Abstract
Signals from cell surface receptors are often relayed via adaptor proteins. NCK1 and NCK2 are Src-Homology (SH) 2 and 3 domain adaptors that regulate processes requiring a remodeling of the actin cytoskeleton. Evidence from gene inactivation in mouse suggests that NCK1 and NCK2 are functionally redundant, although recent reports support the idea of unique functions for NCK1 and NCK2. We sought to examine this question further by delineating NCK1- and NCK2-specific signaling networks. We used both affinity purification-mass spectrometry and BioID proximity labeling to identify NCK1/2 signaling networks comprised of 98 proteins. Strikingly, we found 30 proteins restricted to NCK1 and 28 proteins specifically associated with NCK2, suggesting differences in their function. We report that Nck2−/−, but not Nck1−/− mouse embryo fibroblasts (MEFs) are multinucleated and display extended protrusions reminiscent of intercellular bridges, which correlate with an extended time spent in cytokinesis as well as a failure of a significant proportion of cells to complete abscission. Our data also show that the midbody of NCK2-deficient cells is not only increased in length, but also altered in composition, as judged by the mislocalization of AURKB, PLK1 and ECT2. Finally, we show that NCK2 function during cytokinesis requires its SH2 domain. Taken together, our data delineate the first high-confidence interactome for NCK1/2 adaptors and highlight several proteins specifically associated with either protein. Thus, contrary to what is generally accepted, we demonstrate that NCK1 and NCK2 are not completely redundant, and shed light on a previously uncharacterized function for the NCK2 adaptor protein in cell division.
Keywords: Affinity proteomics, Signal Transduction, Protein-Protein Interactions, SH2/SH3 Domains, Phosphorylation, Cell division, Imaging, Networks, Cell biology, Mass Spectrometry
NCK1 (Noncatalytic region of tyrosine Kinase 1; NCKα)1 and NCK2 (NCKβ, GRB4) adaptors (collectively NCK) are widely expressed proteins that are exclusively composed of Src-Homology (SH) 2 and 3 interaction domains. NCKs contain three SH3 domains, which generally associate with a canonical P-X-X-P motif (1), and a single C-terminal SH2 domain that exhibits a preference for variations of the core pY-D-X-V consensus (2). A comparison of protein sequences revealed a 68% identity that can for the most part be attributed to these SH2-SH3 protein interaction domains (3). The main function of NCK adaptor proteins is to link receptor tyrosine kinase (RTK) activation to effectors involved in cytoskeletal reorganization, such as the Wiscott-Aldrich Syndrome Protein (WASP) and its associated proteins (WIPF1 to −3) (1, 4, 5).
Very few studies reported functions unique to NCK1 or NCK2. For example, activated Platelet-Derived Growth factor Receptor (PDGFRB) associates via two different residues to NCK1 (Y751) and NCK2 (Y1009), and only the latter interaction regulates downstream signaling (6). In addition, NCK2 but not NCK1 was reported to directly bind to EPH family RTKs (EphRs) ligands, B-type ephrins, to initiate reverse signals controlling the actin cytoskeleton (7–9). NCK2 was also found to specifically associate with Paxilin (PXN) in neuronal cells to facilitate neurite outgrowth (10). Finally, NCK2 was recently highlighted as a regulator of adipogenesis, as NCK2- (but not NCK1)-deficient mice display increased adiposity (11). Although simultaneous inactivation of NCK1 and NCK2 genes is embryonic lethal in mouse at mid-gestation (E9.5), single knockout animals did not display any overt phenotype (12). This suggests the existence of a substantial functional overlap between NCK1 and NCK2 functions, at least during early development, despite reports pointing toward a degree of specificity for each protein.
The successful analysis of protein complexes under physiological conditions has been spearheaded by affinity purification followed by mass spectrometry (AP-MS) (13). Although this approach has proven useful to isolate and to characterize intact protein complexes, including those that are dependent on cell stimulation, it still presents limitations in that it requires gentle lysis conditions to preserve interactions, which may not be suitable for proteins with compartmentalized cellular localization. In addition, AP-MS generally fails to capture weak/transient interactions because of the purification process, unless crosslinking is performed. Proximity labeling methods relying on the biotinylation of neighboring proteins by spatially restricted enzymes, for example BioID, were developed to circumvent these issues (14). The covalent labeling of proteins with biotin allows for cell solubilization under stringent conditions and protein isolation without the strict requirement of them being maintained in a complex for the duration of the process. However, classical BioID does not allow for temporal resolution because of long labeling times.
In this study, we used affinity purification-mass spectrometry (AP-MS) and BioID proximity labeling to delineate NCK1 and NCK2 protein networks, which unexpectedly reveal many interactors specific to each adaptor protein. The NCK2 network is enriched for proteins related to the actin cytoskeleton. Strikingly, we find that NCK2, but not NCK1, is localized at the site of furrow ingression and at the midbody during cell division, a process displaying the most extensive reorganization of the actin cytoskeleton. The final stage of cell division, cytokinesis, is a process during which the cytoplasm of the parental cell is partitioned between two daughter cells to terminate their physical separation (15, 16). This process requires the dynamic interplay between components of actin and tubulin cytoskeletons with the plasma membrane to coordinate both the positioning and assembly of an actomyosin ring (17). The mechanical constriction of this ring forces the ingression of the cleavage furrow, leading to the formation of the midbody. This protein-rich structure forms at the site of overlap of antiparallel microtubules derived from the central spindle and serves as an assembly platform and signaling hub for the abscission machinery. Key factors implicated in formation of the cytokinetic furrow are the kinases Polo-like kinase 1 (PLK1) and Aurora-B (AURKB), which are collectively responsible for phosphorylation and recruitment of factors required for furrow ingression (18, 19). Failure in cytokinesis can result in an incomplete separation of the two incipient cells, thus leading to polyploidy or multinucleation (20, 21).
We investigated further the striking compartmentalization of NCK2 at the midbody. We found that Nck2−/− cells delay in cytokinesis, display longer intercellular bridges and often fail to complete abscission. We also found that NCK2 regulation of cytokinesis is dependent on its single SH2 domain, presumably via its interaction with putative tyrosine-phosphorylated targets. Together, our findings demonstrate a clear distinction in NCK1 and NCK2 interaction networks, as well as unique subcellular localization and role for NCK2 during the last stages of cell division, further supporting the notion that NCK1 and NCK2 are not fully redundant in their functions.
EXPERIMENTAL PROCEDURES
Constructs and Sequences
Murine NCK1 (NCBI clone NM010878.3), murine NCK2 (NCBI clone NM010879.3) and human EIF4B (NCBI clone NM001300821.2) were subcloned into pMSCVpuro (Clontech) with a N-terminal 3xFLAG tag, FLAG-BirA*-pcDNA5-FRT-TO (Thermo Fisher Scientific, Waltham, MA), pcDNA3.1 (Thermo Fisher Scientific) with a N-terminal 3xFLAG tag (Sigma-Aldrich, St. Louis, MO), or in pLJM1-EGFP (Addgene, Cambridge, MA, Sabatini lab). Point mutations were introduced using the Q5 mutagenesis kit (New England Biolabs, Ipswich, MA) according to manufacturer instructions. All inserts were fully sequenced and protein expression was verified. siRNA sequences (Integrated DNA Technologies, Coralville, IA) were as follows: (NCK1, Sense: rGrGrCrCrUrUrCrArCrUrCrArCrUrGrGrArArArGrUrUrUGC; Antisense: rGrCrArArArCrUrUrUrCrCrArGrUrGrArGrUrGrArArGrGrCrCrUrA) (NCK2, Sense: rUrArArUrArCrArGrArUrGrArUrUrArArGrGrArArArArCAG; Antisense: rCrUrGrUrUrUrUrCrCrUrUrArArUrCrArUrCrUrGrUrArUrUrArGrU). Nontargeting DsiRNA sequences provided by the manufacturer were used as negative control.
Cell Culture, Transfections, and Virus Infection
Human embryonic kidney 293T cells (HEK293T), Flp-In T-REx HEK293, HeLa cervical carcinoma cells and mouse embryonic fibroblasts (MEF) were cultured in Dulbecco's Modified Eagle's medium (DMEM, Thermo Fisher Scientific) high glucose supplemented with 10% fetal bovine serum (Sigma-Aldrich). HEK293T stable cell lines expressing 3xFLAG-tagged NCK1 or NCK2 were selected with 1.5 μg/ml puromycin (Thermo Fisher Scientific). Flp-In T-REx HEK293 stable lines expressing NCK1 or NCK2 fused with BirA* were generated as described (22), and grown in the presence of 150 μg/ml hygromycin (Thermo Fisher Scientific). Expression was induced with 1 μg/ml tetracycline for 24 h. For the BioID experiments, 50 μm biotin was added at the time of induction. MEFs were generated as previously described (23), then immortalized by the introduction of the SV40 T/t antigens via transfection of the pBSV0.3T/t plasmid (kindly provided by E. W. Khandjian, U. Laval) using jetPRIME (Polyplus, Illkirch, France). HeLa cells were transiently transfected with 3xFLAG constructs using Lipofectamine 2000 (Thermo Fisher Scientific). HeLa cells were synchronized 8 h post-transfection with a double thymidine block as previously described (24), with a relaxation time of 10 h. Transfections of siRNAs in HEK293T were performed using RNAiMAX (Thermo Fisher Scientific). Lentivirus were produced by co-transfection of pLJM1-GFP constructs, pMD2G and psPAX2 (Addgene, Trono lab). Viral supernatants were collected after 48h, filtered through a 0.45 μm membrane and supplemented with 8 μg/ml polybrene (Sigma-Aldrich). Stable clonal lines were obtained by transducing Nck2−/− MEFs and maintaining a 2 μg/ml puromycin selection.
Cell Lysis and Western Blotting
Cells were grown to confluency before lysis. Sodium orthovanadate (Sigma-Aldrich) pre-activated with 30% H2O2 (1: 200) (pervanadate) was added to cell medium at a final concentration of 100 μm for 20 min, as indicated. Cells were washed once with ice cold PBS before lysis. For AP-MS and Western blotting, proteins were extracted by scraping cells in ice-cold lysis buffer as described elsewhere (25, 26). For BioID, cells were lysed in cold RIPA buffer followed by benzonase treatment for 1h (Sigma-Aldrich) and 3 cycles of sonication. Protein concentrations were measured and normalized using a BCA kit (Thermo Fisher Scientific). Lysates were incubated under agitation for 2h at 4 °C with M2 affinity resin (Sigma-Aldrich) for FLAG AP or 3 h at 4 °C with streptavidin agarose beads (Sigma-Aldrich) for BioID. Beads were washed three times with lysis buffer.
For Western blotting, beads were resuspended in 4x Laemmli buffer. A total of 15 μg of whole-cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare). Loading was verified with Ponceau S (Sigma-Aldrich) staining. Western blotting was performed as previously described (27). Primary antibodies were as follows: rabbit anti-NCK1 (Cell Signaling Technology, Danvers, MA, #2319), mouse anti-NCK2 (Abnova, Taipai, Taiwan, #MAB2727), rabbit anti-NCK (gift from T. Pawson, Toronto), rabbit anti-FLAG (Sigma-Aldrich, #F7425), mouse anti-actin (Cell Signaling Technology, #3700), mouse anti-tubulin (DSHB, Iowa City, IA, #E7) and mouse anti-GFP (Abcam, Cambridge, UK, #290). Secondary antibodies were the following: horse anti-mouse IgG (Cell Signaling Technology, #7076), goat anti-rabbit (Cell Signaling Technology, #7074) and streptavidin-HRP (Life Technologies, Carlsbad, CA #434323). Signal was revealed using BioRad (Hercules, CA) Clarity Western ECL substrate and detected with an Amersham Biosciences Imager 600RGB (GE Healthcare).
Immunofluorescence, Cell Labeling, and Microscopy
Cells were fixed with 4% paraformaldehyde (BioShop, Toronto, ON) for 15 min, washed with PBS and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 15 min at room temperature. Cells were blocked in 10% goat serum (Wisent, St. Jean. Baptiste, QC) and 0.1% NP-40 (Sigma-Aldrich) and incubated with the following primary antibodies diluted in blocking solution for 1h: rabbit anti-tubulin (Abcam, #ab18251), rabbit anti-FLAG (Sigma-Aldrich, #F7425), mouse anti-AURKB (BD Biosciences, Franklin Lake, NJ, #611082) and mouse anti-PLK1 (28). Following 3 washes in PBS with 0.1% NP-40 (Sigma-Aldrich), cells were incubated for 1h with the following secondary antibodies: Alexa Fluor 488-goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, #111–545-003), Alexa Fluor 568-goat anti-rabbit IgG (Thermo Fisher Scientific, #A11011), Alexa Fluor 647-goat anti-mouse IgG (Thermo Fisher Scientific, #A21235), Alexa Fluor 647-goat anti-rabbit IgG (Thermo Fisher Scientific, #A21245) and Alexa Fluor 594-sptreptavidin (Thermo Fisher Scientific, #S11227). Actin was visualized using Alexa Fluor 555-phalloidin (Cell Signaling Technology, #8953) or Alexa Fluor 647-phalloidin (Cell Signaling Technology, #8940). Slides were washed 3 times with 0.1% NP-40 (Sigma-Aldrich) in PBS and twice with PBS before being mounted in ProLong Gold Antifade with DAPI (Thermo Scientific). Confocal images were acquired using a FV1000 system (Olympus, Shinjuku, Japan) via the Fluoview 3.0 software.
For fluorogenic labeling and live imaging microscopy, cells were plated into 8-well μ-Slides (Ibidi, Martinsried, Germany) and treated with 90 nm SiR-Actin and 7 μm Verapamil (Cytoskeleton Inc, Denver, CO). Acquisition was performed using an UltraView spinning disc confocal microscope (Perkin-Elmer, Waltham, MA) equipped with an EMCCD camera (Hamamatsu Photonics, Hamamatsu City, Japan), a humidified 5% CO2 thermo-regulated chamber. Images were acquired with the Volocity software (version 6.01, Perkin-Elmer) at 5 min intervals with low laser power at 647 nm for 18–24 h starting at 24 h post-seeding.
For phase contrast live imaging, images were acquired every 5 min for 18–24 h starting at 24 h post-seeding. Images were obtained with a Nikon TE2000 inverted microscope equipped with a humidified 5% CO2 thermo-regulated chamber and a Photometrix Coolsnap FX CCD camera (Roper Scientific, Tucson, AZ) driven by the MetaView software (Version 4.5, Universal Imaging Corp, Bedford Hills, NY).
Experimental Design and Statistical Rationale
Each AP-MS and BioID experiment was performed in biological triplicate (n = 3). Affinity purifications were performed on different days, using cells from successive passages. Controls for each experiment were treated concomitantly to experimental samples. 3xFLAG-GFP was used as a control for 3xFLAG-NCK1/2 in AP-MS experiments, and BirA*-FLAG-GFP for BirA*-FLAG-NCK1/2 BioID experiments. Biological triplicates were required and sufficient to perform SAINT analyses, as reported (29, 30), to distinguish background from bona fide protein associations. Statistical significance for cell imaging-based phenotypes was determined via unpaired t-student test (95% confidence intervals, two tails) or Fisher's exact test (95% confidence intervals, two tails) using Prism 7 software (GraphPad, La Jolla, CA).
Mass Spectrometry Analyses
For MS experiments, beads were additionally washed twice with 20 mm Tris pH 7.4 and proteins were eluted by incubating with agitation at 4 °C with 50 mm H3PO4 before digestion with trypsin, as described (31). A 5600+ TripleTOF mass spectrometer (Sciex, Concord, ON) equipped with a nanoelectrospray ion source and coupled with an Agilent 1200 HPLC system (Agilent, Santa Clara, CA) was used to acquire data for AP-MS experiments. Peptides from tryptic digest were resolved using a New Objective PicoFrit column (15 cm×0.075 mm I.D.) (Scientific Instrument Services, Ringoes, NJ) packed with Jupiter 5 μm C18 (Phenomenex, Torrance, CA) stationary phase. A 65-min linear gradient of a 5–35% mixture of acetonitrile, 0.1% formic acid injected at 300 nL/min was used to elute peptides. Data dependent acquisition mode was used in Analyst version 1.7 (Sciex) to acquire mass spectra. Full scan mass spectrum (400 to 1250m/z) were acquired and followed by collision-induced dissociation of the twenty most intense ions. A period of 20 s and a tolerance of 100 ppm were set for dynamic exclusion. Protein Pilot version 5.0 (Sciex) was used to generate MS/MS peak lists. Mascot (Matrix Science, London, UK; version 2.4.0) and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1) were used to analyze MGF sample files. The Uniprot Homo sapiens reference proteome (March 2014 release, 69150 entries) was used assuming tryptic peptides. Two missed cleavages were allowed. The protein sequences of the GFP and NCK1/2 FLAG-tagged constructs were added to the database to estimate protein sequence coverage. Searches were performed using a fragment ion mass tolerance and a parent ion tolerance of 0.100 Da. Cysteine carbamidomethylation was specified for all samples as a fixed modification. Variable modifications specified were oxidation of methionine, phosphorylation of serine, threonine and tyrosine, conversion of glutamate and glutamine to pyroglutamate, deamidation of asparagine and glutamine.
For BioID, peptide identification was carried out on an Orbitrap Fusion spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Scientific) and coupled to a UltiMate 3000 nanoRSLC (Dionex, Sunnyvale, CA). Data dependent acquisition of mass spectra was performed using XCalibur software version 3.0.63 (Thermo Scientific). Full scan mass spectra (350 to 1800 m/z) were acquired in the orbitrap at a resolution of 120,000 using a maximum injection time of 50 ms and automatic gain control target of 4e5. The quadrupole analyzer allowed for the isolation of selected ions in a window of 1.6 m/z and fragmentation by higher energy collision-induced dissociation (HCD) with 35% energy. The resulting fragments were detected by the linear ion trap at a rapid scan rate. Dynamic exclusion of previously fragmented peptides was set for a period of 20 s and a tolerance of 10 ppm. All MS/MS peak lists were generated using Thermo Proteome Discoverer version 1.4.0.288 (Thermo Scientific). MGF sample files were analyzed using Mascot (Matrix Science, London, UK; version 2.4.0) and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1). Both softwares were programmed to search Uniprot Homo sapiens reference proteome (March 2014 release, 69150 entries) assuming trypsin digestion. Searches were carried out with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10 ppm. Carbamidomethyl of cysteine was specified as a fixed modification. Variable modifications were deamidation of asparagine and glutamine, oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Two miscleavages were allowed.
For both AP-MS and BioID, Scaffold version 4.7.5 (Proteome Sofware Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Criteria for protein identifications were fixed at greater than 99% probability to achieve a FDR less than 1% and required at least 1 peptide to be identified. Peptide and protein probabilities were assigned via the Protein Prophet algorithm (32, 33). Principle of parsimony was applied for proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone. Spectral counts were exported from Scaffold and were formatted according to SAINT algorithm guidelines. SAINT express statistical analyses were carried out including 3xFLAG-GFP controls for AP-MS. For BioID, the maximal spectral count value from Flag-BirA*-GFP (± biotin) for each prey was used as control. Proteins with a SAINT score ≥0.9 were deemed genuine interactors.
Network Modeling and Clustering Analysis
Interaction networks were modeled using Cytoscape v3.1.1. For clustering analysis, ClueGO plug-in v2.1.7 set for GO Biological process and GO Molecular function analysis with upper medium network specificity was used (34). Venn diagrams were prepared using Venn Diagram Plotter (https://omics.pnl.gov/software/venn-diagram-plotter).
RESULTS
AP-MS and BioID Proximity Labeling Delineate NCK1/2 Adaptor Protein Interactomes and Identify Specific Partners for Each NCK1 and NCK2
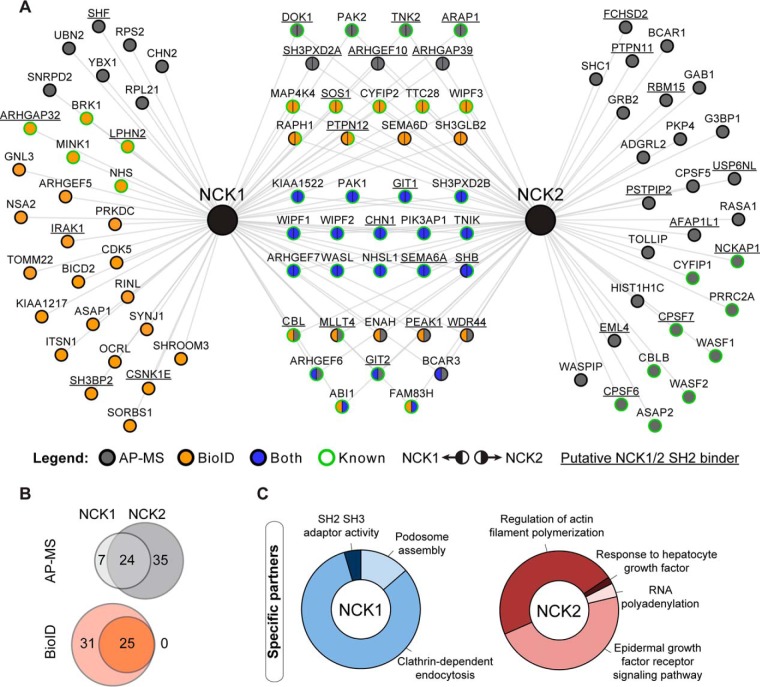
To systematically delineate protein interaction networks nucleated by NCK1 and NCK2 adaptor proteins, we generated clonal HEK293T cells stably expressing 3xFLAG-tagged NCK1 or NCK2 at near endogenous levels (supplemental Fig. S1A). We performed FLAG AP-MS identification of NCK-associated proteins. Asynchronous cells were treated with sodium pervanadate, a phosphatase inhibitor that broadly elevates the total level of endogenous tyrosine phosphorylation, to obtain the full complement of phospho-dependent (SH2) interactions. Using data from three independent AP-MS experiments including a 3xFLAG-GFP line as a negative control, we performed a significance analysis using SAINT to focus on high-confidence interactions (supplemental Table S1–S2) (29, 30). Our analysis identified a total of 66 proteins associated with NCK1 and NCK2 (Fig. 1A–1B).
Fig. 1.
NCK adaptor proteins AP-MS and BioID interactome reveals targets specifically associated with NCK1 or NCK2. A, NCK1/2-associated proteins were identified using AP-MS and BioID and combined to map the NCK1/2 interaction landscape. Previously reported interactions (known) from the BioGRID database were manually annotated. Putative NCK1/2 SH2 binders (underlined) were identified using SH2PepInt. B, Venn diagram representation of the overlap between NCK1- and NCK2-associated proteins found via AP-MS or BioID. C, Gene ontology (GO) and functional pathway analysis via ClueGO of NCK1- and NCK2-specific interactors.
To complement our AP-MS dataset, we further explored NCK1/2 interaction maps via BioID proximity labeling (14). To achieve this, we generated Flp-In T-REx HEK293 stable lines expressing NCK1 or NCK2 fused with BirA*, a promiscuous mutant biotin ligase enzyme that favors covalent biotin binding to adjacent proteins. We validated that the expression levels, function and localization of the chimeras following tetracycline addition were indistinguishable for FLAG-BirA*-NCK1 and FLAG-BirA*-NCK2 (supplemental Fig. S1B–S1D). We affinity-purified biotinylated proteins from cell lysates and identified them by MS, using a FLAG-BirA*-GFP control. We performed biological triplicates and eliminated nonspecific interactions using SAINT (supplemental Table S1, S3) (29, 30). This BioID analysis identified 56 proteins associated with NCK1 and NCK2 (Fig. 1A–1B).
The combination of AP-MS and BioID data revealed a total of 98 proteins associated with NCK1/2. Among them, we found 45 targets previously reported to be NCK1- and/or NCK2-associated proteins, including PAK2 (35), CHN1 (36), CBL (37), WASL (38) and GIT1 (2). In addition, we discovered 53 NCK-associated proteins that were not previously reported in the BioGRID database (Fig. 1A) (39), suggesting previously uncharacterized functions for NCK adaptors. We used the SH2PepInt tool to identify putative NCK1/2 SH2 binding sites on 33 proteins identified in the NCK1/2 network (40, 41), of which only GIT1 (2) and DOK1 (42) were previously reported. As expected, only a subset of NCK-associated proteins was discovered by both AP-MS and BioID. Indeed, 24% of the NCK1 interactome (17/70 proteins) and 23% of the NCK2 interactome (16/68 proteins) were revealed using either approach (Fig. 1A).
To define the specificity landscape of NCK interaction networks, we compared the distinct NCK1 and NCK2 interactions. Strikingly, we found 30 proteins specifically associated with NCK1, and 28 with NCK2 (Fig. 1A). Interestingly, AP-MS revealed more NCK2-specific partners whereas BioID displayed more NCK1-specific interactions. To delineate putative specialized functions for NCK1 or NCK2, we performed gene ontology and pathway term analysis from the proteins specifically associated with NCK1 or NCK2 using the ClueGO (gene ontology) plugin via Cytoscape software (34, 43). This analysis highlighted a strong enrichment for proteins involved in endocytosis for NCK1. In addition, we found that actin polymerization and epidermal growth factor signaling were functions that best represented the NCK2-specific components (Fig. 1C). Together, our findings demonstrate nonredundant protein association profiles for NCK1 and NCK2, supporting the idea that each displays a certain level of specificity. Furthermore, our data highlight several biological processes that may differentially implicate NCK1 or NCK2.
Nck2−/− Cells Are Multinucleated and Display Long Intercellular Bridges
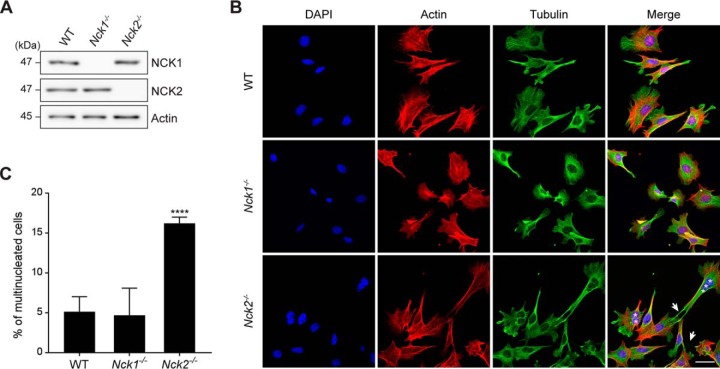
To evaluate the relative contribution of NCK1 and NCK2 to cellular morphology and cytoskeleton dynamics given the discrepancy between their protein networks with regards to actin cytoskeleton regulators, we established primary mouse embryo fibroblasts (MEFs) using animals bearing Nck1 and Nck2 gene targeting (12). Using antibodies that do not cross-react between NCKs, we confirmed that these cell lines specifically lacked NCK1 or NCK2 proteins, respectively (Fig. 2A). Nck1−/− MEFs were morphologically indistinguishable from wild-type cells. Surprisingly, most Nck2−/− MEFs were elongated and displayed extended protrusions that often maintained contact with adjacent cells, reminiscent of intercellular bridges (Fig. 2B). Staining for actin and tubulin did not reveal obvious disparities between the different lines that may explain the phenotype. However, we found that a significant proportion of Nck2−/− MEFs were multinucleated, suggesting a defect in cytokinesis (Fig. 2B–2C). We confirmed our observations using HEK293T cells, which also displayed intercellular bridge-like structures as well as significant multi-nucleation when depleted of NCK2, but not NCK1 using siRNA (supplemental Fig. S2A–S2C). Collectively, these NCK2-specific phenotypes are consistent with a defect in cytokinesis (44), and reinforce the idea that NCK1 and NCK2 adaptors have nonredundant functions.
Fig. 2.
Nck2−/− cells are multi-nucleated and display long protrusions. A, Mouse embryonic fibroblasts (MEFs) bearing Nck1 or Nck2 gene inactivation were analyzed by Western blotting to confirm NCK1/2 protein expression. B, Wild-type, Nck1−/− and Nck2−/− MEFs were analyzed by immunofluorescence for actin (red), tubulin (green) and DAPI (blue) to assess cellular morphology. Multi-nucleation (asterisks) and intercellular bridges (arrows) are indicated. Representative images are presented (scale bar = 40 μm). C, The penetrance of the multi-nucleation phenotype was calculated for each genotype. Mean values and standard deviation from three independent experiments with >100 cells each are presented (WT, n = 374; Nck1−/−, n = 325; Nck2−/−, n = 372) (****p ≤ 0.0001, Fisher's exact test).
NCK2, but Not NCK1, Localizes to the Midbody During Cytokinesis
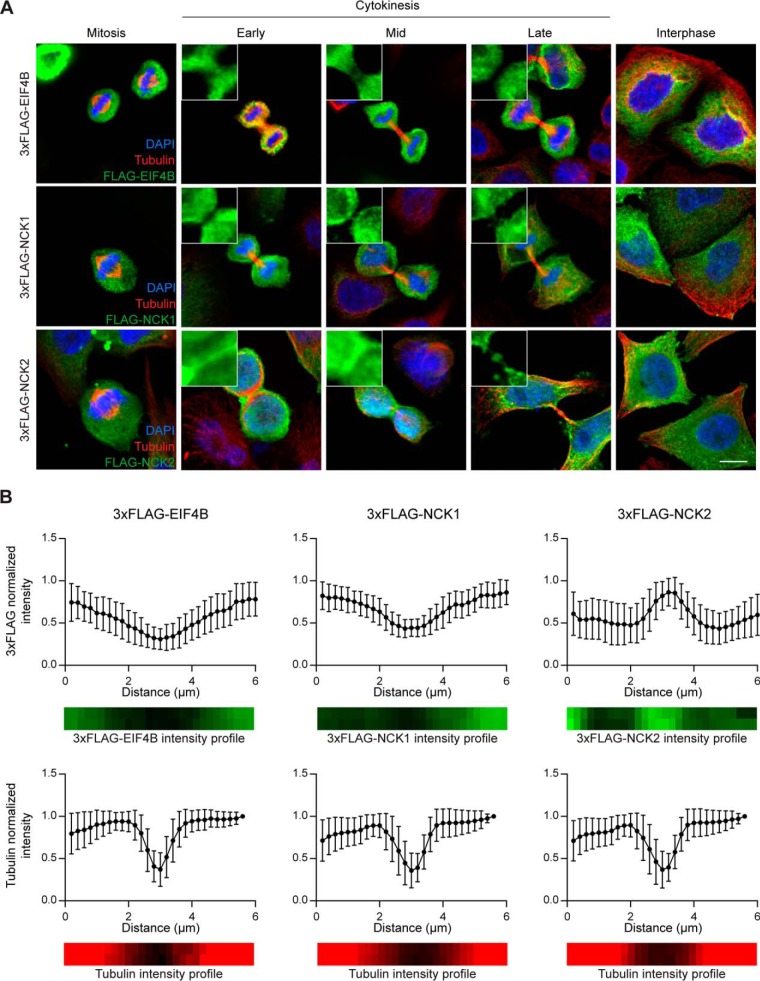
To examine a possible role for NCK during cell division, we first sought to analyze NCK1/2 expression in synchronized HeLa cells undergoing mitosis and cytokinesis. We were unable to detect the endogenous proteins under our fixation conditions (data not shown); we therefore transfected HeLa cells with 3xFLAG-NCK1 or 3xFLAG-NCK2 and assessed their localization during mitosis and cytokinesis. We found that both 3xFLAG-NCK1 and -NCK2 were distributed uniformly in the cytoplasm of daughter cells (Fig. 3A). However, we also detected 3xFLAG-NCK2, but not 3xFLAG-NCK1 or a 3xFLAG-EIF4B negative control, at the midbody in dividing cells (Fig. 3A). This was confirmed by quantifying the 3xFLAG signal across midbody length, the so-called dark zone (45) (Fig. 3B). Exogenous expression of 3xFLAG-NCK1 or -NCK2 did not alter tubulin localization relative to controls (Fig. 3B) and did not lead to cytokinesis defects (data not shown). The observation that NCK2, but not NCK1 localizes at the midbody agrees with the multi-nucleation and extended protrusions phenotypes specific to NCK2-deficient cells. Our observations also represent, to our knowledge, the first evidence of differential subcellular compartmentalization for the two NCK adaptor proteins.
Fig. 3.
NCK2 but not NCK1 localizes to the midbody during cytokinesis. A, HeLa cells were transiently transfected with 3xFLAG-EIF4B (control), 3xFLAG-NCK1 or 3xFLAG-NCK2 and synchronized. Cells were fixed at the cytokinesis stage and immunostained for tubulin (red), 3xFLAG (green) and DAPI (blue) to analyze 3xFLAG-tagged chimeras localization during cell division, as indicated. Representative images are shown (scale bar: 10 μm). B, Quantification of pixel intensity as a function of position within the midbody of images processed as in (A). Distance was measured from a 6 μm line selection over the midbody. Intensity of the 3xFLAG (top panels, green) or tubulin (bottom panels, red) staining is reported following normalization over the most intense pixel. Mean values and standard deviation from three independent experiments with >10 cells each are shown (3xFLAG-EIF4B cells: FLAG staining n = 34, tubulin n = 38; 3xFLAG-NCK1 cells: FLAG n = 33, tubulin n = 40; 3xFLAG-NCK2 cells: FLAG n = 30, tubulin n = 42).
NCK2 Is Required to Complete Cytokinesis and Abscission
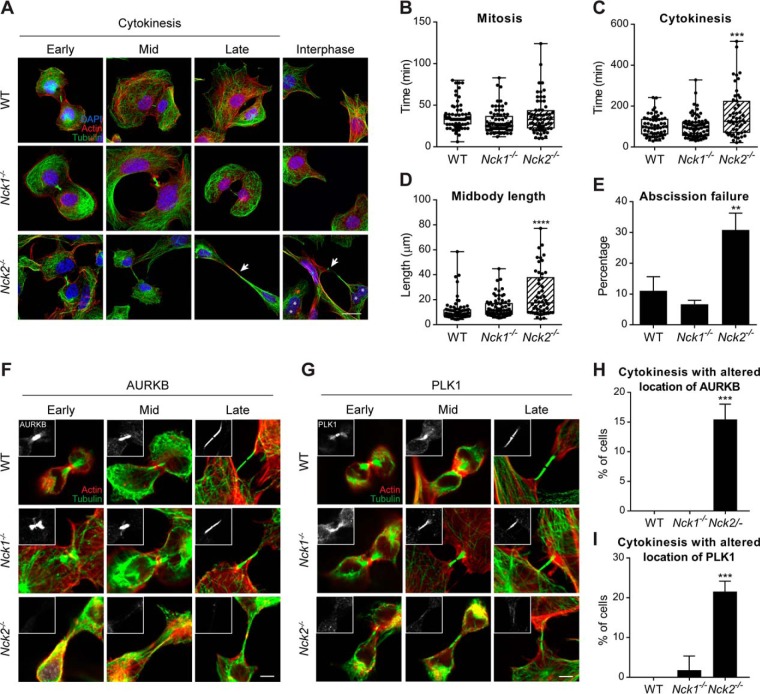
To investigate possible midbody defects in the Nck2−/− MEFs model, we analyzed midbody morphology at different stages of cytokinesis. Although midbody morphology appeared comparable in wild-type and Nck1−/− cells, it was clearly extended in length and persisted between nascent daughter cells for a longer period in Nck2−/− MEFs (Fig. 4A). To quantify this observation, we performed live-cell imaging of mitosis and cytokinesis, using the same lines (supplemental Fig. S3) or in NCK1/2-depleted HEK293T cells (supplemental Fig. S4). We found that Nck2−/− cells (34 ± 7 min) underwent mitosis in a timeframe that was like wild-type (36 ± 4 min) and to Nck1−/− cells (30 ± 9 min) (Fig. 4B). However, we found that Nck2−/− cells required on average 50% more time (156 ± 9 min) to complete cytokinesis compared with wild-type (104 ± 4 min) or Nck1−/− (97 ± 11 min) MEFs (p ≤ 0.001, unpaired t test) (Fig. 4C). In addition, Nck2−/− cells presented on average a significant increase in midbody length (25.8 ± 5.4 μm) during abscission when compared with wild-type (12.2 ± 2.7 μm) and Nck1−/− (13.9 ± 3.6 μm) controls (Fig. 4D). Furthermore, we determined that 30.8% of NCK2-deficient cells failed to fully complete abscission (Fig. 4E). This led to a rise in the proportion of multinucleated cells to 16.2% in Nck2−/− cells, compared with 5.1% in wild-type and 4.7% in Nck1−/− MEFs (p ≤ 0.0001) (Fig. 2C). This also explained the presence of extended intercellular bridges that frequently maintained contact with adjacent cells following division (Fig. 2B).
Fig. 4.
NCK2 is required to complete cytokinesis and abscission. A, Wild-type, Nck1−/− and Nck2−/− MEFs were fixed at different stages of cytokinesis and analyzed by immunofluorescence for actin (red), tubulin (green) and DAPI (blue) to evaluate midbody morphology. Multi-nucleation is indicated with asterisks and extended midbodies with arrows. Representative images are presented (scale bar: 20 μm). B–E, Wild-type, Nck1−/− and Nck2−/− MEFs were stained with SIR-ACTIN and analyzed by live imaging (supplemental Fig. S2). Average time spent in mitosis (B), in cytokinesis (C), average midbody length (D) and abscission failure (E) were calculated for each genotype, from three independent experiments with >20 cells each (WT, n = 70; Nck1−/−, n = 76; Nck2−/−, n = 63) (**p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001; unpaired t test). F–G, Wild-type, Nck1−/− and Nck2−/− MEFs were fixed at different stages of cytokinesis and analyzed by immunofluorescence for actin (red), tubulin (green) and AURKB (insert, panel F) or PLK1 (insert, panel G). Representative images from four experiments are shown (scale bar: 5 μm). H–I, The penetrance of the AURKB/PLK1 mislocalization was calculated for each genotype. Cells in which the AURKB or PLK1 signal at the midbody (identified via the tubulin signal) was barely detectable or not detected at all, relative to WT cells under identical acquisition settings, were considered to display altered staining. Mean values and standard deviation from four independent experiments with >10 cells each are presented (WT, n = 53; Nck1−/−, n = 59; Nck2−/−, n = 81) (*** p ≤ 0.001; Fisher's exact test).
To Further Delineate Midbody Defects in Nck2−/− MEFs, we examined the localization of midbody markers AURKB and PLK1 in our NCK-deficient lines. These kinases are recruited early in cytokinesis to the central spindle and site of furrow ingression and play a crucial role in the recruitment of components of the midbody. In wild-type and Nck1−/− cells undergoing cytokinesis, both proteins localized to the ingressing furrow and midbody in >98% of the cells, as described previously (18, 19). Conversely, we did not detect AURKB (15.4 ± 2.6%) and PLK1 (21.7 ± 2.6%) in a significant proportion of Nck2−/− MEFs, respectively (p ≤ 0.001, Fisher's exact test) (Fig. 4F–4I). PLK1 activity promotes the recruitment of the RhoGEF ECT2 to the central spindle, thereby establishing localized RhoA activation (46). In agreement with the loss of PLK1 at the central spindle and midbody, we observed a concomitant decrease in ECT2 recruitment (supplemental Fig. S5). Collectively, these numbers are consistent with the penetrance of the multi-nucleation and abscission failure phenotypes in Nck2−/− MEFs (Fig. 4C–4E). Together, these findings suggested that NCK2, but not NCK1 is required for the formation of a fully functional midbody in dividing cells.
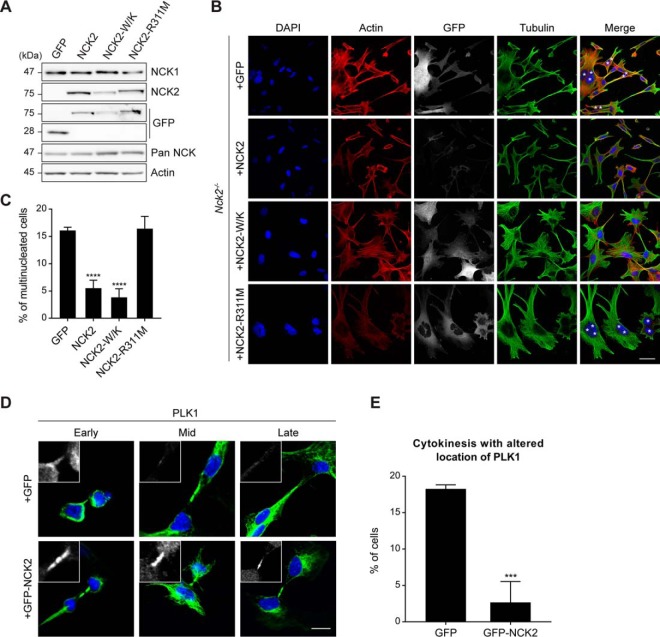
To confirm that our observations were because of the NCK2 loss-of-function, we re-expressed wild-type NCK2 from a stably inserted transgene in Nck2−/− MEFs (Fig. 5A). Expression of GFP-NCK2, but not GFP alone, led to a significant decrease in the proportion of multi-nucleated cells (Fig. 5B–5C). We also discovered that expression of GFP-NCK2 in Nck2−/− cells restored the localization of PLK1 to the midbody during cytokinesis, when compared with cells expressing GFP alone (Fig. 5D–5E). To determine the domain requirement of NCK2 for the regulation of cytokinesis, we introduced point mutations that inactivate the conventional binding of its three SH3 domains (W38K, W148K, W234K; W/K) or single SH2 domain (R311M) and repeated the rescue experiments (47). Interestingly, only the SH3-inactive NCK2 mutant was able to revert multi-nucleation; thus, a functional SH2 domain was required to support NCK2 function during cytokinesis (Fig. 5B–5C).
Fig. 5.
NCK2 regulation of cytokinesis is dependent on its SH2 domain. A, Mouse embryonic fibroblasts (MEFs) bearing Nck2 gene inactivation were infected with GFP, GFP-NCK2 wild-type, SH3-inactive (W38K, W148K, 234K; W/K) or SH2-inactive (R311M) mutants. Lysates were analyzed by Western blotting to confirm GFP or GFP-NCK2 protein expression. B, Nck2−/− MEFs expressing GFP or GFP-NCK2 wild-type and mutants (as indicated) were analyzed by immunofluorescence for actin, tubulin, DAPI and GFP to assess cellular morphology. Multi-nucleation is indicated with asterisks. Representative images are presented (scale bar: 40 μm). C, The penetrance of the multi-nucleation phenotype was calculated for each tentative rescue condition. Mean values and standard deviation from three independent experiments with >100 cells each are presented (+GFP, n = 409; +GFP-NCK2, n = 335; +GFP-NCK2-W/K, n = 328; +GFP-NCK2-R311M, n = 392) (****p ≤ 0.0001; Fisher's exact test). D, Nck2−/− MEFs expressing GFP or GFP-NCK2 wild-type were fixed at different stages of cytokinesis and analyzed by immunofluorescence for tubulin (green), DAPI (blue) and PLK1 (insert panel, white) to assess cellular morphology. Representative images from four experiments are shown (scale bar: 10 μm). E, The penetrance of the PLK1 mislocalization was calculated for each condition. Mean values and standard deviation from four independent experiments with >20 cells each are presented (+GFP, n = 120; +GFP-NCK2, n = 106) (*** p ≤ 0.001; Fisher's exact test).
DISCUSSION
In this work, we report the first high-confidence protein-protein interaction and proximity network for NCK family adaptors. We stressed the complementarity of the AP-MS and BioID approaches by highlighting that <25% of the NCK-associated proteins were revealed by both methods. This observation is consistent with previous reports in which AP-MS and BioID were directly compared (48, 22). We found that the best characterized NCK-associated proteins, such as PAK1/2, CHN1, CBL, WASL and TNIK are common to both NCK1 and NCK2. Interestingly, our AP-MS and BioID data also led to the identification of 30 NCK1- and 28 NCK2-specific partners, the majority of which being new NCK-associated proteins. The large number of NCK1- and NCK2-restricted associations is striking, given that the in vitro binding preference of the respective SH2 domains of NCK1 and NCK2 are virtually indistinguishable (2). However, the analysis of NCK-associated proteins for putative NCK1/2 SH2 binding sites (Y-D/E-X-V/P) revealed several potential targets (Fig. 1A), which suggests that the NCK1 and NCK2 SH2 binding specificities determined in vitro using synthetic peptides might not be reflected in vivo with native proteins in a complex environment. In addition, to our knowledge the binding specificities of corresponding SH3 domains of each NCK have not been explored and may also explain, at least in part, the presence of specific interactions. Differences between the repertoires of NCK1 and NCK2 interactomes are consistent with studies that have described a function specific to one of the two adaptors. NCK2 was previously found to regulate EPH receptor-mediated cell adhesion during early vertebrate development (49) and adipogenesis in a mouse model (11). Yet, no specific NCK2 target was defined in either study. The differential subcellular compartmentalization that we discovered in dividing HeLa cells may also occur in other cell types and/or under different conditions, thus explaining in part the disparities between NCK1- and NCK2-associated proteins. Nonetheless, it is likely that our description of the NCK1/2 signaling network specificity landscape will contribute to providing mechanistic explanations to functional distinctions between NCK1 and NCK2.
We also report for the first time a role for NCK2 in the regulation of cell division that is not redundant with NCK1 function. Interestingly, the function of NCK2 appears to be early during cytokinesis as loss of NCK2 attenuates the recruitment of PLK1 and AURKB to the central spindle, a structure required to position the cleavage plane. Two protein complexes play key roles in central spindle assembly: PRC1 (Protein required for cytokinesis 1) and Centralspindlin (50). The latter consists of a hetero-tetramer of RACGAP1 and KIF23/MKLP1 (51). Their phosphorylation by AURKB and PLK1, respectively, leads to Centralspindlin activation at the onset of anaphase (52, 53). This promotes the recruitment of ECT2 and the subsequent accumulation of active RhoA, which promotes the assembly and stimulates contraction of the actomyosin ring and thus furrow ingression. How the loss of NCK2 prevents PLK1 and AURKB recruitment remains to be explored. Because NCK adaptors play an important role in the assembly and reorganization of the actin cytoskeleton, it is conceivable that they have a similar function during early cytokinesis. Our MS analyses led to the identification of a few proteins previously described to regulate cytokinesis, such as GIT1 (54) and PKP4 (55). Interestingly, the latter was specifically identified in NCK2 complexes, thus suggesting that they may be contribute to NCK2-dependent regulation of the process.
Moreover, considering the requirement for the NCK2-SH2 domain for rescue of the cell division defects, our data suggest that the NCK2 adaptor is a strong candidate for nucleation of essential phospho-tyrosine signaling networks during cytokinesis. Interestingly, the unique Drosophila ortholog of NCK1/2, Dock, localizes at ring canals structures that are assembled from arrested cleavage furrows following incomplete cytokinesis of male germ cells (56). These structures share most of their constituents with contractile rings of dividing cells and display strong phospho-tyrosine signals (57). The subcellular localization of Dock at ring canals was suggested to be dependent on its SH2 domain (56), which supports an evolutionary-conserved SH2-dependent function for NCK adaptors in cytokinesis. Remarkably, a more prominent role for tyrosine phosphorylation during cell division than previously recognized has been recently reported in mammalian cells (58–60). Moreover, tyrosine phosphorylation has been shown to increase at the cleavage furrow of dividing cells (61), suggesting the presence of localized phospho-tyrosine signaling during furrow ingression and cytokinesis. Consistent with this, tyrosine kinases ABL and SRC were shown to localize at the midbody in dividing cells where their functions are required to complete cytokinesis (62–64). The requirement for the tight regulation of phospho-tyrosine signals during cytokinesis is also supported by the findings that PTPN12 and PTPN13 tyrosine phosphatases also contribute to cytokinesis (25, 26). More recently, receptor tyrosine kinase EPHB2-dependent phosphorylation of CIT (Citron kinase) was shown to impair cytokinesis (65). Strikingly, EPHB2 bears a bona fide binding site for NCK adaptors SH2 domains in its juxtamembrane domain when tyrosine phosphorylated (66), thus suggesting a possible mechanism for cytokinesis regulation.
In summary, the NCK1 and NCK2 interaction profiles revealed a plethora of new candidates that may pave the way for the mechanistic dissection of NCK1 and NCK2 contribution to biological processes. The data led to the discovery that NCK2 but not NCK1 is present at the midbody in dividing cells, where it is required for proper cytokinesis and abscission. This supports the very few previous reports showing that NCK1 and NCK2 functions are not completely redundant.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (67), with the data set identifier PXD008824.
Supplementary Material
Acknowledgments
We thank members of the Bisson laboratory for discussions, J. Ruston, J. Du and T. Pawson for the pan-NCK antibody as well as Nck1−/− and Nck2−/− MEFs. Mass spectrometry was performed at the CHU de Québec – Université Laval Proteomics Platform.
Footnotes
* This work was funded by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) (418615-2012, 2018-06293 to N.B. and 2016-05841 to S.E.), and Operating Grants from the Canadian Institutes for Health Research (CIHR) (130335 to N.B., 312588 to S.E.). N.B. was also supported by Leader's Opportunity Funds from the Canada Foundation for Innovation (30308, 34963), holds a Canada Research Chair (Tier 2) in Cancer Proteomics, and previously a Junior 1 salary award from the Fonds de Recherche du Québec-Santé (FRQ-S) with funds from the Quebec Breast Cancer Foundation. S.E. holds a Junior 2 salary award from the Fonds de Recherche du Québec-Santé (FRQ-S). K.J. and S.L.B. held PROTEO scholarships.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- NCK
- non-catalytic region of tyrosine kinase
- AP-MS
- affinity purification-mass spectrometry
- AURKB
- Aurora-B
- CIT
- citron kinase
- DMEM
- Dulbecco's Modified Eagle's medium
- EphR
- EPH family receptors
- HEK293T
- human embryonic kidney 293T cells
- MEF
- mouse embryonic fibroblasts
- PRC1
- protein required for cytokinesis 1
- PXN
- paxillin
- PDGFRB
- platelet-derived growth factor receptor beta
- PLK1
- polo-like kinase 1
- RT
- room temperature
- RTK
- receptor tyrosine kinase
- SH
- Src homology
- WASP
- Wiscott-Aldrich syndrome protein.
REFERENCES
- 1. Buday L., Wunderlich L., and Tamas P. (2002) The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal 14, 723–731 [DOI] [PubMed] [Google Scholar]
- 2. Frese S., Schubert W. D., Findeis A. C., Marquardt T., Roske Y. S., Stradal T. E., and Heinz D. W. (2006) The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236–18245 [DOI] [PubMed] [Google Scholar]
- 3. Chen M., She H., Davis E. M., Spicer C. M., Kim L., Ren R., Le Beau M. M., and Li W. (1998) Identification of Nck family genes, chromosomal localization, expression, and signaling specificity. J. Biol. Chem. 273, 25171–25178 [DOI] [PubMed] [Google Scholar]
- 4. Li W., Fan J., and Woodley D. T. (2001) Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene 20, 6403–6417 [DOI] [PubMed] [Google Scholar]
- 5. Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., and Mayer B. J. (2001) Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276, 26448–26452 [DOI] [PubMed] [Google Scholar]
- 6. Guan S., Fan J., Han A., Chen M., Woodley D. T., and Li W. (2009) Non-compensating roles between Nckalpha and Nckbeta in PDGF-BB signaling to promote human dermal fibroblast migration. J. Invest. Dermatol. 129, 1909–1920 [DOI] [PubMed] [Google Scholar]
- 7. Bong Y. S., Park Y. H., Lee H. S., Mood K., Ishimura A., and Daar I. O. (2004) Tyr-298 in ephrinB1 is critical for an interaction with the Grb4 adaptor protein. Biochem. J. 377, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowan C. A., and Henkemeyer M. (2001) The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature 413, 174–179 [DOI] [PubMed] [Google Scholar]
- 9. Su Z., Xu P., and Ni F. (2004) Single phosphorylation of Tyr304 in the cytoplasmic tail of ephrin B2 confers high-affinity and bifunctional binding to both the SH2 domain of Grb4 and the PDZ domain of the PDZ-RGS3 protein. Eur. J. Biochem. 271, 1725–1736 [DOI] [PubMed] [Google Scholar]
- 10. Guan S., Chen M., Woodley D., and Li W. (2007) Nckbeta adapter controls neuritogenesis by maintaining the cellular paxillin level. Mol. Cell Biol. 27, 6001–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dusseault J., Li B., Haider N., Goyette M. A., Cote J. F., and Larose L. (2016) Nck2 Deficiency in mice results in increased adiposity associated with adipocyte hypertrophy and enhanced adipogenesis. Diabetes 65, 2652–2666 [DOI] [PubMed] [Google Scholar]
- 12. Bladt F., Aippersbach E., Gelkop S., Strasser G. A., Nash P., Tafuri A., Gertler F. B., and Pawson T. (2003) The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol. Cell Biol. 23, 4586–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gingras A. C., Gstaiger M., Raught B., and Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 14. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Green R. A., Paluch E., and Oegema K. (2012) Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 [DOI] [PubMed] [Google Scholar]
- 16. Pollard T. D. (2010) Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller A. L. (2011) The contractile ring. Curr. Biol. 21, R976–R978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carmena M., and Earnshaw W. C. (2003) The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 19. Takaki T., Trenz K., Costanzo V., and Petronczki M. (2008) Polo-like kinase 1 reaches beyond mitosis–cytokinesis, DNA damage response, and development. Curr. Opin. Cell Biol. 20, 650–660 [DOI] [PubMed] [Google Scholar]
- 20. Barr F. A., and Gruneberg U. (2007) Cytokinesis: placing and making the final cut. Cell 131, 847–860 [DOI] [PubMed] [Google Scholar]
- 21. Pollard T. D., and Wu J. Q. (2010) Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11, 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lambert J. P., Tucholska M., Go C., Knight J. D., and Gingras A. C. (2015) Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics 118, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bisson N., Tremblay M., Robinson F., Kaplan D. R., Trusko S. P., and Moss T. (2008) Mice lacking both mixed-lineage kinase genes Mlk1 and Mlk2 retain a wild type phenotype. Cell Cycle 7, 909–916 [DOI] [PubMed] [Google Scholar]
- 24. Ma H. T., and Poon R. Y. (2017) Synchronization of HeLa Cells. Methods Mol. Biol. 1524, 189–201 [DOI] [PubMed] [Google Scholar]
- 25. Angers-Loustau A., Cote J. F., Charest A., Dowbenko D., Spencer S., Lasky L. A., and Tremblay M. L. (1999) Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J. Cell Biol. 144, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herrmann L., Dittmar T., and Erdmann K. S. (2003) The protein tyrosine phosphatase PTP-BL associates with the midbody and is involved in the regulation of cytokinesis. Mol. Biol. Cell 14, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beigbeder A., Chartier F. J. M., and Bisson N. (2017) MPZL1 forms a signalling complex with GRB2 adaptor and PTPN11 phosphatase in HER2-positive breast cancer cells. Sci. Rep. 7, 11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaguchi T., Goto H., Yokoyama T., Sillje H., Hanisch A., Uldschmid A., Takai Y., Oguri T., Nigg E. A., and Inagaki M. (2005) Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J. Cell Biol. 171, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi H., Liu G., Mellacheruvu D., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2012) Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr. Protoc. Bioinformatics Chapter 8, Unit8 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beigbeder A., Velot L., James D. A., and Bisson N. (2016) Sample preparation for mass spectrometry analysis of protein-protein interactions in cancer cell lines and tissues. Methods Mol. Biol. 1458, 339–347 [DOI] [PubMed] [Google Scholar]
- 32. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 33. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 34. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W. H., Pages F., Trajanoski Z., and Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bokoch G. M., Wang Y., Bohl B. P., Sells M. A., Quilliam L. A., and Knaus U. G. (1996) Interaction of the Nck adapter protein with p21-activated kinase (PAK1). J. Biol. Chem. 271, 25746–25749 [DOI] [PubMed] [Google Scholar]
- 36. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., and Pawson T. (2006) A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 37. Rivero-Lezcano O. M., Sameshima J. H., Marcilla A., and Robbins K. C. (1994) Physical association between Src homology 3 elements and the protein product of the c-cbl proto-oncogene. J. Biol. Chem. 269, 17363–17366 [PubMed] [Google Scholar]
- 38. Rivero-Lezcano O. M., Marcilla A., Sameshima J. H., and Robbins K. C. (1995) Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol. Cell Biol. 15, 5725–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N. K., O'Donnell L., Oster S., Theesfeld C., Sellam A., Stark C., Breitkreutz B. J., Dolinski K., and Tyers M. (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kundu K., Costa F., Huber M., Reth M., and Backofen R. (2013) Semi-supervised prediction of SH2-peptide interactions from imbalanced high-throughput data. PLoS ONE 8, e62732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kundu K., Mann M., Costa F., and Backofen R. (2014) MoDPepInt: an interactive web server for prediction of modular domain-peptide interactions. Bioinformatics 30, 2668–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu B. A., Engelmann B. W., Jablonowski K., Higginbotham K., Stergachis A. B., and Nash P. D. (2012) SRC Homology 2 domain binding sites in insulin, IGF-1 and FGF receptor mediated signaling networks reveal an extensive potential interactome. Cell Commun. Signal 10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Normand G., and King R. W. (2010) Understanding cytokinesis failure. Adv. Exp. Med. Biol. 676, 27–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu C. K., Coughlin M., and Mitchison T. J. (2012) Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 23, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su K. C., Takaki T., and Petronczki M. (2011) Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell 21, 1104–1115 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka M., Gupta R., and Mayer B. J. (1995) Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol. Cell Biol. 15, 6829–6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Couzens A. L., Knight J. D., Kean M. J., Teo G., Weiss A., Dunham W. H., Lin Z. Y., Bagshaw R. D., Sicheri F., Pawson T., Wrana J. L., Choi H., and Gingras A. C. (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal 6, rs15. [DOI] [PubMed] [Google Scholar]
- 49. Bisson N., Poitras L., Mikryukov A., Tremblay M., and Moss T. (2007) EphA4 Signaling Regulates Blastomere Adhesion in the Xenopus Embryo by Recruiting Pak1 to Suppress Cdc42 Function. Mol. Biol. Cell 18, 1030–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mierzwa B., and Gerlich D. W. (2014) Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell 31, 525–538 [DOI] [PubMed] [Google Scholar]
- 51. Pavicic-Kaltenbrunner V., Mishima M., and Glotzer M. (2007) Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol. Biol. Cell 18, 4992–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guse A., Mishima M., and Glotzer M. (2005) Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15, 778–786 [DOI] [PubMed] [Google Scholar]
- 53. Mishima M., Pavicic V., Gruneberg U., Nigg E. A., and Glotzer M. (2004) Cell cycle regulation of central spindle assembly. Nature 430, 908–913 [DOI] [PubMed] [Google Scholar]
- 54. Hagemann N., Ackermann N., Christmann J., Brier S., Yu F., and Erdmann K. S. (2013) The serologically defined colon cancer antigen-3 interacts with the protein tyrosine phosphatase PTPN13 and is involved in the regulation of cytokinesis. Oncogene 32, 4602–4613 [DOI] [PubMed] [Google Scholar]
- 55. Wolf A., Keil R., Gotzl O., Mun A., Schwarze K., Lederer M., Huttelmaier S., and Hatzfeld M. (2006) The armadillo protein p0071 regulates Rho signalling during cytokinesis. Nat. Cell Biol. 8, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 56. Abdallah A. M., Zhou X., Kim C., Shah K. K., Hogden C., Schoenherr J. A., Clemens J. C., and Chang H. C. (2013) Activated Cdc42 kinase regulates Dock localization in male germ cells during Drosophila spermatogenesis. Dev. Biol. 378, 141–153 [DOI] [PubMed] [Google Scholar]
- 57. Hime G. R., Brill J. A., and Fuller M. T. (1996) Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 109 (Pt 12), 2779–2788 [DOI] [PubMed] [Google Scholar]
- 58. Caron D., Byrne D. P., Thebault P., Soulet D., Landry C. R., Eyers P. A., and Elowe S. (2016) Mitotic phosphotyrosine network analysis reveals that tyrosine phosphorylation regulates Polo-like kinase 1 (PLK1). Sci. Signal 9, rs14. [DOI] [PubMed] [Google Scholar]
- 59. Elowe S. (2017) Tyr(less) kinase signaling during mitosis. Cell Cycle 16, 746–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. St-Denis N., Gupta G. D., Lin Z. Y., Gonzalez-Badillo B., Veri A. O., Knight J. D. R., Rajendran D., Couzens A. L., Currie K. W., Tkach J. M., Cheung S. W. T., Pelletier L., and Gingras A. C. (2016) Phenotypic and Interaction Profiling of the Human Phosphatases Identifies Diverse Mitotic Regulators. Cell Rep. 17, 2488–2501 [DOI] [PubMed] [Google Scholar]
- 61. Ng M. M., Chang F., and Burgess D. R. (2005) Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell 9, 781–790 [DOI] [PubMed] [Google Scholar]
- 62. Chen S., and Tang D. D. (2014) c-Abl tyrosine kinase regulates cytokinesis of human airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 50, 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kasahara K., Nakayama Y., Nakazato Y., Ikeda K., Kuga T., and Yamaguchi N. (2007) Src signaling regulates completion of abscission in cytokinesis through ERK/MAPK activation at the midbody. J. Biol. Chem. 282, 5327–5339 [DOI] [PubMed] [Google Scholar]
- 64. Soeda S., Nakayama Y., Honda T., Aoki A., Tamura N., Abe K., Fukumoto Y., and Yamaguchi N. (2013) v-Src causes delocalization of Mklp1, Aurora B, and INCENP from the spindle midzone during cytokinesis failure. Exp. Cell Res. 319, 1382–1397 [DOI] [PubMed] [Google Scholar]
- 65. Jungas T., Perchey R. T., Fawal M., Callot C., Froment C., Burlet-Schiltz O., Besson A., and Davy A. (2016) Eph-mediated tyrosine phosphorylation of citron kinase controls abscission. J. Cell Biol. 214, 555–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Holland S. J., Gale N. W., Gish G. D., Roth R. A., Songyang Z., Cantley L. C., Henkemeyer M., Yancopoulos G. D., and Pawson T. (1997) Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 16, 3877–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (67), with the data set identifier PXD008824.