Abstract
Accurate segmentation of hippocampus from infant magnetic resonance (MR) images is very important in the study of early brain development and neurological disorder. Recently, multi-atlas patch-based label fusion methods have shown a great success in segmenting anatomical structures from medical images. However, the dramatic appearance change from birth to 1-year-old and the poor image contrast make the existing label fusion methods less competitive to handle infant brain images. To alleviate these difficulties, we propose a novel multi-atlas and multi-modal label fusion method, which can unanimously label for all voxels by propagating the anatomical labels on a hypergraph. Specifically, we consider not only all voxels within the target image but also voxels across the atlas images as the vertexes in the hypergraph. Each hyperedge encodes a high-order correlation, among a set of vertexes, in different perspectives which incorporate 1) feature affinity within the multi-modal feature space, 2) spatial coherence within target image, and 3) population heuristics from multiple atlases. In addition, our label fusion method further allows those reliable voxels to supervise the label estimation on other difficult-to-label voxels, based on the established hyperedges, until all the target image voxels reach the unanimous labeling result. We evaluate our proposed label fusion method in segmenting hippocampus from T1 and T2 weighted MR images acquired from at 2-week-old, 3-month-old, 6-month-old, 9-month-old, and 12-month-old. Our segmentation results achieves improvement of labeling accuracy over the conventional state-of-the-art label fusion methods, which shows a great potential to facilitate the early infant brain studies.
1 Introduction
The human brain undergoes a rapid physical growth and fast functional development during the first year of life. In order to characterize such dynamic changes in vivo, accurate segmentation of anatomical structures from the MR images is very important in imaging-based brain development studies. Since hippocampus plays an important role in learning and memory function, many studies aim to find the imaging markers around hippocampus. Unfortunately, the poor image contrast and dramatic appearance change make the segmentation of hippocampus from the infant images in the first year of life very challenging, due to the dynamic white matter myelination progress [1]. Since different modalities convey diverse imaging characteristics at different brain development phases, integration of multi-modal imaging information can significantly improve the segmentation accuracy[2]. However, the modulation of each modality is hard-coded and subjective to the expert’s experience.
In medical image analysis area, there has been a recent spike in multi-atlas patch based segmentation methods [3–5], which first register all atlas images to the underlying target image and then propagate the labels from the atlas domain to the target image. The assumption behind is that two voxels should bear the same anatomical label if their local appearances are similar. However, a critical issue in current multi-atlas patch based label fusion methods is that the labels are determined separately at each voxel. As a result, there is no guarantee that labeled anatomical structures are spatially consistent. On the other hand, conventional graph-cut based methods [6] can jointly segment the entire ROI by finding a minimum graph cut. However, only the image information within the target image is utilized during this segmentation.
To combine the power of multi-atlas and graph-based approaches, we propose a novel multi-atlas and multi-modal label fusion method for segmenting the hippocampus from the infant brain images in the first year of life. Hypergraph has shown its superiority in image retrieval [7, 8] and recognition [9]. Our idea is to use hypergraph to leverage (1) the information integration from multiple atlases and multiple imaging modalities, and (2) the label fusion for all target image voxels under consideration.
Specifically, we regard all voxels within the target image and the atlas images as the vertexes in the graph. In hypergraph, we generalize the concept of conventional graph edge (only connects two vertexes at a time) to the hyperedge (groups a set of vertexes simultaneously), in order to reveal the high-order correlations for more than two voxels. In general, our hyperedges in the multi-atlas scenario encode three types of voxels correlations: (1) feature affinity: only the vertexes with similar appearance are connected by hyperedge, (2) spatial coherence: the vertexes located in a certain neighborhood from target image belong to the same hyperedge, and (3) atlas correspondence: each vertex from target image and its corresponding vertexes across all atlas images form the hyperedge. It is worth noting that the hypergraph is very flexible to incorporate the multi-modal information by constructing the above three types of hyperedges w.r.t. different imaging modalities. After constructing the hypergraph, the vertexes from both the atlas image voxels and the target image voxels with high labeling confidence are considered bearing the known labels. Thus, our label fusion method falls into the semi-supervised hypergraph learning framework, i.e., the latent labels on the remaining voxels in the target image are influenced by the connected vertexes with known labels. The principle to propagate the labels is that the vertexes sitting in the same hyperedge should have the same label, with the minimal discrepancy of labels on the vertexes with existing labels before and after label propagation.
Our proposed method has been comprehensively evaluated on segmenting hippocampus from T1 and T2 weighted MR images at 2-week-old, 3-month-old, 6-month-old, 9-month-old and 12-month-old. The segmentation result shows a great improvement compared to the state-of-the-art method [3–5] in terms of labeling accuracy.
2 Method
Given the atlases (including T1, T2 weighted MR images and the ground truth label of hippocampus), the goal is to segment the hippocampus from the unlabeled target image. Since the T1 and T2 weighted images are acquired from the same subject at the same time, it is not difficult to register them to the same space. Due to the dynamic appearance change and poor image resolution, it is challenging to apply deformable image registration for the infant images in the first year of life. Thus, only linear registration is used to map each atlas to the underlying target image domain. After that, our label fusion method consists of two main steps: 1) hypergraph construction (Section 2.1) and 2) label propagation (Section 2.2), as detailed below.
2.1 Hypergraph Construction
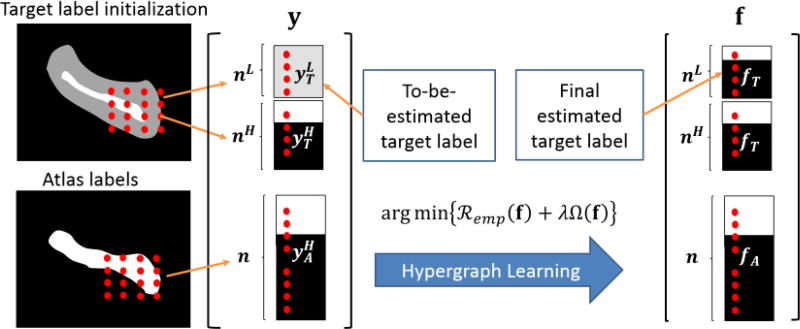
We assume that N atlas images, each with T1 and T2 weighted MR images, are used to label the target image. The hypergraph is constructed to accommodate the complete information from both the multi-atlas and multi-modalities images, where the hypergraph is denoted as 𝒢 = (𝒱, ℰ, W) with the vertex set 𝒱, the hyperedge set ℰ and its weight W. In order for computation efficiency, only the voxels within a bounding box (shown in the left column of Fig. 1 which covers the hippocampus with enough margin) are used as the vertexes, instead of using the voxels from the entire image.
Fig. 1.
The framework of hypergraph learning based label propagation.
Hypergraph Initialization
Assume the number of voxels inside the bounding box is q. The hypergraph vertexes are built on every voxel from the bounding boxes of the target image and the atlas images. For each atlas image, we vectorize the voxels in the bounding box into a column vector 𝒱s with the length of q, where s = 1, …, N. With the same vectorising order, we can obtain the column vector of graph vertexes from target image, denoted by 𝒰. Thus, the vertex set 𝒱 is eventually the combination of voxels from the target image and all atlas images, i.e., 𝒱 = {𝒰, 𝒱1, …, 𝒱N}. It is worth noting that each element in 𝒱s bears the known label and the label on each element of 𝒰 is currently unknown.
It is clear that the voxels along the interface of multiple structures are more difficult to determine the labels than other points. In light of this, we go one step further to classify the elements in 𝒰 into two categories based on their difficulties in labeling. To achieve it, we use an existing label fusion method, such as the majority voting, to predict the label of each element in 𝒰 as well as the confidence value in terms of the voting predominance. If the influence for voting one label dominates the other labels, we regard the labeling result on the underlying target image voxel has high confidence to determine its label. Otherwise, we regard the underlying target image voxel needs other heuristics to find its label. Thus, we can divide 𝒰 into two parts, 𝒰L (low confidence) and 𝒰H (high confidence), where the number of elements in 𝒰L and 𝒰H is qL and qH (q = qL + qH), respectively.
The advantage of separate 𝒰 to 𝒰L and 𝒰H is that it allows the label propagation from not only the atlas images but also the some reliable regions of the target image, which are more specific to label fusion of underlying image. Specifically, for each element in 𝒰H and {𝒱s|s = 1, …, N}, we assign the label with either 0 (background) or 1 (hippocampus). For each element in 𝒰L, the label value is assigned to 0.5 since the label is uncertain. Following the same order of 𝒱, we stack all the label values into a label vector y. As shown in the left bracket in Fig. 1, the first part of y is the uncertain labels for voxels 𝒰L (in gray), followed by the known labels from 𝒰H and {𝒱s|s = 1, …, N} (hippocampus in white and background in black). Thus, our hypergraph based label fusion turns into a semi-supervised learning scenario, i.e., optimize for a new label vector f (in the right of Fig. 1) which should be 1) as close to y as possible and 2) propagate the known labels from 𝒰H and {𝒱s|s = 1, …, N} to the difficult-to-label vertexes 𝒰L. The leverage of label propagation is a set of hyperedges, as detailed next.
Hyperedge Construction
Since the goal is to estimate the latent labels for the difficult-to-label voxels in the target image, we only construct hyperedges centered at each vertex u ∈ 𝒰L in the low confidence voxel set. In total, we constructed 2 × 3 types of hyperedges w.r.t. two imaging modalities and three measurements. Specifically, for each modality, we construct the following three hyperedges on each vertex u:
Feature Affinity (FA) hyperedge eFA. It consists of K nearest vertexes (v ∈ 𝒱), where their patchwise similarities w.r.t. the underlying vertex u are the highest K hits in the feature space.
Local Coherence (LC) hyperedge eLC. It consists of all the vertexes v ∈ 𝒰 from the target image and being located within a spatial neighborhood of u.
Atlas Correspondence (AC) hyperedge eAC. It consists of all vertexes v ∈ {𝒱s|s = 1, …, N} across the atlas images at the corresponding spatial locations w.r.t. u.
Since each of the above hyperedge is centered by u, we call u as the owner of hyperedge e (we omit the superscript of e hereafter).
Incidence Matrix Construction
After the construction of hyperedges for each u, we build a incidence matrix H with row representing the vertexes 𝒱 and column representing the hyperedges ℰ, which encode all the information within the hypergraph 𝒢. Each entry H(v, e) in H measures the affinity between the each vertex v and the owner u of the hyperedge e ∈ ℰ:
| (1) |
where the ‖․‖2 is the L2 norm computed between intensity image patch p(v) and p(u) for vertex v and the hyperedge owner u. σ is the averaged patchwise similarity between u and all vertexes within the hyperedge e. For simplicity, each hyperedge is initialized with an equal weight, W(e) = 1. The degree of a vertex v is defined as d(v) = Σe∈ℰ W(e)H(v, e), and the degree of hyperedge is defined as δ(e) = Σv∈𝒱H(v, e). Thus, two diagonal matrices Dv and De can be formed with each entry along the diagonal using the vertex degree and hyperedge degree, respectively.
According to this formulation, the construction of the hypergraph captures the high-order relationship among the all the vertices across different image modalities, feature affinities, local neighborhood coherences and the atlas correspondences. And any two vertexes within the same hyperedge should have similar labels. Following gives the detailed hypergraph learning for label propagation.
2.2 Labels Propagation via Hypergraph Learning
Given the initialization on the hypergraph, we employed a semi-supervised learning method to perform the label fusion on the constructed hypergraph. The objective function [10] is defined as:
| (2) |
where f is the likelihood of hippocampus on each vertex v ∈ 𝒱. The first term ℛemp(f) is an empirical loss, which prevent the dramatic change for those the high probability values in f. The second term Ω(f) is a regularization term on the hypergraph, which restrict the labels at similar range for those vertexes within a same hyperedge. The λ is a positive weighing parameter between the two terms.
The first empirical loss term ℛemp(f) is defined by
| (3) |
The empirical loss term is designed for the minimization of the differences before and after label fusion.
The second term Ω(f) is a regularizer on the hypergraph, which defined as follows:
| (4) |
where , and Δ can be viewed as normalized hypergraph Laplacian matrix. Here, the regulation term Ω(f) constrains the labels of vertexes in the same hyperedge should held a similar value during the label propagation. Thus, the objective function, Equation (2), can be rewritten as:
| (5) |
By differentiating the objective function with respect to f, the optimal f can be computed iteratively as below:
| (6) |
Given the estimated f, the anatomical label on each difficult-to-label target voxel u ∈ 𝒰L can be determined by
| (7) |
It should be noted that using the semi-supervised learning to perform the binary classification on the hypergraph allows prediction of the entire target image voxels simultaneously, while combining several correlations across all the voxels between the target image and atlas images.
3 Experiments
3.1 Date acquisition and preprocessing
In the experiments, MR images of 10 healthy infant subjects are acquired from a Siemens head-only 3T scanner. For each subject, both T1- and T2-weighted MR images were acquired in five data sets at 2 weeks, 3 months, 6 months, 9 months and 12 months of age. T1-weighted MR images were acquired with 144 sagittal slices at a resolution of 1 × 1 × 1mm3, while T2-weighted MR images were acquired with 64 axis slices at resolution of 1.25 × 1.25 × 1.95mm3. For each subject, the T2-weighted MR image is linearly aligned to the T1-weighted MR image at the same age and then further resampled to 1 × 1 × 1mm3. Standard preprocessing was performed including skull stripping [11], intensity inhomogeneity correction [12]. The manual segmentations of the hippocampal regions for all 10 subjects are available and used as ground-truth for evaluation.
3.2 Evaluation of the proposed method
For all the experiments, the patch size is set as 5 × 5 × 5 voxels. The number of nearest neighborhood vertexes K is 10 in constructing eFA. The spatial neighborhood in constructing eLC and eAC is set to 3 × 3 × 3 voxel. Parameter λ in equation (2) is 10.
To evaluate the performance of the proposed method, we adopted the leave-one-out cross-validation. In each cross-validation step, one subject is used as target images and the remaining 9 subjects were used as the atlas images. Our proposed hypergraph patch labeling (HPL) method is compared with three state-of-the-art multi-atlas patch-labeling methods: local-weighted majority voting (LMV) [3], non-local mean (NLM) [4] and sparse patch labeling (SPL) [5].
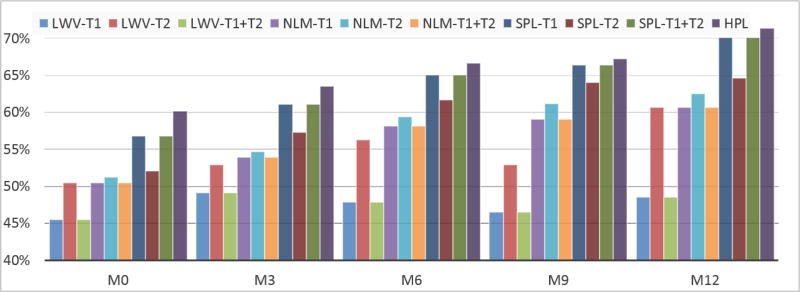
Table 1 gives the average dice ratio of four comparison methods for 2-week, 3-month, 6-month, 9-month and 12-month data respectively. It can be seen that our method can achieve better segmentation accuracy than all other three counterpart methods. Fig. 2 further shows the impact of using our proposed method for segmenting the infant hippocampus compared with other different labeling methods under different modalities.
Table 1.
The average dice ratio and standard deviation of four comparison methods: local-weighted majority voting, non-local mean, sparse patch labeling and hypergraph patch labeling for 2 week, 3 month, 6 month, 9 month and 12 month data.
| LMV | NLM | SPL | HPL | |
|---|---|---|---|---|
| 2 week | 0.45 ± 0.12* | 0.51 ± 0.12 | 0.57 ± 0.10 | 0.60 ± 0.11 |
| 3 month | 0.49 ± 0.09* | 0.54 ± 0.10* | 0.61 ± 0.04 | 0.64 ± 0.04 |
| 6 month | 0.48 ± 0.09* | 0.58 ± 0.11 | 0.65 ± 0.06 | 0.67 ± 0.07 |
| 9 month | 0.46 ± 0.07* | 0.59 ± 0.08* | 0.66 ± 0.04 | 0.67 ± 0.04 |
| 12 month | 0.48 ± 0.10* | 0.61 ± 0.05* | 0.70 ± 0.04 | 0.71 ± 0.04 |
(* indicates the significant improvement of HPL over other compared methods (p<0.05), T1+T2).
Fig. 2.
Comparison of the average dice ratio of the ten infant subjects between different labeling methods across five time points: 2-week (M0), 3-month (M3), 6-month (M6), 9-month (M9) and 12-month (M12)
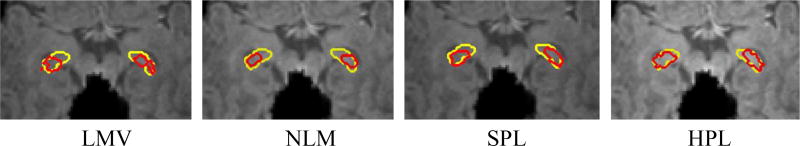
Finally, Fig. 3 shows the final segmentation results by the four methods and their corresponding manually segmentations for one typical subject at 2-week-old. Through visual inspection, our estimated labeling results (bottom line) are closer to the ground truth (yellow contours).
Fig. 3.
Infant hippocampus segmentation comparison between the automatic segmentation using the four methods (red circles) and the ground truth (yellow circles)
4 Discussion and Conclusion
With our proposed method, we achieved a better segmentation result with fewer atlases compared with the state-of-the-art patch-based method. The reasons are due to the followings: (1) the label propagation based on the hypergraph can adaptively combine the complementary information both from T1- and T2- weighed images; (2) for each image modality, three types of voxels correlations are considered, i.e. feature affinity, local spatial neighborhood and atlas corresponding spatial regions, which extensively exploits the high order correlations among a group of voxels; (3) Instead of labeling each voxel independently as in the existing methods, our semi-supervised hypergraph learning method performs a global optimization to predict the anatomical labels for all the voxels in the target image simultaneously. (4) Through the label propagation process, we allow the high confidence voxels to guide the nearby low confidence voxels, which provides valuable heuristics to overcome the uncertainty in label fusion.
In this paper, we propose a multi-atlas and multi-modal label fusion method for the segmentation of the hippocampus from infant MR images. We combine the advantages of conventional multi-atlas and graph-based approaches by encoding the feature affinity within the multi-modal feature space, spatial coherence, and the atlas information via hypergraph. Then, the whole label fusion procedure falls into the semi-supervised hypergraph learning framework, where the estimation of the latent anatomical labels is adaptively influenced by the connected counterparts in the hypergraph. The experiment results show more accurate hippocampus labeling results from MR images in the different phases of first year of life, using our proposed method with comparison to the state-of-the-art methods.
References
- 1.Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH. A structural MRI study of human brain development from birth to 2 years. J. Neurosci. Off. J. Soc. Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Shi F, Yap P-T, Gilmore JH, Lin W, Shen D. 4D multi-modality tissue segmentation of serial infant images. PloS One. 2012;7:e44596. doi: 10.1371/journal.pone.0044596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, van Ginneken B. Multi-atlas-based segmentation with local decision fusion--application to cardiac and aortic segmentation in CT scans. IEEE Trans. Med. Imaging. 2009;28:1000–1010. doi: 10.1109/TMI.2008.2011480. [DOI] [PubMed] [Google Scholar]
- 4.Coupé P, Manjón JV, Fonov V, Pruessner J, Robles M, Collins DL. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D, Guo Q, Wu G, Shen D. Sparse Patch-based Label Fusion for Multi-atlas Segmentation; Proceedings of the Second International Conference on Multimodal Brain Image Analysis; 2012. pp. 94–102. [Google Scholar]
- 6.Shi J, Malik J. Normalized Cuts and Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 2000;22:888–905. [Google Scholar]
- 7.Gao Y, Ji R, Cui P, Dai Q, Hua G. Hyperspectral image classification through bilayer graph-based learning. IEEE Trans. Image Process. 2014;23(7):2769–2778. doi: 10.1109/TIP.2014.2319735. [DOI] [PubMed] [Google Scholar]
- 8.Gao Y, Wang M, Zha Z-J, Shen J, Li X, Wu X. Visual-textual joint relevance learning for tag-based social image search. IEEE Trans. Image Process. 2013;22(1):363–376. doi: 10.1109/TIP.2012.2202676. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Wang M, Tao D, Ji R, Dai Q. 3-D object retrieval and recognition with hypergraph analysis. IEEE Trans. Image Process. 2012;21(9):4290–4303. doi: 10.1109/TIP.2012.2199502. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Huang J, Schölkopf B. Learning with Hypergraphs: Clustering, Classification, and Embedding. Proceedings of NIPS. 2006;19:1601–1608. [Google Scholar]
- 11.Shi F, Wang L, Dai Y, Gilmore JH, Lin W, Shen D. LABEL: Pediatric brain extraction using learning-based meta-algorithm. NeuroImage. 2012;62:1975–1986. doi: 10.1016/j.neuroimage.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]