Abstract
Microtubules (MTs) and associated proteins can self-organize into complex structures such as the bipolar spindle, a process in which RanGTP plays a major role. Addition of RanGTP to M-phase Xenopus egg extracts promotes the nucleation and self-organization of MTs into asters and bipolar-like structures in the absence of centrosomes or chromosomes. We show here that the complex proteome of these RanGTP-induced MT assemblies is similar to that of mitotic spindles. Using proteomic profiling we show that MT self-organization in the M-phase cytoplasm involves the non-linear and non-stoichiometric recruitment of proteins from specific functional groups. Our study provides for the first time a temporal understanding of the protein dynamics driving MT self-organization in M-phase.
Keywords: Mitosis, Cell division, Cell biology, Protein complex analysis, Protein-Protein Interactions, Microtubule, Proteomic profiling, RanGTP, Self-organization, Spindle
The ability to self-organize molecular machines and higher order complex forms is an essential feature of living systems (1, 2). Self-organization occurs at different scales from animals to tissues, from cells to molecules. Microtubules (MTs)1 polymerize through a self-assembly process of tubulin dimers and typically alternate between phases of growth and shrinkage, an intrinsic property termed dynamic instability (3). In vitro MTs form patterns, but in combination with molecular motors they self-organize into defined structures such as asters, bundles, and networks (4–6). These basic properties may provide some principles underlying their self-organization into more complex cellular structures such as the bipolar spindle (1, 6, 7). Although, there is an extensive list of spindle components obtained by proteomics (8–11), genetic screening (12, 13), and targeted functional studies, how all these components drive the robust self-assembly of the bipolar spindle is not fully understood yet. Moreover, it is unclear how all these components contribute or drive the assembly of such a dynamic structure.
The Xenopus laevis egg extract system has been instrumental for understanding the molecular basis of M-phase MT assembly in the proximity of chromosomes and define a pathway essential for spindle assembly both in meiosis and mitosis (14). This pathway is triggered by the GTP bound form of the small GTPase Ran (15, 16). Indeed, the addition of RanGTP to M-phase Xenopus egg extracts is sufficient to drive MT nucleation and organization into asters and bipolar-like structures, called mini-spindles. The mechanism at play involves the release of a class of Nuclear Localization Signal (NLS) containing proteins from inhibitory interactions with importins. These so-called Spindle Assembly Factors (SAFs) have essential roles in RanGTP-dependent MT nucleation, stabilization, and organization, and thereby in bipolar spindle assembly (17).
Here, using mass spectrometry-based proteomics analysis we first validate the RanGTP-dependent MT assemblies as complex spindle-like structures. We then exploit this system to look for general principles driving MT and spindle self-organization. Our data suggest the non-linear recruitment of specific functional protein groups to MTs as they self-organize.
EXPERIMENTAL PROCEDURES
Xenopus Egg Extracts and RanGTP Dependent MTs
Xenopus laevis female and male frogs were purchased from Nasco and were used at an age between 1 and 3 years. All experiments involving animals were performed according to standard protocols approved by the ethical committee of the Parc de Recerca Biomédica of Barcelona, Barcelona, Spain. Fresh cytosolic-factor-arrested Xenopus egg extracts (CSF extract) were prepared as previously described (18) with the following modifications: Cytochalasin D was added to a final concentration of 13.3 μg/ml (instead of 20 μg/ml) and 1.5–2 ml of CSF extract were clarified by a second centrifugation step in 2 ml tubes for 15 min at 15,000 × g. After centrifugation, the clarified cytoplasmic layer was carefully retrieved using a 2 ml syringe and a 1.2 × 40 mm needle taking care not to disturb the black pellet and the lipid top layer. Clarified extracts quality was assessed by checking spindle assembly around exogenously added demembranated Xenopus sperm nuclei as described (18) and MT aster and mini spindle assembly on addition of RanQ69L·GTP.
RanQ69L·GTP Purification
His-RanQ69L was expressed in BL21 (DE3) Rep4 E. Coli for 3 h at 30 °C after induction with 2 mm IPTG. Cells were pelleted and washed using cold PBS. Cells were lysed by incubation with Lysozyme (100 μg/ml, 15 min at room temperature under constant stirring) in lysis buffer (PBS, 500 mm NaCl, 0.1 mm MgCl2, 0.5% Triton X-100, 1 mm DTT, 0.1 mm PMSF, 0.1 mm GTP). Cell lysates were cleared first by centrifugation (30 min, 27,000 rcf at 4 °C) and then by filtering using a low binding filter 0.45 μm (Millex - HP Filter Unit, Fast Flow & Low Binding Millipore, Burlington, MA, ref SLHP033RS). The protein was purified by affinity chromatography using a 1 ml HisTRAP HP column (GE Healthcare, Chicago, IL, ref 17–5247-01) on FPLC-AKTA purifier. After incubation of the cell lysate, the column was washed with 10 column volumes of washing buffer (PBS, 500 mm NaCl, 0.1 mm MgCl2, 0.1 mm GTP, 40 mm Imidazole, pH 7.7). Elution was performed by collecting 500 μl fractions while mixing in gradually increasing concentrations of Elution buffer (PBS, 500 mm NaCl, 0.1 mm MgCl2, 0.1 mm GTP, 500 mm Imidazole) with Wash buffer. Fractions corresponding to the peak (around 150 mm Imidazole) were pooled and dialyzed against CSF-XB. GTP was loaded onto RanQ69L by incubating the purified protein with 1 mm GTP for 2 h on ice. After incubation, 20 mm MgCl2 were added drop by drop to avoid precipitation of the protein. RanQ69L·GTP was concentrated using Amicon Ultracel-3k membrane (Millipore, ref UFC900324) at the minimum concentration of 150 μm.
RanGTP Asters and Mini-spindle Assembly in Xenopus Egg Extracts
RanQ69L·GTP was added to the clarified egg extract at 15 μm (19), together with rhodamine-labeled tubulin. The reactions were incubated in a water bath at 20 °C and the formation of MT asters followed by squashing 1 μl of the reaction mixture and 3 μl of fix solution (11% formaldehyde, 48% glycerol, 1 μg/ml Hoechst in CSF-XB buffer (10 mm Hepes, 100 mm KCl, 0,1 mm CaCl2, 2 mm MgCl2, 50 mm sucrose, 5 mm EGTA)) under a coverslip. Samples were taken at 15, 20, 30, and 50 min and analyzed using an inverted DMI-6000-B Leica (Wetzlar, Germany) wide-field fluorescent microscope.
For the purification of RanGTP dependent MT assemblies, 200 μl of clarified extract was supplemented with RanQ69L·GTP and incubated in a water bath 20 °C for 15, 20, 30, or 50 min. Meanwhile 5 ml of aster cushion (25% glycerol, 1× BRB80, in water) were added to thin wall, ultra-clear centrifuge tubes 16 × 102 mm (Beckman Coulter, Brea, CA, ref 344061). After the indicated time, 1 ml of aster dilution (10% glycerol, 1x BRB80, 0,1% TritonX-100, 1 mm GTP) was vigorously added to the egg extract reaction and further mixed by inversion 5 times. The reaction was then laid on top of the 5 ml aster cushion and centrifuged for 20 min, at 3200 × g at room temperature. Cushion was aspirated and the pellet resuspended in 50 μl of aster dilution buffer supplemented with 20 μm Taxol (T7402, Sigma, St Louis, MO). The resuspended pellet was carefully laid on top of 800 μl of aster cushion supplemented with 1 mm GTP and 20 μm Taxol in a 2 ml tube. Samples were centrifuged for 20 min, at 4800 × g at room temperature. Cushion was aspirated and the pellet resuspended in 35 μl of Laemmli buffer, transferred to a clean tube and boiled for 10 min at 95 °C. Five sixth of the resulting samples were run on a precasted gradient gel (4- 15% Criterion TGX, 12 + 2 wells, 45 μl, BioRad, Hercules, California, ref 567–1083) for 45 min at 60mA. Gels were stained over night using colloidal blue (Invitrogen Life Technologies, Carlsbad, CA, LC6025). Gels were cut into 6 bands and processed for proteomic analysis.
Sample Preparation for Mass Spectrometry Analysis
Gel bands were destained with 40% acetonitrile (ACN) +100 mm ammonium bicarbonate, reduced with dithiothreitol (10 mm, 30 min, 56 °C), alkylated in the dark with iodoacetamide (55 mm, 30 min, 25 °C), dehydrated with ACN and digested with 0.7 μg of trypsin (Promega, Madison, MN, cat # V5113) overnight at 37 °C. After digestion, peptides were extracted and cleaned up on a homemade Empore C18 column (3M, St. Paul, MN) (20).
Samples were analyzed using a LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to an EasyLC (Thermo Fisher Scientific). Peptides were loaded directly onto the analytical column at 1.5–2 μl/min using a wash-volume of 4 to 5 times injection volume and separated by reverse-phase chromatography using a 12-cm column with an inner diameter of 75 μm, packed with 5 μm C18 particles (Nikkyo Technos Co., Ltd., Tokyo, Japan). Chromatographic gradients started at 97% buffer A and 3% buffer B with a flow rate of 300 nl/min, and gradually increased to 93% buffer A and 7% buffer B in 1 min, and to 65% buffer A/35% buffer B in 90 min. After each analysis, the column was washed for 10 min with 10% buffer A/90% buffer B. Buffer A: 0.1% formic acid in water. Buffer B: 0.1% formic acid in acetonitrile.
The mass spectrometer was operated in positive ionization mode with nanospray voltage set at 2.2 kV and source temperature at 250 °C. Ultramark 1621 for the FT mass analyzer was used for external calibration before the analyses. Moreover, an internal calibration was also performed using the background polysiloxane ion signal at m/z 445.1200. The instrument was operated in DDA mode and full MS scans with 1 micro scans at resolution of 60000 were used over a mass range of m/z 350–2000 with detection in the Orbitrap. Auto gain control (AGC) was set to 1E6, dynamic exclusion (60 s) and charge state filtering disqualifying singly charged peptides was activated. In each cycle of DDA analysis, following each survey scan the top ten most intense ions with multiple charged ions above a threshold ion count of 5000 were selected for fragmentation at normalized collision energy of 35%. Fragment ion spectra produced via collision-induced dissociation (CID) were acquired in the Ion Trap, AGC was set to 5e4, isolation window of 2.0 m/z, activation time of 0.1ms and maximum injection time of 100 ms was used. All data were acquired with Xcalibur software v2.2.
Protein Identification and Quantification
The MaxQuant software suite (v1.5.5.1) was used for peptide identification and label-free protein quantitation (21). The data were searched against an in-house generated database containing all proteins corresponding to phrog database (22) and the corresponding decoy entries (release January 2016, 79214 entries). A precursor ion mass tolerance of 4.5 ppm at the MS1 level was used, and up to three missed cleavages for trypsin were allowed. The fragment ion mass tolerance was set to 0.5 Da. Oxidation of methionine, protein acetylation at the N-terminal defined as variable modification; whereas carbamidomethylation on cysteines was set as a fix modification. The minimum number of razor and unique peptides (“min. razor + unique”) for a protein group to be considered as identified was set to 1. Identified peptides and proteins were respectively filtered using a 1% FDR. Protein areas were calculated using the average of the three most intense unique peptides per the protein group. In cases in which two or more protein groups mapped to the same human reference gene, a consensus area was calculated with the sum of the different protein groups.
Protein Dynamics Analysis
The average of the intensity areas obtained in the three replicas at a given time point was calculated for each protein. The relative protein variation was obtained by calculating the difference of the average intensity of a given protein in a certain time point with the median of the protein intensity (from all the samples) resulting in a dynamics profile of each protein. We normalized the profile of each protein using the relative variation of the total tubulin that was detected. By doing so, the profile of the total tubulin became flat (centered on zero), and the values for each protein corresponded to the difference between the relative abundance of the protein and the relative abundance of the total tubulin. As an example and quality control of the normalization process, the integrity of specific protein complexes was monitored, as the TUBGCPs (supplemental Fig. S5A) and SKA proteins.
A protein was considered to bind nonstiochiometrically to MTs and defined as dynamic when its deviation from the tubulin average at any given time point was higher than the error of the assay. The error of the assay (0.9) was defined by the maximum difference between the “total of tubulin alpha” and “total of tubulin beta” after normalization.
Clustering Analysis
The Perseus (version 1.6.0.7) software was used for the unbiased clustering analysis using the relative abundance of the dynamics proteins. Hierarchical clustering was done using Pearson correlation as the distance parameter and average as the linkage. To remove the randomly fluctuating proteins 25 clusters were defined in the first clustering. All clusters showing random fluctuations of the relative protein abundance profiles were eliminated before the last clustering analysis using the same parameters that resulted in seven clusters.
Gene Ontology Enrichment Analysis
The online CleverGO tool (http://www.tartaglialab.com/GO_analyser/universal) was used to determine the Cellular Content of the RanMT proteome and obtain the corresponding graphical representation. The results were obtained using the following parameters in the software: Similarity strength threshold, 0.75; Minimal precision, 0.5; Minimal Lvl, 4; p value cutoff, 0.01. The AmiGO 2 software was used (http://amigo.geneontology.org/) to obtain the list of proteins having GOs related to spindle, microtubules, centrosomes, or nucleus.
Network Analysis
The online tool NetworkAnalyst (http://www.networkanalyst.ca/) was used to visualize protein-protein interactions networks based on STRING (confidence cutoff 400 and with experimental evidence) generating zero-order networks. The lists of one step interactions were obtained by using the online tool NetworkAnalyst (http://www.networkanalyst.ca/). Interactions with experimental evidence were selected using STRING with a confidence cutoff of 400. The proteins used as nodes to assemble the 4 functional networks were selected based on available information from the literature.
Cell Culture
HeLa cells were grown at 37 °C in a 5% CO2 humid atmosphere in DMEM 4,5g/L Glucose, supplemented with Ultraglutamine (Lonza, Basel, Switzerland), 10% fetal bovine serum (Invitrogen), penicillin, and streptomycin. HeLa cells were regularly controlled for mycoplasma contamination.
The siRNA targeting human CBX3 was a SMARTpool ON-TARGETplus from Dharmacon (ref l-010033–00), siRNA targeting human DnaJB6 was a unique siRNA sequence from Dharmacon, Lafayette, CO (5′-CUAUGAAGUUCUAGGCGUG-3′) and the CTR was a scrambled siRNA is (5′-CGUACGCGGAAUACUUCGAUU-3′). siRNAs were transfected with Lipofectamine RNAiMAX (Invitrogen, ref. 13778030) using 100pmols per well in six-wells plates according to the manufacturer protocol. Cells were analyzed 72 h or 48 h after transfection for CBX3 and DnaJB6 respectively.
The inhibitor Qz was purchased from Millipore (218717–50MG) and resuspended in DMSO to a final concentration of 10 mm. Qz was added to cells 30 min before Nocodazole wash out at the final concentration of 5 μm or 30 μm.
Plasmid transfections were carried out using 2.5 μg of DNA per well in six-well plates with TransIT-HeLaMONSTER by Mirus, Madison, WI, according to manufacturer instructions.
Microtubule Regrowth Assays
For MT regrowth experiments, cells were plated 1 day before transfection when silencing was required or 2 days before the experiment. 2 μm Nocodazole (Sigma) were added to the medium for 3 h and washed out three times with PBS and one time with media at 37 °C. Cells were fixed in methanol at −20 °C immediately before Nocodazole washout (time 0) and at the different time points after nocodazole washout. Samples were processed for immunofluorescence as described and the number of MT asters present in mitotic cells recorded.
Statistical Analysis
Statistical analysis of MT regrowth experiments was performed using Prism 6 (Graphpad, La Jolla, CA). The data for control and CBX3 silenced cells showed a normal distribution (D'Agostino & Pearson omnibus normality test) and equal variance (F test). Therefore in this case the statistical analysis was performed by unpaired two tailed t test. The data for CK2 and DnaJB6 showed non-normal distribution and unequal variance, therefore the statistical analysis was carried out by unpaired two tailed Mann Whitney test.
Antibodies
Rabbit anti-CK2α was a gift of Claudia Götz (University of the Saarland, Germany) and was used at 1:200 as suggested by the original laboratory. Homemade produced anti-DnaJB6 was used at a final concentration of 5 μg/ml for immunofluorescence and 1 μg/ml for Western blotting. The following commercial antibodies were used: mouse anti-α-tubulin (DM1A, Sigma) at 1:10,000 for Western blotting and at 1:1000 for immunofluorescence, rabbit anti-β-tubulin (ab6046, Abcam, Cambridge, UK) at 1:700, rabbit anti-CBX3 (abnova, Taiwan, PAB15582) 1:400. Secondary antibodies anti-rabbit and anti-mouse conjugated to Alexa-488, 568 were used at 1:1000 for immunofluorescence and 680 (Invitrogen), or IRdye 800 CW (Li-cor, Lincoln, Nebraska) at 1:10000 for Western blotting.
Anti-DnaJB6 Antibody Production
The human DnaJB6 cDNA fragment (DnaJB6-CT) was obtained by reverse transcription on total human RNA and PCR with specific primers:
FW: 5′-(EcoRI) AATTCCGGGACATCTATGACAAATATGGC-3′.
R: 5′-AAGTCGACCAAAGGCAATCACTAGCTAGA (XbaI).
The resulting cDNA was inserted using restriction enzymes into pMAL-C2 vector for expression in bacteria. MBP-DnaJB6-CT peptide was expressed in BL21ril bacterial cells and purified. Home made polyclonal anti-DnaJB6 antibodies were obtained by injecting rabbits with MBP-DnaJB6-CT. The serum was affinity-purified on MBP-DnaJB6 covalently bound to HiTrap columns (GE Healthcare Life Sciences) following the company's protocol after passing the serum through an MBP column.
Immunofluorescence
Cells were grown on coverslips and fixed in freeze cold methanol for 10 min. Blocking and antibody dilution buffer was 2% BSA (Sigma), 0.1% Triton-X100 (Sigma) in PBS 1×. In the case of DnaJB6, cells were fixed in 4% PFA for 7 min after a pre-extraction step of 6 s (in BRB80–1X, 0,5% triton and 1 mm DSP) and blocking and antibody dilution buffer was 0.5% BSA, 0.1% tritonX100 in PBS1X. Coverslips were mounted in 10% Mowiol (Calbiochem, Merck) in 0.1 m TrisHCl at pH 8.2, 25% glycerol (Merck, Kenilworth, NJ). Fixed cells were visualized and imaged with 63x or 100x objectives on an inverted DMI-6000 Leica wide-field fluorescent microscope. Pictures were acquired with the Leica Application Suite software as 211.3 × 211.3 micron images with 2048 × 2048 pixels and were processed using Adobe Photoshop CS5.1 (Adobe) and mounted using Adobe Illustrator CS5.1 (Adobe, San Jose, CA).
Western Blots
Blots were developed using Alexa Fluor 680 (Invitrogen) and IRdye 800CW (Li-cor) labeled antibodies and analyzed through the Odyssey Infrared imaging system (Li-Cor) (149.68 pixels/inch image resolution). For CBX3 transfer using semi-dry device, the transfer time should not be longer than 1 h from a 1.5 mm protein gel.
RESULTS
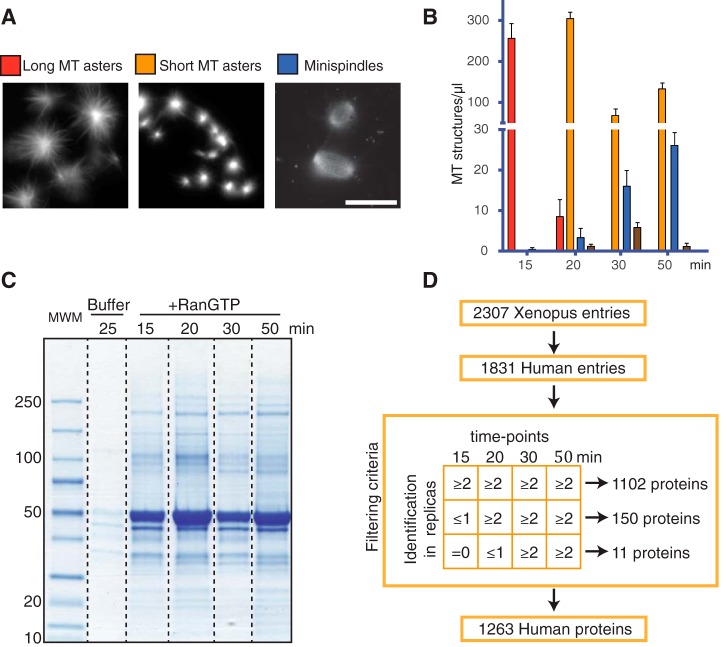
To characterize RanGTP dependent MT self-organization in egg extract we quantified the type of MT structures at different incubation times (Fig. 1A, 1B). Very few MTs formed during the first 10 min of incubation with RanGTP. At 15 min, extracts contained relatively long MTs organized into asters. At 20 min, the number of asters increased, and their morphology changed: asters had shorter MTs and higher tubulin fluorescence signal, suggesting an increase of the number of MTs per aster. At 30 min, the number of asters decreased whereas more complex bipolar-like MT structures (mini-spindles) appeared. The proportion of mini-spindles increased after longer incubation times (50 min) (Fig. 1B). These data indicate that RanGTP promotes a MT self-organization process that transits through intermediate steps involving changes in MT dynamics and organization.
Fig. 1.
RanGTP-induced MT self-organization in Xenopus egg extracts. A, Representative fluorescence images of RanGTP induced MT assemblies in egg extract upon incubation with RanGTP. Scale bar = 20 μm. B, Quantification of the different types of MT assemblies in 1 μl of egg extract at different times of incubation as indicated. Red bars, MT asters with long MTs; Orange bars, MT asters with short MTs; Blue bars, minispindles; Brown bars, aggregates of MT assemblies. Data obtained from four independent experiments. C, Colloidal blue stained SDS-PAGE of the MT pellets obtained at different time of incubation of egg extracts with RanGTP (as indicated). Molecular weight markers are indicated on the left in kDa. D, Schematic representation of the conversion and filtering process of the Xenopus proteins identified by mass spectrometry to obtain the final RanMT proteome of human proteins.
Identification of the M-phase RanGTP-induced MT Proteome
To study RanGTP-dependent MT self-organization, we first aimed at identifying all proteins associated to these MTs through label-free mass spectrometry based proteomics. We optimized a protocol (see methods section) to obtain clean RanGTP-induced MT pellets from egg extracts at different time points of incubation (15, 20, 30, and 50 min). As control, we processed in parallel egg extract supplemented with buffer. SDS-PAGE analysis of the samples showed that they contained a wide range of proteins of different molecular weights (Fig. 1C). As expected, a major band at the molecular weight of tubulin was present in all the MT pellets. Similar results were obtained in three experimental replicas. All the samples (three replicas, all the time points) were then processed for protein identification by nano LC-MS/MS (see experimental procedures section).
Altogether, 2307 Xenopus laevis proteins were identified (False Discovery Rate, FDR 1%) using the protein contigs from a reference database (22). Because Xenopus laevis is a allotetraploid organism (23) these data include isoforms for many proteins. Before further analysis, we converted frog protein entries into human protein entries. This resulted in a proteome of 1831 human protein entries (supplemental Table S1).
As a first quality control we checked whether the proteome included RanGTP regulated SAFs described so far in different systems. Indeed, most of them were identified (17 out of 22) (17), indicating that our experimental approach was successful (supplemental Table S2).
We then took advantage of the three independent experiments to filter out inconsistent data and thus reduce the number of contaminants and potential false positives. We assumed that proteins specifically associated to the MT assemblies should be consistently identified in multiple independent experiments at any given time point and recruited in a time consistent manner. Following these criteria (see Fig. 1D) we defined a RanMT proteome consisting of 1263 human proteins (1102 present at all time points, 150 first identified at 20 min and 11 first identified at 30 min) (supplemental Table S3).
The RanMT Proteome
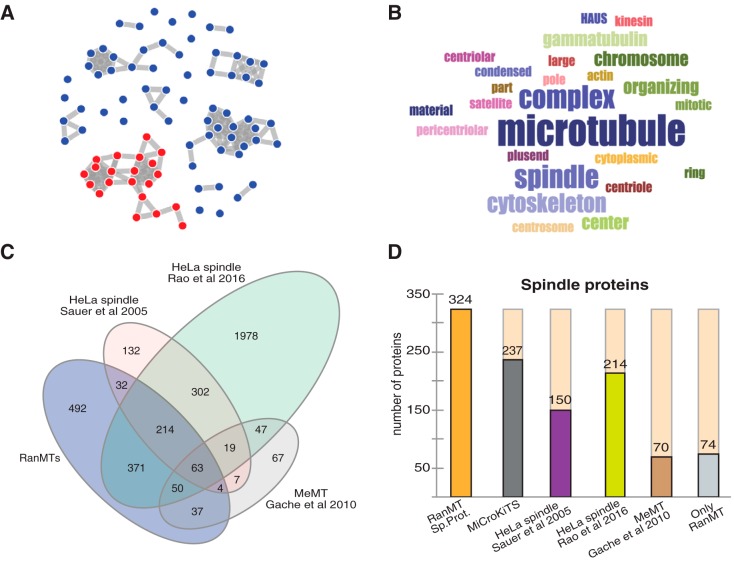
To obtain a global view on the RanMT proteome, we performed an enrichment analysis of gene ontology (GO) cellular component using the online CleverGO tool (http://www.tartaglialab.com/GO_analyser/universal). This revealed four major GO clusters (Fig. 2A): one including GO terms related to spindle and MTs (254 proteins; Fig. 2B). Another including GO terms related to peroxisomes, and other membrane bound organelles (322 proteins; supplemental Fig. S1A), another one including GO terms related to nucleus and nucleoplasm (391 proteins; supplemental Fig. S1B) and the last one including GO terms related to ribosomes and mitochondria (78 proteins; supplemental Fig. S1C).
Fig. 2.
The RanMT proteome. A, Distribution of the GO terms of the RanMT proteome obtained with the online CleverGO tool (analysis conditions: Similarity strength threshold, 0.75; Minimal precision, 0.5; Minimal Lvl, 4; p value cutoff, 0.01). GO terms (represented as dots) more closely related are connected with a gray line. Four major clusters include more than 8 GO terms. The cluster that contains GO terms related to spindles is highlighted in red. B, Word cloud showing the words represented at the GO terms included in the cluster shown in red in (A). Word size is proportional to its frequency in the cluster GO terms. C, Venn-diagram showing the number of proteins identified in our study (RanMT blue) and the overlaps with the HeLa spindle proteomes described by Sauer et al. (8) (pink) and Rao et al. (10) (green) and the MeMT proteome described by Gache et al. (11) (gray). D, Bar graph showing the number of spindle proteins identified in our study (RanMT Sp. Prot., orange bar) and the overlaps with MiCroKiTS (dark gray bar), the HeLa spindle proteomes described by Sauer et al. (8) (magenta bar) and Rao et al. (10) (green bar) and the MeMT proteome described by Gache et al. (11) (brown bar). The number of spindle proteins that are unique to our study is shown on the right (Only RanMT, light gray bar).
We also searched the database MiCroKITS (24) to look for spindle associated proteins in the RanMT proteome. We found that 237 proteins were present in this database which accounts for 18.76% of the RanMT proteome. Altogether, the GO and MiCroKITS analysis defined 324 proteins as spindle proteins in the RanMT proteome (25.6% of the proteome) (Fig. 2D; supplemental Table S3).
As expected, we identified more than half of the proteins previously found to associate with taxol-stabilized MTs in Xenopus egg extracts (MeMT) (154 out of the 267 in the MeMT) (11) (Fig. 2C; supplemental Table S3). The RanMT proteome is in fact richer in spindle or MT-related proteins as it contains 154 proteins in this category not present in the MeMT proteome (Fig. 2D). Interestingly, some of these proteins are particularly relevant for spindle assembly, such as the components of the augmin (or HAUS) complex that drives MT amplification in M-phase (25) and many proteins involved in MT organization. Altogether these data suggest that the higher complexity of the RanMT proteome may correlate with the higher complexity of MT self-organization triggered by RanGTP.
The physiological relevance of the RanMT proteome is reinforced by its high overlap with HeLa cell spindle proteomes (8, 10) (supplemental Table S3). We identified 40% of the proteins included in one of them (313 out of the 773 proteins) (8) and 22,9% of proteins included in the other one (698 proteins out of 3046) (10) (Fig 2C). Moreover, a high number of the 324 spindle proteins is also present in the two HeLa spindle proteomes (149 and 214 respectively) (Fig 2D).
Interestingly, spindle proteins are more represented in the RanMT proteome (25.6%) than in the HeLa spindle proteomes (as described by the authors, 11.9% in Rao et al. (10) 20.2% in Sauer et al. (8). Moreover, we identified 74 spindle proteins absent in all the other reported proteomes (8–11) (Fig 2D).
It is also noteworthy that the RanMT proteome is highly enriched in phosphatases and kinases. We indeed identified 12 phosphatases (4% of the 251 human phosphatases, http://hupho.uniroma2.it) and 49 kinases (9% of the 518 human kinases (26)). Interestingly, 20 of these kinases are included in the group of 324 spindle proteins and 5 of them are not present in the MeMT and HeLa spindle proteomes (MAPK1, NEK1, NME7, PLK3, PRKACB). In total, 20 of the 49 identified kinases are not present in any of these proteomes (supplemental Table S3). Because only some of the 49 kinases have been shown to play a role in mitosis (27), it will be interesting to explore whether more have a role in spindle assembly.
In total, we identified 431 proteins not found in any of the previously reported spindle- and MeMT proteomes (8–11). Interestingly, 74 (17.2%) of these proteins are spindle proteins (as defined above) and some have been functionally characterized such as HAUS2 and 3, HOOK3, DCTN3 and 5, DYNLL2, KATNA1, CENPJ, Cep120, Cep152, Cep164, Cep63 and CETN3. This indicates that the RanMT proteome includes novel spindle factors that remain to be identified.
Overall, our data suggest that the RanGTP-induced MT assemblies are highly complex and physiologically relevant structures thereby providing further validation for their use as a model system to study various aspects of spindle assembly (28–32).
Identification of Novel Spindle Proteins
To explore whether the RanMT proteome indeed includes novel spindle factors, we selected several proteins with no previously described function in spindle assembly, trying to cover a wide range of features and considering the availability of reagents.
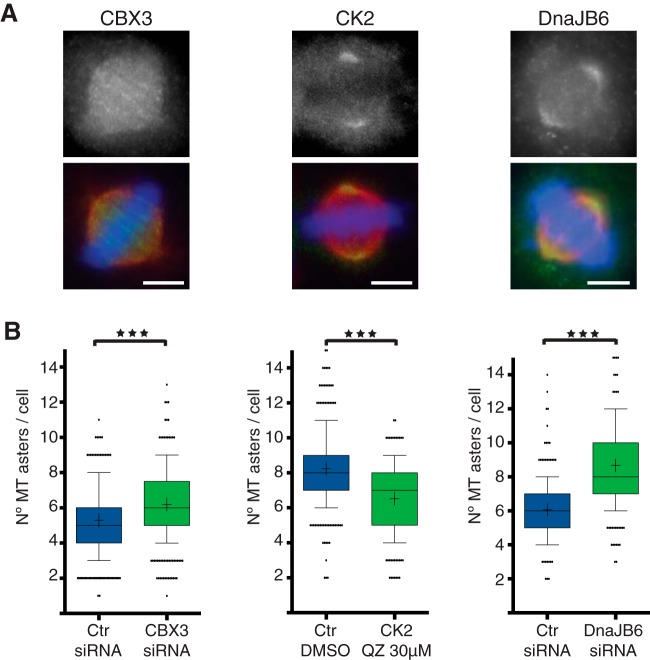
We checked the localization of the candidates in mitotic HeLa cells by immunofluorescence using antibodies or by expressing fluorescently tagged proteins. Three candidates showed interesting spindle localization (supplemental Table S4). The signals observed with the anti-CK2α, anti-CBX3, and anti-DnaJB6 antibodies suggested an association of these proteins with spindle MTs: CBX3 along the spindle MTs, and CK2α and DnaJB6 enriched at the spindle poles (Fig. 3A). We further tested whether any of these three proteins could have a role in chromosome/RanGTP dependent MT assembly using a MT regrowth assay (33). Cells were either silenced for CBX3 or DnaJB6 or incubated with Quinalizarin (QZ), a specific inhibitor of CK2, at concentrations previously described to be both specific and effective (5 μm and 30 μm) (34–36). We found that the number of MT asters present at 3 min after nocodazole washout was significantly different upon silencing or inhibition of each of the three candidates compared with controls. CK2 inhibition using QZ significantly reduced the number of asters at the two concentrations tested (Fig. 3B; 5 μm not shown), with a stronger effect in cells incubated with 30 μm QZ (average number of MT asters per cell of 6.5 ± 0.1 S.E. (standard error of the mean) in control cells and 5.7 ± 0.1 S.E. in 30 μm QZ. p value <0.0001 in Mann Whitney test). In contrast, CBX3 and DnaJB6 silenced cells formed significantly more asters than controls (Fig. 3B). CBX3 silencing increased the average number of asters per cell from 5.3 (± 0.1 S.E., control cells) to 6.2 (± 0.1 S.E., CBX3 silenced cells) (p value <0.0001 in t test) and silencing of DnaJB6 increased the average number of asters per cell from 6.5 (± 0.1 S.E., control cells) to 8.26 (± 0.2 S.E., DnaJB6 silenced cells). Altogether, these data suggest that the three proteins have a role in the chromosome/RanGTP-dependent MT assembly pathway. In addition, these data indicate that the RanMT proteome indeed contains novel players in spindle assembly and putative novel RanGTP regulated SAFs.
Fig. 3.
Identification of novel SAFs. A, Immunofluorescence images from HeLa cells showing the localization of three proteins identified by mass spectrometry, CBX3, CK2, and DnaJB6 as indicated. Upper row, Immunofluorescence signal detected with antibodies against each of the proteins as indicated. Lower row, overlaid images (in green, signal for each protein as shown above, in red tubulin, and in blue DNA). Scale bar = 5 μm. B, Quantifications of MT regrowth assays in HeLa cells for Control and CBX3 or DnaJB6 silenced cells and for cells incubated with DMSO or with the CK2 inhibitor Quinalizarin (QZ) 30 μm. The graphs show the number of MT asters in mitotic cells fixed 3 min after nocodazole washout. n = 3, experiments performed in different days (total sample size, 413 control and 409 CBX3 silenced cells; 308 control DMSO and 323 QZ incubated cells; 220 control and 223 DnaJB6 silenced cells). Box and whiskers plot, boxes show values between the 25th and the 75th percentiles, with a line at the median and a + at the mean, whiskers extend from the 10th to the 90th percentile and dots correspond to outliers. p value <0.0001 in all cases, obtained by unpaired two tailed t test (for CBX3) or Mann Whitney test (for CK2 and DnaJB6).
Functional Networks in the RanMT Proteome
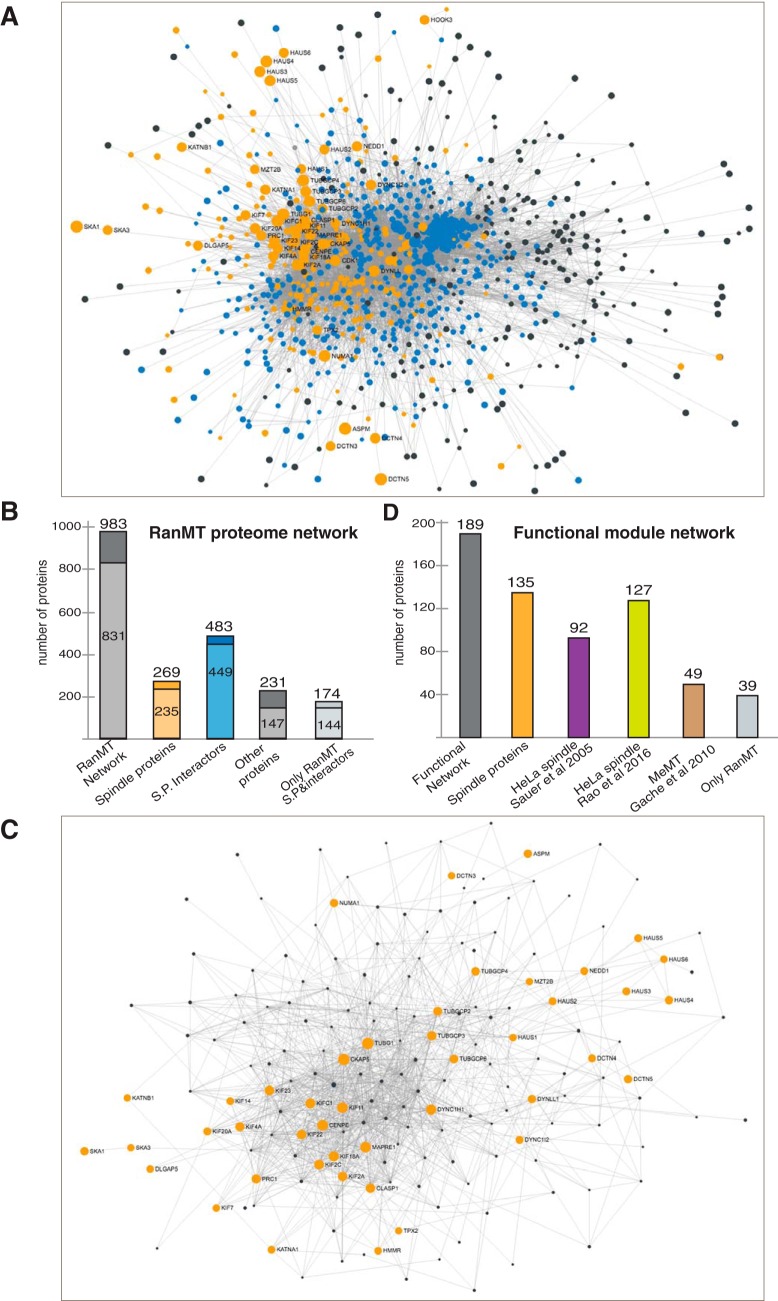
To evaluate the complexity and connectivity in the RanMT proteome, we generated a protein-protein interaction network using STRING (37), considering only experimentally validated data (Fig. 4A). The resulting network included 983 proteins (77.8% of the RanMT proteome). Interestingly 831 (84.5%) had a high degree of connectivity (more than 2 connections) (Fig. 4B). The network included 269 of the 324 spindle proteins. These proteins were connected with 483 proteins. Altogether this generated a subnetwork of 752 proteins. Interestingly these proteins have more connectivity than the other proteins of the general network (90% with more than 2 connections for proteins in the spindle subnetwork versus 63% for the others) (Fig. 4B). This analysis together with our previous one defines a group of 807 proteins (324 spindle proteins and 483 interactors) as the central core of the RanMT proteome (63.9% of the RanMT proteome).
Fig. 4.
Protein-protein interaction networks in the RanMT proteome. A, Network showing experimentally validated protein-protein interactions within the RanMT proteome. Orange nodes represent spindle proteins (defined in the text). Blue nodes represent their first neighbor interactors. Gray dots represent the rest of proteins. Dot size is directly proportional to the number of connections. Gray lines correspond to experimentally validated protein interactions. Most of the proteins are highly connected at the center of the network. B, Bar graph showing the number of proteins identified in our study that are present in the network shown in (A) (RanMT dark gray), the spindle proteins present within the network (orange bar), the proteins connected to the spindle proteins (blue bar), the rest of the proteins (dark gray bar) and the proteins in the network only identified in our study (light gray bar). Within each bar the number of proteins having high connectivity (more than 2 connections) is shown in lighter color. C, Network showing experimentally validated protein-protein interactions within the functional modules as defined in the text (Fig. 3). The orange nodes correspond to the selected key proteins. Gray nodes correspond to their first neighbor interactors. D, Bar graph showing the number of proteins in the Functional module network shown in (B) (gray bar) and the overlaps with the spindle proteins identified in our study (Spindle proteins, orange bar), the HeLa spindle proteomes described by Sauer et al. (8) (magenta bar), and Rao et al. (10) (green bar), the MeMT proteome described by Gache et al. (11) (brown bar). The number of proteins in this network unique to our study is shown on the right (Only RanMT, light gray bar).
We then focused on the key functions involved in MT self-organization. We defined 4 modules for MT nucleation, MT dynamics regulation (MT stabilization and destabilization), and MT organization. We selected in total 49 key proteins within RanMT proteome with essential functions in any of the four functional modules (supplemental Table S3). Using these proteins as nodes we generated protein-protein interaction networks for each functional module considering first-neighbor connections (supplemental Fig. S3A–S3D). Interestingly, although most of the key proteins used to build the networks are unique to each module, the resulting networks included several proteins present in more than one network. These data suggested that the functional modules are not independent but highly interconnected (supplemental Fig. S3A). We therefore decided to build a single core functional network. This network included 189 proteins (47 nodes and 142 interactors) (Fig. 4C, supplemental Fig. S3A, supplemental Table S3) including 135 spindle proteins (as defined above). Interestingly, 39 proteins are absent from previously reported spindle- and MeMT proteomes (Fig. 4D). Altogether, these data defined a core functional interactome for the main modules that drive MT self-organization.
Our data suggest that, although the core functions associated to MT self-organization are usually assigned to a few key players, many other spindle and/or non-spindle proteins are most certainly required to account for the whole process.
The Dynamics of the RanMT Proteome
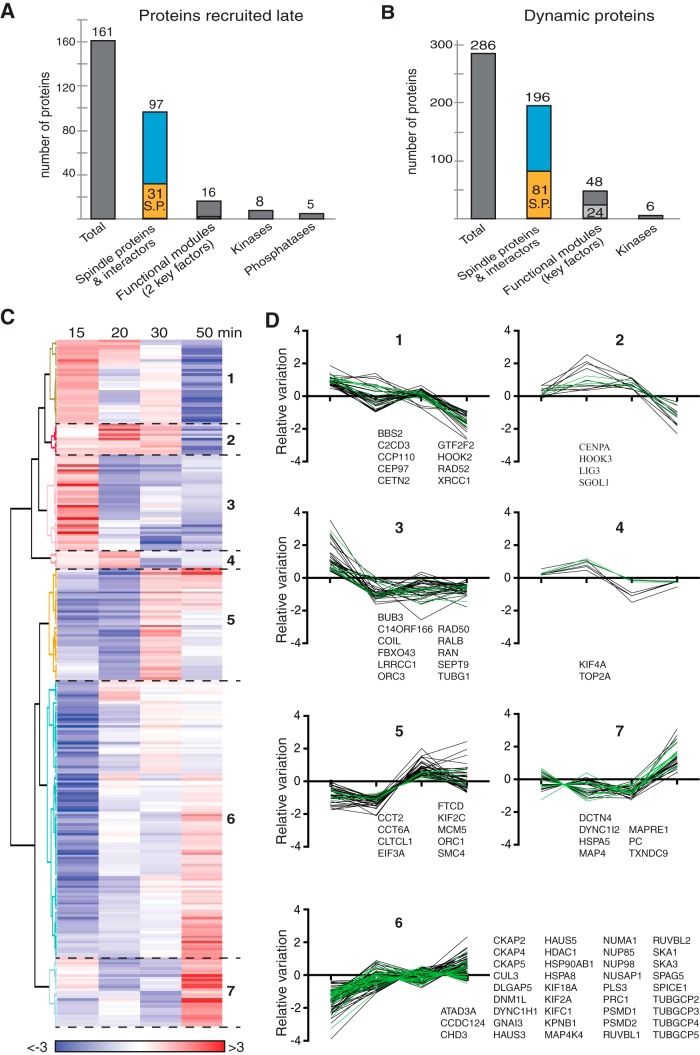
One of our major aims was to study the dynamics of MT self-organization through proteomic profiling of the RanGTP-induced MT assemblies in egg extracts. As described above, a large part of the proteome was identified throughout the time course of the experiment. However, 161 proteins were not present at the earlier time points: 150 proteins were recruited at 20 min and 11 were recruited at 30 min (in total 12.7% of the proteome) (Fig. 1D). Interestingly, these proteins include 31 spindle proteins and 66 spindle protein interactors, accounting for 60% of these proteins. In addition, 16 are included in the RanMT core functional network suggesting that they have a role in the main MT functional modules that we defined above (Fig. 5A). This indicated that the proteome is indeed dynamic and its composition may correlate with changes in MT dynamics and organization.
Fig. 5.
Dynamics of the RanMT proteins that associate non-stochiometrically with MTs along the time course of the experiment. A, Bar graph showing the number of proteins that are recruited from 20 or 30 min after initiation of the incubation of egg extracts with RanGTP (Proteins recruited late, left gray bar). The second bar shows the overlap with the RanMT spindle proteins (orange) and their interactors (blue). The third bar shows the proteins present in the Functional module network. The two last bars show the numbers of kinases and phosphatases. B, Bar graph showing the number of RanMT proteins present in all the time-points but binding non-stochiometrically to the MTs (Dynamic proteins, dark gray bar). The second bar shows the overlap with the RanMT spindle proteins (orange) and their interactors (blue). The third bar shows the proteins present in the Functional module network. The number of selected key factors is indicated in light gray. The number of kinases is shown on the left bar. C, Heatmap showing the relative changes in abundance of the 286 dynamic proteins as shown in (B), throughout the time course of the experiment (time of incubation of the EE with RanGTP shown at the top). The unsupervised clustering analysis generated 7 clusters numbered on the right. The color code is indicated at the bottom. D, Profiles of relative abundance of the proteins in the cluster shown in (C). The 0 in the Y axis corresponds to the relative variation of the total tubulin (normalization factor). Each line corresponds to the relative abundance variation of one protein compared with tubulin (see Experimental Procedures). Spindle proteins are shown with green lines. The gene names for the spindle proteins are reported for each graph.
We then investigated whether other proteins could also contribute to introducing variations in the time specific RanMT proteomes. We obtained the relative abundance profiles for all the proteins over time by calculating for each of them the deviation of intensity at each time point from the calculated average intensity value throughout the experiment. We then normalized all the protein profiles on the profile of the total tubulin. 508 proteins did not show significant variations from the experimental error (maximum difference observed between the total alpha and beta tubulins, see methods for details). This indicated that their association with MTs is stoichiometric throughout the experiment. We defined these proteins as 'non-dynamic'. The remaining 595 proteins were defined as “dynamic” (supplemental Table S3). Importantly, the changes in the relative abundance of these dynamic proteins over time were not associated with an overall lower abundance compared with non-dynamic proteins indicating that the data are reliable (supplemental Fig. S4). We discarded from further analysis dynamic proteins showing random fluctuations over time and performed unsupervised clustering analysis of the reaming 286 dynamic proteins (Fig. 5B, 5C). The resulting 7 clusters defined 2 main types of profiles: one of relative protein decrease over time (clusters 1 to 4) and another one of relative protein enrichment over time (clusters 5 to 7) (Fig. 5C, 5D, supplemental Table S5).
As expected, proteins known to be in complex were in the same cluster. This was the case for members of the yTURC (Cluster 6, but TubG1 is in Cluster1), the augmin/Haus complex (cluster 6), pontin and reptin (RUVBL1 and RUVBL2, cluster 1), and the SKA complex (SKA1 and SKA3, Cluster 6). Interestingly, these dynamic proteins include 81 spindle proteins and 115 spindle protein interactors. They also include 49 proteins present in the RanMT core functional network including 24 selected key factors (Fig. 5B) that became enriched over time, albeit with different timings and profiles.
The two main complexes involved in MT nucleation (the yTURC and the augmin/HAUS complex) are actively recruited at early time points of incubation reaching a plateau after 20 min (Fig. 5D and supplemental Fig. S5A). This profile is consistent with the differences of MT assemblies observed at 15 min (asters with relatively few and long MTs) and 20 min (asters with more and shorter MTs) and with the larger MT pellet obtained at 20 min (Fig. 1). Interestingly, these data agree with spindle tubulin density profiles published previously (38).
Different classes of proteins involved in the regulation of MT dynamics and organization were also enriched over time, (Fig. 5D, supplemental Table S3, S5 and supplemental Fig. S5B, S5C), including the MT depolymerases KIF2a and KIF2c, the MT polymerase CKAP5 (chTOG/XMAP215), and the MT organization factors KIFC1 (HSET), PRC1 and NuMA. Interestingly, two members of the dynein-dynactin complex (DCTN4 and DYNC1I2) strongly enriched at the last time point when the proportion of mini spindles is the highest.
Interestingly, 14 of the kinases present in the RanMT proteome are dynamic. 12 kinases have an enrichment profile and 2 decrease over time (supplemental Table S3, S5). In addition, 8 kinases and 5 phosphatases are recruited at 20 min for the first time (kinases: CDK8, CSNK2A2, GAK, NEK1, NME3, PRPS1, STK3, and STK36; phosphatases: GAK, NUDT9, PPP1CA, PPP2R5C, and PPP6C). These data suggest that the equilibrium of protein phosphorylation-dephosphorylation is temporally regulated and may have a major role in defining the dynamics of the RanMT proteome and RanGTP-induced MT self-organization.
Overall, we have addressed the complex and dynamic self-organization of MTs in M-phase through the characterization of their associated proteome. We showed that this proteome is indeed dynamic. The dynamics of the proteome provides novel insights into the temporal regulation of MT associated proteins activities that may drive MT self-organization.
DISCUSSION
The spindle is a complex self-organized molecular machine. Our understanding of spindle assembly has been mostly based on functional studies on individual proteins or small functional groups in various experimental systems and some modeling approaches (39–41). Frog egg extracts have been instrumental for defining the self-organization properties of MTs and associated proteins in a physiological cell cytoplasm (42, 43). In vitro experiments and computer modeling approaches established that MT self-organization arises from the collective activities of motors and MTs and the energy dissipation mostly from the motors moving along the MTs (1, 44). Bipolar spindle assembly, however, involves a much larger variety of activities and molecules. Therefore, this complex self-organization process has been difficult to address experimentally as a whole. Addition of RanGTP to M-phase Xenopus egg extracts is sufficient to trigger MT nucleation and organization into mini-spindle like structures in the absence of chromosomes and centrosomes (16). Here, we describe the proteome composition of RanGTP-induced MT assemblies. We show that the complexity and the composition of the RanMT proteome is comparable to that of mitotic spindles assembled in tissue culture cells (8–10) (Fig. 2C, 2D). Our data therefore provide further validation for the RanGTP-induced MT assemblies as a model system to study general principles in spindle assembly. The similarities between the RanMT and mitotic spindle proteomes also indicate that meiotic and mitotic spindles may not be so different in terms of properties and general organization principles because the RanMT proteome was obtained from egg extracts arrested in meiosis.
Although there is a significant overlap between the RanMT and mitotic spindle proteomes we identified more spindle proteins (8–10). This is probably because of intrinsic advantages of the egg extract system that provides a good source of relatively abundant and clean preparations of RanGTP induced MT assemblies. Moreover they can be purified without extensive and long manipulation steps, such as those required for mitotic spindle preparations (8, 10). Interestingly, the RanMT proteome also includes 431 proteins that are absent in any of the reported spindle proteomes (8–11) and 74 of these proteins are spindle proteins included in MiCroKITS. This suggests that the RanMT proteome most certainly includes novel SAFs. In support of this idea, we identified novel putative spindle proteins some of them with a function in chromosome dependent MT assembly (Fig. 3). Interestingly, two of them (DnaJB6 and CBX3) have predicted NLS and are therefore novel putative RanGTP regulated SAFs.
We used a network-based approach to try to reach a global understanding of MT and spindle self-organization. We could place most of the proteome into a single protein-protein interaction network based on existing experimentally validated data (Fig. 4A). However, several spindle proteins and validated SAFs (55 in total) are absent from the network. This suggests that some connecting proteins may still be missing in the RanMT proteome and additional work on the unconnected spindle proteins is required to generate a full spindle network. Interestingly, the network we assembled seems to be robust because its topology does not change significantly when considering only the spindle proteins and their first neighbor interactors (Fig. 4A). This also suggests that the number of false positives in the RanMT proteome is low. Interestingly, we defined a “core functional network” including 189 proteins that accounts for the main activities involved in MT self-organization. 54 proteins in this core functional network have no yet been reported to function in spindle assembly and may therefore be interesting novel players.
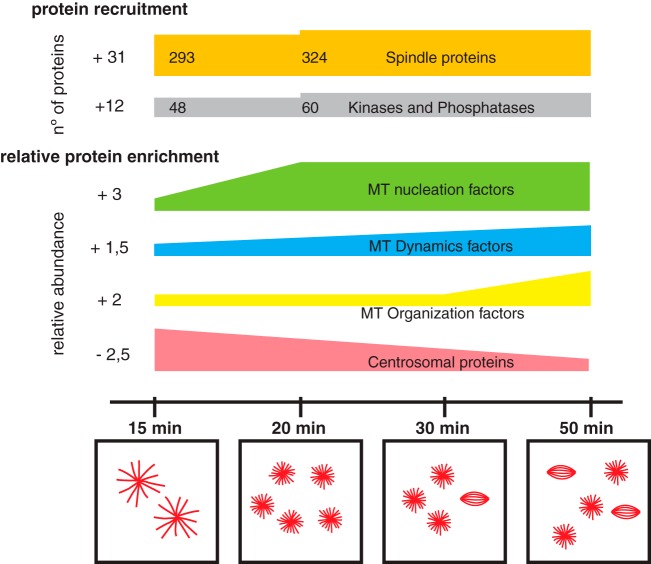
To unravel general principles in spindle self-organization, we used RanGTP-induced MT assemblies to capture temporal snapshots of MT self-organization. We found that the proteomes associated with the diverse MT assemblies change over time both qualitatively and quantitatively. In addition to the 161 proteins recruited to MT assemblies after the earliest time points, our data show that 286 proteins are dynamic and bind to MTs non-stochiometrically throughout the time course of the experiment. The unsupervised clustering analysis of these dynamic proteins indicates that we did grasp functionally relevant enrichment patterns throughout the process of MT self-organization. Our results suggests an earlier recruitment of MT nucleation factors, followed by the progressive enrichment of factors regulating MT dynamics and a final peak of motors involved in MT organization particularly pole formation (Fig. 6, supplemental Fig. S5A–S5C). Interestingly, the relative abundance of some centrosomal core components recruited initially decrease overtime.
Fig. 6.
Schematic representation of the dynamics of the RanMT proteome. At the top schematic representation of proteins recruitment along the experiment, the number of recruited proteins is indicated at the left side. Spindle proteins are shown in orange and kinases and phosphatases in gray. Below, temporal enrichment general profiles of key functionalities involved in MT self-organization are shown as indicated (total enrichment shown at the left side), as well as the dynamic centrosomal proteins. Temporal scale is shown at the bottom together with representative drawings of characteristic MT assemblies observed at different time points of incubation of egg extracts with RanGTP.
A simple form of MT self-organization that has been recapitulated in vitro with purified components is the formation of asters with different topologies depending on the type of motor (5). In egg extracts this is also the first type of MT organization occurring upon addition of taxol or RanGTP. However more complex spindle-like structures only assemble upon RanGTP addition. The augmin complex was not described as associated to the taxol stabilized MTs in egg extract (11) nor was it included in other in vitro experiments (5, 44) suggesting that it is not required for the formation of MT asters. Our data however show that the augmin complex becomes enriched when MT asters have formed. This enrichment agrees with the previously proposed presence of a positive feedback loop in which MTs nucleated in a RanGTP dependent manner recruit more MT-nucleation factors promoting microtubule branching (45). Altogether and in agreement with previous reports (45, 46) this suggests that MT branching through the augmin dependent pathway may play a role in the self-organization of more complex spindle like structures. Our data also points to the importance of MT dynamics regulation in this process. This regulation appears to occur in a time dependent fashion that involves the enrichment of both stabilizing and destabilizing activities detected with the proteomics approach. This is interesting because MT dynamics is defined by the balance of stabilizing and destabilizing factors as previously shown in vitro (47, 48). Our proteomic data also correlate well with the change in morphology of the MT asters that transit from having relatively long MTs at the earlier time points to shorter ones.
MT organization is highly dependent on motor activities as shown in vitro with purified components. Our proteomics data show that the RanGTP MT assemblies have a large number of associated motors (14 kinesins and dynein-dynactin components) with several of them becoming enriched over time. The collective activities of these motor proteins together with the specific properties of the MTs (length, number and dynamics) define their organization potential (44).
Another interesting data from our proteomic analysis is the temporally regulated recruitment of kinases and phosphatases (Fig. 6, green bar). This could suggest the existence of feedback loops and/or amplification mechanisms for driving the enrichment of some proteins within specific functional groups. Because no mitotic function has been clearly established for several of the enzymes identified, it will be interesting to address their potential role in mitosis in spindle assembly.
Altogether, our results validate our approach to address MT self-organization using mass spectrometry based proteomic profiling. Overall, MT self-organization transits through phases that we have captured and characterized in terms of specific proteome composition and dynamics. Indeed, we identified non-stoichiometric MT binders and by performing unsupervised clustering analysis we found a temporal coordination of main MT dependent activities. Our results suggest a very strong functional crosstalk between the regulation of MT number, length and dynamics that in turns define the specific functional properties of a large number of motor proteins that collectively drive MT self-organization. This is in agreement with some of the principles proposed to participate in MT self-organization, but our experimental approach provides for the first time a system level analysis.
In summary, our work provides for the first time a global and precise view of the highly complex process of MT self-organization in M-phase.
DATA AVAILABILITY
Data from this work are available via ProteomeXchange (49) with identifier PXD008306 and annotated spectra of identified peptides can be browsed at MSViewer with accession number q6sv4oww94 (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer).
Supplementary Material
Acknowledgments
We thank C. Götz (University of the Saarland) for the gift of the anti-CK2α antibodies and N. Mallol and L. Avila for excellent technical support.
Footnotes
* This work was supported by grants from the Spanish Ministry of Economy (MINECO) I+D grants BFU2012-37163 and BFU2015-68726-P. The CRG/UPF Proteomics Unit is part of the “Plataforma de Recursos Biomoleculares y Bioinformáticos (ProteoRed)” supported by grant PT17/0019 of Instituto de Salud Carlos III from the Spanish Government. We acknowledge support from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership, “Programa CERCA Generalitat de Catalunya”, and “Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement” (2017SGR595 to ES and 2017SGR478 to IV). We also acknowledge support from the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013–2017”, SEV-2012-0208. T. C. and M. R. were supported by FPI fellowships from the Spanish Ministry of Economy BES-2010-031355 and BES-2013-064601, respectively.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- MTs
- microtubules
- CSF extract
- cytosolic factor arrested Xenopus laevis egg extract
- EE
- egg extract
- FDR
- false discovery rate
- GO
- gene ontology
- MeMT
- Taxol-stabilized MTs in Xenopus egg extracts
- NLS
- nuclear localization signal
- RanMT
- RanGTP induced microtubules
- RT
- room temperature
- SAFs
- spindle assembly factors
- SEM
- standard error of the mean.
REFERENCES
- 1. Karsenti E. (2008) Self-organization in cell biology: a brief history. Nature Reviews Mol. Cell Biol. 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 2. Vignaud T., Blanchoin L., and Thry M. (2012) Directed cytoskeleton self-organization. Trends Cell Biol. 22, 671–682 [DOI] [PubMed] [Google Scholar]
- 3. Mitchison T., and Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 4. Cortes S., Glade N., Chartier I., and Tabony J. (2006) Microtubule self-organisation by reaction-diffusion processes in miniature cell-sized containers and phospholipid vesicles. Biophys. Chem. 120, 168–177 [DOI] [PubMed] [Google Scholar]
- 5. Surrey T., Nedelec F. J., Leibler S., and Karsenti E. (2001) Physical properties determining self-organization of motors and microtubules. Science 292, 1167–1171 [DOI] [PubMed] [Google Scholar]
- 6. Nedelec F. J. (2002) Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 158, 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nedelec F. J., Surrey T., and Karsenti E. (2003) Self-organisation and forces in the microtubule cytoskeleton. Curr. Opinion Cell Biol. 15, 118–124 [DOI] [PubMed] [Google Scholar]
- 8. Sauer G., Körner R., Hanisch A., Ries A., Nigg E. A., and Silljé H. H. (2005) Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics 4, 35–43 [DOI] [PubMed] [Google Scholar]
- 9. Bonner M. K., Poole D. S., Xu T., Sarkeshik A., Yates J. R. 3rd, and Skop A. R. (2011) Mitotic Spindle Proteomics in Chinese Hamster Ovary Cells. PLoS ONE 6, e20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rao S. R., Flores-Rodriguez N., Page S. L., Wong C., Robinson P. J., and Chircop M. (2016) The Clathrin-dependent Spindle Proteome. Mol. Cell. Proteomics 15, 2537–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gache V., Waridel P., Winter C., Juhem A., Schroeder M., Shevchenko A., and Popov A. V. (2010) Xenopus meiotic microtubule-associated interactome. PLoS ONE 5, e9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neumann B., Held M., Liebel U., Erfle H., Rogers P., Pepperkok R., and Ellenberg J. (2006) High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods 3, 385–390 [DOI] [PubMed] [Google Scholar]
- 13. Neumann B., and Walter T. (2010) and Hrich. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke P. R., and Zhang C. (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9, 464–477 [DOI] [PubMed] [Google Scholar]
- 15. Nakamura M., Masuda H., Horii J., Kuma Ki., Yokoyama N., Ohba T., Nishitani H., Miyata T., Tanaka M., and Nishimoto T. (1998) When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 143, 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., and Mattaj I. W. (1999) Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 [DOI] [PubMed] [Google Scholar]
- 17. Cavazza T., and Vernos I. (2015) The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 3, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desai A., Murray A. W., Mitchison T. J., and Walczak C. E. (1999) The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385–412 [DOI] [PubMed] [Google Scholar]
- 19. Caudron M., Bunt G., Bastiaens P., and Karsenti E. (2005) Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309, 1373–1376 [DOI] [PubMed] [Google Scholar]
- 20. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 21. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22. Wühr M., Freeman R. M., Presler M., Horb M. E., Peshkin L., Gygi S., and Kirschner M. W. (2014) Deep Proteomics of the Xenopus laevis Egg using an mRNA-Derived Reference Database. Curr. Biol. 24, 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S. J., Quigley I., Heinz S., Ogino H., Ochi H. (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Z., Ma L., Wang Y., Pan Z., Ren J., Liu Z., and Xue Y. (2015) MiCroKiTS 4.0: a database of midbody, centrosome, kinetochore, telomere and spindle. Nucleic Acids Res. 43, D328–D334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petry S., Groen A. C., Ishihara K., Mitchison T. J., and Vale R. D. (2013) Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 27. Ma H. T., and Poon R. Y. (2011) How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 435, 17–31 [DOI] [PubMed] [Google Scholar]
- 28. Zhang X., Ems-McClung S. C., and Walczak C. E. (2008) Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell 19, 2752–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitchison T. J., Nguyen P., Coughlin M., and Groen A. C. (2013) Self-organization of stabilized microtubules by both spindle and midzone mechanisms in Xenopus egg cytosol. Mol. Biol. Cell 24, 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Groen A. C., Coughlin M., and Mitchison T. J. (2011) Microtubule assembly in meiotic extract requires glycogen. Mol. Biol. Cell 22, 3139–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilde A, Lizarraga S. B., Zhang L., Wiese C., Gliksman N. R., Walczak C. E., and Zheng Y. (2001) Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3, 221–227 [DOI] [PubMed] [Google Scholar]
- 32. Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., and Zheng Y. (2003) A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 33. Meunier S., and Vernos I. (2011) K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 13, 1406–1414 [DOI] [PubMed] [Google Scholar]
- 34. Cozza G., Mazzorana M., Papinutto E., Bain J., Elliott M., di Maira G., Gianoncelli A., Pagano M. A., Sarno S., Ruzzene M., Battistutta R., Meggio F., Moro S., Zagotto G., and Pinna L. A. (2009) Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem. J. 421, 387–395 [DOI] [PubMed] [Google Scholar]
- 35. Wilhelm N., Kostelnik K., Gtz C., and Montenarh M. (2012) Protein kinase CK2 is implicated in early steps of the differentiation of pre-adipocytes into adipocytes. Mol. Cell. Biochem. 365, 37–45 [DOI] [PubMed] [Google Scholar]
- 36. Franchin C., Cesaro L., Salvi M., Millioni R., Iori E., Cifani P., James P., Arrigoni G., and Pinna L. (2015) Quantitative analysis of a phosphoproteome readily altered by the protein kinase CK2 inhibitor quinalizarin in HEK-293T cells. Biochim. Biophys. Acta 1854, 609–623 [DOI] [PubMed] [Google Scholar]
- 37. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dinarina A. Pugieux C., Corral M. M., Loose M., Spatz J., Karsenti E., and Nédélec F. (2009) Chromatin shapes the mitotic spindle. Cell 138, 502–513 [DOI] [PubMed] [Google Scholar]
- 39. Prosser S. L., and Pelletier L. (2017) Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187–201 [DOI] [PubMed] [Google Scholar]
- 40. Pavin N., and Tolic I. M. (2016) Self-organization and forces in the mitotic spindle. Annu. Rev. Biophys. 45, 279–298 [DOI] [PubMed] [Google Scholar]
- 41. Schaffner S. C., and José J. V. (2006) Biophysical model of self-organized spindle formation patterns without centrosomes and kinetochores. Proc. Natl. Acad. Sci. U.S.A. 103, 11166–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verde F., Berrez J. M., Antony C., and Karsenti E. (1991) Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol. 112, 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karsenti E., and Vernos I. (2001) The mitotic spindle: a self-made machine. Science 294, 543–547 [DOI] [PubMed] [Google Scholar]
- 44. Dogterom M., and Surrey T. (2013) Microtubule organization in vitro. Curr. Opin. Cell Biol. 25, 23–29 [DOI] [PubMed] [Google Scholar]
- 45. Oh D., Yu C. H., and Needleman D. J. (2016) Spatial organization of the Ran pathway by microtubules in mitosis. Proc. Natl. Acad. Sci. U.S.A. 113, 8729–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petry S., Pugieux C., Nedelec F. J., and Vale R. D. (2011) Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. U.S.A. 108, 14473–14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tournebize R., Popov A., Kinoshita K., Ashford A. J., Rybina S., Pozniakovsky A., Mayer T. U., Walczak C. E., Karsenti E., and Hyman A. A. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nature Cell Biol. 2, 13–19 [DOI] [PubMed] [Google Scholar]
- 48. Kinoshita K., Arnal I., Desai A., Drechsel D. N., and Hyman A. A. (2001) Reconstitution of physiological microtubule dynamics using purified components. Science. 294, 1340–1343 [DOI] [PubMed] [Google Scholar]
- 49. Deutsch E. W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D. S., Bernal-Llinares M., Okuda S., Kawano S., Moritz R. L., Carver J. J., Wang M., Ishihama Y., Bandeira N., Hermjakob H., and Vizcaíno J. A. (2017) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from this work are available via ProteomeXchange (49) with identifier PXD008306 and annotated spectra of identified peptides can be browsed at MSViewer with accession number q6sv4oww94 (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer).