SUMMARY
Histone deacetylase inhibitors (HDACis) have fascinated researchers in almost all fields of oncology for many years owing to their pleiotropic effects on nearly every aspect of cancer biology. Since the approval of the first HDACi vorinostat for the treatment of cutaneous T-cell leukemia in 2006, more than a hundred clinical trials have been initiated with a HDACi as a single agent or in combination therapy. Although a number of epigenetic and nonepigenetic molecular mechanisms of action have been proposed, biomarkers for response prediction and patient selection are still lacking. One of the inherent problems in the field of HDACis is their ‘reverse’ history of drug development: these compounds reached clinical application at an early stage, before the biology of their targets, HDAC1–11, was sufficiently understood. This review summarizes the current knowledge on the human family of HDACs as drug targets in pediatric and adult brain tumors, the efficacy and molecular action of HDACis in preclinical models, as well as the current status of the clinical development of these compounds in the field of neuro-oncology.
Practice Points.
Expression of histone deacetylases (HDACs) is frequently dysregulated in brain tumors compared with normal brain tissue.
Expression patterns differ depending on histology and tumor grade.
Mutation rate of HDAC genes in brain tumors is low to nonexistent.
HDACs have been validated as potential targets for the treatment of brain tumors in preclinical models.
Small-molecule HDAC inhibitors can induce apoptosis, differentiation and cell-cycle arrest in brain tumor cells.
Biomarkers for response prediction to HDAC inhibitor treatment are missing.
HDAC inhibitors might reveal their full anticancer potential in combination therapy approaches.
HDAC inhibitors currently used in ongoing clinical trials are vorinostat, panobinostat, entinostat and valproic acid.
Initial results from completed trials do not show efficacy of HDAC inhibitors as monotherapy.
Individual patients do show responses to HDAC inhibitor treatment.
Development of biomarkers for response to HDAC inhibition is crucial for the successful translation of the promising preclinical findings into the clinic.
Currently, HR23B is the most prominent predictive biomarker, and histone acetylation of peripheral blood mononuclear cells the most widely used pharmacodynamic biomarker for HDAC inhibition.
Critical points for the successful progression of research exploring the targeting of HDACs in brain tumors are elucidation of the function of HDAC isoenzymes, development of inhibitors with a class- or isoenzyme-specific inhibitory profile, development of biomarkers for response prediction and investigation of rational combination therapies.
The human histone deacetylases (HDACs) are divided into the families of ‘classical’ HDAC1–11 and of sirtuins (also referred to as class III HDACs) [1]. Classical HDAC inhibitors (HDACis) act through complexing the catalytically critical zinc ion of HDAC1–11 at the base of the enzymatic pocket. By contrast, sirtuins do not share this catalytical mechanism, they are NAD+ dependent and thus are not affected by classical HDACis [1]. This review focuses on the classical HDACs and their small-molecule inhibitors.
The 11 classical HDACs are subdivided into class I, IIa/b and IV according to their homology to yeast orthologs [2]. Class I HDAC1, 2, 3 and 8 are predominantly located in the nucleus and there seems to be a tissue-specific subcellular distribution; however, as has been reported for smooth muscle cells, HDAC8 is mainly found in the cytoplasm [3]. Class IIa HDAC4, 5, 7 and 9 can shuttle between the nucleus and the cytoplasm, while class IIb HDAC6 and 10 are predominantly located in the cytoplasm [4]. Much less is known regarding the single class IV HDAC11. It is now clear from knockout mice experiments that HDACs have nonredundant functions during embryonal development, resulting in distinct phenotypes ranging from early embryonal death to postnatal heart defects, growth plate and endothelial cell dysfunctions, and craniofacial defects [5]. In cancer biology, distinct functions of individual HDAC family members have also been described. For example, class I HDAC1–3 have been found in multiprotein complexes with oncogenic fusion transcripts, such as PML–RARa and AML-1–ETO, driving dedifferentiation of leukemic cells [6,7]. In addition, class I HDAC1–3 are frequently overexpressed in adult solid cancers [8]. In pediatric neuroblastoma, class I HDAC8 is associated with advanced-stage disease and poor clinical outcome and plays a distinct role in differentiation [9]. Much less is known regarding the function of class II HDACs in cancer biology. Class IIa HDAC5 and 9 are overexpressed in subgroups of pediatric medulloblastoma tumors and functional analysis showed that they are involved in proliferation of medulloblastoma cells [10]. Only scarce information on class IV HDAC11 in cancer biology is available. Very recently, HDAC11 was found to be a promising selective drug target in carcinomas [11].
Historically, the first substrates known to be deacetylated by HDACs were histone proteins, hence their name; however, it is now clear that there is a great number of nonhistone nuclear as well as cytoplasmatic substrate proteins that are deacetylated. Thus, many researchers are in favor of the term ‘lysine deacetylases’ to indicate the fact that HDACs are a more general acting class of enzymes removing acetyl groups from ε-amino-lysine residues of many different proteins [12]. Among the nonhistones substrates of HDACs there are key proteins, such as p53, STAT3, HSP70 and tubulin that are regulated in their biological function by reversible acetylation [1]. In fact, it has been estimated by high-resolution mass spectrometry that more than 3600 acetylation sites in 1750 proteins are regulated after treating a given cell type with a HDACi, giving rise to the term ‘acetylome’ [13]. It has been proposed that the acetylome, regulated by HDACs and HATs, functions as a regulatory signaling network in cells similar to the phosphoproteome network controlled by kinases and phosphatases [14]. A useful acetylome database covering a wide range of protein acetylation information is the CPLA [201].
HDACs can be inhibited by several small-molecule chemical compounds that fall into five basic structural classes: hydroxamic acids, benzamides, cyclic tetrapeptides, short-chain fatty acids and electrophilic ketones [15]. The compounds currently in clinical use are unselective, and, therefore, they target several HDACs simultaneously. Vorinostat (suberoylanilide hydroxamic acid) was the first HDACi to be approved for clinical use in cutaneous T-cell lymphoma in 2006, followed by depsipeptide (romidepsin, FK228) in 2009 [16]. Although vorinostat and many of the other hydroxamic acid compounds (i.e., TSA, ITF 2357 and panobinostat) have been described as being pan-HDACis, this view was recently challenged through a chemical phylogenetics approach suggesting that these compounds are in fact class I HDACis [17].
Of great clinical importance in neuro-oncology is the ability of compounds to cross the blood–brain barrier (BBB) (Table 1). The ability of HDACis to penetrate the BBB is not well studied. Some preclinical mouse models and clinical trials suggest that vorinostat is able to pass the BBB [18–20]. Data from PET-based experiments performed by Hooker et al. suggest that MS-275 (entinostat) does not penetrate the BBB of nonhuman primates at clinically relevant concentrations [21], although it has been described as a potent brain region selective HDACi in rodents [22]. Valproic acid (a short-chain fatty acid) is an HDACi with extensively clinically proven BBB penetration, as it has long been used for the treatment of epilepsy. Phenylbutyrate, another short-chain fatty acid-type HDACi, has been developed for the treatment of urea cycle disorders. Both compounds are weak HDACis in that they require 1–5 mM concentrations to exert HDAC inhibitory activity in vitro. However, these concentrations can hardly be reached in patients (Table 1) and phenylbutyrate must be given as continuous intravenous infusion or ingested in large amounts (several grams) of tablets per day, which still results in submillimolar plasma concentrations (Table 1).
Table 1. . Clinically achievable concentrations of HDAC inhibitors.
| Compound | Cmax (mol/l) | Class | Penetration of BBB | Ref. |
|---|---|---|---|---|
| VPA | 3.41E-04 | Short-chain fatty acid | Yes [114] | [115] |
| Phenylbutyrate | 3.00E-04 | Short-chain fatty acid | Yes [116] | [117] |
| Vorinostat (SAHA) | 4.49E-06 | Hydroxamic acid | Yes [118] | [119] |
| Panobinostat (LBH589) | 4.09E-08 | Hydroxamic acid | Yes [120] | [121] |
| Belinostat (PXD-101) | 1.00E-07 | Hydroxamic acid | Little in nonhuman primates [122] | [123] |
| Abexinostat (PCI-24781) | 3.39E-07 | Hydroxamic acid | N/A | [124] |
| Dacinostat (LAQ824) | 2.17E-06 | Hydroxamic acid | N/A | [125] |
| Givinostat (ITF2357) | 4.98E-07 | Hydroxamic acid | N/A | [126] |
| Entinostat (MS-275) | 3.90E-07 | Benzamide | No/little in nonhuman primates [22]; yes [116] | [127] |
| Romidepsin (depsipeptide) | 8.74E-07 | Cyclic tetrapeptide | No/little [101] | [128] |
BBB: Blood–brain barrier; Cmax: Maximum concentration; N/A: Not applicable; SAHA: Suberoylanilide hydroxamic acid; VPA: Valproic acid.
All HDACis currently in clinical use are associated with class-specific toxicities, the most frequent being fatigue, electrolyte disturbances, and gastrointestinal and hematological side effects, which limit the applicable dosages and, therefore, impede the realization of HDACis full anticancer potential. It is currently under discussion which of the HDAC family members are mediating these limiting toxicities. As a consequence, several groups are trying to develop truly isoform-selective inhibitors with a broader therapeutic window [23]. However, this approach requires exact knowledge regarding the molecular function and mechanism of single HDAC isoforms in tumor tissue and normal organs. One of the few current promising examples is the development of HDAC8-selective inhibitors that are at least 200-fold more active against HDAC8 compared with other HDAC family members [24,25] and the identification of HDAC8 as a selective drug target in neuroblastoma [9]. Finally, a recent publication describes novel class IIa selective HDACis [26]; however, true class IIa isoenzyme-selective HDACis are still missing.
Expression of HDACs in brain tumor tissue
Dysregulation of the expression of HDAC genes has been reported for numerous cancer entities [27]. However, literature systematically analyzing the expression of all 11 classical HDAC isoenzymes in cancer cells and the correlation of expression patterns with stage of the disease or patient survival remains sparse. This holds true for the expression of HDACs in brain tumor tissue in particular. Lucio-Eterovic et al. demonstrated that class II and IV HDACs are downregulated in glioblastoma multiforme (GBM) in comparison with low-grade astrocytoma and normal brain tissue at the mRNA level [28]. Campos et al. restricted their HDAC expression analysis to HDAC1–3 in astrocytic gliomas and found that HDAC1 and 2 are strongly expressed in gliomas independent of tumor grade. HDAC3, however, showed an inverse correlation with grade, with strong HDAC3 expression correlating with increased survival probabilities [29]. Milde et al. found a correlation of high HDAC5 and 9 expression and an unfavorable prognosis for patients with medulloblastoma [10].
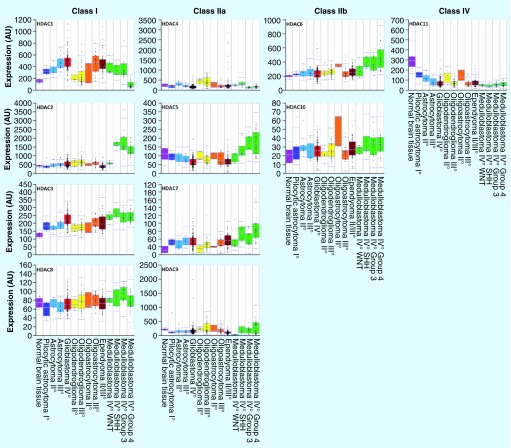
In order to systematically display HDAC1–11 expression patterns across a wide spectrum of different brain tumor entities of different grade, as well as normal brain tissue, we analyzed publicly available data via the R2 microarray analysis and visualization platform (Figure 1) [202]. For this analysis we used the probesets with the highest average present signal. Other probesets for the same HDAC did not show considerable differences in observed trends in expression levels.
Figure 1. Histone deacetylase mRNA expression in brain tumors, as analyzed by gene-expression microarrays, visualized by R2.
Dataset GSE16011 [129]: age at diagnosis: 11–81 years; normal brain tissue: n = 8; pilocytic astrocytoma WHO I°: n = 8; astrocytoma WHO II°: n = 13; astrocytoma WHO III°: n = 16; glioblastoma multiforme WHO IV°: n = 159; oligodendroglioma WHO II°: n = 8; oligodendroglioma WHO III°: n = 44; oligoastrocytoma WHO II°: n = 3; and oligoastrocytoma WHO III°: n = 25. Dataset GSE21687 [130]: age: 0–59 years; ependymoma WHO II° and III°: n = 83. Dataset GSE37418 [131]: age: 3–16 years; medulloblastoma WHO IV° WNT: n = 8; medulloblastoma WHO IV° SHH: n = 10; medulloblastoma WHO IV° group 3: n = 16; and medulloblastoma WHO IV° group 4: n = 39. Probesets: HDAC1: 201209_at; HDAC2: 201833_at; HDAC3: 216326_s_at; HDAC4: 204225_at; HDAC5: 202455_at; HDAC6: 206846_s_at; HDAC7: 217937_s_at; HDAC8: 223345_at; HDAC9: 205659_at; HDAC10: 226672_s_at; and HDAC11: 227679_at.
As shown in Figure 1, many brain tumor entities show considerable alterations in their HDAC expression compared with normal brain tissue. In addition, the expression of the same HDAC isoenzyme may differ within the same tumor entity depending on tumor grade or molecular subgroup. The expression data presented in Figure 1 suggests a correlation of increased HDAC1 and 3 expression and tumor grade in astrocytoma (data available for astrocytoma [WHO II° and III°] and GBM [WHO IV°]). Medulloblastomas, in particular, show a diverse HDAC expression pattern. While upregulated in all examined brain tumor entities, HDAC1 is relatively downregulated in only one molecular subgroup of medulloblastoma (group 4). HDAC2, however, showing no alteration of expression in all the other brain tumor entities, is considerably overexpressed in three out of four molecular subgroups in medulloblastoma (groups 3, 4 and SHH, relative to WNT). A similar trend can be observed for HDAC5 and 6. Intriguingly, all brain tumor entities show an upregulation of class I HDAC1 and 3 and a downregulation of class IV HDAC11 compared with normal brain tissue.
In pediatric brain tumors, next-generation sequencing technologies have identified mutations in genes directly or indirectly involved in epigenetic regulation as a common theme [30–35]. However, little information is available on mutations in HDAC genes. HDAC2 mutations have been discovered in group 4 medulloblastoma [35]; other publications describing the sequencing of large medulloblastoma cohorts, however, did not describe HDAC mutations [32,34]. Therefore, the mutation rate of HDAC genes in medulloblastoma seems to be very low.
While epigenetically effective mutations in the chromatin component H3.3 have been described in GBM [30,31], data on HDAC mutations again is scarce.
We analyzed the publicly available The Cancer Genome Atlas data set on GBM [203] via the open access cBio Cancer Genomics Portal [36,204]. At the time of manuscript submission, data of whole-exome sequencing for 276 GBM tumor samples was available. We found that only very few GBM tumors have mutations in or copy number alterations of HDAC genes (amplification or homozygous deletion) (Table 2). According to MutSig analysis [205], no mutation of HDAC genes in GBM is considered a significantly mutated driver candidate.
Table 2. . Rate of mutation rates and copy number alterations in glioblastoma.
| Gene | Somatic mutations | Copy number alterations | |||
|---|---|---|---|---|---|
| Cases (n) | % | Cases (n) | % | ||
| HDAC9 | 4 | 1.4 | 7 | 2.5 | |
| HDAC2 | 3 | 1.1 | 4 | 1.4 | |
| HDAC5 | 3 | 1.1 | 0 | 0.0 | |
| HDAC6 | 3 | 1.1 | 0 | 0.0 | |
| HDAC8 | 2 | 0.7 | 0 | 0.0 | |
| HDAC1 | 1 | 0.4 | 1 | 0.4 | |
| HDAC3 | 1 | 0.4 | 0 | 0.0 | |
| HDAC4 | 1 | 0.4 | 2 | 0.7 | |
| HDAC10 | 0 | 0.0 | 4 | 1.4 | |
| HDAC11 | 0 | 0.0 | 1 | 0.4 | |
| HDAC7 | 0 | 0.0 | 1 | 0.4 | |
Taken together, the mutation frequency of HDACs in all brain tumors currently sequenced seems to be very low to nonexisting. Therefore, it seems likely that the expression or functional status within multiprotein complexes determines the oncogenic potential of HDAC family members.
Functional validation of HDACs & efficacy of HDACis in preclinical models
As mentioned above, HDAC expression is frequently dysregulated in brain tumors compared with normal brain tissue. Considering that HDACs act as important epigenetic, as well as nonepigenetic, regulators of key signaling pathways involved in many, if not, all hallmarks of cancer [15], and since, unlike genetic mutations, epigenetic modifications are thought to be reversible, it is tempting to speculate that targeting of HDACs has the potential to reverse the malignant phenotype of cancer cells. This renders HDACs highly interesting targets for anticancer therapy.
In preclinical investigations much effort has been made to evaluate the functional relevance of HDACs in brain tumor cells. Currently, the most frequent experimental approaches of modulating HDAC activity include interference with HDACs either by si/shRNA-mediated knockdown or by enzymatic inhibition with small-molecule inhibitors. While knockdown experiments have the advantage of exclusively targeting single HDAC isoenzymes individually, established HDACis usually target a variety of HDAC family members, impeding the attribution of an observed effect to a specific enzyme. However, knockdown experiments are technically much more challenging, more difficult to control and the pharmacology of si/shRNA-mediated knockdown in in vivo models remains complex [37], while experiments with HDACis promise a quick translation of interesting findings from bench to bedside, if applied in clinically relevant concentrations. Finally, since HDACs usually function within multiprotein complexes, knockdown of a particular HDAC can cause disruption of the function of a whole protein complex. Thus, a given biological effect cannot necessarily be attributed to enzymatic function, and, therefore, be translatable into a strategy involving HDACis, but could also be explained by HDACs functioning as bromodomain proteins, that is, readers of epigenetic marks acting independently from their catalytic activity.
As reviewed below, HDACis have a broad range of effects on brain tumor cells. Most of the research has focused on the two most frequent devastating malignancies in neuro-oncology: GBM in adults and medulloblastoma in pediatrics. It is important to note that in all investigations, the observed effect of HDAC inhibition was dose dependent and relevant anti-tumoral effects were only observed above a distinct threshold level. In many investigations, final inhibitor concentrations are far from clinically achievable dosages, which renders these findings untranslatable into clinical applications. This review only comments on investigations working with clinically achievable concentrations of inhibitors as summarized in Table 3. Two reasons for applying inhibitors at higher concentrations were considered valid to make exceptions:
The clinically achievable concentration of the applied inhibitor is unknown, but the investigators demonstrated in control experiments that the inhibitor does not affect normal brain tissue or cells at the highest concentration used;
High concentrations of the inhibitor are required to be able to investigate molecular mechanisms involved in functional consequences of HDAC inhibition. Elucidation of these mechanisms might offer rationales for intelligent combination therapies that might allow lowering the dosage of the inhibitor.
Table 3. . Ongoing clinical trials using histone deacetylase inhibitors in brain tumors.
| HDAC inhibitor | Combination treatment | Histology | Phase | Age (years) | Clinicaltrials identifier/status |
|---|---|---|---|---|---|
| Panobinostat | Stereotactic radiation | Recurrent glioma, high-grade meningioma, brain metastasis | I | >18 | NCT01324635; recruiting |
| Valproic acid | Radiation therapy, temozolomide | High-grade glioma | II | 18–90 | NCT00302159; recruiting |
| Vorinostat | Stereotactic radiation | Anaplastic astrocytoma, anaplastic oligodendroglioma, glioblastoma, gliosarcoma, mixed glioma | I | >18 | NCT01378481; recruiting |
| Vorinostat | Bevacizumab | Recurrent glioblastoma multiforme, malignant glioma | II | >18 | NCT01738646; recruiting |
| Vorinostat | Bevacizumab, temozolomide | Recurrent malignant glioma | I/II | >18 | NCT00939991; active, not recruiting |
| Vorinostat | Erlotinib, temozolomide | Recurrent glioblastoma multiforme | I/II | >18 | NCT01110876; recruiting |
| Vorinostat | Bevacizumab | Recurrent glioblastoma | I/II | >18 | NCT01266031; active, not recruiting |
| Vorinostat | Temozolomide | Relapsed primary brain tumors and spinal chord tumors, gliomas, AT/RT, germ cell tumors, MB, PNET, meningioma | I | 1–21 | NCT01076530; completed |
| Vorinostat | Temozolomide | Malignant glioma | I | >18 | NCT00268385; active, not recruiting |
| Vorinostat | Temozolomide, bevacizumab, radiation therapy | Newly diagnosed high-grade glioma | Randomized Phase II/III trial | 3–21 | NCT01236560; recruiting |
| Vorinostat | Radiation | Newly diagnosed pontine glioma | I/II | 3–21 | NCT01189266; recruiting |
| Valproic acid | Etoposide | Neuronal tumors and brain metastases | I | ND | NCT00513162; completed |
| Vorinostat | Bevacizumab, irinotecan | Recurrent glioblastoma | I | >18 | NCT00762255; active, not recruiting |
| Phenylbutyrate | – | Relapsed brain tumors | II | 2–21 | NCT00006450; completed |
Details of each trial can be found on www.clinicaltrials.gov and by searching for the relevant trial number.
AT/RT: Atypical teratoid rhabdoid tumor; HDAC: Histone deacetylase; MB: Medulloblastoma; ND: Not defined; PNET: Primitive neuroectodermal tumor.
It appears that, in preclinical models of all investigated brain tumor entities, small-molecule HDACis of different chemical classes were able to considerably change the cellular phenotype. Although very promising, these findings are far from satisfying. The constantly observed response of brain tumor cells to HDAC inhibition, especially in contrast with the missing response of normal brain tissue [38], suggests a certain degree of tumor addiction to the catalytic activity of HDACs. Tumor addiction to a targetable enzyme, opens up a therapeutic window. However, cell lines of the same entity show big differences in their response to the same HDACi: some go into apoptosis while others experience only a cell-cycle arrest, half maximal inhibitory concentration values for induction of apoptosis differ dramatically from cell line to cell line across all entities. Why are some tumor cells more susceptible to HDACi treatment than others? This is a key question to be answered as it could ultimately result in the development of biomarkers for response prediction in the clinical setting. This is of particular clinical interest since a limited number of individual patients show a dramatic response, while for the majority of patients no benefit of HDACi treatment could be demonstrated.
As HDACs have a broad variety of substrates apart from histones, such as transcription factors p53 and HSP90, HDAC inhibition will ultimately result in a rather difficult-to-survey global change in acetylation patterns of cellular proteins. The challenge of current HDAC research is to identify those HDAC-dependent acetylation patterns and to elucidate molecular mechanisms of tumor addiction to functional consequences of the acetylation status of different proteins.
When it comes to histones, the most prominent target of HDACs, the functional consequences of the catalytic activity of HDACs are anything but easy to predict. Mechanistically speaking, it is known that an altered histone acetylation pattern will change the availability of DNA for transcription. Functionally speaking, this will ultimately result in a change of gene expression. However, to functionally link the action of a specific HDAC isoenzyme to the altered expression of a specific (set of) gene(s) is much more difficult. Unlike transcription factors, HDACs do not interact directly with DNA. This renders them incapable of directing their regulatory effect on transcription towards a defined region of the DNA. Therefore, HDACs are dependent on other proteins to recruit them to specific regions of the DNA where they will then effectuate their histone deacetylating activity. HDAC expression alone is, therefore, not sufficient to predict a cancer cell‘s susceptibility to HDACi treatment. The given set of recruitment proteins (e.g., transcription factors and repressive complexes), which will vary considerably from cancer cell to cancer cell, determines the gene expression-modifying effect of HDACs on the histone level. However, it appears reasonable that, if both HDAC isoenzyme and the appropriate DNA recruitment protein(s) are present in a cancer cell, the HDAC can be targeted towards a region of the DNA encoding for tumor suppressor pathways and/or inhibition of oncogenic pathways. This could partially explain tumor addiction to HDACs and has been shown for several tumor suppressor genes in different brain tumor entities (see the ‘Modulation of tumor suppressor pathways’ section).
Inhibition of HDACs results in a change of expression in up to 20% of the genes of the total genome in a given cell [39]. Depending on the genetic and epigenetic make-up of the cell and HDACi concentration applied, the following mechanisms of action have been implicated in the response of brain tumor cells to HDAC inhibition:
Modulation of tumor suppressor pathways and/or of oncogenic pathways;
Inhibition of cell cycle and induction of differentiation;
Induction of cell death mechanisms;
Sensitization towards irradiation and other chemotherapeutic agents;
Interference with cell–matrix interaction: invasion and angiogenesis;
Other mechanisms: telomerase activity and an increase in genomic instability;
Crosstalk between regulative acetylation and methylation mechanisms.
▪ Modulation of tumor suppressor pathways
As HDAC activity is linked to increased chromatin condensation and subsequent silencing of gene expression, a tempting strategy of exploiting molecular alterations in cancer involves the reactivation of tumor suppressor genes by inhibition of HDAC activity. Indeed, several groups have shown that HDACis might be of use when it comes to reinducing tumor suppressor genes in brain tumors. Schmidt et al. identified the Ras-related protein on chromosome 22 (RRP22) as an important tumor suppressor gene in gliomas [40]. RRP22 expression is negatively correlated with tumor grade and overall survival. This group demonstrated that upon treatment with TSA, RRP22 is re-expressed. Gao et al. described NECL1 as a frequently lost tumor suppressor gene in GBMs. NECL1 expression could be reinduced by TSA treatment [41]. Vibhakar et al. showed that DKK1, a Wnt antagonist and tumor suppressor gene in medulloblastoma, is frequently downregulated compared with normal tissue of the cerebellum and could be reinduced by TSA treatment [42]. Tamannai et al. identified ING1 as a tumor suppressor gene in GBM, and described the reinduction of ING1 expression through TSA treatment [39]. Spiller et al. have found that treatment with retinoic acid can induce the transcription of BMP2, leading to apoptosis, and concurrent treatment with vorinostat potentiated the effect of retinoic acid by facilitating accessibility for transcription, resulting in a dramatic increase in apoptosis [43].
▪ Inhibition of cell cycle & differentiation
Cell cycle
For uncontrolled proliferation, cancer cells have to find a way to escape their intrinsic control machinery during the cell cycle. Mutations and dysregulations of proteins essential for controlling check points of the cell cycle are, therefore, constitutively found in cancer cells [44]. A frequently observed effect of HDAC inhibition in brain tumor cells, is the induction of cell-cycle arrest. Accumulation of cells in either the G1 or G2/M phase of the cell cycle after HDAC inhibition has been reported, and several mechanistic explanations for the HDACi-mediated cell-cycle arrest have been proposed.
Concerning G1 arrest, the majority of published data point to the involvement of p21. Treatment of GBM cells with vorinostat has been shown to increase the acetylation of the histones in the promoter region of CDKN1A, inducing the expression of p21 [45], leading to G1 cell-cycle arrest. Yin et al. also observed an increase in expression of p27, and decreased expression of CDK2, CDK4, CCND1 and CCND2 upon treatment with vorinostat [45]. An upregulation of p21 associated with G1 cell-cycle arrest was also found in GBM cells after treatment with TSA [46], MS-275 [47], LAQ824 and sodium butyrate [48]. TSA also induced G1 cell-cycle arrest by upregulation of p21 in a medulloblastoma cell line [49]. However, it has been shown that both 4-PB and TSA can induce G1 cell-cycle arrest in GBM cells independently of p21 [49,50]. With regard to G2/M arrest, it has been demonstrated that vorinostat in clinically relevant concentrations leads to G2/M arrest in GBM cells [45,51,52] and TSA causes G2/M arrest in medulloblastoma cells [49,53].
The role of p53 in HDACi-mediated cell-cycle arrest remains unclear. While Jin et al. demonstrated that HDACi-induced cell-cycle arrest is dependent on p53 in GBM cells [54], other investigations came to the conclusion that cell-cycle arrest in GBM cells was independent from p53 [45,51]. Further clarification of these observations is of great interest since p53 is frequently mutated in GBM.
Differentiation
While HDACis such as TSA and 4-PB suppress differentiation of neural stem cells into astrocytes and oligodendrocytes in the physiological setting [55], several groups report differentiation effects in brain tumor cells after treatment with HDACi. For both medulloblastoma [56] and GBM [57,58], induction of differentiation has been reported after treatment with HDACis. Of note, the differentiated phenotype became irreversible when HDACis were applied for long periods of time, for example, 28 days [56,58]. Milde et al. demonstrated that treatment with vorinostat induced neuronal differentiation with loss of stem cell properties in an ependymoma stem cell model, including inhibition of self-renewal as shown by a reduction of neurosphere formation [59]. Several groups propose that HDAC inhibition leads to the activation of transcription factors that control differentiation programs. Morita et al. suggest that in glioma cells HDACis lead to an elevation of BDNF gene expression [57], induced by an increase of neuroactive 5α-reduced steroid metabolites. Her et al. demonstrated that this increase is caused by a TSA-mediated increase in 5α-R expression [60]. Taylor et al. showed that HDAC1 and 2 are complexed with REST in medulloblastoma, and HDACi treatment ultimately results in expression of the REST target gene SYN1, leading to neuronal differentiation [61]. In a very interesting approach, Wei et al. demonstrated in a rat glioma model that metabolites, such as alanine, lactate, inositol, N-acetylaspartate and creatinine, which usually show a diverse picture in tumor tissue, are restored to the levels of normal brain tissue after treatment with vorinostat, indicating differentiation [62].
▪ Induction of cell death mechanisms
Apoptosis
The overwhelming body of published data creates the impression that the application of an HDACi can induce apoptosis in all investigated brain cancer entities. However, many published experiments did not use clinically achievable concentrations and applying HDACi at relevant concentrations provides a more diverse picture when it comes to the induction of apoptosis. Premkumar et al. have found that in several different GBM cell lines clinically achievable concentrations of vorinostat monotherapy, while inhibiting cell proliferation, do not induce apoptosis [52]. Others have shown a slight increase in apoptotic GBM cells after treatment with relatively low doses of TSA or 4-PB [39,48,58]. In medulloblastoma, vorinostat, TSA and MS-275 have been demonstrated to have a proapototic effect as single agents [43,51,63,64]. However, while TSA induced apoptosis in ependymoma cells [65], clinically achievable concentrations of vorinostat did not lead to increased cell death but to differentiation of ependymoma cells [66].
HDACis can disturb the equilibrium of pro- and anti-apoptotic proteins present in the cell, resulting in apoptosis. For example, Sawa et al. showed that romidepsin induces apoptosis in several GBM cell lines partially by the increased expression of the proapoptotic protein Bad and reduced transcription of antiapoptotic proteins Bcl-xL and Bcl-2 [67]. HDAC inhibition was also found to increase the transcription and acetylation of p53, while simultaneously negatively regulating the IKK–NF-κB pathway and survivin expression, promoting the mitochondrial apoptosis pathway in GBM [54,68]. In a GBM in vivo model, Ugur et al. showed that vorinostat increased Caspase 3 expression [69]. Rahman et al. also identified Caspase 3 as essential for TSA-induced apoptosis in medulloblastoma, primitive neuroectodermal tumor (PNET) and ependymoma [49]. Häcker et al. found that HDAC inhibition with MS-275 leads to acetylation of Ku70 in medulloblastoma, which leads to its dissociation from the proapoptotic Bax protein, the latter being activated by simultaneously upregulated p53 [70]. Upregulation of Caspase 8 upon HDACi treatment has been reported for medulloblastoma [71], atypical teratoid rhabdoid tumor [63], as well as GBM cells [46], while Sonnemann et al. found an activation of Caspase 9 and 3 upon HDACi treatment in medulloblastoma cells [64].
Sensitiziation to extrinsic & intrinsic apoptotic pathways
Disturbing the equilibrium of pro- and anti-apoptotic proteins with HDACis, even if not necessarily directly inducing apoptosis, appears to reliably sensitize tumor cells to extrinsic and intrinsic apoptotic pathways. Several groups report that HDACi pretreatment of medulloblastoma [71] and GBM cells in vitro [64], as well as GBM cells in an in vivo model [71], sensitizes towards TRAIL-induced apoptosis. Bangert et al. showed that treatment of GBM cells with MS-275, leads to the downregulation of cFLIPL and cFLIPS, partially by MS-275-induced upregulation of cMYC, which mediates the increased sensitivity towards TRAIL-induced apoptosis [72]. Sharma et al. found that the observed induction of apoptosis in glioma cells after treatment with scriptaid was concurrent with a dramatic increase in Ras activity, while apoptosis induction could be prevented by inhibiting JNK [73].
▪ Sensitization towards oxidative stress, irradiation & other chemotherapeutic agents
Oxidative stress and DNA damage are known to induce apoptosis via the intrinsic (mitochondrial) pathway. As HDACis have been reported to result in reactive oxygen species generation [74], and to induce and sensitize cancer cells towards DNA damage [75,76], combining HDACis with proteasome inhibitors, irradiation or DNA-damaging chemotherapeutics is a rational approach to try to increase the induction of apoptosis in cancer cells.
Proteasome inhibition
Proteasomes are an important part of the survival machinery of cancer cells. Yu et al. have shown that the combination of the proteasome inhibitor bortezomib with different HDACis (TSA, LBH589 and LAQ824) results in dramatic induction of apoptosis in GBM cells [77]. Both agents synergistically lead to mitochondrial injury with loss of mitochondrial membrane potential and consecutive release of cyctochrome C, an increase in the generation of reactive oxygen species and an increasing level of proapoptotic Noxa and MCL-1 cleavage [52].
HDACis & radiation therapy
Heterochromatization of DNA protects DNA from radiation-induced DNA damage [78]. Leading to the loosening of the chromatin structure, HDACis bear the potential to facilitate radiation-induced DNA double-strand breaks (DSB) in cancer therapy. For both GBM [51,75,79,80] and medulloblastoma [64,81], increases in radiation-induced apoptosis upon concurrent HDACi treatment has been described. Shabason et al. found that the radiation-sensitizing effect of HDACis is most pronounced when the HDACis are applied both before and after irradiation [79]. This may be due to the fact that HDACis not only promote radiation-induced DSB by euchromatization of the genome, but also negatively regulate the expression of DNA repair proteins, such as Rad51 and DNA-PK, which are crucial to nonhomologous end joining and homologous recombination [52,82]. However, data from Manova et al. showed that chromatin acetylation itself does not affect DSB repair by nonhomologous end joining in GBM cells [83].
DNA-damaging chemotherapeutic agents
Several groups have tested whether HDACis can increase apoptosis induced by clinically applied DNA-damaging chemotherapeutic agents. It has been reported, that MS-275 and vorinostat enhance apoptosis in both GBM [47] and medulloblastoma [64] induced by doxorubicin, etoposid and cisplatin. An interesting finding by Kitange et al. demonstrates the synergistic effect of combining vorinostat with temozolomide on induction of cell apoptosis in a GBM mouse model despite upregulation of MGMT, which is considered a mechanism of acquired temozolomide resistance [76].
▪ Interference with cell–matrix interaction: invasion & angiogenesis
HDACis reveal great potential in influencing the interaction of brain tumor tissue with its environment. Osuka et al. showed in in vitro experiments that treatment with the HDACi valproic acid significantly reduced the secretion of the VEGF in several GBM cell lines under normoxic conditions [84]. A reduction of angiogenesis upon treatment with vorinostat was also observed in in vivo models of GBM [62,63].
With increasing degrees of malignancy, brain tumors present a more invasive phenotype. Invasiveness and malignancy show a positive correlation with expression of the matrix metalloproteases MMP-2 and -9 in GBM [85]. Chen et al. found that the tumor suppressor gene RECK, which is downregulated in high-grade gliomas and negatively regulates MMP-2 and -9, can be reinduced in glioma cells by treatment with the HDACi valproic acid [86]. In addition, Papi et al. demonstrated that HDAC inhibition in GBM cells leads to the upregulation of the protein level of TIMP [87], while Konduri et al. showed that HDAC inhibition reinduces the expression of TFPI-2, a broad-range proteinase inhibitor in glioma cells [88]. An et al. found that the observed reduction of invasiveness in glioma cells upon treatment with vorinostat was not related to the activity of MMPs, but to an increase in cell–cell adhesion [89]. Together these findings suggest that HDACis block the invasiveness of GBM cells.
▪ Other mechanisms: telomerase activity & an increase in genomic instability
Telomerase
Telomerase activity (or an alternative lengthening of telomeres) is crucial for the immortalization of cells [90]. While generally repressed in somatic cells, the telomerase reverse transcriptase hTERT, encoding the catalytic domain of the human telomerase, is reactivated in approximately 85% of all malignancies, including malignant brain tumors [91]. Although the repression of hTERT in the physiological setting is at least partially mediated by HDAC1 and 2 [91], it has been shown that HDAC inhibition by TSA decreases hTERT expression at the mRNA level in medulloblastoma and GBM cell lines [53], as well as in CNS PNET and ependymoma cell lines [49]. Sharma et al. showed that scriptaid decreases telomerase activity in glioma cells [73], which could lead to telomere damage and consecutively contribute to induction of apoptosis [92].
Increase in genomic instability
While most research in the field of HDACs has been focusing on the change of gene expression after HDAC inhibition for many years, several groups recently have shown that part of the cytotoxicity of HDACis can be attributed to an increase in genomic instability following increased histone acetylation [53,82]. DNA DSBs were evidenced by H2AX activation after treatment with TSA in high-grade childhood CNS PNET, medulloblastoma and ependymoma cell lines, suggesting an activated DNA damage response [49]. As a consequence, HDACis were combined with other DNA-damaging chemotherapeutics. Combining vorinostat with the topoisomerase inhibitor SN38 in GBM cell lines was shown to have a synergistic effect on DNA damage [93]. MS-275 potentiated the DSB-inducing effect of doxorubicine in GBM cell lines and triggered the acetylation of the DNA repair protein Ku70, attenuating its damage response activity [70].
▪ Crosstalk between regulative acetylation & methylation mechanisms
The tight coordination of histone acetylation with other regulative epigenetic mechanisms, including histone and DNA methylation, has been described in several cancer entities [55]. While hypermethylation of CpG islands, mediated by DNMT, already leads to transcriptional repression of genes in cancer cells [63], proteins with a binding domain for methylated DNA can ultimately serve as recruitment proteins for HDACs. Concurrent histone deacetylation and DNA hypermethylation results in long-term silencing of these genes [47]. Espada et al. could show that loss of the DNMT enzyme leads to an alteration of the histone acetylation patterns [94]. On the histone level, Singh et al. could show that treating GBM cells with the HDACi PCI-24781 induces hypermethylation of histones [39]. The methylation status of histones (e.g., dimethylation of lysine 4 on histone H3, leading to transcriptional activation) is generally coordinated by the enzyme LSD1, residing in a complex with HDAC1 and 2 [95]. Simultaneously inhibiting LSD1 with tranylcypromine and HDACs with PCI-24781 strongly induced apoptosis in GBM cells [38]. Finally, Ecke et al. demonstrated that combination therapy of the DNMT inhibitor 5-aza-dC and the HDACi valproic acid effectively diminishes tumor formation in a ptch medulloblastoma in vivo model [96].
Clinical trials of HDACis in neuro-oncology
Early case reports applying HDACis in individual patients with brain tumors suggested activity in clinical neuro-oncology [97,98]. Owing to its good safety profile and easy clinical availability, valproic acid was applied as the first HDACi in maintenance therapy for pediatric patients with high-grade glioma [99]. Recently, a retrospective analysis of 544 adult GBM patients receiving antiepileptic drug treatment during radiation therapy suggests improved overall survival of patients receiving valproic acid [100]. Phenylbutyrate is another weak short-chain fatty acid HDACi that has been developed for the treatment of urea cycle disorders with limited clinical experience (Table 3) due to the short half-life and high doses required. The first prospective clinical trial applying a hydroxamic acid-type HDACi in neuro-oncology was reported in 2009 [20]. In this Phase II study, vorinostat was given as a single agent to 66 patients with recurrent GBM. The trial met the prospectively defined primary efficacy end point, with nine of the first 52 patients being progression free at 6 months, suggesting modest activity. Biopsy in pre- and post-treated GBMs showed increased histone acetylation in immunohistochemical staining in tumor samples, suggesting BBB penetration of vorinostat and on-target effects in GBM tissue. However, owing to the small number of samples and technical limitations of quantification of immunohistochemistry, this observation has to be interpreted with caution. A Phase I/II trial of single-agent romidepsin included 35 patients with recurrent GBM. No objective response was seen in the Phase II trial and the authors concluded that single-agent romidepsin in its standard dose was not effective. Besides low antiglioma activity of romidepsin as a single agent these disappointing results might partially be due to insufficient BBB penetration of the compound or ABCB1-mediated increase of drug efflux in the tumor tissue [101].
Since HDACis show antiangiogenic, chemotherapy- and radiation-sensitizing activity in preclinical models, current trials combine HDACis with bevacizumab, chemotherapy and radiation therapy. The HDACis applied include the hydroxamic acids vorinostat and panobinostat, the benzamide MS-275 (entinostat) and the short-chain fatty acid valproic acid.
Drappatz et al. reported a Phase I study of panobinostat in combination with bevacizumab, for recurrent high-grade glioma. The trial determined a dose recommendation of panobinostat 30 mg three-times per week, every other week, in combination with bevacizumab 10 mg/kg every other week. There were some promising signals of efficacy with three patients showing partial responses and seven stable diseases, and a Phase II trial of this combination is underway [102]. In another Phase I trial, vorinostat was combined with bevacizumab and irinotecan in patients with recurrent GBM [103]. The authors report a high toxicity of this regimen and the need for frequent dose reductions of irinotecan. Of interest, the authors noted that high-dose vorinostat was associated with an improved progression-free and overall survival when compared with low-dose vorinostat. In a plasma proteomic-based antibody array platform, the authors identified low IGFBP-5 pretreatment levels as significant predictors of progression-free survival and decreasing PDGF-AA plasma levels as predictors of tumor recurrence.
Another rational combination partner of HDACis is the proteasome inhibitor bortezomib, since both compounds target critical components of the proteasomal machinery. In a Phase II trial, Friday et al. assessed the combination of vorinostat and bortezomib in patients with recurrent GBM. However, the trial was closed at the predetermined interim analysis with none out of 34 patients being progression-free at 6 months, suggesting that this combination is not effective in recurrent GBM [104].
In addition, several early clinical trials are currently recruiting patients with brain tumors, mainly patients in relapse, but also upfront (Table 3). In these trials, the HDACis are combined with standard therapy, including radiation and temozolomide, but also with other targeted compounds, such as bevacizumab, erlotinib and bortezomib (Table 3). Of note, a Phase II/III randomized trial is currently testing vorinostat plus radiation versus temozolomide plus radiation versus bevacizumab plus radiation as first-line treatment in young patients (3–21 years) with high-grade glioma (NCT01236560) (Table 3).
At this point, it is too early to make a firm statement on the use of HDACis in the treatment of brain tumors. The first Phase I/II completed trials indicate that single-agent HDACis have no or only modest efficacy. However, some trials indicate that combination treatment might be more effective and there seems to be a dose-dependent effect of HDACis. Since single patients do show responses, the greatest challenge still is to identify reliable biomarkers for patient selection in future clinical trials.
Predictive & pharmacodynamic biomarkers for HDACis in clinical trials
As with all molecularly defined therapeutic strategies, development of predictive markers allowing for stratification of patients in clinical trials will be crucial to the success of HDACis in the clinic. Data on predictive markers, however, are sparse, the most prominent marker today being HR23B, a protein involved in nucleotide excision repair, as well as the ubiquitin-mediated proteolytic pathway. First described by Khan et al. in cutaneous T-cell lymphoma, it has been shown that HR23B protein expression serves as a predictive marker for responsiveness to vorinostat, with strong HR23B protein expression predicting a good response to vorinostat [105]. Moreover, HR23B can easily be assessed in formalin-fixed paraffin-embedded samples, making it ideally suited for routine clinical application. HR23B has since been validated as a predictive marker for response to belinostat in hepatocellular carcinoma (HCC) by Yeo et al., indicating that this marker could possibly be valid for more than single tumor entities [106]. However, it remains to be shown if HR23B can serve as a predictive marker in the field of neuro-oncology. Other biomarkers predictive of poor response to HDAC inhibition include nuclear accumulation of STAT1 and high levels of nuclear pSTAT3 in malignant cells in skin samples of cutaneous T-cell lymphoma patients [107], and overexpression of 17 antioxidant genes in acute myeloid leukemia patient‘s peripheral blood cells [108].
Preclinical data from HCC in vivo studies indicate that the methylation status of a set of tumor suppressor genes is a predictive marker for HDAC class I inhibition, in general, and HDAC1 inhibition, in particular, with strong methylation predicting poor response [109]. Moreover, low ID2 gene expression has been proposed as a predictive marker in HCC [110]. Almost no data on HDAC expression as predictive markers exist. In brain tumor cell lines, the expression pattern of HDACs was shown not to be predictive for the responsiveness of these cell lines to HDACis [111].
Histone acetylation of peripheral blood mononuclear cells (PBMCs) has been proposed as a pharmocodynamic biomarker in the clinical setting since PBMCs can be collected easily and repetitively in patients during clinical trials. Indeed, histone H4 acetylation has been shown to be a strong positive predictor of response in a clinical trial applying vorinostat in breast cancer [112]. However, determination of histone acetylation in PBMCs strongly depends on the type of acetylation assay used, as well as the time point of PBMC collection, and is associated with a large variability. Pretreatment HDAC2 protein expression levels in PBMCs have been proposed as predictive markers for vorinostat response, since they correlated with the extent of the histone H4 acetylation change in a Phase I study [113].
Further studies on predictive and pharmacodynamic biomarkers in brain tumors and their molecular subgroups are, therefore, urgently required, if HDACis are to be successful in the clinic.
Conclusion & future perspective
In summary, the success of HDACis in the treatment of brain tumors will be largely dependent on:
Elucidation of the molecular function of single HDAC isoenzymes;
Development of class- or (possibly more desirably) isoenzyme-specific HDACis;
Patient selection by application of valid predictive markers and/or entity-specific isoenzyme inhibition;
Development of synergistic combination therapies.
Although preclinical and some clinical data suggest a promising action of unselective HDACis in brain tumors, we are still lacking a thorough understanding of individual HDAC isoenzyme function(s) in brain tumors and normal tissues. However, the understanding of molecular functions of HDAC isoenzymes is crucial if we are to improve cancer therapy by offering drugs that are more effective with a broad therapeutic window. This can be achieved with HDACis that have either higher specificity against cancer cells or higher selectivity for single HDAC isoenzymes. The basis for this is the development of class-and/or isoenzyme-selective HDACis; although very promising first steps have been undertaken, the panel of inhibitors applicable in the clinical setting needs to be significantly expanded.
As has been reported in numerous publications, expression of HDAC isoenzymes is highly variable, and is dependent on the tumor entity, and clinical and/or molecular subgroup within one entity. Not surprisingly, HDAC isoenzyme expression is often correlated with clinical course. Conversely, our knowledge on HDAC isoenzyme expression profiles and their predictive value for response to HDACis remains largely unexplored. Success and failure of clinical trials testing HDACis will, however, be highly dependent on selecting the right patient population that is clearly defined by molecular markers. Further efforts are required in the development of predictive and pharmacodynamic biomarkers, be it based on HDAC isoenzymes themselves, their downstream targets, or functionally linked or surrogate markers.
Since HDACi monotherapy will probably not be successful in the treatment of cancer patients, rational and synergistic combination therapies need to be fully characterized. This will require employment of adequate preclinical in vitro and in vivo models for the corresponding tumor entities and their molecular subgroups.
Footnotes
Financial & competing interests disclosure
The authors are supported by the Talents in Medicine Program of Heidelberg University Hospital to J Ecker. O Witt has scientific cooperations with MSD, Pharmacyclics, Bayer Healthcare, and consultant agreements with Novartis and Merck/USA. T Milde is supported by a grant from the B Braun foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: ▪ of interest ▪▪ of considerable interest
- 1.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 2.Yang X-J, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9(3):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waltregny D, Leval LD, Glénisson W, et al. Expression of histone deacetylase 8, a class I histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. Am. J. Pathol. 2004;165(2):553–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marks PA. Histone deacetylase inhibitors: a chemical genetics approach to understanding cellular functions. Biochim. Biophys. Acta. 2010;1799(10–12):717–725. doi: 10.1016/j.bbagrm.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Detailed review on histone deacetylase (HDAC) isoenzyme functions.
- 6.Atsumi A, Tomita A, Kiyoi H, Naoe T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML–RARα as a component of the N-CoR co-repressor complex to repress transcription in vivo . Biochem. Biophys. Res. Commun. 2006;345(4):1471–1480. doi: 10.1016/j.bbrc.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 7.Amann JM, Nip J, Strom DK, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell Biol. 2001;21(19):6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weichert W, Röske A, Gekeler V, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9(2):139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 9.Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin. Cancer Res. 2009;15(1):91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 10.Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin. Cancer Res. 2010;16(12):3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 11.Deubzer HE, Schier MC, Oehme I, et al. HDAC11 is a novel drug target in carcinomas. Int. J. Cancer. 2013;132(9):2200–2208. doi: 10.1002/ijc.27876. [DOI] [PubMed] [Google Scholar]
- 12.Giannini G, Cabri W, Fattorusso C, Rodriquez M. Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med. Chem. 2012;4(11):1439–1460. doi: 10.4155/fmc.12.80. [DOI] [PubMed] [Google Scholar]
- 13.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 14.Smith KT, Workman JL. Introducing the acetylome. Nat. Biotechnol. 2009;27(10):917–919. doi: 10.1038/nbt1009-917. [DOI] [PubMed] [Google Scholar]
- 15.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 16.Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics. 2013;7:47–60. doi: 10.2147/BTT.S29965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradner JE, West N, Grachan ML, et al. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ Thorough investigation of the selectivity profiles of HDAC inhibitors (HDACis).
- 18.Hockly E, Richon VM, Woodman B, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington‘s disease. Proc. Natl Acad. Sci. USA. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahnen E, Eyupoglu IY, Brichta L, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J. Neurochem. 2006;98(1):193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 20.Galanis E, Jaeckle KA, Maurer MJ, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a North Central Cancer Treatment Group study. J. Clin. Oncol. 2009;27(12):2052–2058. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooker JM, Kim SW, Alexoff D, et al. Histone deacetylase inhibitor, MS-275, exhibits poor brain penetration: PK studies of [C]MS-275 using positron emission tomography. ACS Chem. Neurosci. 2010;1(1):65–73. doi: 10.1021/cn9000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonini MV. From the cover: the benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc. Natl Acad. Sci. USA. 2006;103(5):1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280(2):211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22(5):1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 25.Krennhrubec K, Marshall BL, Hedglin M, Verdin E, Ulrich SM. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorg. Med. Chem. Lett. 2007;17(10):2874–2878. doi: 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 26.Lobera M, Madauss KP, Pohlhaus DT, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol. 2013;9(5):319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 27.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Lucio-Eterovic AKB, Cortez MaA, Valera ET, et al. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer. 2008;8(1):243. doi: 10.1186/1471-2407-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos B, Bermejo JL, Han L, et al. Expression of nuclear receptor corepressors and class I histone deacetylases in astrocytic gliomas. Cancer Sci. 2011;102(2):387–392. doi: 10.1111/j.1349-7006.2010.01792.x. [DOI] [PubMed] [Google Scholar]
- 30.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 31.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 32.Jones DT, Jager N, Kool M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones DT, Northcott PA, Kool M, Pfister SM. The role of chromatin remodeling in medulloblastoma. Brain Pathol. 2013;23(2):193–199. doi: 10.1111/bpa.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Northcott PA, Shih DJ, Peacock J, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugh TJ, Weeraratne SD, Archer TC, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Introduces a very useful cancer genomics online platform.
- 37.Kim C, Shah BP, Subramaniam P, Lee K-B. Synergistic induction of apoptosis in brain cancer cells by targeted codelivery of siRNA and anticancer drugs. Mol. Pharm. 2011;8(5):1955–1961. doi: 10.1021/mp100460h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh MM, Manton CA, Bhat KP, et al. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. Neuro Oncol. 2011;13(8):894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamannai M, Farhangi S, Truss M, et al. The inhibitor of growth 1 (ING1) is involved in trichostatin a-induced apoptosis and Caspase 3 signaling in p53-deficient glioblastoma cells. Oncol. Res. 2010;18(10):469–480. doi: 10.3727/096504010x12704916124828. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt N, Windmann S, Reifenberger G, Riemenschneider MJ. DNA hypermethylation and histone modifications downregulate the candidate tumor suppressor gene RRP22 on 22q12 in human gliomas. Brain Pathol. 2012;22(1):17–25. doi: 10.1111/j.1750-3639.2011.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J, Chen T, Liu J, et al. Loss of NECL1, a novel tumor suppressor, can be restored in glioma by HDAC inhibitor-trichostatin A through Sp1 binding site. Glia. 2009;57(9):989–999. doi: 10.1002/glia.20823. [DOI] [PubMed] [Google Scholar]
- 42.Vibhakar R, Foltz G, Yoon J-G, et al. Dickkopf-1 is an epigenetically silenced candidate tumor suppressor gene in medulloblastoma. Neuro Oncol. 2007;9(2):135–144. doi: 10.1215/15228517-2006-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiller SE, Ditzler SH, Pullar BJ, Olson JM. Response of preclinical medulloblastoma models to combination therapy with 13-cis retinoic acid and suberoylanilide hydroxamic acid (SAHA) J. Neurooncol. 2008;87(2):133–141. doi: 10.1007/s11060-007-9505-1. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36(3):131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin D, Ong JM, Hu J, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor: effects on gene expression and growth of glioma cells in vitro and in vivo . Clin. Cancer Res. 2007;13(3):1045–1052. doi: 10.1158/1078-0432.CCR-06-1261. [DOI] [PubMed] [Google Scholar]
- 46.Komata T, Kanzawa T, Nashimoto T. Histone deacetylase inhibitors, N-butyric acid and trichostatin A, induce Caspase-8- but not Caspase-9-dependent apoptosis in human malignant glioma cells. Int. J. Oncol. 2005;26(5):1345–1352. doi: 10.3892/ijo.26.5.1345. [DOI] [PubMed] [Google Scholar]
- 47.Bangert A, Häcker S, Cristofanon S, Debatin K-M, Fulda S. Chemosensitization of glioblastoma cells by the histone deacetylase inhibitor MS275. Anticancer Drugs. 2011;22(6):494–499. doi: 10.1097/CAD.0b013e32834631e0. [DOI] [PubMed] [Google Scholar]
- 48.Egler V, Korur S, Failly M, et al. Histone deacetylase inhibition and blockade of the glycolytic pathway synergistically induce glioblastoma cell death. Clin. Cancer Res. 2008;14(10):3132–3140. doi: 10.1158/1078-0432.CCR-07-4182. [DOI] [PubMed] [Google Scholar]
- 49.Rahman R, Osteso-Ibanez T, Hirst RA, et al. Histone deacetylase inhibition attenuates cell growth with associated telomerase inhibition in high-grade childhood brain tumor cells. Mol. Cancer Ther. 2010;9(9):2568–2581. doi: 10.1158/1535-7163.MCT-10-0272. [DOI] [PubMed] [Google Scholar]
- 50.Lopez CA, Feng FY, Herman JM, Nyati MK, Lawrence TS, Ljungman M. Phenylbutyrate sensitizes human glioblastoma cells lacking wild-type P53 function to ionizing radiation. Int. J. Radiat. Oncol. Biol. Phys. 2007;69(1):214–220. doi: 10.1016/j.ijrobp.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 51.Xu J, Sampath D, Lang FF, et al. Vorinostat modulates cell cycle regulatory proteins in glioma cells and human glioma slice cultures. J. Neurooncol. 2011;105(2):241–251. doi: 10.1007/s11060-011-0604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Premkumar DR, Jane EP, Agostino NR, Didomenico JD, Pollack IF. Bortezomib-induced sensitization of malignant human glioma cells to vorinostat-induced apoptosis depends on reactive oxygen species production, mitochondrial dysfunction, Noxa upregulation, Mcl-1 cleavage, and DNA damage. Mol. Carcinog. 2013;52(2):118–133. doi: 10.1002/mc.21835. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Develops rationales for intelligent combination therapies: HDACis and proteasome inhibitors.
- 53.Khaw AK, Silasudjana M, Banerjee B, Suzuki M, Baskar R, Hande MP. Inhibition of telomerase activity and human telomerase reverse transcriptase gene expression by histone deacetylase inhibitor in human brain cancer cells. Mutat. Res. 2007;625(1–2):134–144. doi: 10.1016/j.mrfmmm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Jin H, Liang L, Liu L-F, Deng W-P, Liu J-W. HDAC inhibitor DWP0016 activates p53 transcription and acetylation to inhibit cell growth in U251 glioblastoma cells. J. Cell. Biochem. 2012;114(7):98–09. doi: 10.1002/jcb.24491. [DOI] [PubMed] [Google Scholar]
- 55.Khan Z, Akhtar M, Ekström TJ. HDAC inhibitor 4-phenylbutyrate preserves immature phenotype of human embryonic midbrain stem cells: implications for the involvement of DNA methyltransferase. Int. J. Mol. Med. 2011;28(6):977–983. doi: 10.3892/ijmm.2011.791. [DOI] [PubMed] [Google Scholar]
- 56.Li XN, Parikh S, Shu Q, et al. Phenylbutyrate and phenylacetate induce differentiation and inhibit proliferation of human medulloblastoma cells. Clin. Cancer Res. 2004;10(3):1150–1159. doi: 10.1158/1078-0432.ccr-0747-3. [DOI] [PubMed] [Google Scholar]
- 57.Morita K, Gotohda T, Arimochi H, Lee M-S, Her S. Histone deacetylase inhibitors promote neurosteroid-mediated cell differentiation and enhance serotonin-stimulated brain-derived neurotrophic factor gene expression in rat C6 glioma cells. J. Neurosci. Res. 2009;87(11):2608–2614. doi: 10.1002/jnr.22072. [DOI] [PubMed] [Google Scholar]
- 58.Svechnikova I, Almqvist PM, Ekström TJ. HDAC inhibitors effectively induce cell type-specific differentiation in human glioblastoma cell lines of different origin. Int. J. Oncol. 2008;32(4):821–827. [PubMed] [Google Scholar]
- 59.Milde T, Lodrini M, Savelyeva L, et al. HD-MB03 is a novel group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. J. Neurooncol. 2012;110(3):335–348. doi: 10.1007/s11060-012-0978-1. [DOI] [PubMed] [Google Scholar]
- 60.Her S, Lee M-S, Morita K. Trichostatin A stimulates steroid 5α-reductase gene expression in rat C6 glioma cells via a mechanism involving Sp1 and Sp3 transcription factors. J. Mol. Neurosci. 2010;41(2):252–262. doi: 10.1007/s12031-009-9284-6. [DOI] [PubMed] [Google Scholar]
- 61.Taylor P, Fangusaro J, Rajaram V, et al. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Mol. Cancer Ther. 2012;11(8):1713–1723. doi: 10.1158/1535-7163.MCT-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei L, Hong S, Yoon Y, et al. Early prediction of response to Vorinostat in an orthotopic rat glioma model. NMR Biomed. 2012;25(9):1104–1111. doi: 10.1002/nbm.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furchert SE, Lanvers-Kaminsky C, Juürgens H, Jung M, Loidl A, Frühwald MC. Inhibitors of histone deacetylases as potential therapeutic tools for high-risk embryonal tumors of the nervous system of childhood. Int. J. Cancer. 2007;120(8):1787–1794. doi: 10.1002/ijc.22401. [DOI] [PubMed] [Google Scholar]
- 64.Sonnemann J, Kumar KS, Heesch S, et al. Histone deacetylase inhibitors induce cell death and enhance the susceptibility to ionizing radiation, etoposide, and TRAIL in medulloblastoma cells. Int. J. Oncol. 2006;28(3):755–766. [PubMed] [Google Scholar]
- 65.Rahman R, Osteso-Ibanez T, Hirst RA, et al. Histone deacetylase inhibition attenuates cell growth with associated telomerase inhibition in high-grade childhood brain tumor cells. Mol. Cancer Ther. 2010;9(9):2568–2581. doi: 10.1158/1535-7163.MCT-10-0272. [DOI] [PubMed] [Google Scholar]
- 66.Milde T, Kleber S, Korshunov A, et al. A novel human high-risk ependymoma stem cell model reveals the differentiation-inducing potential of the histone deacetylase inhibitor Vorinostat. Acta Neuropathol. 2012;122(5):637–650. doi: 10.1007/s00401-011-0866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sawa H, Murakami H, Kumagai M, et al. Histone deacetylase inhibitor, FK228, induces apoptosis and suppresses cell proliferation of human glioblastoma cells in vitro and in vivo . Acta Neuropathol. 2004;107(6):523–531. doi: 10.1007/s00401-004-0841-3. [DOI] [PubMed] [Google Scholar]
- 68.Hsu Y-F, Sheu J-R, Hsiao G, et al. p53 in trichostatin A induced C6 glioma cell death. Biochim. Biophys. Acta. 2011;1810(5):504–513. doi: 10.1016/j.bbagen.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Ugur HC, Ramakrishna N, Bello L, et al. Continuous intracranial administration of suberoylanilide hydroxamic acid (SAHA) inhibits tumor growth in an orthotopic glioma model. J. Neurooncol. 2007;83(3):267–275. doi: 10.1007/s11060-007-9337-z. [DOI] [PubMed] [Google Scholar]
- 70.Häcker S, Karl S, Mader I, et al. Histone deacetylase inhibitors prime medulloblastoma cells for chemotherapy-induced apoptosis by enhancing p53-dependent Bax activation. Oncogene. 2011;30(19):2275–2281. doi: 10.1038/onc.2010.599. [DOI] [PubMed] [Google Scholar]; ▪ Develops rationales for intelligent combination therapies: HDACis and chemotherapeutics.
- 71.Häcker S, Dittrich A, Mohr A, et al. Histone deacetylase inhibitors cooperate with IFN-γ to restore Caspase-8 expression and overcome TRAIL resistance in cancers with silencing of Caspase-8. Oncogene. 2009;28(35):3097–3110. doi: 10.1038/onc.2009.161. [DOI] [PubMed] [Google Scholar]
- 72.Bangert A, Cristofanon S, Eckhardt I, et al. Histone deacetylase inhibitors sensitize glioblastoma cells to TRAIL-induced apoptosis by c-myc-mediated downregulation of cFLIP. Oncogene. 2012;31(44):4677–4688. doi: 10.1038/onc.2011.614. [DOI] [PubMed] [Google Scholar]; ▪ Develops rationales for intelligent combination therapies: HDACis and TRAIL.
- 73.Sharma V, Koul N, Joseph C, Dixit D, Ghosh S, Sen E. HDAC inhibitor, scriptaid, induces glioma cell apoptosis through JNK activation and inhibits telomerase activity. J. Cell. Mol. Med. 2010;14(8):2151–2161. doi: 10.1111/j.1582-4934.2009.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ungerstedt JS. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA. 2005;102(3):673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camphausen K. Enhanced radiation-induced cell killing and prolongation of H2AX foci expression by the histone deacetylase inhibitor MS-275. Cancer Res. 2004;64(1):316–321. doi: 10.1158/0008-5472.can-03-2630. [DOI] [PubMed] [Google Scholar]
- 76.Kitange GJ, Mladek AC, Carlson BL, et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin. Cancer Res. 2012;18(15):4070–4079. doi: 10.1158/1078-0432.CCR-12-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu C, Friday BB, Yang L, et al. Mitochondrial Bax translocation partially mediates synergistic cytotoxicity between histone deacetylase inhibitors and proteasome inhibitors in glioma cells. Neuro Oncol. 2008;10(3):309–319. doi: 10.1215/15228517-2007-063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ljungman M. The influence of chromatin structure on the frequency of radiation-induced DNA strand breaks: a study using nuclear and nucleoid monolayers. Radiat. Res. 1991;126(1):58–64. [PubMed] [Google Scholar]
- 79.Shabason JE, Tofilon PJ, Camphausen K. Grand rounds at the National Institutes of Health: HDAC inhibitors as radiation modifiers, from bench to clinic. J. Cell. Mol. Med. 2011;15(12):2735–2744. doi: 10.1111/j.1582-4934.2011.01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chinnaiyan P, Cerna D, Burgan WE, et al. Postradiation sensitization of the histone deacetylase inhibitor valproic acid. Clin. Cancer Res. 2008;14(17):5410–5415. doi: 10.1158/1078-0432.CCR-08-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar KS, Sonnemann J, Hong le TT, et al. Histone deacetylase inhibitors, but not vincristine, cooperate with radiotherapy to induce cell death in medulloblastoma. Anticancer Res. 2007;27(1A):465–470. [PubMed] [Google Scholar]
- 82.Chinnaiyan P, Vallabhaneni G, Armstrong E, Huang S-M, Harari PM. Modulation of radiation response by histone deacetylase inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2005;62(1):223–229. doi: 10.1016/j.ijrobp.2004.12.088. [DOI] [PubMed] [Google Scholar]
- 83.Manova V, Singh SK, Iliakis G. Processing of DNA double strand breaks by alternative non-homologous end-joining in hyperacetylated chromatin. Genome Integr. 2012;3(1):4. doi: 10.1186/2041-9414-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Osuka S, Takano S, Watanabe S, Ishikawa E, Yamamoto T, Matsumura A. Valproic acid inhibits angiogenesis in vitro and glioma angiogenesis in vivo in the brain. Neurol. Med. Chir. (Tokyo) 2012;52(4):186–193. doi: 10.2176/nmc.52.186. [DOI] [PubMed] [Google Scholar]
- 85.Wang M, Wang T, Liu S, Yoshida D, Teramoto A. The expression of matrix metalloproteinase-2 and -9 in human gliomas of different pathological grades. Brain Tumor Pathol. 2003;20(2):65–72. doi: 10.1007/BF02483449. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, Tsai Y-H, Tseng S-H. Valproic acid affected the survival and invasiveness of human glioma cells through diverse mechanisms. J. Neurooncol. 2012;109(1):23–33. doi: 10.1007/s11060-012-0871-y. [DOI] [PubMed] [Google Scholar]
- 87.Papi A, Ferreri AM, Rocchi P, Guerra F, Orlandi M. Epigenetic modifiers as anticancer drugs: effectiveness of valproic acid in neural crest-derived tumor cells. Anticancer Res. 2010;30(2):535–540. [PubMed] [Google Scholar]
- 88.Konduri SD, Srivenugopal KS, Yanamandra N, et al. Promoter methylation and silencing of the tissue factor pathway inhibitor-2 (TFPI-2), a gene encoding an inhibitor of matrix metalloproteinases in human glioma cells. Oncogene. 2003;22(29):4509–4516. doi: 10.1038/sj.onc.1206695. [DOI] [PubMed] [Google Scholar]
- 89.An Z, Gluck CB, Choy ML, Kaufman LJ. Suberoylanilide hydroxamic acid limits migration and invasion of glioma cells in two and three dimensional culture. Cancer Lett. 2010;292(2):215–227. doi: 10.1016/j.canlet.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Meyerson M. Role of telomerase in normal and cancer cells. J. Clin. Oncol. 2000;18(13):2626–2634. doi: 10.1200/JCO.2000.18.13.2626. [DOI] [PubMed] [Google Scholar]
- 91.Rahman R, Grundy R. Histone deacetylase inhibition as an anticancer telomerase-targeting strategy. Int. J. Cancer. 2011;129(12):2765–2774. doi: 10.1002/ijc.26241. [DOI] [PubMed] [Google Scholar]
- 92.Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2012;39(5):444–456. doi: 10.1016/j.ctrv.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 93.Sarcar B, Kahali S, Chinnaiyan P. Vorinostat enhances the cytotoxic effects of the topoisomerase I inhibitor SN38 in glioblastoma cell lines. J. Neurooncol. 2010;99(2):201–207. doi: 10.1007/s11060-010-0127-7. [DOI] [PubMed] [Google Scholar]
- 94.Espada J, Ballestar E, Fraga MF, et al. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279(35):37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 95.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Ecke I, Petry F, Rosenberger A, et al. Antitumor effects of a combined 5-aza-2´deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in ptch mutant mice. Cancer Res. 2009;69(3):887–895. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 97.Witt O, Schweigerer L, Driever PHI, Wolff J, Pekrun A. Valproic acid treatment of glioblastoma multiforme in a child. Pediatr. Blood Cancer. 2004;43(2):181. doi: 10.1002/pbc.20083. [DOI] [PubMed] [Google Scholar]
- 98.Deboer R, Batjer H, Marymont M, et al. Response of an adult patient with pineoblastoma to vorinostat and retinoic acid. J. Neurooncol. 2009;95(2):289–292. doi: 10.1007/s11060-009-9921-5. [DOI] [PubMed] [Google Scholar]
- 99.Wolff JEA, Kramm C, Kortmann R-D, et al. Valproic acid was well tolerated in heavily pretreated pediatric patients with high-grade glioma. J. Neurooncol. 2008;90(3):309–314. doi: 10.1007/s11060-008-9662-x. [DOI] [PubMed] [Google Scholar]
- 100.Barker CA, Bishop AJ, Chang M, Beal K, Chan TA. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int. J. Radiat. Oncol. Biol. Phys. 2013;86(3):504–509. doi: 10.1016/j.ijrobp.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iwamoto FM, Lamborn KR, Kuhn JG, et al. A Phase I/II trial of the histone deacetylase inhibitor romidepsin for adults with recurrent malignant glioma: North American Brain Tumor Consortium Study 03–03. Neuro Oncol. 2011;13(5):509–516. doi: 10.1093/neuonc/nor017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Drappatz J, Lee EQ, Hammond S, et al. Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma. J. Neurooncol. 2012;107(1):133–138. doi: 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- 103.Chinnaiyan P, Chowdhary S, Potthast L, et al. Phase I trial of vorinostat combined with bevacizumab and CPT-11 in recurrent glioblastoma. Neuro Oncol. 2011;14(1):93–100. doi: 10.1093/neuonc/nor187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Friday BB, Anderson SK, Buckner J, et al. Phase II trial of vorinostat in combination with bortezomib in recurrent glioblastoma: a north central cancer treatment group study. Neuro Oncol. 2012;14(2):215–221. doi: 10.1093/neuonc/nor198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khan O, Fotheringham S, Wood V, et al. HR23B is a biomarker for tumor sensitivity to HDAC inhibitor-based therapy. Proc. Natl Acad. Sci. USA. 2010;107(14):6532–6537. doi: 10.1073/pnas.0913912107. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ First clinical description of HR23B as a predictive marker for HDACi treatment.
- 106.Yeo W, Chung HC, Chan SL, et al. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter Phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J. Clin. Oncol. 2012;30(27):3361–3367. doi: 10.1200/JCO.2011.41.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fantin VR, Loboda A, Paweletz CP, et al. Constitutive activation of signal transducers and activators of transcription predicts vorinostat resistance in cutaneous T-cell lymphoma. Cancer Res. 2008;68(10):3785–3794. doi: 10.1158/0008-5472.CAN-07-6091. [DOI] [PubMed] [Google Scholar]
- 108.Garcia-Manero G, Yang H, Bueso-Ramos C, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111(3):1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 109.Wong JC, Tang G, Wu X, et al. Pharmacokinetic optimization of class-selective histone deacetylase inhibitors and identification of associated candidate predictive biomarkers of hepatocellular carcinoma tumor response. J. Med. Chem. 2012;55(20):8903–8925. doi: 10.1021/jm3011838. [DOI] [PubMed] [Google Scholar]
- 110.Tsunedomi R, Iizuka N, Harada S, Oka M. Susceptibility of hepatoma-derived cells to histone deaaylase inhibitors is associated with ID2 expression. Int. J. Oncol. 2013;42(4):1159–1166. doi: 10.3892/ijo.2013.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Graham C, Tucker C, Creech J, et al. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo . Clin. Cancer Res. 2006;12(1):223–234. doi: 10.1158/1078-0432.CCR-05-1225. [DOI] [PubMed] [Google Scholar]
- 112.Munster PN, Thurn KT, Thomas S, et al. A Phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br. J. Cancer. 2011;104(12):1828–1835. doi: 10.1038/bjc.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Munster PN, Marchion D, Thomas S, et al. Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br. J. Cancer. 2009;101(7):1044–1050. doi: 10.1038/sj.bjc.6605293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hrebackova J, Hrabeta J, Eckschlager T. Valproic acid in the complex therapy of malignant tumors. Curr. Drug Targets. 2010;11(3):361–379. doi: 10.2174/138945010790711923. [DOI] [PubMed] [Google Scholar]
- 115.Voso MT, Santini V, Finelli C, et al. Valproic acid at therapeutic plasma levels may increase 5-azacytidine efficacy in higher risk myelodysplastic syndromes. Clin. Cancer Res. 2009;15(15):5002–5007. doi: 10.1158/1078-0432.CCR-09-0494. [DOI] [PubMed] [Google Scholar]
- 116.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat. Rev. Neurosci. 2006;7(10):784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 117.Gore SD, Weng L-J, Figg WD, et al. Impact of prolonged infusions of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin. Cancer Res. 2002;8(4):963–970. [PubMed] [Google Scholar]
- 118.Palmieri D, Lockman PR, Thomas FC, et al. Vorinostat inhibits brain metastatic colonization in a model of triple-negative breast cancer and induces DNA double-strand breaks. Clin. Cancer Res. 2009;15(19):6148–6157. doi: 10.1158/1078-0432.CCR-09-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fakih MG, Fetterly G, Egorin MJ, et al. A Phase I, pharmacokinetic, and pharmacodynamic study of two schedules of vorinostat in combination with 5-fluorouracil and leucovorin in patients with refractory solid tumors. Clin. Cancer Res. 2010;16(14):3786–3794. doi: 10.1158/1078-0432.CCR-10-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garbes L, Riessland M, Holker I, et al. LBH589 induces up to 10-fold SMN protein levels by several independent mechanisms and is effective even in cells from SMA patients non-responsive to valproate. Hum. Mol. Genet. 2009;18(19):3645–3658. doi: 10.1093/hmg/ddp313. [DOI] [PubMed] [Google Scholar]
- 121.Rathkopf D, Wong BY, Ross RW, et al. A Phase I study of oral panobinostat alone and in combination with docetaxel in patients with castration-resistant prostate cancer. Cancer Chemother. Pharmacol. 2010;66(1):181–189. doi: 10.1007/s00280-010-1289-x. [DOI] [PubMed] [Google Scholar]
- 122.Warren KE, Mccully C, Dvinge H, et al. Plasma and cerebrospinal fluid pharmacokinetics of the histone deacetylase inhibitor, belinostat (PXD101), in non-human primates. Cancer Chemother. Pharmacol. 2008;62(3):433–437. doi: 10.1007/s00280-007-0622-5. [DOI] [PubMed] [Google Scholar]
- 123.Steele NL, Plumb JA, Vidal L, et al. A Phase 1 pharmacokinetic and pharmacodynamic study of the histone deacetylase inhibitor belinostat in patients with advanced solid tumors. Clin. Cancer Res. 2008;14(3):804–810. doi: 10.1158/1078-0432.CCR-07-1786. [DOI] [PubMed] [Google Scholar]
- 124.Evens AM, Ai W, Balasubramanian S, et al. 51st Ash Annual Meeting and Exposition. New Orleans, LA, USA: 5–8 December 2009. Phase I analysis of the safety and pharmacodynamics of the broad spectrum histone deacetylase inhibitor (HDACi) PCI-24781 in relapsed and refractory lymphoma. Presented at. [Google Scholar]
- 125.Bono JSD, Kristeleit R, Tolcher A, et al. Phase I pharmacokinetic and pharmacodynamic study of LAQ824, a hydroxamate histone deacetylase inhibitor with a heat shock protein-90 inhibitory profile, in patients with advanced solid tumors. Clin. Cancer Res. 2008;14(20):6663–6673. doi: 10.1158/1078-0432.CCR-08-0376. [DOI] [PubMed] [Google Scholar]
- 126.Furlan A, Monzani V. Pharmacokinetics, safety and inducible cytokine responses during a Phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat) Mol. Med. 2011;17(5–6):353–362. doi: 10.2119/molmed.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kummar S, Gutierrez M, Gardner ER, et al. Phase I trial of MS-275, a histone deacetylase inhibitor, administered weekly in refractory solid tumors and lymphoid malignancies. Clin. Cancer Res. 2007;13(18):5411–5417. doi: 10.1158/1078-0432.CCR-07-0791. [DOI] [PubMed] [Google Scholar]
- 128.Sandor V, Bakke S, Robey RW, et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin. Cancer Res. 2002;8:718–728. [PubMed] [Google Scholar]
- 129.Gravendeel LaM, Kouwenhoven MCM, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 130.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robinson G, Parker M, Kranenburg TA, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
▪ Websites
- 201.http://cpla.biocuckoo.org/ GPS The Cuckoo Workgroup. CPLA introduction.
- 202.http://r2.amc.nl R2: microarray analysis and visualization platform.
- 203.https://tcga-data.nci.nih.gov/tcga/ The Cancer Genome Atlas Data Portal. TCGA Data Portal Overview.
- 204.www.cbioportal.org/ Memorial Sloan–Kettering Cancer Center. cBioPortal for Cancer Genomics.
- 205.www.broadinstitute.org/cancer/cga/mutsig Cancer Genome Analysis. MutSig.