Abstract
Nicotinamide adenine dinucleotide (NAD) plays an essential role in all aspects of human life. NAD levels decrease as humans age, and supplementation with NAD precursors plays a protective role against aging and associated disease. Less is known about the effects of decreased NAD on cellular processes, which is the basis for understanding the relationship between cellular NAD levels and aging-associated disease. In the present study, cellular NAD levels were decreased by overexpression of CD38, a NAD hydrolase, or by treating cells with FK866, an inhibitor of nicotinamide phosphoribosyltransferase (NAMPT). Quantitative proteomics revealed that declining NAD levels downregulated proteins associated with primary metabolism and suppressed cell growth in culture and nude mice. Decreased glutathione synthesis caused a 4-fold increase in cellular reactive oxygen species levels, and more importantly upregulated proteins related to movement and adhesion. In turn, this significantly changed cell morphology and caused cells to undergo epithelial to mesenchymal transition (EMT). Secretomic analysis also showed that decreased NAD triggered interleukin-6 and transforming growth factor beta (TGFβ) secretion, which activated integrin-β-catenin, TGFβ-MAPK, and inflammation signaling pathways to sustain the signaling required for EMT. We further revealed that decreased NAD inactivated sirtuin 1, resulting in increased signal transducer and activator of transcription 3 (STAT3) acetylation and phosphorylation, and STAT3 activation. Repletion of nicotinamide or nicotinic acid inactivated STAT3 and reversed EMT, as did STAT3 inhibition. Taken together, these results indicate that decreased NAD activates multiple signaling pathways to promote EMT and suggests that age-dependent decreases in NAD may contribute to tumor progression. Consequently, repletion of NAD precursors has potential benefits for inhibiting cancer progression.
Keywords: Aging, Cell adhesion, Lung cancer, Omics, SILAC, Secretome, Phosphoproteome, Epithelial-Mesenchymal Transition
Nicotinamide adenine dinucleotide (NAD)1 is essential to life and participates in all major biological processes (1–3). The NAD pool consists of NAD, reduced nicotinamide adenine dinucleotide (NADH), nicotinamide adenine dinucleotide phosphate (NADP), and reduced nicotinamide adenine dinucleotide phosphate (NADPH). As co-factors for a variety of oxidoreductase enzymes, NAD(H) mainly functions in biodegradation reactions and energy generation, whereas NADP(H) functions in fatty acid and cholesterol synthesis. NADPH is also an essential element in the cellular defense system and maintains redox homeostasis, in which both glutaredoxin (GRX) and thioredoxin reductase (TrxR) are acceptors of NADPH electrons for reduction of disulfide bonds. NAD is synthesized in cells via de novo biosynthesis and salvage pathways. Tryptophan is the precursor for de novo NAD synthesis, in which kynureninase catalyzes hydrolysis of 3-hydroxy-l-kynurenine to generate 3-hydroxy-anthranilic acid. Nicotinamide phosphoribosyltransferase (NAMPT) is the rate-limiting enzyme in the NAD salvage pathway, which is essential for maintaining cellular NAD levels (4). Moreover, inhibiting NAMPT is a therapeutic approach in cancer treatment (5).
Besides its roles in cell metabolism, NAD is a substrate for various adenosine diphosphate (ADP)-ribosyltransferases in regulation of cellular signal transduction, including NAD-dependent deacetylases (sirtuins), poly(ADP-ribose) polymerases(PARPs), mono(ADP-ribosyl)-transferases (ARTs), tRNA 2′-phosphotransferases, and ADP-ribosylcyclases (CD38 and CD157). NAD functions as both a cofactor and substrate for sirtuins to remove acetyl groups from acetyl-lysine residues of proteins (6–10). Consequently, NAD plays a key role in a broad range of cellular processes performed by sirtuins (10–20). NAD is a substrate of PARP for ADP-ribosylation of nuclear proteins in cellular stress responses (21, 22) that regulate cell proliferation, death, differentiation, and senescence. All these findings suggest that NAD is essential for cellular metabolism, mitochondrial function, stress response, and genomic integrity. More importantly, recent studies have shown that NAD levels decline as humans age. Indeed, it was estimated that NAD levels are ∼5 ng/mg (protein) in teenagers but decrease to 3 ng/mg (protein) by the age of 50. Reduced levels of NAD metabolism are observed in aged rats and mice (23, 24), whereas increasing NAD levels is an effective approach for prolonging lifespan (25–28).
Cellular NAD levels are regulated through NAD biosynthesis, NAD metabolism, and NAD-dependent nonredox enzymatic reactions. ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 (CD38) is a transmembrane glycoprotein mainly expressed in B cells. It functions as a NAD glycohydrolase as well as an ADP-ribosyl cyclase that generates cyclic ADP-ribose to initiate calcium efflux (29, 30). NADase activity of CD38 is 100-times higher than its ADP-ribosyl cyclase activity, therefore CD38 is an efficient tool for modulating cellular NAD levels (31, 32). Increased NAD levels in Parp1−/− or Cd38−/− mice and mice treated with NAD precursors improve mitochondrial function and protect mice against metabolic damage induced by high-fat diets (24, 33–35). More importantly, a recent study found that CD38 increases with age and dictates age-dependent NAD decline (36).
However, the effect of decreased NAD on biological processes has not been well characterized, although it is important for understanding the relationship between cellular NAD levels and aging-associated disease. In our previous study (37), we established a stable cell line that overexpresses CD38. We found that a 35% decrease in NAD levels in 293T cells caused significant changes in proteostasis and reactive oxygen species (ROS) homeostasis, with downregulation of glycolytic enzymes and antioxidant proteins leading to decreased cell proliferation rates and increased cell susceptibility to oxidative stress. Thus, the purpose of the present study was to examine the effect of decreased NAD on cellular processes in other cell lines. Herein, we established stable CD38-expressing cell lines to comprehensively determine how cellular NAD levels affect protein expression, cell proliferation, and cellular responses to oxidative stress. Our findings demonstrate that decreased cellular NAD levels cause cells to undergo epithelial-mesenchymal transition (EMT), whereas signal transducer and activator of transcription 3 (STAT3) inhibition reversed EMT, indicating that STAT3 is a vital regulator in NAD-mediated EMT.
EXPERIMENTAL PROCEDURES
Cell Culture and SILAC Labeling
The human lung cancer cell line, A549, human liver cancer cell, HepG2 and human embryonic kidney cell line, 293T, were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cells were grown in RPMI 1640 medium or Dulbecco's Modified Eagle's Medium (Wisent, Montreal, QC, Canada). The medium was supplemented with 10% FBS (Wisent) and 1% penicillin/streptomycin (Wisent). For stable isotope labeling using amino acids (SILAC), cells were washed twice with PBS (Wisent) before replacing the culture medium with SILAC. A549 cells were grown in SILAC medium for 7 passages and tested for full incorporation before proteomic analysis. SILAC RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA) was supplemented with 10% dialyzed-FBS (Wisent), 1% penicillin/streptomycin, 40 mg/L isotope labeling l-13C615N2 lysine·HCl (Thermo Fisher Scientific) and 200 mg/L isotope labeling l-13C6 arginine·HCl (Thermo Fisher Scientific).
Establishment of a Stable CD38 Overexpressing Cell Line
Human CD38 cDNA was obtained from the U266 cell line. A FLAG tag was added at the C terminus and DNA sequence encoding amino acids 2–43 deleted from the N terminus. This recombinant human CD38 DNA was then cloned into PLVX-IRES-ZsGreen1 lentiviral transfer vector. The 293T cell line was transfected with PLVX-IRES-ZsGreen1 or PLVX-CD38-IRES-ZsGreen1 using lentiviral packaging vectors and polyethylenimine (Sigma, St Louis, MO). Supernatants were harvested after 48 h and concentrated with PEG6000. Precipitated lentiviral particles were resuspended in PBS. A549 and HepG2 cells were infected in the presence of 5 μg/ml polybrene (Sigma). Infected cells were sorted by flow cytometry. A single green fluorescent protein (GFP)-positive cell was seeded into a single well of a 96-well plate to select a clone with intense and uniform GFP expression for further analysis.
NAD and NADH Content Assay
NAD and NADH content were measured by NAD/NADH assay (BioAssay Systems, Hayward, CA), according to the manufacturer's instruction. Briefly, cells were washed with PBS and 4 × 105 cells counted for each cell line. NAD or NADH extraction buffer were added, and extracts heated at 60 °C for 5 min. Samples and NAD standards were then reacted with a working reagent. Absorbance at 565 nm was measured to quantify NAD and NADH concentrations.
Cell Proliferation Assay Determined by CCK-8
Cells were seeded in 96-well plates with 2000 cells/well. Cell proliferation rate was determined by the Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan). CCK-8 reagent was added to treated cells and incubated at 37 °C for 2 h. Optical density (OD) was measured at 450 nm with a microplate reader (Bio-Rad, Hercules). Relative cell number was represented as the ratio between absorbance in 450 nm at a particular hour and 0 h.
Experimental Design and Statistical Rationale
All experiments were performed at least three times to ensure statistical power > 80%. Proteomic analysis was performed using biological triplicates. Mock-infected control cells (designated CD38(−) cells) were cultured in SILAC culture media with 13C615N2 lysine and 13C6 arginine. CD38-transfected cells were cultured in normal RMPI-1640 media and designated as CD38(+) cells. A Venn diagram was used to evaluate the number of overlapping proteins identified from proteomic studies using three biological replicates. Significantly changed proteins were screened by volcano plot analysis using R (V.3.3.2). Histograms of SILAC ratios showed protein fold-changes in proteomics data and demonstrated normal data distribution. Proteins with fold-change > 2 or < 0.5 and p values < 0.05 from t test statistics were considered significantly changed between CD38(−) and CD38(+) cells. Calculation of Pearson correlation was performed to ensure reproducibility of protein quantification.
Sample Preparation
Protein (200 μg) was extracted from CD38(−) and CD38(+) cells and mixed. Protein disulfides were reduced by dithiothreitol at 5 mm for 60 min at room temperature. Next, free cysteine residues were alkylated with iodoacetamide at 12 mm for 45 min in the dark at room temperature. Protein samples were digested with trypsin for 14 h at 37 °C. Peptides were desalted with Sep-Pak C18 cartridges (Thermo-Pierce Biotechnology, Rockford, IL). Eluents were centrifuged by speedvac followed by HPLC separation. Collected eluents were combined into 12 fractions and analyzed by LC-MS/MS.
LC-MS/MS Analysis
For LC-MS/MS analysis, SILAC labeled peptides were separated by 135 min gradient elution at a flow rate of 0.3 μl/min with a Thermo-Dionex Ultimate 3000 HPLC system that was directly interfaced with a Thermo Orbitrap Fusion Lumos mass spectrometer. The analytical column was a homemade fused silica capillary column (75 μm inner-diameter, 150 mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 Å, 5 μm; Varian, Lexington, MA). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The Thermo Orbitrap Fusion Lumos mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 4.0.27.10 software. A single full-scan mass spectrum was done in the Orbitrap (300–1500 m/z, 120,000 resolution). The spray voltage is 2500 V and the AGC target is 200,000. This was followed by 3 s data-dependent MS/MS scans in an ion routing multipole at 30% normalized collision energy (HCD). The charge state screening of ions was set at 2–7. The exclusion duration was set at 15 s. Mass window for precursor ion selection was set at 1.6 m/z. The MS/MS resolution was 30,000. The MS/MS maximum injection time was 60 ms and the AGC target was 200,000.
Peptide and Protein Identification
Peak lists from LC-MS/MS analysis were generated with the SEQUESTTM searching algorithm using Proteome Discoverer software (version 2.1; Thermo Fisher Scientific). Spectra were searched against the UniProt human reference proteome (release on March 17, 2017, containing 21,042 entries) using an in-house Proteome Discoverer Searching Algorithm (version 2.1; Thermo Fisher Scientific). The search criteria were: full tryptic specificity was required; two missed cleavage sites were allowed; oxidation of methionine, 13C(6)15N(2) at lysine, 13C(6) at arginine, and acetylation of protein N terminus were set as variable modifications; carbamidomethylation of cysteine was set as the fixed modification; and precursor ion mass tolerance was set at 10 ppm for all MS and 20 mmu for all MS2 spectra. Peptide false discovery rate (FDR) was estimated using the percolator function provided by Proteome Discoverer, with a cutoff score of 1% based on decoy database searching.
Protein Quantification
Proteome Discoverer Searching Algorithm was used for protein quantification. Briefly, relative protein expression ratios were calculated using the peak area of Lys0Arg0 divided by the peak area of Lys8Arg6. Only peptides assigned to a given protein group were considered unique. Proteins with two or more unique peptide matches were regarded as confident identifications and further quantified. Differences in protein abundance between sample and control groups were calculated by averaging unique peptide ratios for that protein. Quantitative precision was expressed as protein ratio variability. Proteins with fold-change > 2 or < 0.5 and p values < 0.05 by t test statistic were considered significantly changed.
MS proteomic data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRoteomics IDEntifications (PRIDE) partner repository with the data set identifiers, PXD010284, PXD010286, PXD009916, and PXD009917.
Detection of Cellular ROS
ROS levels in CD38(−) and CD38(+) A549 cells were detected using CellROX® Deep Red Reagents (Invitrogen, Grand Island, NY), according to the manufacturer's protocol. Briefly, cells were seeded into 6-well cell culture plates. CellROX® Deep Red Reagents were added to a final concentration of 5 μm and incubated at 37 °C for 30 min. Fluorescence was measured with a BD FACSAria II Flow Cytometer (BD Biosciences, San Jose, CA).
Detection of Cellular Glutathione (GSH) Levels
Cells were washed twice with ice-cold PBS and extracted with 80% precooled methanol. Samples were dried by speedvac and redissolved in 80% methanol for LC-MS/MS analysis. Quantitative analysis of metabolites extracted from CD38(−) and CD38(+) A549 cells was performed using the Q-Exactive Mass Spectrometer. Atlantis HILIC silica columns (2.1 × 100 mm, 3 μm; Waters, Milford, MA) were used for positive separation. Metabolites were identified based on retention time on LC analysis. Accurate mass was measured with < 5 ppm mass accuracy. TraceFinder was used to identify peaks and extract quantitative information.
Susceptibility of CD38(−) and CD38(+) A549 Cells to Hydrogen Peroxide, and Cisplatin
CD38(−) and CD38(+) A549 cells were seeded into 96-well plates with 4000 cells/well. After 36 h incubation, cells were treated with hydrogen peroxide (Aladdin, Shanghai, China) and cisplatin (Selleck, Houston, TX) in triplicate for 24 h. Cell counting kit-8 (CCK-8) reagent was added to treated cells and incubated at 37 °C for 2 h. Optical density (OD) was measured at 450 nm with a microplate reader (Bio-Rad). Cell viability was represented as the percentage of viable cells compared with untreated cells.
Phosphoproteomic Analysis
CD38(−) A549 cells were cultured in SILAC culture media and CD38(+) A549 cells were cultured in normal RMPI-1640 media. Cells were harvested and lysed for 30 min on ice with RIPA lysis buffer (Solarbio, Beijing, China) supplemented with 1% Protease Inhibitor Mixture and phosphatase inhibitor (Thermo Fisher Scientific). Supernatants were collected after centrifugation and four-volumes of ice-cold acetone added, followed by overnight precipitation at −20 °C. The supernatant was discarded after 7600 rpm centrifugation at 4 °C for 10 min, and pellets dissolved in 8 m urea (in 10 mm HEPES, pH8.0). Protein concentration was determined using a BCA protein assay kit (Solarbio). Protein (5 mg) was used for the following experiment. In-solution digestion was performed. Protein disulfides were reduced by dithiothreitol at 1 mm for 60 min at room temperature, and free cysteine residues alkylated with iodoacetamide at 5 mm for 60 min in the dark at room temperature. Protein samples were digested with trypsin for 14 h at 37 °C. Peptides were desalted with Sep-Pak C18 cartridges and fractioned by HPLC separation. Collected eluents were combined into 12 fractions and dried by speedvac. Each fraction was dissolved with 10 ml 80% acetonitrile and 5%TFA. TiO2 beads (SHIMADZU, Kyoto, Japan) were pre-incubated in 2,5-Dihydroxybenzoic acid (Aladdin) in 80% acetonitrile and 1%TFA (5 μl/mg of beads) for 20 min. 2 mg of beads was added to every fraction and incubated on rotator for 30 min. Beads were washed twice with 30% acetonitrile and 1%TFA, loaded into C8 STAGE-TIP, and centrifuged at 500g for 5 min. Then, beads were washed on-tip with 50% acetonitrile and 1%TFA followed by 80% acetonitrile and 1%TFA. Peptides were eluted with 20 μl 5% ammonium hydroxide, 20 μl 10% ammonium hydroxide, and 10% ammonium hydroxide with 25% acetonitrile. Reduced the volume by speedvac and analyzed by LC-MS/MS. The MS/MS spectra from LC-MS/MS analysis were processed to generate the peak list and searched against the UniProt human reference proteome (release on March 17, 2017, containing 21,042 entries) using an in-house Sequest HT Algorithm in Proteome Discoverer software (PD, version 1.4; Thermo Fisher Scientific). The search criteria were: full tryptic specificity was required; two missed cleavage sites were allowed; oxidation of methionine, 13C(6)15N(2) at lysine, 13C(6) at arginine, deamidation at asparagine and glutamine, phosphorylation at serine, threonine and tyrosine were set as variable modifications; carbamidomethylation of cysteine was set as the fixed modification; and precursor ion mass tolerance was set at 20 ppm for all MS and 20 mmu for all MS2 spectra. Peptide false discovery rate (FDR) was estimated using the percolator function provided by Proteome Discoverer, with a cutoff score of 1% based on decoy database searching.
Secretomic Analysis
Cells were cultured in medium supplemented with 10% FBS until they reached 70% confluency. Cells were washed with PBS three times, and then incubated in serum and phenol red free medium at 37 °C for 12 h. Medium was collected and centrifuged at 10,000 rpm for 10 min to remove large debris. Supernatants were collected, and four-volumes of ice-cold acetone added, followed by overnight precipitation at −20 °C. The supernatant was discarded after 10,000 rpm centrifugation at 4 °C for 10 min, and pellets dissolved in 8 m urea. Supernatants were collected after centrifugation at 12,000 rpm for 20 min at 4 °C. Protein concentration was determined using a BCA protein assay kit (Solarbio). Equal protein amounts were used for the following experiment. In-solution digestion was performed. Protein disulfides were reduced by dithiothreitol at 5 mm for 60 min at room temperature, and free cysteine residues alkylated with iodoacetamide at 12 mm for 45 min in the dark at room temperature. Protein samples were digested with trypsin for 14 h at 37 °C. Peptides were desalted with Sep-Pak C18 cartridges, labeled using TMT reagents (Thermo Fisher Scientific) according to the manufacturer's instructions, and fractioned by HPLC separation. Collected eluents were combined into 12 fractions and analyzed by LC-MS/MS. The MS/MS spectra from LC-MS/MS analysis were processed to generate the peak list and searched against the UniProt human reference proteome (release on March 17, 2017, containing 21,042 entries) using an in-house Sequest HT Algorithm in Proteome Discoverer software (PD, version 1.4; Thermo Fisher Scientific). The search criteria were: full tryptic specificity was required; two missed cleavage sites were allowed; oxidation of methionine, deamidation at asparagine and glutamine were set as variable modifications; carbamidomethylation of cysteine, TMT6plex at lysine, and TMT6plex at protein N terminus were set as the fixed modifications; and precursor ion mass tolerance was set at 20 ppm for all MS and 20 mmu for all MS2 spectra. Peptide false discovery rate (FDR) was estimated using the percolator function provided by Proteome Discoverer, with a cutoff score of 1% based on decoy database searching.
Western Blot Analysis
Cells were harvested and lysed for 30 min on ice with RIPA lysis buffer (Solarbio) supplemented with 1% Protease Inhibitor Mixture (Thermo Fisher Scientific). Supernatants were collected after centrifugation at 12,000 rpm for 20 min at 4 °C. Protein concentration was determined using a BCA protein assay kit (Solarbio). Proteins were separated on 12% SDS-PAGE gels and transferred onto PVDF transfer membranes. Western blot analysis followed a standard procedure. Actin antibody was purchased from Abmart (Shanghai, China) and used as an internal control. FLAG antibody was purchased from Sigma. Antibodies against β-catenin, E-cadherin, N-cadherin, Snail1, pStat3 Y705 and S727, and Ac-Stat3 K685 were purchased from Cell Signaling Technology (Danvers, MA). Cytokeratin 18 antibody was purchased from Millipore (Boston, MA). ZO-1, vimentin, and Stat3 antibodies were obtained from Proteintech (Chicago, IL).
Quantitative Real-time PCR (qPCR)
Total RNA was extracted from cells using the RNAprep Pure Cell Kit (Tiangen, Beijing, China). cDNA was synthesized from 3 μg total RNA using the TIANScript RT kit (Tiangen). Quantitative real-time PCR was performed using the Roche LightCycler® 480II Detection System with SYBR green (Tiangen), according to the manufacturer's instructions, and β-actin used as an internal control. Primers were acquired from Primer Bank (http://pga.mgh.harvard.edu/primerbank/). The primers are listed in supplemental Table S1.
Invasion Assay
Invasion assays were performed using 8-μm-pore transwell chambers (Corning, New York, NY). Briefly, cells were starved for 24 h before harvest. Cells were trypsinized, washed twice with PBS, and resuspended in serum-free medium. Next, 1.3 × 105 cells were counted and seeded into transwell chambers, with serum-containing media added to the lower chamber. Cells were incubated at 37 °C with 5% CO2. Cells that invaded through Matrigel (Corning)-coated membrane were trypsinized, and cell counting kit-8 (CCK-8) reagent added to treated cells and incubated at 37 °C for 3 h. Optical density (OD) was measured at 450 nm with a microplate reader.
Xenograft Experiments
Animal studies were approved by the Animal Research Ethics Committee of the Tsinghua University. For xenograft experiments, 5 × 106 CD38(−) and CD38(+) A549 cells were trypsinized, washed three times with PBS, and resuspended in 200 μl PBS before injecting subcutaneously into 5-week-old female nude mice (Vital River Company, Beijing, China). Tumor volumes were measured at 37 days after injection.
Statistical Methods
GraphPad Prism 5.0 software (La Jolla, CA) was used for statistical analysis. Significant differences were determined by Student's t test. p values < 0.05 were considered significant.
RESULTS
Decreased NAD Levels by CD38 Overexpression or FK866 Treatment Reduces Cell Proliferation Rates
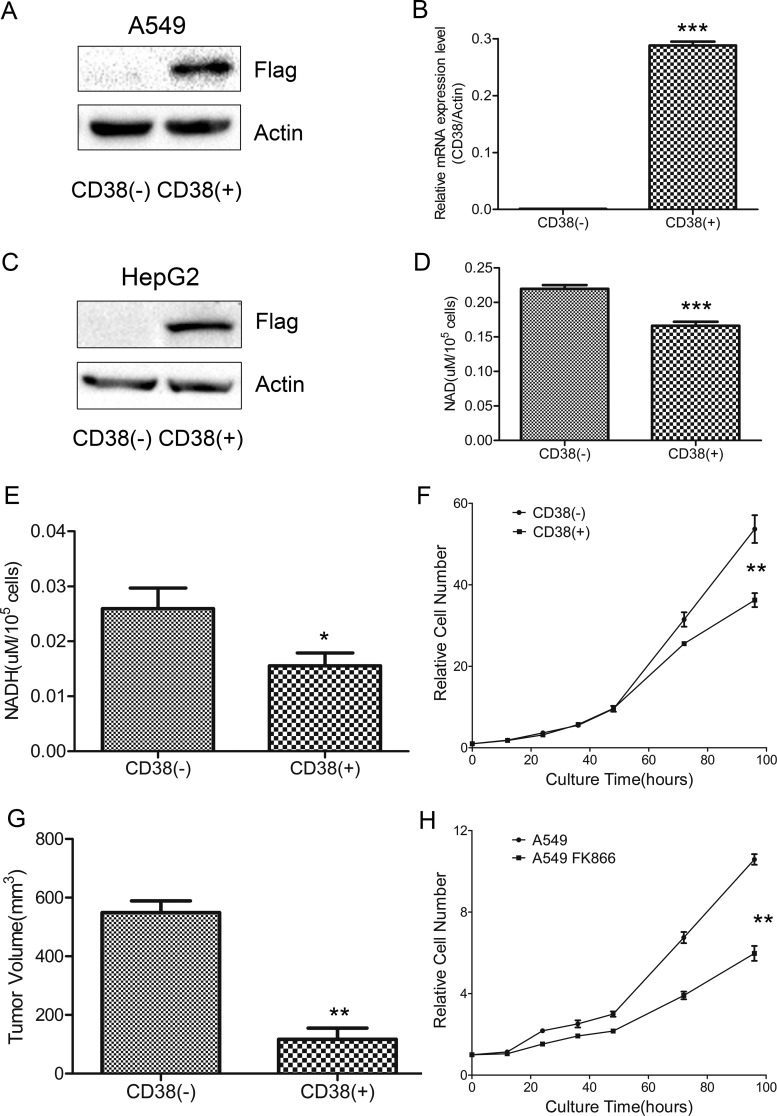
CD38 is an ectoenzyme that anchors to the cell membrane. To express intracellular CD38, the N-terminal helical domain was deleted, and a FLAG-tagged sequence added to the C terminus to generate recombinant human CD38 DNA (supplemental Fig. S1A). Lentiviral particles containing CD38 vector were transfected into A549 and HepG2 cells to produce stable CD38-overexpressing (CD38(+)) cell lines. CD38 overexpression was examined by Western blotting and qPCR (Fig. 1A–1C, supplemental Fig. S2A). CD38 overexpression decreased cellular NAD levels, as determined by NAD assay, with a 25% decrease in NAD levels achieved in A549 CD38(+) cells (Fig. 1D). Similarly, NADH levels were 40% lower in CD38(+) cells than control cells (A549 CD38(−)) (Fig. 1E). In HepG2 CD38(+) cells, the NAD and NADH levels were also decreased compared with HepG2 CD38(−) cells (supplemental Figs. S2B, S2C). The NAD/NADH ratio was nearly unchanged between CD38(+) and CD38(−) A549 cells (supplemental Fig. S2D).
Fig. 1.
Lowering cellular NAD levels decreases cell growth rate. A, Western blotting confirmed CD38 overexpression in A549 cells. B, qPCR analysis confirmed overexpressed CD38 mRNA levels in A549 cells (n = 3). C, Western blotting confirmed CD38 overexpression in HepG2 cells. D, E, Cellular NAD and NADH levels in CD38(−) and CD38(+) A549 cells (n = 3). F, Growth curves of CD38(−) and CD38(+) A549 cells determined by CCK-8 assay (n = 3). G, Tumor samples harvested from mice 37 days after injection (n = 3). Tumor volumes (mm3) were measured using digital calipers and calculated using the formula: π/6 × length (mm) × width2 (mm). H, Growth curves of untreated- and 10 nm FK866-treated A549 cells determined by CCK-8 assay (n = 3). Data were analyzed using Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001. *p < 0.05 was considered statistically significant. All values represent mean from at least three biological replicates ± S.E.
Proliferation rates for CD38(−) and CD38(+) cells were determined by CCK-8 assay (Fig. 1F) and by cell counting assay (supplemental Fig. S2E), showing that CD38(+) cells grew more slowly than CD38(−) cells. At 96 h, the number of CD38(+) cells was ∼40% less than CD38(−) cells. Subcutaneous injection of CD38(+) cells into immunocompromised mice gave rise to exponentially growing tumors, and control cells grew to higher volumes compared with CD38(+) cells (Fig. 1G). Xenograft experiment demonstrated that CD38 overexpression significantly inhibited tumor growth of A549 cells in nude mice, providing further evidence that CD38 overexpression inhibits cell proliferation. Similarly, FK866 treatment decreased cell growth (Fig. 1H).
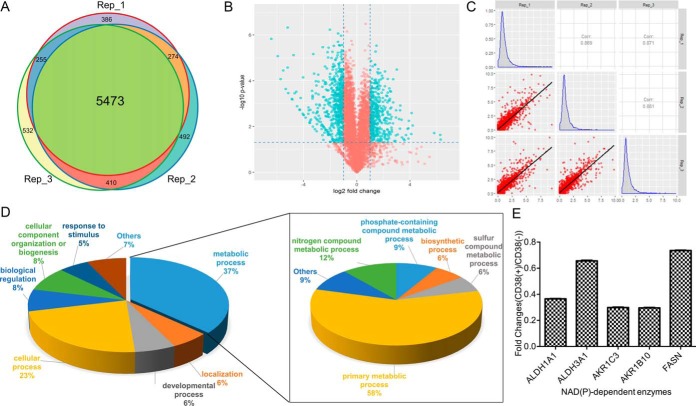
Next, proteomic analysis was performed in CD38(+) and CD38(−) cells. Equal amounts of CD38(+) and CD38(−) cell protein were digested in solution and then fractionated by HPLC into twelve fractions. Each fraction was analyzed by nano-LC-MS/MS to identify differentially expressed proteins. Quantitation values were determined based on SILAC ratios. Experiments were performed in biological triplicates, with ∼6500 proteins identified in each experiment (supplemental Table S2). The false-positive rate was estimated to be < 1%. Based on SILAC ratios (> 2 or < 0.5), 973 proteins were differentially expressed between CD38(+) and CD38(−) A549 cells, with 562 proteins downregulated and 411 upregulated (supplemental Tables S3 and S4). Venn diagram showed the number of overlapping proteins identified in proteomic studies from three biological replicates (Fig. 2A), demonstrating that our proteomic data are highly reproducible. Volcano plots revealed significantly changed proteins between CD38(+) and CD38(−) cells. Blue dots in upper left and upper right sections represent proteins with fold-change < 0.5 or > 2 and p values < 0.05 from t test statistics (Fig. 2B). Histograms revealed fold-changes of proteomic data, which showed normal Gaussian distributions (Fig. 2C). Pearson correlation was used to determine protein quantification reproducibility. High Pearson correlation coefficient indicates high correlation in fold-change between three independent experiments (Fig. 2C). To understand the biological relevance of differentially expressed proteins, Gene Ontology (GO) was used to cluster molecular functions and biological processes. Overall, 562 downregulated proteins were classified into several significant groups of biological processes according to their molecular functions, including energy and carbohydrate metabolism, RNA processing and protein synthesis, DNA repair, chaperone, and cell cycle regulation.
Fig. 2.
Proteomic analysis of CD38(−) and CD38(+) cells. A, Venn diagram showed that proteomic data were highly reproducible. B, Volcano plots showed significantly changed proteins (labeled in blue) between CD38(+) and CD38(−) A549 cells with fold-change > 2 or < 0.5 and adjusted p value < 0.05 from t test statistics. C, Pearson correlation diagram embedded with a histogram revealed that protein fold-changes in our proteomic data showed normal distributions. High Pearson correlation coefficient indicates high correlation of protein fold-changes between three independent experiments. D, Functional classification of downregulated proteins between CD38(−) and CD38(+) A549 cells by PANTHER (http://www.pantherdb.org). E, SILAC ratios for NAD(P)-dependent enzymes.
Gene list annotations were summarized in a pie chart using the PANTHER bioinformatics platform (http://www.pantherdb.org/). More than one-third of downregulated proteins were associated with cellular metabolism (Fig. 2D), in which half the proteins participated in primary metabolic processes including carbohydrate metabolic process, cellular amino acid metabolic process, lipid metabolic process, nucleobase-containing compound metabolic process, and protein metabolic process. Among abundant proteins with a high number of peptide spectrum matches (> 300), we noticed that the NAD(P)-dependent enzymes, retinal dehydrogenase (ALDH1A1), aldehyde dehydrogenase-dimeric NADP-preferring (ALDH3A1), aldo-keto reductase family 1 member C3 and B10 (AKR1C3/B10), and fatty acid synthase (FASN) were autonomously downregulated (Fig. 2E) and participated in co-factor metabolism. These results suggest that lowering NAD levels decreased primary metabolism as well as cofactor and lipid synthesis, with an overall resultant effect of decreased proliferation rates in CD38(+) cells.
CD38 Overexpression Increases Cellular ROS Levels and Susceptibility to Oxidative Stress
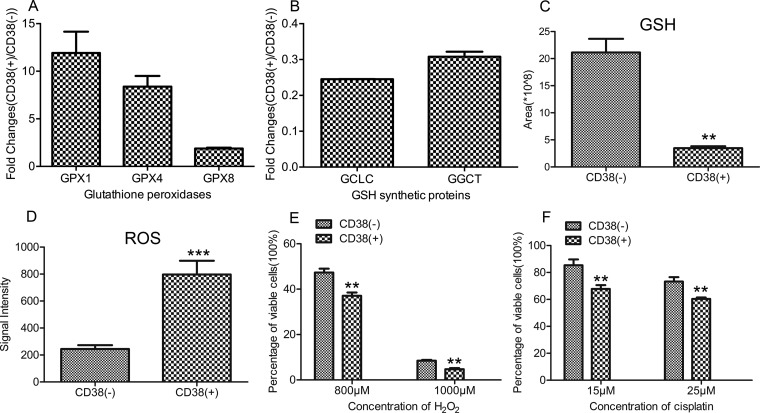
Among upregulated proteins, GSH peroxidase expression (GPX1, 4, 8) displayed the high fold-changes in CD38(+) A549 cells (Fig. 3A), which was derived from the MS spectra (supplemental Fig. S3A). GPXs reduce hydrogen peroxide (H2O2) or lipid hydroperoxide to water and alcohol while generating oxidized GSH (GSSG). However, our proteomic analysis did not show a significant change in GSH reductase (GSR) expression between CD38(+) and CD38(−) cells. Furthermore, two enzymes involved in GSH synthesis, glutamate-cysteine ligase catalytic subunit (GCLC) and gamma-glutamylcyclotransferase (GGCT), were downregulated in CD38(+) cells (Fig. 3B), which was derived from the MS spectra (supplemental Fig. 3B). These results suggest that lowered NAD decreases cellular GSH concentration. This was confirmed by metabolomic analysis showing a 5-fold decrease in GSH levels in CD38(+) cells (Fig. 3C). Cellular ROS levels were measured using a CellROX® Deep Red kit, which showed that CD38(+) cells exhibited much stronger fluorescence with a 4-fold increase in ROS levels (Fig. 3D). Further, we determined the GSH and ROS levels in untreated- and NAC-treated A549 CD38(+) cells and found that the GSH levels were unchanged in the NAC-treated A549 CD38(+) cells but the ROS levels were significantly decreased in NAC-treated A549 CD38(+) cells, suggesting that CD38 overexpression caused a defect in GSH synthesis (supplemental Figs. S3C, S3D).
Fig. 3.
CD38 overexpression increases ROS levels and susceptibility to oxidative stress. A, SILAC ratios for three GSH peroxidases. B, SILAC ratios for GSH synthetic proteins. C, Graphical representation of relative concentrations of GSH in CD38(−) and CD38(+) A549 cells by metabolomic analysis (n = 3). D, Graphical representation of cellular ROS levels in CD38(−) and CD38(+) A549 cells (n = 3). E, Percentage of viable cells between CD38(−) and CD38(+) A549 cells treated with different concentration of H2O2 (n = 3). F, Percentage of viable cells between CD38(−) and CD38(+) A549 cells treated with different concentration of cisplatin (n = 3). Data were analyzed using Student's t test. **p < 0.01. *p < 0.05 was considered statistically significant. All values represent mean from at least three biological replicates ± S.E.
Downregulation of antioxidant proteins (peroxiredoxin [PRDX]-1 and PRDX6) and decreased GSH suggests that CD38 overexpression increases susceptibility to oxidative stress. CD38(−) and CD38(+) A549 cells were treated with different concentrations of H2O2 or cisplatin for 24 h to determine susceptibility to oxidative stress. Dose-dependent effects of H2O2 or cisplatin are represented as percentage of viable cells after 24 h treatment, as measured by CCK-8 assay. The percentage of viable cells was 37 and 47% for CD38(+) and CD38(−) cells, respectively, after treatment with 800 μm H2O2 for 24 h. The number decreased to 5% when CD38(+) cells were treated with 1000 μm H2O2 for 24 h (Fig. 3E), demonstrating that CD38(+) cells are extremely sensitive to H2O2 treatment. The percentage of viable cells was 60 and 70% for CD38(+) and CD38(−) cells, respectively, after cells were treated with 25 μm cisplatin for 24 h (Fig. 3F).
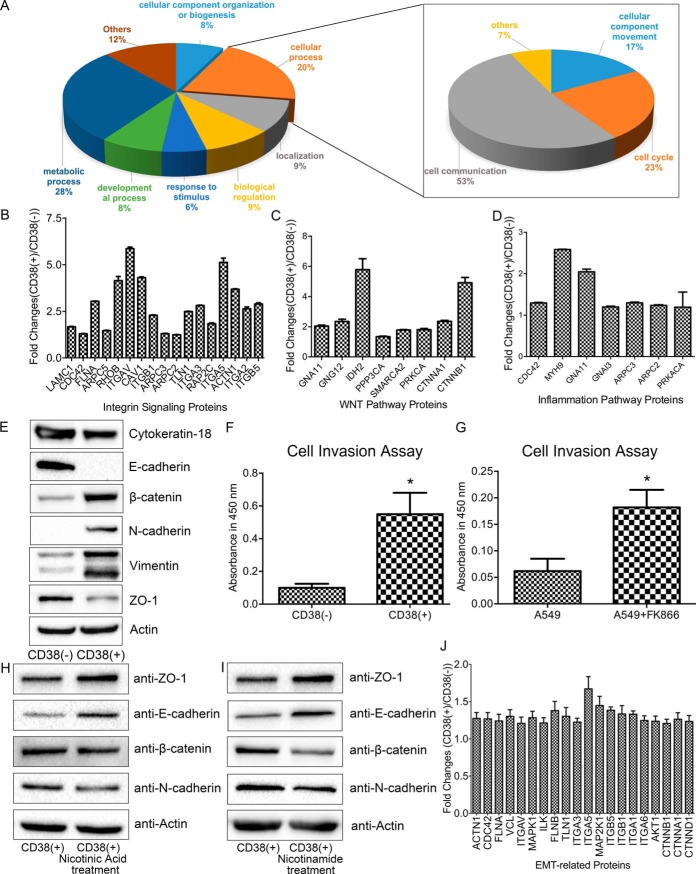
Decreased NAD Activates Multiple EMT Associated Pathways
Upregulated proteins were summarized by a pie chart using the PANTHER bioinformatics platform (Fig. 4A). Accordingly, 10% of proteins were associated with cell communication including integrin signaling, the Wnt pathway, and inflammatory process (Figs. 4B–4D). Ingenuity Pathway Analysis (IPA) identified significantly changed canonical pathways associated with differentially expressed proteins between CD38(−) and CD38(+) A549 cells such as integrin signaling, PI3K/AKT signaling, ERK/MAPK signaling, and ephrin receptor signaling, which suggests that CD38 overexpression causes cells to undergo EMT (supplemental Fig. S4A). Indeed, CD38(+) A549 cells exhibited significant morphological changes compared with CD38(−) cells, showing transformation of epithelial cells to elongated and spindle-like cells of a similar shape as EMT intermediates (supplemental Figs. S5A, S5B). Western blotting with EMT markers confirmed that CD38 overexpression mediated the EMT phenotype (Fig. 4E). Overall, CD38(+) cells exhibit significantly higher expression of the mesenchymal markers, vimentin, N-cadherin, and β-catenin, and lower expression of the epithelial markers, ZO-1, E-cadherin, and cytokeratin 18. Using a cell invasion assay, CD38(+) cells were found to display higher migration rates (Fig. 4F). Thus, altogether we show that CD38 overexpressing cells undergo EMT in A549 cells.
Fig. 4.
Lowering cellular NAD levels activates multiple EMT-associated pathways and causes cells to undergo EMT. A, Functional classification of upregulated proteins between CD38(−) and CD38(+) A549 cells by PANTHER (http://www.pantherdb.org). B, SILAC ratios of upregulated proteins related to the integrin signaling pathway. C, SILAC ratios of upregulated proteins related to the Wnt signaling pathway. D, SILAC ratios of upregulated proteins related to the inflammation signaling pathway. E, Western blotting of expression levels of β-catenin, vimentin, cytokeratin-18, E-cadherin, N-cadherin, ZO-1, and β-actin in CD38(−) and CD38(+) A549 cells. F, Migration rates of CD38(−) and CD38(+) A549 cells determined by cell invasion assay (n = 4). G, Migration rates of untreated-, and 10 nm FK866-treated cells determined by cell invasion assay (n = 3). H, I, Western blotting of expression levels of β-catenin, E-cadherin, N-cadherin, ZO-1, and β-actin in untreated- and nicotinic acid (500 μm)- or nicotinamide (1 mm)-treated CD38(+) cells. J, Ratios of upregulated proteins related to the EMT process in CD38(−) and CD38(+) HepG2 cells. *p < 0.05 was considered statistically significant. All values represent mean from at least three biological replicates ± S.E.
To confirm that lower NAD levels in CD38(+) cells are the driving force for cells to undergo EMT, we treated A549 cells with 10 nm FK866 for 72 h. This resulted in increased invasiveness, like CD38 overexpression (Fig. 4G). Proteomic analysis was performed to identify differentially expressed proteins between FK866-treated and untreated A549 cells. We revealed that differentially expressed proteins associated with EMT were like those found in CD38 overexpressing cells (supplemental Figs. S6A). Repletion of nicotinic acid or nicotinamide reversed EMT, leading to increased ZO-1 and E-cadherin expression but lowered β-catenin and N-cadherin expression. These results suggest that decreased NAD-induced EMT is a reversible process (Fig. 4H, 4I). Proteomic analysis also confirmed that EMT associated pathways were upregulated in CD38 overexpressing HepG2 cells (Fig. 4J), indicating that the NAD-mediated EMT process is a universal event.
Secretome and Phosphoproteomic Analyses Support Activation of Multiple EMT-associated Pathways in CD38(+) Cells
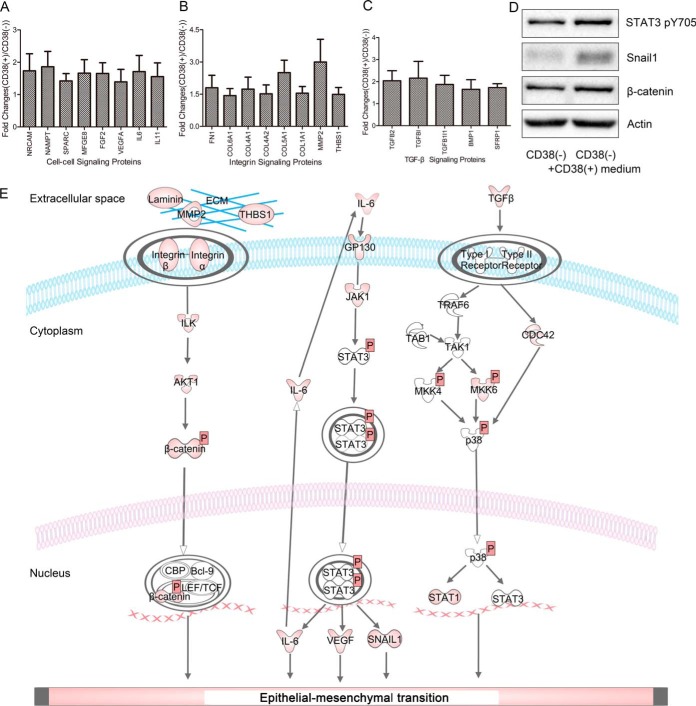
Secretome analysis confirmed that EMT-associated pathways were activated in CD38(+) A549 cells (supplemental Table S5). Here, proteins associated with interleukin (IL)-6-, integrin-, and transforming growth factor beta (TGFβ)-signaling pathways were highly secreted compared with CD38(−) A549 cells (Fig. 5A–5C). Among known extracellular proteins, both TGFβ1 and TGFβ2 were 2-fold higher in CD38(+) cells compared with CD38(−) cells, suggesting that CD38 overexpression activates the TGFβ pathway in A549 cells. Proteomic analysis revealed that TGFβ-induced protein ig-h3 (TGFBI) and TGFβ-1-induced transcript 1 protein (TGFB1I1) were increased 4-times in CD38(+) cells. Further, phosphoproteomic analysis showed that TGFβ1-induced transcript 1 protein (TGFB1I1) was increased in CD38(+) cells (supplemental Table S6). It is known that TGFβ activates the mothers against decapentaplegic homolog (Smad) signaling pathway, but both expression and phosphorylation levels of Smad proteins were not altered in CD38(+) cells, suggesting that a TGFβ/Smad-independent signaling pathway is activated. This was confirmed by increased phosphorylation of mitogen-activated protein kinase kinase (MKK)-4, MKK6, and mitogen-activated protein kinase 14 (p38).
Fig. 5.
Secretomic analysis reveals that CD38(+) cells have an EMT-phenotype. A–C, Ratios of secreted proteins associated with the EMT process. D, Western blotting of expression levels of β-catenin, Snail1, Stat3 pY705, and β-actin in CD38(−) and CD38(+) cell medium-treated CD38(−) cells. E, Schematic diagram showing activated pathways in CD38(+) A549 cells.
Secretome analysis also revealed that CD38(+) A549 cells release higher levels of laminin, matrix metallopeptidase 2 (MMP2), and thrombospondin 1 (THBS1), which are ligands for integrins. Combined with proteomic results showing that integrin α-2, -3, -5, -V, integrin β-1, -5, and β-catenin were all upregulated in CD38(+) cells, we concluded that CD38 overexpression also activates the integrin pathway. Secretome analysis further detected higher IL6 in the medium of CD38(+) cells, whereas phosphoproteomic analysis showed that STAT3 S727 phosphorylation was increased (supplemental Fig. S7A). Combined with proteomic results showing upregulated JAK1 in CD38(+) cells, we concluded that CD38 overexpression also activates inflammatory pathways. To determine whether secreted proteins from CD38(+) A549 cells promote the EMT process, we collected cell culture medium and used it for culturing CD38(−) A549 cells. After two-days of culture, cells were collected and probed by Western blot analysis. We found upregulated Snail1 and β-catenin and activated STAT3 (Fig. 5D), demonstrating that secreted proteins from CD38(+) cells cause cells to undergo the EMT process. Pathways activated by CD38 overexpression are shown in Fig. 5E.
Inhibition of STAT3 Reverses EMT in CD38(+) Cells
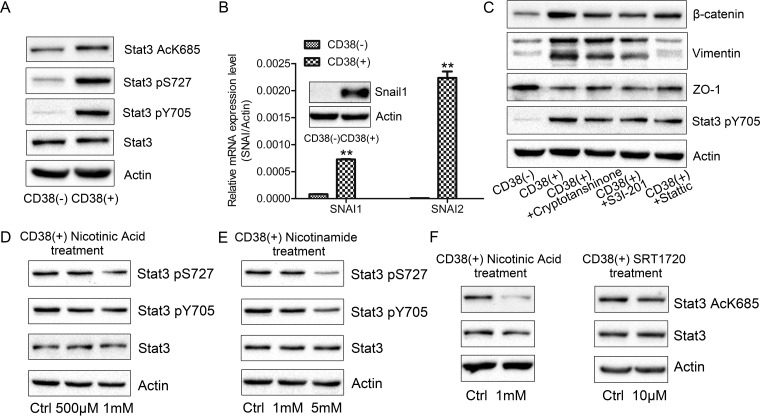
Secretome and proteomic analyses suggested that CD38 overexpression activates the IL6-JAK1-STAT pathway. To further confirm STAT3 pathway activation, we performed phosphoproteomic analysis. Phosphorylation of the S727 residue of STAT3 was increased in CD38(+) A549 cells, which was further validated by Western blotting. The increase in Y705 phosphorylation of STAT3 was also validated by Western blotting. Phosphorylation of S727 promotes STAT3 transcriptional activity (38, 39), whereas phosphorylation of Y705 induces STAT3 dimerization, nuclear translocation, and subsequent DNA binding (40, 41). Further, K685 acetylation is important for STAT3 dimerization, which leads to subsequent nuclear translocation and transcriptional regulation (42, 43). In the present study, we found increased acetylation of the STAT3 K685 residue (Fig. 6A). STAT3 is a vital downstream target of TGFβ-, integrin-, and inflammatory pathways, which induce Snail expression (44, 45). Indeed, SNAI1 and SNAI2 mRNA levels were increased in CD38(+) A549 cells and increased Snail1 protein expression levels were validated by Western blotting (Fig. 6B). Inhibition of STAT3 with cryptotanshinone, S3I-201, and stattic in CD38(+) A549 cells reversed EMT, resulting in increased ZO-1 and decreased vimentin and β-catenin expression (Fig. 6C). More strikingly, nicotinic acid or nicotinamide supplementation of cell culture medium resulted in decreased phosphorylation of S727 and Y705 in STAT3, suggesting that intracellular NAD levels mediate the STAT3 activation state (Fig. 6D, 6E). Moreover, supplementation with nicotinic acid and SRT1720, a SIRT1 activator, decreased STAT3 K685 acetylation (Fig. 6F), indicating that SIRT1 plays a role in NAD-mediated STAT3 activation.
Fig. 6.
STAT3 is a vital regulator in the NAD-mediated EMT process. A, Western blotting of expression levels of Stat3 pS727, Stat3 pY705, Stat3 AcK685, Stat3, and β-actin in CD38(−) and CD38(+) A549 cells. B, mRNA and protein expression levels of SNAI1 and SNAI2 in CD38(−) and CD38(+) A549 cells (n = 3). C, Western blotting of β-catenin, vimentin, ZO-1, Stat3 pY705, and β-actin expression levels in untreated- and cryptotanshinone (15 μm, 48 h)-, S3I-201 (150 μm, 48 h)-, and stattic (1.5 μm, 48 h)-treated CD38(+) cells. D, E, Western blotting of Stat3 pS727, Stat3 pY705, Stat3, and β-actin expression levels in untreated- and nicotinic acid (500 μm, 48 h)- or nicotinamide (1 mm, 48 h)-treated CD38(+) cells. F, Western blotting of Stat3 AcK685, Stat3, and β-actin expression levels in untreated- and nicotinic acid (500 μm, 48 h)- or SRT1720 (10 μm, 8h)-treated CD38(+) cells. **p < 0.01. *p < 0.05 was considered statistically significant. All values represent mean from at least three biological replicates ± S.E.
DISCUSSION
Cellular NAD levels decline with age, whereas supplementation with NAD precursors exert a protective role to slow down aging and prevent metabolic and neurodegenerative disease. NAD homeostasis is maintained by both NAD synthetic and consumption pathways. NAD is largely consumed via nonredox NAD-dependent enzymes such as PARPs, ADP-ribose transferases, NADase, and sirtuins, which use NAD and generate nicotinamide, whereas NAMPT is the rate-limiting enzyme to convert nicotinamide back to NAD. Previous studies have established that CD38 can be used for tuning cellular NAD levels and studying NAD function without interfering with major cellular processes (31, 32). As demonstrated, increased NAD in Cd38−/− mice improves mitochondrial function and protects mice against metabolic damage induced by a high-fat diet (24, 34, 35).
As we showed in a previous study, CD38 overexpression in 293T cells downregulates all glycolytic enzymes and slows cell proliferation (37). Thus, we examined the effect of CD38 overexpression on cellular processes in A549 and HepG2 cells. CD38 overexpression in A549 and HepG2 cells was confirmed by Western blotting and qPCR. Consequently, NAD and NADH level were decreased by 25 and 40%, respectively, in CD38(+) A549 cells (Fig. 1D, 1E). A SILAC-based quantitative proteomic study was performed to identify differentially expressed proteins in CD38(+) and CD38(−) cells. Unlike the results observed in 293T cells, in which CD38 overexpression downregulates glycolytic enzymes and decreases cell proliferation, CD38 overexpression did not alter expression of glycolytic enzymes in A549 cells. Instead, proteins that participated in co-factor metabolism were downregulated in CD38(+) cells, suggesting that lowered cellular NAD levels decrease primary metabolism, and lead to a decreased proliferation rate in CD38(+) cells and xenograft experiments. This also suggests that decreased NAD promotes cell-specific responses. Moreover, GPXs were upregulated whereas proteins involved in GSH biosynthesis were downregulated, resulting in decreased GSH and elevated ROS levels in CD38(+) cells. Consequently, downregulation of antioxidant proteins, PRDX1 and PRDX6, and decreased GSH increased susceptibility to oxidative stress in CD38(+) cells.
More strikingly, a combination of proteomic, secretomic, and phosphoproteomic analyses identified activation of multiple pathways related to EMT in CD38(+) cells. Accordingly, cells were transformed from a cobblestone-shaped epithelial population to an elongated and spindle-like morphology, like EMT intermediates. The EMT phenotype was further confirmed by Western blotting and cell invasion assays (Fig. 4E, 4F). Vimentin was 2-fold downregulated in CD38(+) 293T cells compared with CD38(−) cells, whereas its expression was upregulated in CD38(+) A549 cells. As a major intermediate filament protein, vimentin is ubiquitously and highly expressed in epithelial cancer cells, and regulates accelerated tumor growth, invasion, poor prognosis, and drug resistance (46–48). Decreased NAD-mediated EMT was observed in multiple cell lines, including A549 and HepG2. As EMT promotes tumor cell metastasis, our results suggest that decreased NAD contributes to tumor progression in addition to its roles in maintaining mitochondrial integrity. Secretion of IL6, integrin, and TGFβ can induce EMT in nearby cells, indicating that decreased NAD also mediates the complex interactions between cells and their adjacent microenvironment. More importantly, treatment with NAD precursors reverses EMT in CD38(+) cells. This further confirms that nicotinamide and nicotinic acid inhibit cancer metastasis in xenograft models (49).
STAT3 is a downstream target of TGFβ-, integrin-, and inflammatory pathways, which induce Snail expression (38, 39). STAT3 inhibition reversed EMT by downregulating vimentin and β-catenin, and upregulating ZO-1 expression (Fig. 6C). Treating CD38(+) cells with NAD precursors decreased phosphorylation of the S727 and Y705 residues in STAT3, indicating that increased NAD inactivates transcriptional activity of STAT3 (40–43). SIRT1, a NAD-dependent deacetylase, inactivates STAT3 (50). Here, we demonstrate that decreased NAD decreases SIRT1 activity and increases STAT3 acetylation. Supplementation with nicotinic acid or treatment with SRT1720, a SIRT1 activator, decreased acetylation of STAT3 K685, which inactivated STAT3. Taken together, we demonstrate that STAT3 plays a vital role in NAD-mediated EMT.
In summary, we demonstrate that decreased NAD activates multiple EMT-associated pathways to enhance cell invasiveness, in which STAT3 is a vital regulator. Moreover, supplementation with NAD precursors reverses EMT by inactivating STAT3 activity and providing a potential strategy to halt cancer cell progression.
DATA AVAILABLITY
The MS proteomic data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRoteomics IDEntifications (PRIDE) partner repository with the data set identifiers, PXD010284, PXD010286, PXD009916, and PXD009917.
Supplementary Material
Acknowledgments
We thank the Protein Chemistry Facility at the Center for Biomedical Analysis of Tsinghua University for sample analysis. We thank Rachel James from Edanz Group China for editing the English text of this manuscript.
Footnotes
* This work was supported by the National Key Research and Development Program of China (Grant 2017YFA0505103), NSFC 31270871 (H.D.), the Chinese Ministry of Science and Technology (2014CBA02005) and the Science and Technology Pillar Program of Sichuan (2016JZ0015).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- NAD
- nicotinamide adenine dinucleotide
- CD38
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1
- SILAC
- stable isotope labeling with amino acids in cell culture
- PRDX1
- peroxiredoxin-1
- PRDX6
- peroxiredoxin-6
- JAK1
- janus kinase1
- STAT3
- signal transducer and activator of transcription 3
- TGFβ1
- transforming growth factor beta-1
- TGFβ2
- transforming growth factor beta-2
- Smad
- mothers against decapentaplegic homolog
- MKK6
- dual specificity mitogen-activated protein kinase kinase 6
- MKK4
- dual specificity mitogen-activated protein kinase kinase 4
- p38
- mitogen-activated protein kinase 14
- IL6
- interleukin-6
- NAMPT
- nicotinamide phosphoribosyltransferase.
REFERENCES
- 1. Verdin E. (2015) NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 [DOI] [PubMed] [Google Scholar]
- 2. Chiarugi A., Dolle C., Felici R., and Ziegler M. (2012) The NAD metabolome–a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 [DOI] [PubMed] [Google Scholar]
- 3. Yang Y., and Sauve A. A. (2016) NAD+ metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 1864, 1787–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Samal B., Sun Y., Stearns G., Xie C., Suggs S., and McNiece I. (1994) Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell. Biol. 14, 1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garten A., Petzold S., Korner A., Imai S., and Kiess W. (2009) Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol. Metab. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imai S., Armstrong C. M., Kaeberlein M., and Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 7. Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., and Schramm V. L. (2001) Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40, 15456–15463 [DOI] [PubMed] [Google Scholar]
- 8. Frye R. A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 9. Borra M. T., Langer M. R., Slama J. T., and Denu J. M. (2004) Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 10. Imai S., and Guarente L. (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi M., and Guarente L. (2016) mTORC1 and SIRT1 Cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166, 436–450 [DOI] [PubMed] [Google Scholar]
- 12. Mouchiroud L., Houtkooper R. H., Moullan N., Katsyuba E., Ryu D., Canto C., Mottis A., Jo Y. S., Viswanathan M., Schoonjans K., Guarente L., and Auwerx J. (2013) The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoon M. J., Yoshida M., Johnson S., Takikawa A., Usui I., Tobe K., Nakagawa T., Yoshino J., and Imai S. (2015) SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 21, 706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gomes A. P., Price N. L., Ling A. J., Moslehi J. J., Montgomery M. K., Rajman L., White J. P., Teodoro J. S., Wrann C. D., Hubbard B. P., Mercken E. M., Palmeira C. M., de Cabo R., Rolo A. P., Turner N., Bell E. L., and Sinclair D. A. (2013) Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155, 1624–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin J., He B., Zhang X., Lin H., and Wang Y. (2016) SIRT2 Reverses 4-oxononanoyl lysine modification on histones. J. Am. Chem. Soc. 138, 12304–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown K. D., Maqsood S., Huang J. Y., Pan Y., Harkcom W., Li W., Sauve A., Verdin E., and Jaffrey S. R. (2014) Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 20, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D. T., and Chen D. (2013) SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang W., Nagasawa K., Munch C., Xu Y., Satterstrom K., Jeong S., Hayes S. D., Jedrychowski M. P., Vyas F. S., Zaganjor E., Guarani V., Ringel A. E., Gygi S. P., Harper J. W., and Haigis M. C. (2016) Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell 167, 985–1000 e1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhardwaj A., and Das S. (2016) SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc. Natl. Acad. Sci. U.S.A. 113, E538–E547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W., Li Y., Yang J., Hao Y., Yuan T., Yuan G., Wang H., Ju Z., Mao Z., Li J., Qu J., Tang F., and Liu G. H. (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 26, 190–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo X., and Kraus W. L. (2012) On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson B. A., and Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 23. Braidy N., Guillemin G. J., Mansour H., Chan-Ling T., Poljak A., and Grant R. (2011) Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS ONE 6, e19194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Yoshino J., Mills K. F., Yoon M. J., and Imai S. (2011) Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonkowski M. S., and Sinclair D. A. (2016) Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mills K. F., Yoshida S., Stein L. R., Grozio A., Kubota S., Sasaki Y., Redpath P., Migaud M. E., Apte R. S., Uchida K., Yoshino J., and Imai S. I. (2016) Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metab. 24, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P., D'Amico D., Ropelle E. R., Lutolf M. P., Aebersold R., Schoonjans K., Menzies K. J., and Auwerx J. (2016) NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 [DOI] [PubMed] [Google Scholar]
- 28. Li J., Bonkowski M. S., Moniot S., Zhang D., Hubbard B. P., Ling A. J., Rajman L. A., Qin B., Lou Z., Gorbunova V., Aravind L., Steegborn C., and Sinclair D. A. (2017) A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science 355, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sauve A. A., Munshi C., Lee H. C., and Schramm V. L. (1998) The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry 37, 13239–13249 [DOI] [PubMed] [Google Scholar]
- 30. Dong M., Si Y. Q., Sun S. Y., Pu X. P., Yang Z. J., Zhang L. R., Zhang L. H., Leung F. P., Lam C. M., Kwong A. K., Yue J., Zhou Y., Kriksunov I. A., Hao Q., and Lee H. C. (2011) Design, synthesis and biological characterization of novel inhibitors of CD38. Org. Biomol. Chem. 9, 3246–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aksoy P., White T. A., Thompson M., and Chini E. N. (2006) Regulation of intracellular levels of NAD: a novel role for CD38. Biochem. Biophys. Res. Commun. 345, 1386–1392 [DOI] [PubMed] [Google Scholar]
- 32. Chini E. N. (2009) CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr. Pharm. Des. 15, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bai P., Canto C., Oudart H., Brunyanszki A., Cen Y., Thomas C., Yamamoto H., Huber A., Kiss B., Houtkooper R. H., Schoonjans K., Schreiber V., Sauve A. A., Menissier-de Murcia J., and Auwerx J. (2011) PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 13, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barbosa M. T., Soares S. M., Novak C. M., Sinclair D., Levine J. A., Aksoy P., and Chini E. N. (2007) The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 21, 3629–3639 [DOI] [PubMed] [Google Scholar]
- 35. Canto C., Houtkooper R. H., Pirinen E., Youn D. Y., Oosterveer M. H., Cen Y., Fernandez-Marcos P. J., Yamamoto H., Andreux P. A., Cettour-Rose P., Gademann K., Rinsch C., Schoonjans K., Sauve A. A., and Auwerx J. (2012) The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camacho-Pereira J., Tarrago M. G., Chini C. C. S., Nin V., Escande C., Warner G. M., Puranik A. S., Schoon R. A., Reid J. M., Galina A., and Chini E. N. (2016) CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 23, 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu Y., Wang H., Wang Q., and Deng H. (2014) Overexpression of CD38 decreases cellular NAD levels and alters the expression of proteins involved in energy metabolism and antioxidant defense. J. Proteome Res. 13, 786–795 [DOI] [PubMed] [Google Scholar]
- 38. Darnell J. E. Jr, Kerr I. M., and Stark G. R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 39. Ihle J. N. (1995) Cytokine receptor signalling. Nature 377, 591–594 [DOI] [PubMed] [Google Scholar]
- 40. Wen Z., Zhong Z., and Darnell J. E. Jr. (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 41. Yokogami K., Wakisaka S., Avruch J., and Reeves S. A. (2000) Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10, 47–50 [DOI] [PubMed] [Google Scholar]
- 42. Wang R., Cherukuri P., and Luo J. (2005) Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 280, 11528–11534 [DOI] [PubMed] [Google Scholar]
- 43. Yuan Z. L., Guan Y. J., Chatterjee D., and Chin Y. E. (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 44. Saitoh M., Endo K., Furuya S., Minami M., Fukasawa A., Imamura T., and Miyazawa K. (2016) STAT3 integrates cooperative Ras and TGF-beta signals that induce Snail expression. Oncogene 35, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 45. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Satelli A., and Li S. (2011) Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol. Life Sci. 68, 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verrills N. M., Walsh B. J., Cobon G. S., Hains P. G., and Kavallaris M. (2003) Proteome analysis of vinca alkaloid response and resistance in acute lymphoblastic leukemia reveals novel cytoskeletal alterations. J. Biol. Chem. 278, 45082–45093 [DOI] [PubMed] [Google Scholar]
- 48. Urbani A., Poland J., Bernardini S., Bellincampi L., Biroccio A., Schnolzer M., Sinha P., and Federici G. (2005) A proteomic investigation into etoposide chemo-resistance of neuroblastoma cell lines. Proteomics 5, 796–804 [DOI] [PubMed] [Google Scholar]
- 49. Santidrian A. F., Matsuno-Yagi A., Ritland M., Seo B. B., LeBoeuf S. E., Gay L. J., Yagi T., and Felding-Habermann B. (2013) Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J. Clin. Invest. 123, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nie Y., Erion D. M., Yuan Z., Dietrich M., Shulman G. I., Horvath T. L., and Gao Q. (2009) STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.