0.852±0.054 |
| Zhang [8] area error (cm2) | 46.8±27.7 | – | 46.6±22.1 | 44.0±21.7 |
| Proposed v.s. Zhang [8] Dice p-value | 3.3×10−5 | – | 2.4×10−5 | 5.6×10−8 |
| Proposed v.s. Zhang [8] area error p-value | 3.8×10−6 | – | 7.6×10−7 | 1.6×10−6 |
4 Conclusions
We presented an automated workflow to segment the skeletal muscle, psoas muscle, visceral adipose tissue and subcutaneous adipose tissue from clinically acquired abdominal CT scans. The results on 40 subjects from two separate clinical datasets demonstrated that the proposed framework was able to achieve 0.854, 0.740, 0.887 and 0.933 Dice similarity coefficient for the skeletal muscle, psoas muscle, VAT and SAT, respectively. The future work can be the improvement of the proposed method by using shape prior in psoas muscle segmentation.
Acknowledgments
This research was supported by NIH 1R03EB012461, NIH 2R01EB006136, NIH R01EB006193, NIH P30 CA068485, NIH R01 HL 098445 (PI - Carr), and AUR GE Radiology Research Academic Fellowship, and in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by VISE/VICTR VR3029 and the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This research was supported in part by the National Natural Science Foundation of China (Grant No. 91630311) and the Fundamental Research Funds for the Central Universities (Grant No. 2017XZZX007-02).
Footnotes
References
- 1.Yip C, Dinkel C, Mahajan A, Siddique M, Cook GJ, Goh V. Imaging body composition in cancer patients: visceral obesity, sarcopenia and sarcopenic obesity may impact on clinical outcome. Insights into imaging. 2015;6:489–497. doi: 10.1007/s13244-015-0414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SY, Gallagher D. Assessment methods in human body composition. Current opinion in clinical nutrition and metabolic care. 2008;11:566–572. doi: 10.1097/MCO.0b013e32830b5f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popuri K, Cobzas D, Esfandiari N, Baracos V, Jagersand M. Body Composition Assessment in Axial CT Images Using FEM-Based Automatic Segmentation of Skeletal Muscle. IEEE transactions on medical imaging. 2016;35:512–520. doi: 10.1109/TMI.2015.2479252. [DOI] [PubMed] [Google Scholar]
- 4.Idoate F, Cadore EL, Casas-Herrero A, Zambom-Ferraresi F, Marcellán T, de Gordoa AR, Rodriguez-Mañas L, Bastarrika G, Marques MC, Martínez-Velilla N. Adipose tissue compartments, muscle mass, muscle fat infiltration, and coronary calcium in institutionalized frail nonagenarians. European radiology. 2015;25:2163–2175. doi: 10.1007/s00330-014-3555-5. [DOI] [PubMed] [Google Scholar]
- 5.Yao J, Sussman DL, Summers RM. SPIE Medical Imaging. Orlando, Florida: SPIE; 2011. Fully automated adipose tissue measurement on abdominal CT. 79651Z. [Google Scholar]
- 6.Inoue T, Kitamura Y, Li Y, Ito W, Ishikawa H. International MICCAI Workshop on Medical Computer Vision. Springer; 2015. Psoas major muscle segmentation using higher-order shape prior; pp. 116–124. [Google Scholar]
- 7.Chung H, Cobzas D, Birdsell L, Lieffers J, Baracos V. SPIE Medical Imaging. International Society for Optics and Photonics; 2009. Automated segmentation of muscle and adipose tissue on CT images for human body composition analysis. 72610K-72610K-72618. [Google Scholar]
- 8.Zhang W, Liu J, Yao J, Summers RM. SPIE Medical Imaging. International Society for Optics and Photonics; 2013. Segmenting the thoracic, abdominal and pelvic musculature on CT scans combining atlas-based model and active contour model. 867008-867008-867006. [Google Scholar]
- 9.Xu Z, Conrad BN, Baucom RB, Smith SA, Poulose BK, Landman BA. Abdomen and spinal cord segmentation with augmented active shape models. Journal of Medical Imaging. 2016;3:036002–036002. doi: 10.1117/1.JMI.3.3.036002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine. 2010;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. IEEE transactions on pattern analysis and machine intelligence. 2013;35:611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan J, Bae E, Tai XC. Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on. IEEE; 2010. A study on continuous max-flow and min-cut approaches; pp. 2217–2224. [Google Scholar]
- 13.Yuan J, Ukwatta E, Tai X, Fenster A, Schnoerr C. A fast global optimization-based approach to evolving contours with generic shape prior submission. IEEE TPAMI, also UCLA Tech Report CAM. 2012;12:38. [Google Scholar]


