Abstract
Background:
Artificial intelligence is advancing at an accelerated pace into clinical applications, providing opportunities for increased efficiency, improved accuracy, and cost savings through computer-aided diagnostics. Dermatopathology, with emphasis on pattern recognition, offers a unique opportunity for testing deep learning algorithms.
Aims:
This study aims to determine the accuracy of deep learning algorithms to diagnose three common dermatopathology diagnoses.
Methods:
Whole slide images (WSI) of previously diagnosed nodular basal cell carcinomas (BCCs), dermal nevi, and seborrheic keratoses were annotated for areas of distinct morphology. Unannotated WSIs, consisting of five distractor diagnoses of common neoplastic and inflammatory diagnoses, were included in each training set. A proprietary fully convolutional neural network was developed to train algorithms to classify test images as positive or negative relative to ground truth diagnosis.
Results:
Artificial intelligence system accurately classified 123/124 (99.45%) BCCs (nodular), 113/114 (99.4%) dermal nevi, and 123/123 (100%) seborrheic keratoses.
Conclusions:
Artificial intelligence using deep learning algorithms is a potential adjunct to diagnosis and may result in improved workflow efficiencies for dermatopathologists and laboratories.
Keywords: Artificial intelligence, computational pathology, computer-aided diagnosis, deep learning algorithm, dermatopathology, digital pathology, whole slide images
INTRODUCTION
Digital pathology systems were used in 32% of anatomic pathology laboratories in the United States in 2016 with another 7% of laboratories planning to add a system in 2017.[1] Advantages of digital pathology over traditional microscopy include improving efficiencies in educational training, tumor boards, research, frozen tissue diagnosis, permanent archiving, teleconsultation, access to published medical data, quantitative image analysis, quality assurance testing, establishment of biorepositories, and pharmaceutical research.[2,3,4,5,6,7,8,9] Acceptance of digital pathology for use in primary diagnosis was accelerated by validation of diagnosis of whole slide images (WSI) as equivalent to diagnosis of glass slides.[10,11,12,13,14,15,16] In April 2017, the US Food and Drug Administration (FDA) approved the first whole slide scanner system for use in primary diagnosis and other vendors are likely to follow.[17,18]
A digital workflow enhances opportunities for integration of diagnoses to a patient's electronic medical record, sharing of images between clinicians and patients, improved storage capabilities, and increased workflow efficiencies.[19,20]
Dermatopathology is ideally suited for the implementation of digital workflow and WSI for primary diagnosis. Specimens are small and the number of slides per case is low, although the workflow in academic centers, where the dermatopathology section is in the pathology department, may yield a different case mix and increased slide numbers related to large complex excisions and accompanied sentinel lymph node materials. Dermatopathology case volume is often high and while there is controversy among pathologists regarding workflow efficiency of the computer monitor or tablet versus the microscope, data suggest that with training and exposure, equivalence can be reached.[15,21,22,23,24]
Since dermatopathology is an image-intense subspecialty and low-power recognition is often an important first step for accurate diagnosis,[25,26] application of deep learning algorithms to cutaneous WSI allows for triage capabilities and computer-assisted diagnosis. Pathologists remain critical for rendering a final and accurate diagnosis.
Once digital pathology is utilized by a laboratory, there is a myriad of new software-driven opportunities that increase workflow efficiency and facilitate precision diagnoses through quantitative applications. These applications, enabled by artificial intelligence and deep learning, have shown to reduce error rates,[27] and increase efficiency in key disciplines of pathology. The evolution of artificial intelligence's role in laboratory medicine is a direct product of technological advances in hardware for image capture and massive computational advances in computer vision and machine learning.
The first indication of the benefit of computer-assisted diagnosis to laboratories was in 1996 with Hologic's ThinPrep system,[28] a revolutionary tool for cytopathology laboratories. Anatomic pathology laboratories are currently seeing error reduction and increased efficiency with quantitative immunohistochemistry, which utilizes more primitive computer vision techniques than deep learning, to quantify protein expression.[29] For example, the FDA approved automated detection, counting, and computer-generated analysis of HER2 gene for therapeutic determination in breast cancer.[30,31] Computer-aided counting of mitoses in breast cancer WSI represents another step on the artificial intelligence continuum.[32]
Dermatologists are also realizing the value of artificial intelligence and deep learning and have used algorithms to aid in diagnosis of clinical images.[33]
The combined effect of the introduction of deep learning, the democratization of powerful graphics processing unit (GPU) computing capacity, and increasing acceptance and use of digital pathology has created an unprecedented opportunity to explore the power of deep learning. Coupled with the acknowledged premise that fundamental dermatopathological diagnoses often rely on low power and pattern recognition,[25,26] we hypothesized that dermatopathology was an ideal starting point for the development of deep learning algorithms for case triage and computer-aided diagnosis.
In this study, we utilize expertly annotated “ground truth” WSI and the clinical and medical expertise of dermatopathologists in combination with the experience of an artificial intelligence team, to develop computer software that teaches itself the ability to make binary classifications as to whether test WSI represents ground truth diagnosis or not. Initial results from algorithmic testing of three common dermatopathology diagnoses, nodular basal cell carcinoma (BCC), dermal nevus, and seborrheic keratosis, indicate proof of concept and is the framework to allow additional development of other relevant dermatopathologic predictive models.
METHODS
Before the study, Institutional Review Board (IRB) approval was sought from and exempted by the IRB at Boonshoft School of Medicine, Wright State University, Dayton, Ohio (IRB # 06194). All hematoxylin and eosin-stained glass slides used in this study were de-identified of patient information before scanning into WSI and stored in our biorepository.
Data collection and annotation
Algorithmic development and testing were partitioned into three sequential studies, to include training and testing of each individual algorithm, and allow for assessment and correction of study design elements. Table 1 shows the total number and diagnoses of WSI used for training and testing the algorithms [Table 1].
Table 1.
Number and diagnoses of whole slide images used in algorithm training and testing
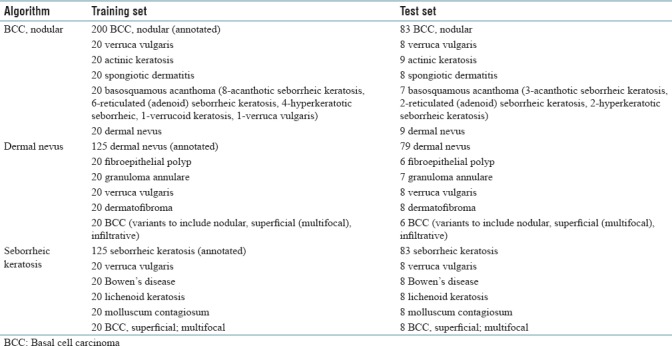
In the first study, a training set of 200 previously diagnosed nodular BCC glass slides and 100 distractor glass slides were deidentified and scanned into WSI at ×40 magnification (Aperio AT2 Scanscope Console, Leica Biosystems). The distractor set for nodular BCC consisted of 20 slides each of verrucae vulgares, actinic keratoses, spongiotic dermatitis, dermal nevi, and basosquamous acanthomas. (The term basosquamous acanthoma reflects different types of seborrheic keratoses, verrucoid keratoses, and verrucae and is an epithelial hyperplasia/hypertrophy that histologically manifests a thickening of the spinous layer to include papillations and acanthosis and a variable scale/crust and inflammation). These five distractor diagnoses were selected to include common neoplasms and an inflammatory condition.
In the second study, the training set was reduced to 125 previously diagnosed dermal nevi glass slides and 100 distractor glass slides, 20 slides each of fibroepithelial polyps, granuloma annulare, verrucae vulgares, dermatofibromas, and variants of BCCs. All deidentified glass slides were scanned into WSI at ×40 magnification.
In the third study, 125 previously diagnosed seborrheic keratoses glass slides and 100 distractor glass slides, 20 slides each of superficial BCCs, molluscum contagiosum, lichenoid keratoses, Bowen's disease, and verrucae vulgares, were deidentified and scanned into WSI at ×40 magnification.
All WSI were uploaded to the Proscia Pathology Cloud into separately labeled folders. Positive training examples were then annotated according to criteria as determined by the dermatopathologists and computer scientists. Figure 1 shows multiple sections of annotated nodular BCC [Figure 1].
Figure 1.
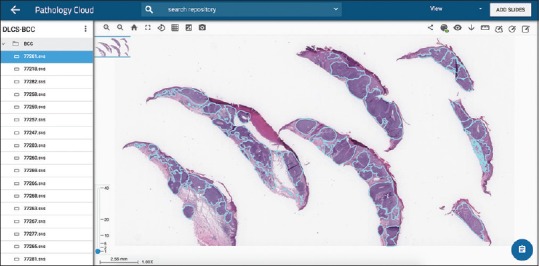
Nodular basal cell carcinoma. Multiple H and E-stained sections annotated for artificial intelligence training phase (×1 magnification)
For each algorithm, randomly selected new test sets of hematoxylin and eosin stained previously diagnosed slides, as shown in Table 1, consisting of approximately 70% lesion of interest and 30% distractors, were de-identified, scanned at ×40 magnification into WSI, and uploaded into the Proscia Pathology Cloud.
Algorithm development and training
For each individual dermatological diagnosis, corresponding positive and negative image sets were preprocessed and decomposed to extract features relevant to the differential diagnosis. Each WSI was parsed in hierarchical fashion to access regions of the slide at different magnification levels.[34] WSI, containing available resolutions from ×1 to ×40 magnification, contained on average over 70% unnecessary background space which slowed down image processing; therefore, tissue samples on WSI were detected and isolated. The WSI was then decomposed into many smaller tiles representing square regions of tissue. Each individual pixel within the tile was assigned a corresponding label, and a binary mask was created to indicate delineation of normal and neoplastic or inflammatory tissue, according to the expert annotations. This process was performed in parallel to generate a training set containing image data and labeled annotations from 200 nodular BCCs and 125, respectively, for dermal nevi and seborrheic keratoses. Distractors had no pixels with corresponding positive labels and image data was tiled and produced a binary mask where all pixels were homogeneously labeled.
Once data collection was complete, we used custom developed versions of fully convolutional neural networks (CNNs) to identify high probability regions of each dermatological lesion. CNNs are trained end-to-end, pixels-to-pixels, and represent a breakthrough in deep learning-based semantic segmentation. This technology adapts the contemporary classification networks into fully convolutional networks and transfers their learned representations by fine tuning to the segmentation task.[35] The CNN employed for this study is a derivative of a typical Visual Geometry Group (VGG) network architecture introduced by Simonyan and Zisserman.[36] A VGG network consists of multiple layers of neural nodes which, with each succeeding layer, encodes a progressively higher level pattern. The first layer helps in extracting information relevant to the edges and gradients present in the WSI tile. The second layer feeds on this information and outputs feature maps which represent patterns formed by the connections between the edges and gradients. Further, the third layer extracts objects formed by these patterns. The final few layers translate these maps into a set of feature maps which, once optimized, provide the maximum distinction between the diagnoses. Our optimization was performed using stochastic gradient descent with momentum and regularized the model with a convergent loss function to ensure prevention of overfitting.[37]
Algorithm testing
For double-blind testing, a WSI with no pixel labels was deconstructed into tiles of the same size as the training exemplars, and each tile assigned a probability by the semantic segmentation model, which involves assigning one semantic class to each pixel of the input image. CNNs were used to model the class likelihood of pixels directly from either the WSI or the image patches. The WSI was reconstructed with a third dimension class likelihood which is a value on a uniform probability density function representing likelihood of that pixel belonging to the given lesion. This third dimension is visualized by generating a heat map where each pixel is assigned a value on the probability continuum and assigned a corresponding red, green, and blue color value to represent that class likelihood.
The resulting heat map [Figure 2] is a representation of the model's ability to distinguish between benign and neoplastic or inflammatory cells within a given WSI. For example, visually investigative results indicate that the traditional basaloid nesting pattern in the dermis is continually identified on images where BCC is present. As an additional regularization technique, we implemented a rule-based discrimination system to eliminate false positives by identifying each contiguous region or nest of the lesion identified in any image and generated a feature vector representing size, shape, and texture of that region. These independent variables were used to train a classification model to determine if each individual region was the lesion of interest or not and we used this classification system as the final arbiter of truth [Figure 2].
Figure 2.
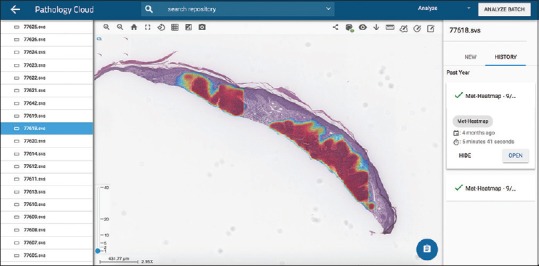
Nodular basal cell carcinoma. Probability heat map visualization generated by deep learning model indicates high probability regions of nodular basal cell carcinoma (×3 magnification)
Slide level evaluation was a composition of all individual patterns available, identified by the postprocessing system on the slide, and the final label indicating the presence of the lesion or not on a given WSI. Each slide output was compared against established ground truth diagnosis and results evaluated to determine model performance.
RESULTS
Timing profile
Our models were implemented using Caffe[38] on a machine equipped with a 32-Core processor, 60GB RAM, and four NVIDIA K520 GPUs. For each algorithm, the training and validation was conducted over a period of 4 days. During the testing phase, we used the trained algorithm to predict the probability of the presence of the target pathology in the input tiles. The average compute time for the testing phase was found to be 40 s/image.
Receiver operating characteristics analysis
The classifying model was examined with different probability values (0.0–1.0) as threshold. This analysis is illustrated in Figures 3–5, by receiver operating characteristics curves for nodular BCC, dermal nevus, and seborrheic keratosis, respectively. The area under the curve correlates with the detection accuracy. The optimal threshold value was recognized to provide the best sensitivity and specificity [Figures 3–5].
Figure 3.
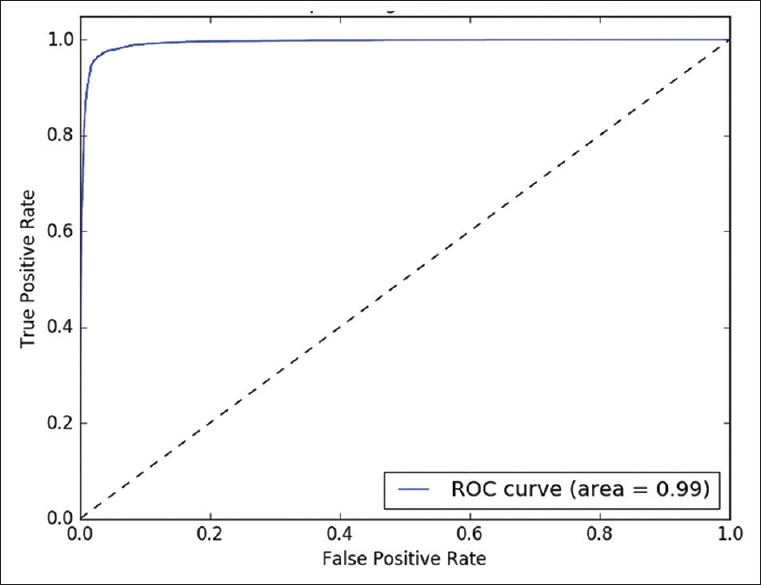
Nodular basal cell carcinoma. Receiver operating characteristics curve for binary detection
Figure 5.
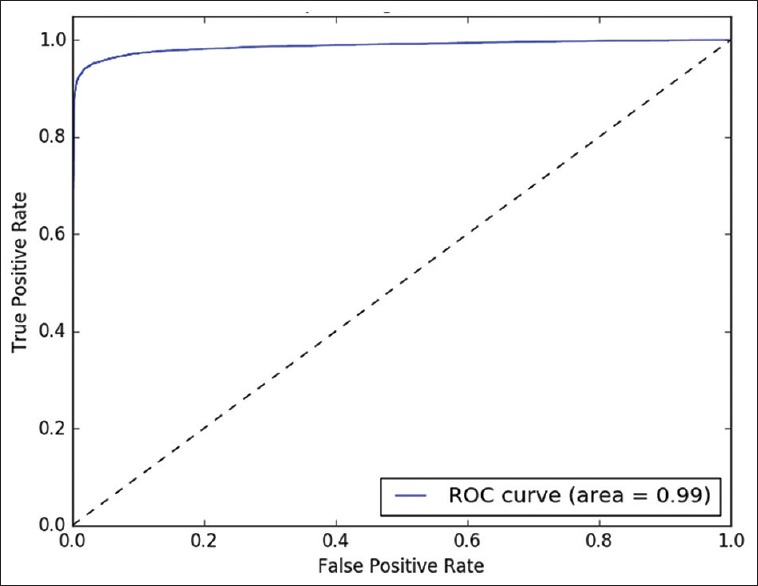
Seborrheic keratosis. Receiver operating characteristics curve for binary detection
Figure 4.
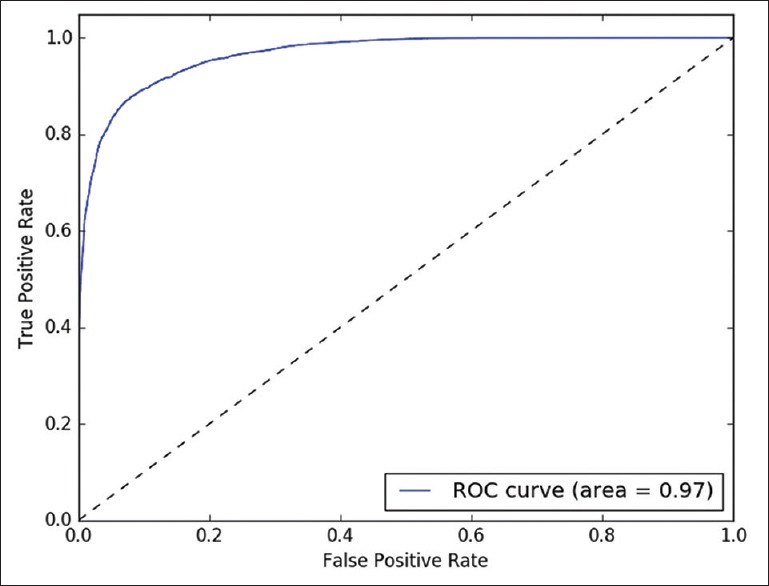
Dermal nevus. Receiver operating characteristics curve for binary detection
Sensitivity and specificity analysis
The computer algorithm for nodular BCC correctly classified 123 of 124 test WSI (99.45% balanced accuracy) as nodular BCC or not. The computer algorithm for dermal nevi also yielded a high percentage of accurate results as it correctly classified 113 of 114 test WSI (99.4% balanced accuracy) as dermal nevus or not. The computer algorithm correctly classified 100% of the test WSI as seborrheic keratosis or not on all 123 cases. Table 2 shows the results of the binary classification by diagnosis [Table 2].
Table 2.
Results of algorithm testing
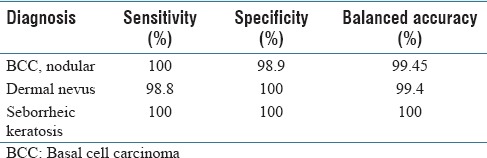
Balanced accuracy is the average of the sensitivity and specificity testing. Sensitivity testing includes only the results obtained from testing on the target pathology. For example, to validate the sensitivity of the nodular BCC detection model, a dataset of 83 randomly selected nodular BCC images was subjected to prediction by the algorithm. All of the 83 images were correctly labeled as nodular BCC by the algorithm, exhibiting a sensitivity of 100% on this dataset.
Specificity testing describes the testing conducted with distractors. For example, the specificity of the nodular BCC algorithm was tested with 41 randomly chosen images from different pathologies that are visually similar or commonly found in the laboratory workflow. Of the 41 distractor images, 40 were accurately labeled as non-BCC by the algorithm, resulting in a 98.9% specificity. Thus, the balanced accuracy for the nodular BCC algorithm was 99.45%.
DISCUSSION
Artificial intelligence, and specifically deep learning, is now being applied to the fields of clinical dermatology and pathology.[39,40,41,42,43,44,45,46,47,48] Both specialties are image intense, rely on the integration of visual skills for diagnosis, and are a natural segue for algorithmic development. Esteva et al. trained a CNN dataset, consisting of approximately 129,000 clinical images and 2000 diseases, to recognize nonmelanoma skin cancer versus benign seborrheic keratosis, and melanoma versus benign nevi, equal to the clinical diagnostic impressions of 21 board-certified dermatologists. The authors project their results to a practical application of incorporating deep neural networks into clinician's mobile devices to yield diagnoses beyond the confines of the office/clinic.[33]
Bejnordi et al. tested the performance of seven deep learning algorithms to detect lymph node metastasis in women with breast cancer against a panel of 11 participating pathologists with results reflecting equivalence of the algorithm versus the pathologists for the WSI classification task.[46]
Dermatopathology is an image-intense specialty and our WSI biorepository provided an optimum source for development and testing of this technology. We investigated application to three common diagnoses in skin pathology, representing approximately 20% of specimens diagnosed in our dermatopathology laboratory; nodular BCC, dermal nevus, and seborrheic keratosis.
Algorithmic diagnosis of nodular BCC was successful in 123 of 124 cases with a sensitivity and specificity of 100% and 98.9%, respectively. The single false positive was a dermal nevus incorrectly classified as nodular BCC. Although speculative as to the algorithm's misclassification of this WSI, the specimen reflected, particularly in the papillary and upper reticular dermis, nested aggregates of nevocytes with similarity in size to nests of BCC. Nevocytes were more diffusely arranged in the deeper reticular dermis.
Dermal nevi also reflected a high degree of accuracy with 113 out of 114 cases being correctly diagnosed by the algorithm with sensitivity of 98.8% and specificity of 100%. The one false-negative WSI [Figure 6] was a dermal nevocellular nevus and although unknown why this was misclassified by the algorithm, the dermatopathologist noted the specimen having a diffuse arrangement of nevocytes rather than a combination of nests and diffuse arrangements, which are seen in many dermal nevi [Figure 6].
Figure 6.
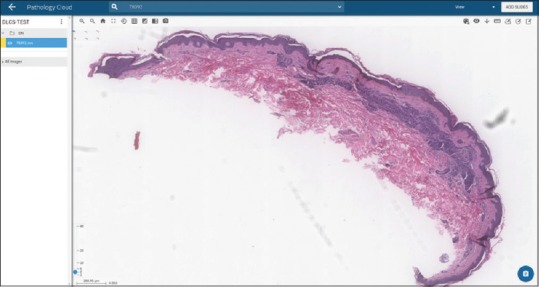
Dermal nevus. H and E-stained whole slide images not identified by algorithm (false negative) (×2 magnification)
Seborrheic keratoses were the highest yield with the algorithm making the diagnosis in 123 out of 123 cases. This was particularly significant since the seborrheic keratosis training and test set was more rigorous and included a blend of acanthotic (72), reticulated (24), hyperkeratotic (12), and other variants (17) of seborrheic keratoses.
The distractor diagnoses for each particular lesion were variably rigorous yet followed machine learning sequential training, starting with more obvious distractors and moving to the more difficult ones, respectively. In addition, the distractors selected mirrored the more typical diagnoses in a dermpath laboratory workflow. Thus, distractors for nodular BCC only included one dermal process, and namely, a dermal nevus. The other four distractors were epithelial proliferations and dermatitis (verruca, actinic keratosis, basosquamous acanthoma, and spongiotic dermatitis). No other basaloid neoplasms, such as trichoepithelioma, spiradenoma, nodular hidradenoma, or Merkel cell carcinoma and lymphoma, were included for this proof of concept study.
Distractors for dermal nevi included four out of five dermal processes and/or neoplasms. Fibroepithelial polyps were included because of their polypoid morphology, similar in silhouette morphology to many dermal nevi, yet an absence of nevocytes in the stroma. Considerations for more rigorous distractors in the training process might include syringomas, mastocytomas, Spitz nevi, and potentially, a nevoid melanoma.
Distractors for seborrheic keratosis all reflected varying involvement of the epithelium and significantly, a superficial (multifocal) variant of BCC. Since seborrheic keratoses represent a proliferation of an intermediate keratinocyte, particularly in the acanthotic variant of seborrheic keratosis, the low-power impression can mimic the basaloid proliferation of superficial (multifocal) BCC. The rigor of this grouping of distractors was at a different level than the nodular BCC and dermal nevi.
Future data collection to enhance rigor of the algorithms, utilizing more simulant distractors similar to a dermatopathologist deciding between two or more lesions, will be critical.
Our workflow did not involve an explicit stain normalization step but fine-tuning models with datasets developed using different staining conventions is expected to produce similar results. We leverage the fact that deep learning models, when trained with datasets that encompass all the variations introduced by scanners and staining conventions, tend to base their decisions on features or patterns that explicitly define the diagnosis, thereby eliminating any bias.
A significant limitation of our study was that the statistical analysis was a binary classification system. Organization of the study follows typical medical image analysis machine learning protocol,[49] with a train-test split showing generalizability of the model and performance on out-of-sample observations. Development of a multivariate classification system integrating the individual algorithms into a holistic system would enhance the future studies.
CONCLUSIONS
Results from this study provide proof of concept that can serve as a framework for refinement and expansion of algorithmic development for common diagnoses in a dermatopathology laboratory. Exposing the algorithmic library to a digital workflow of all dermatopathology specimens is essential to collection of meaningful results that allow build out and refinement of these and further algorithms. With rapid advancements occurring in both medicine and technology, clinicians, and computer scientists must work collaboratively to take full advantage of the unprecedented opportunities that are transforming medicine.[50] Integration of refined and fully developed computer algorithms into digital pathology workflow could facilitate efficient triaging of cases and aid in diagnosis, resulting in significant cost savings to the health-care system.
Financial support and sponsorship
DLCS-Clearpath LLC and Proscia Inc. provided funding support for this study.
Conflicts of interest
Dr. Thomas Olsen is an owner at DLCS-Clearpath, LLC.
Acknowledgments
The authors wish to acknowledge the following individuals: Joel Crockett, MD, for expertise in annotation of images; Michael A. Bottomley, MS, Statistical Programmer Analyst, Statistical Consulting Center, Department of Mathematics and Statistics, Wright State University, Dayton, Ohio, for review of data results; and Julianna Ianni, Ph.D., from Proscia, Inc. for review and edits to the final manuscript.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2018/9/1/32/242369
REFERENCES
- 1.Klipp J. Poughkeepsie NY: Laboratory Economics; 2017. The US Anatomic Pathology Market: Forecasts and Trends 2017-2020; pp. 79–80. [Google Scholar]
- 2.Zembowicz A, Ahmad A, Lyle SR. A comprehensive analysis of a web-based dermatopathology second opinion consultation practice. Arch Pathol Lab Med. 2011;135:379–83. doi: 10.5858/2010-0187-OA.1. [DOI] [PubMed] [Google Scholar]
- 3.Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36–45. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen PS, Riber-Hansen R, Raundahl J, Steiniche T. Automated quantification of MART1-verified Ki67 indices by digital image analysis in melanocytic lesions. Arch Pathol Lab Med. 2012;136:627–34. doi: 10.5858/arpa.2011-0360-OA. [DOI] [PubMed] [Google Scholar]
- 5.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: Current status and future perspectives. Histopathology. 2012;61:1–9. doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 6.Al Habeeb A, Evans A, Ghazarian D. Virtual microscopy using whole-slide imaging as an enabler for teledermatopathology: A paired consultant validation study. J Pathol Inform. 2012;3:2–6. doi: 10.4103/2153-3539.93399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero Lauro G, Cable W, Lesniak A, Tseytlin E, McHugh J, Parwani A, et al. Digital pathology consultations-a new era in digital imaging, challenges and practical applications. J Digit Imaging. 2013;26:668–77. doi: 10.1007/s10278-013-9572-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei BR, Simpson RM. Digital pathology and image analysis augment biospecimen annotation and biobank quality assurance harmonization. Clin Biochem. 2014;47:274–9. doi: 10.1016/j.clinbiochem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekins S. The next era: Deep learning in pharmaceutical research. Pharm Res. 2016;33:2594–603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooney E, Hood AF, Lampros J, Kempf W, Jemec GB. Comparative diagnostic accuracy in virtual dermatopathology. Skin Res Technol. 2011;17:251–5. doi: 10.1111/j.1600-0846.2010.00493.x. [DOI] [PubMed] [Google Scholar]
- 11.Al-Janabi S, Huisman A, Vink A, Leguit RJ, Offerhaus GJ, Ten Kate FJ, et al. Whole slide images for primary diagnostics in dermatopathology: A feasibility study. J Clin Pathol. 2012;65:152–8. doi: 10.1136/jclinpath-2011-200277. [DOI] [PubMed] [Google Scholar]
- 12.Bauer TW, Schoenfield L, Slaw RJ, Yerian L, Sun Z, Henricks WH, et al. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch Pathol Lab Med. 2013;137:518–24. doi: 10.5858/arpa.2011-0678-OA. [DOI] [PubMed] [Google Scholar]
- 13.Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: A systematic review. Arch Pathol Lab Med. 2017;141:151–61. doi: 10.5858/arpa.2016-0025-RA. [DOI] [PubMed] [Google Scholar]
- 14.Shah KK, Lehman JS, Gibson LE, Lohse CM, Comfere NI, Wieland CN, et al. Validation of diagnostic accuracy with whole-slide imaging compared with glass slide review in dermatopathology. J Am Acad Dermatol. 2016;75:1229–37. doi: 10.1016/j.jaad.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 15.Kent MN, Olsen TG, Feeser TA, Tesno KC, Moad JC, Conroy MP, et al. Diagnostic accuracy of virtual pathology vs. traditional microscopy in a large dermatopathology study. JAMA Dermatol. 2017;153:1285–91. doi: 10.1001/jamadermatol.2017.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JJ, Jedrych J, Pantanowitz L, Ho J. Validation of digital pathology for primary histopathological diagnosis of routine, inflammatory dermatopathology cases. Am J Dermatopathol. 2018;40:17–23. doi: 10.1097/DAD.0000000000000888. [DOI] [PubMed] [Google Scholar]
- 17.FDA Allows Marketing of First Whole Slide Imaging System for Digital Pathology. Food and Drug Administration. 2017. Apr 12, [Last accessed on 2018 Apr 30]. Available from: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm552742.htm .
- 18.Whole Slide Imaging for Primary Diagnosis: Now it is Happening. CAP Today. 2017. May, [Last accessed on 2018 Apr 30]. Available from: http://www.captodayonline.com/whole-slide-imaging-primary-diagnosis-now-happening/
- 19.Kent MN. Abstract presented at: Pathology Visions 2016. San Diego, CA: 2016. Oct 24, Practical Implementation of Digital Pathology in Dermatopathology Practice. [Google Scholar]
- 20.Kent MN. Detroit, MI: 2017. Dec 8-9, Deploying a Digital Pathology Solution in a Dermatopathology Reference Lab. Abstract presented at: Association for Pathology Informatics Digital Pathology Workshop, Henry Ford Hospital. [Google Scholar]
- 21.Nielsen PS, Lindebjerg J, Rasmussen J, Starklint H, Waldstrøm M, Nielsen B, et al. Virtual microscopy: An evaluation of its validity and diagnostic performance in routine histologic diagnosis of skin tumors. Hum Pathol. 2010;41:1770–6. doi: 10.1016/j.humpath.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Randell R, Ruddle RA, Mello-Thoms C, Thomas RG, Quirke P, Treanor D, et al. Virtual reality microscope versus conventional microscope regarding time to diagnosis: An experimental study. Histopathology. 2013;62:351–8. doi: 10.1111/j.1365-2559.2012.04323.x. [DOI] [PubMed] [Google Scholar]
- 23.Thorstenson S, Molin J, Lundström C. Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: Digital pathology experiences 2006-2013. J Pathol Inform. 2014;5:14. doi: 10.4103/2153-3539.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vodovnik A. Diagnostic time in digital pathology: A comparative study on 400 cases. J Pathol Inform. 2016;7:4–8. doi: 10.4103/2153-3539.175377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackerman AB. Philadelphia, PA: Lea & Febinger; 1978. Histologic Diagnosis of Inflammatory Skin Diseases: A Method Based on Pattern Analysis. [Google Scholar]
- 26.Ackerman AB. 3rd ed. Baltimore, MD: Williams & Wilkins; 1997. Histologic Diagnosis of Inflammatory Skin Diseases: An Algorithmic Method Based on Pattern Analysis. [Google Scholar]
- 27.Zhang X, Zou J, He K, Sun J. Accelerating very deep convolutional networks for classification and detection. IEEE Trans Pattern Anal Mach Intell. 2016;38:1943–55. doi: 10.1109/TPAMI.2015.2502579. [DOI] [PubMed] [Google Scholar]
- 28.Chivukula M, Saad RS, Elishaev E, White S, Mauser N, Dabbs DJ, et al. Introduction of the thin prep imaging system (TIS): Experience in a high volume academic practice. Cytojournal. 2007;4:6. doi: 10.1186/1742-6413-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Food and Drug Administration. US Department of Health and Human Services 510(k) summary k061602. 2007. Jan 23, [Last accessed on 2018 Apr 30]. Available from: http://www.accessdata.fda.gov/cdrh_docs/pdf6/k061602.pdf .
- 31.Food and Drug Administration. US Department of Health and Human Services 510(k) Summary k080909. [Last accessed on 2018 Apr 30]. Available from: http://www.accessdata.fda.gov/cdrh_docs/pdf8/k080909.pdf .
- 32.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–48. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M. OpenSlide: A vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:640–51. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 36.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition.arXiv preprint arXiv: 1409.1556. 2014 [Google Scholar]
- 37.Ashiquzzaman A, Tushar AK, Islam R, Kim JM. Reduction of overfitting in diabetes prediction using deep learning neural network. In: Kim K, Kim H, Baek N, editors. IT Convergence and Security 2017. Lecture Notes in Electrical Engineering. Singapore: Springer; 2018. [Google Scholar]
- 38.Yangqing J, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, et al. Caffe: Convolutional architecture for fast feature embedding. Proceedings of the 22nd ACM International Conference on Multimedia. ACM. 2014 [Google Scholar]
- 39.Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, González Osorio FA. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med Image Comput Comput Assist Interv. 2013;16:403–10. doi: 10.1007/978-3-642-40763-5_50. [DOI] [PubMed] [Google Scholar]
- 40.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016;33:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Song Y, Xia F, Zhu C, Zhang Y, Song W, et al. Rapid and accurate intraoperative pathological diagnosis by artificial intelligence with deep learning technology. Med Hypotheses. 2017;107:98–9. doi: 10.1016/j.mehy.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Radiya-Dixit E, Zhu D, Beck AH. Automated classification of benign and malignant proliferative breast lesions. Sci Rep. 2017;7:9900. doi: 10.1038/s41598-017-10324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, et al. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18:281. doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, et al. Deep learning for classification of colorectal polyps on whole-slide images. J Pathol Inform. 2017;8:30. doi: 10.4103/jpi.jpi_34_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller DD, Brown EW. Artificial intelligence in medical practice: The question to the answer? Am J Med. 2018;131:129–33. doi: 10.1016/j.amjmed.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 48.Golden JA. Deep learning algorithms for detection of lymph node metastases from breast cancer: Helping artificial intelligence be seen. JAMA. 2017;318:2184–6. doi: 10.1001/jama.2017.14580. [DOI] [PubMed] [Google Scholar]
- 49.Wei Q, Dunbrack RL Jr. The role of balanced training and testing data sets for binary classifiers in bioinformatics. PLoS One. 2013;8:e67863. doi: 10.1371/journal.pone.0067863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudin RS, Bates DW, MacRae C. Accelerating innovation in health IT. N Engl J Med. 2016;375:815–7. doi: 10.1056/NEJMp1606884. [DOI] [PMC free article] [PubMed] [Google Scholar]


