Abstract
Chromatin condenses several folds to form mitotic chromosomes during cell division and decondenses post-mitotically to reoccupy their nuclear territory and regain their specific transcriptional profile in a precisely lineage specific manner. This necessitates that the features of nuclear architecture and DNA topology persist through mitosis. We compared the proteome of nuclease and high salt resistant fraction of interphase nucleus known as nuclear matrix (NuMat) and an equivalent biochemical fraction in the mitotic chromosome known as mitotic chromosome scaffold (MiCS). Our study elucidates that as much as 67% of the NuMat proteins are retained in the MiCS indicating that the features of nuclear architecture in interphase nucleus are retained on the mitotic chromosomes. Proteins of the NuMat/MiCS have large dynamic range of MS signal and were detected in sub-femtomolar amounts. Chromatin/RNA binding proteins with hydrolase and helicase activity are highly enriched in NuMat as well as MiCS. Although several transcription factors involved in functioning of interphase nucleus are present exclusively in NuMat, protein components responsible for assembly of membrane-less nuclear bodies are uniquely retained in MiCS. Our study clearly indicates that the features of nuclear architecture, in the structural context of NuMat, are retained in MiCS and possibly play an important role in maintenance of cell lineage specific transcriptional status during cell division and thereby, serve as components of cellular memory.
Keywords: Drosophila, Cell cycle, Chromatin function or biology, Subcellular analysis, Gene Expression, Structural Biology, Cellulra memory, Drosophila S2 cell, Mitotic chromosome scaffold, Nuclear Matrix
Genetic information that is encoded in a linear DNA sequence, needs to be organized in three-dimensional space of the nucleus in the form of chromatin for appropriate expression of genes (1–3). During interphase, interior of the nucleus is organized on a ribonucleo-protein network known as nuclear matrix (NuMat)1 (4). Specific attachment of chromatin fibers to NuMat create a higher order structure made up of discrete topological loop domains (5, 6). NuMat, along with facilitating the packaging of DNA, organizes the nuclear microenvironment in a way that transcription, replication, splicing and repair are executed in a coordinated manner over the highly compacted DNA. However, during open mitosis the three-dimensional organization of the nucleus breaks down as the chromosomes condense to enable faithful segregation of duplicated chromatids into daughter cells. Most genes are thought to be silenced during cell division but are precisely re-activated in daughter cells to achieve gene expression pattern that is indistinguishable from the mother cell (7, 8). One of the mechanisms by which this is accomplished is by bookmarking of genes mediated by locus specific retention of transcription factors and deposition of heritable epigenetic marks (9–11). Several studies suggest that along with this, the information for 3D organization of chromatin in the nucleus is also stably inherited in daughter cells. For example, chromosome positions are transmitted through mitosis and non-random higher order chromatin arrangements can be stably propagated over many cell cycles (12, 13). Early evidence for transmission of information for higher order chromatin organization through mitosis came from the pioneering work by Laemmli and co-workers, who showed that in histone depleted mitotic chromosomes, DNA loops are attached to a proteinaceous scaffold. This arrangement of DNA loops in mitotic chromosomes is similar to the DNA loops that are attached to NuMat in interphase nucleus (14, 15).
Considering that NuMat and mitotic chromosome scaffold (MiCS) are comparable biochemical structures, it is reasonable to expect that at least certain components of the NuMat are retained in MiCS (16, 17). However, our knowledge of the precise relationship between composition of NuMat and MiCS remains limited. Hence, it is important to identify the key structural and functional components of NuMat that are represented in MiCS. In this study, we have carried out a detailed proteomic analysis of NuMat and MiCS in Drosophila S2 cells.
Our results indicate that the bulk of MiCS proteins are a subset of NuMat proteome whereas only a small fraction of MiCS proteins is unique. These findings suggest that NuMat proteins are packaged into MiCS as chromosomes condense. We hypothesize that during mitotic exit, as the chromosomes decondense, these proteins provide the information to organize each chromosome into their territories and functional domains in the daughter nuclei. Thus, the NuMat and MiCS share much of the components responsible for hierarchical organization of chromatin that are likely to be, at least partially, specific to a cell type.
EXPERIMENTAL PROCEDURES
Cell Culture
S2 cells were cultured in Schneider's Media (GIBCO, Gaithersburg, MD) with 10% heat inactivated FBS at 25 °C. Cells were arrested at metaphase with colcemid (Sigma, St. Louis, MO) at a concentration of 20 μg/ml for 20–24 h.
Transfection of S2 Cells and Preparation of Stable Cell Lines
S2 cell transfections were done using effectene transfection reagent (Qiagen, Netherlands, 301425) according to manufacturer instructions. Briefly, 106 cells/ml in S2 cell culture medium were plated in each well of the 6-well plate 24 h before transfection. For each well, 1 μg/2 μl of plasmid DNA was mixed with 94.8 μl of enhancer buffer and 3.2 μl of enhancer in a microfuge tube. The solution was mixed by pipetting and incubated at room temperature for 5 mins. 4 μl of effectene reagent was added to the tube, mixed by pipetting, and incubated at room temperature for 10 mins. 500 μl of fresh S2 medium was added to each reaction mixture. In the meanwhile, 500 μl of the medium was aspirated out from each well. The reaction mixture thus prepared was added dropwise into each well. The 6-well plates were incubated in 25 °C incubator for 48 h. 100 μg/ml hygromycin was added to each well and the plates were further incubated in 25 °C incubator for 48 h. Following this the media was changed every 48 h with fresh media containing 100 μg/ml hygromycin. After 3 weeks when cell death was negligible the cells were transferred to T25 flasks and maintained in media containing 100 μg/ml hygromycin. The expression of proteins was induced by adding 500 μm CuSO4 in the media and incubating the cells in 25 °C incubator for 24 h. All the steps were carried out in laminar flow hood.
Nuclei Purification
Pelleted cells were suspended in nuclear isolation buffer (NIB) with 0.25 m sucrose (NIB - 15 mm Tris, pH 7.4, 40 mm KCl, 1 mm EDTA, 0.1 mm EGTA, 0.1 mm PMSF, 0.25 mm spermidine, 0.1 mm spermine). Cell suspension was passed 3 times through a 22-gauge needle to rupture cell membrane. Homogenate thus obtained was spun at 600 × g for 2 mins to remove unbroken cells and debris. Supernatant was spun at 3000 × g to collect crude nuclear pellet. The crude nuclear pellet was suspended in 1 m sucrose in NIB, and centrifuged at 6000 × g to get pure nuclear pellet. The pellet was washed twice before being used for NuMat preparation.
Chromosome Purification
Chromosomes were purified according to Mg-Hex buffer method (modified from Wray and Stubblefield (18) by Lewis and Laemmli (14)). Briefly, arrested S2 cells were pelleted, washed and resuspended in RSB (10 mm Tris pH 7.4, 10 mm NaCl, 5 mm Mgcl2). After incubation for 15 mins at room temperature, cells were pelleted down at 1400 × g and resuspended for swelling in Mg-Hexelene buffer (1 m Hexelene glycol, 0.1 mm PIPES, 1 mm MgCl2, 0.1 mm and 1% thiodiglycol). Swollen cells were pelleted and resuspended in Mg-Hex buffer with 0.5% Na-DOC and homogenized with 20–25 strokes at 400 rpm with Teflon pestle in a tight-fitting glass tube. Homogenate was spun at 600 × g for 5 mins to remove nuclei and unbroken cells. Supernatant was collected and spun at 1400 × g for 30 mins to collect chromosomes.
An aliquot of nuclei/chromosomes was lysed in 0.5% SDS and absorbance was measured at 260 nm to find out the DNA content. Based on the DNA content, nuclear and chromosomal yield was calculated in OD units.
NuMat/MiCS Preparation
For NuMat preparation, 10 ODUs of nuclei were incubated with 100 μg/ml DNase I in digestion buffer (DB) (DB - 20 mm Tris, pH 7.4, 20 mm KCl, 70 mm NaCl, 10 mm MgCl2, 0.125 mm spermidine, 0.05 mm spermine, 0.1 mm PMSF, 0.5% Triton X-100) at 4 °C for 1 h. Digestion was followed by extraction in extraction buffer (10 mm Hepes, pH 7.5, 4 mm EDTA, 0.25 mm spermidine, 0.1 mm PMSF, 0.5% Triton X-100) containing 0.4 m NaCl for 5 min and an additional 5 min in extraction buffer containing 2 m NaCl to obtain NuMat. The NuMat pellet was washed twice with wash buffer (5 mm Tris, 20 mm KCl, 1 mm EDTA, 0.25 mm spermidine, 0.1 mm PMSF) and stored at −70 °C.
For MiCS preparation, 10 ODUs of isolated chromosomes were digested with 10 μg of DNaseI as well as 15 units of MNase, at RT for 30 mins in digestion buffer (15 mm Tris pH 7.4, 15 mm NaCl, 20 mm KCl, 2 mm CaCl2, 5 mm MgCl2, 0.1% NP 40, 0.1% Trition-X-100 and 0.1 mm PMSF). Digested chromosomes were pelleted down at 900 × g and resuspended in extraction buffer (2 m NaCl, 10 mm Tris-Cl pH 9.0, 10 mm EDTA, 0.1% Trition-X-100, 0.1 mm PMSF) for removal of histones. 10 ODU were extracted in 10 ml of extraction buffer, for 15 mins at RT. The MiCS pellet was washed twice in wash buffer (10 mm Tris-Cl pH 9.0, 50 mm NaCl, 10 mm EDTA, 0.1% Trition-X-100, 0.1 mm PMSF) and stored in −70 °C till further use.
The NuMat and MiCS pellets were used for 1D and 2D PAGE, Western blotting and LC-MS/MS.
SDS-PAGE, Western Blotting, and DIGE
NuMat/MiCS protein samples were resolved on 12% SDS-PAGE gel and were either silver stained for 1D profile or transferred to PVDF for western hybridization with Lamin Dm0, topoisomerase 2, histone or GFP antibody.
For DIGE, 150 μg of protein was labeled with Cy Dyes from Ettan DIGE (GE Healthcare, Chicago, IL) and applied to 11 cm (3–10) strips from BioRad in rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% Pharmalyte). Focusing was done at 4000 V with linear ramp for 40,000 V Hrs. After focusing, the strips were equilibrated in buffer (Tris-Cl 50 mm pH 8.8, 6 m urea, 30% v/v glycerol, 2% SDS) containing 0.01g/ml DTT and 0.025 g/ml iodoacetamide. The focused proteins were resolved for second dimension on 12% SDS-PAGE. Gels were scanned using Typhoon scanner, documented and analyzed for spot pattern through Image Master 2D Platinum v7.05 DIGE software.
In-Gel Digestion and LC-MS/MS
For tryptic digestion, 2–4 μg of protein were resolved on 12% SDS-PAGE. Gel slices were cut and dehydrated twice using 50% ACN followed by 100% ACN and the protein in the gel slices were digested with 100 μl-200 μl of Trypsin solution (10 μg/ml, Promega, Madison, WI) in 25 mm ammonium bicarbonate. Digestion was carried out at 37 °C overnight. Digested peptides were eluted with 50% ACN and 5% TFA, vacuum dried and stored at −70 °C till loaded on to the mass spectrometer.
Tryptic peptides were loaded on a 50 cm long column, packed with 1.8 μm C18 beads. Column had been heated at 48 °C in a homemade oven. The peptides were separated using linear gradient from 5% to 35% of buffer B (95% acetonitrile and 0.5% acetic acid) with flow rate of 75 nl/min followed by a wash up to 95% of buffer B. The length of the gradient was adjusted to 480 mins.
The Proxeon LC system was directly connected with Thermo fisher scientific LTQ Orbitrap Velos instrument using Proxeon nanoelectrospray source. The nano source was operated at 2.2 kV and the ion transfer tube at 200 °C without sheath gas. The mass spectrometer was programmed to acquire in a data dependent mode. The scans were acquired with resolution 60,000 at m/z 400 in orbitrap mass analyzer with lock mass option enabled for the 445.120024 ion. The 25 most intense peaks containing double or higher charge state were selected for sequencing and fragmentation in the ion trap by using collision induced dissociation with normalized collision energy of 40%, activation q = 0.25, activation time of 10 ms and one micro scan. Dynamic exclusion was activated for all sequencing events to minimize repeated sequencing. Peaks selected for fragmentation more than once within 30 s were excluded from selection for next 90 s and the maximum number of excluded peak was 500.
Data Processing and Analysis
The raw spectra obtained were processed with Andromeda search engine (MaxQuant software version 1.1.0.39). Search was performed against D. melanogaster protein database (FlyBase assembly version Dmel_5.37 that contains 23605 entries in the forward database). Trypsin/P was specified as the cleavage enzyme and up to two missed cleavages were allowed. The initial precursor mass tolerance was set at 10 ppm and fragment mass deviation was set at 0.25 Da. The search included cysteine carbamidomethylation as fixed and oxidation of methionine as variable modification. The identification has been done keeping 1% false discovery rate at the peptide and protein level. Proteins identified with multiple peptides as well as one unique one unique peptide with high confidence were listed and analyzed further.
Experimental Design and Statistical Rationale
Three biological replicates from independent biological preparations were analyzed for each of NuMat and MiCS. Only proteins identified in all three biological replicates with two or more peptides, were considered as true NuMat and MiCS components and were used for further analysis. NuMat and MiCS thus identified were compared and the common and unique proteins were listed (supplementary Table S3). For estimation of sensitivity of single run analysis, total summed peptide intensities of each protein group in NuMat and MiCS were taken as proxies to estimate the protein amount. Summed peptide intensities were normalized to the molecular weight of proteins. Spearman Rank correlation was used to judge the reliability of quantification measurements between biological replicates. Further process was followed from the earlier report (19).
Bioinformatics Analysis
Annotation of the proteins was derived using clusterProfiler, an R package. Enrichment of protein domains in NuMat and MiCS proteins were analyzed based on InterPro database where analysis was done using DAVID annotation tool (v6.8 (http://david.abcc.ncifcrf.gov)).
In Situ NuMat/MiCS Preparation in Drosophila Embryo and Immunostaining
Drosophila embryos (0–2 h old) were collected, de-chorionated and washed thoroughly with running water to remove sodium hypochlorite. Embryos (0.1 g) were then rinsed in distilled water and pretreated in 4:1 heptane:PBS for 2 min. Fixation was carried out in 0.8 ml heptane, 0.1 ml 10× PBS with 4% formaldehyde for 20 min at room temperature with continuous mixing. After fixation, heptane and aqueous phase was removed. A mixture of 1:1 (v/v) ice cold methanol/heptane was added to devitellinize the embryos. The tube was shaken vigorously, and the embryos were allowed to settle to the bottom. This step is repeated several times. These fixed embryos were brought to aqueous medium by several washes in PBS + 0.1% Triton-X-100 (PBT) and used directly for immuno-staining as control embryos. To prepare NuMat, the fixed embryos were brought to aqueous medium by several washes in DB (composition same as in MuMat/MiCS preparation) without DNase I. Digestion was carried out with DNase I (200 μg/ml) in DB for 30 min at room temperature. Embryos were then washed two times in extraction buffer without NaCl by suspending and then allowing to settle down in the tube because of gravity. Extraction was carried out with 2 m NaCl in extraction buffer or 20 min. The embryos were finally washed several times in PBS with 0.5% Triton-X-100 (PBT). These embryos with in situ mitotic chromosome scaffolds were used for immuno-staining.
For antibody detection, anti-Lamin Dm0 (mouse), anti-fibrillarin (rabbit) and an in house developed anti-NuMat (rat) antibody were used alone or in combination. The primary antibodies were used at a dilution of 1:100 in PBT. Incubation was carried out for 1 h at 37 °C or at 4 °C overnight. Primary antibody was followed by secondary antibody treatment (anti-mouse/rabbit/rat Cy3/FITC at 1:300 dilution) for another hour. Embryos were then washed and mounted in Vectasheild with DAPI. Confocal laser scanning was carried out on a Zeiss LSM 510 META (Carl Zeiss Inc., Germany) with excitation at 488 nm, 543 nm, 633 nm (Ar-ion and HeNe lasers) and 760 nm at a pinhole of 1 AU. The scanning was done in the multi-track mode using a 10×, 63× and 100× 1.4NA objective. The emission of FITC was acquired using a 500–530 BP filter, that of Cy3 with a 565–615 BP filter and that of Cy5 with 650–710 BP filter set. Optical sections were taken 0.35 μm intervals. Individual optical sections were projected to give information in 3D using the Zeiss LSM software version 3.2 SP2. Later the images were assembled using Adobe Photoshop 6.0.
Plasmids
The vectors carrying the cDNA sequences of Orc2(GH13824), Orc5(RE16687), Mcm2 (LD47441) and Mcm5(RE67590) proteins were obtained from DGRC (Drosophila Genome Research Center). eGFP cDNA was amplified from pEGFP-C1 plasmid using appropriate primers with kpn1 and spe1 restriction enzyme sites introduced in the forward and reverse primers respectively. The Orc2, Orc5, Mcm2 and Mcm5 full length cDNAs were amplified from the DGRC clones using appropriate primers harboring BamH1(for Orc2 and Orc5) or EcoR1(for Mcm2) or Xho1(for Mcm5) restriction enzyme sites in the forward primers and Kpn1 restriction enzyme site in the reverse primers. The amplified cDNA sequences of Orc2, Orc5, Mcm2, and Mcm5 were digested with Kpn1 and ligated with Kpn1 digested eGFP amplicon. The resulting product was further amplified using the respective forward primers for each of the genes and the reverse primer for GFP. The products were digested with appropriate restriction enzymes and cloned into pMk33-cTAP(SG) vector, between BamH1 (for Orc2 and Orc5) or EcoR1 (for Mcm2) or Xho1 (for Mcm5) and Spe1 sites, such that the proteins are C-terminally tagged with GFP. For empty vector control GFP was amplified from pEGFP-C1 plasmid using a different forward primer harboring a BamH1 restriction enzyme site instead of Kpn1. This was subsequently digested and cloned between BamH1 and Spe1 sites into pMk33-cTAP(SG) vector. The pMk33-cTAP(SG) plasmid contains an inducible metallothionine promoter(MT) and a hygromycin selection marker.
RESULTS
NuMat and MiCS Preparation and Quality Control
NuMat is the salt and nuclease resistant fraction of the interphase nucleus, whereas MiCS is an equivalent biochemical fraction from mitotic chromosomes. S2 cell nuclei were purified according to published protocol and were found to be free of cytosolic contaminants when observed under fluorescence microscope (Fig. 1A, a). The enlarged pictures in inset shows that the nuclei were free of adhering membranes. The isolated nuclei were also assessed by western hybridization with antibodies against proteins known to localize to other sub-cellular compartments (Fig. 1B). Aliquots were analyzed at different steps of nuclei isolation and analyzed by Western blot analysis. Western blot analysis showed that endoplasmic reticulum, golgi and cytosolic contaminants were efficiently removed from the isolated nuclei. S2 cells were arrested in metaphase with microtubule depolymerizing drug colcemid and highly purified mitotic chromosomes were obtained using Mg-Hexelene buffer method, adapted from Laemmli et al. (14). Fluorescence microscopy after DAPI staining showed that the chromosomal fraction was homogenous, and no contaminating nuclei were detected (Fig. 1A, b). Nuclei and chromosomes were digested with nucleases and subjected to high salt extraction to yield NuMat and MiCS respectively. The representative silver stained SDS-PAGE pictures showed that NuMat/MiCS were enriched in high molecular weight proteins (Fig. 2, Lanes 2 and 6). Digestion with nucleases removes bulk of chromatin and associated proteins (Fig. 2, Lanes 3 and 7) whereas stepwise salt and detergent extraction further removes the tightly associated proteins like histones (Lanes 4 and 8). Quality of the preparations was assessed by Western blot analysis with known NuMat and MiCS protein (Fig. 2, Lower panels). A known NuMat protein, lamin Dm0, was used as positive control for NuMat preparations and topoisomerase II that localizes in MiCS was used as a positive control for chromosome scaffold preparations. These proteins, as expected, were enriched in the respective insoluble fractions of NuMat/MiCS. During preparation, histones were eluted out by salt extraction and thus western with antibody against H3 served as a negative control to assure that the preparation is of good quality. Interestingly, lamin Dm0 was also seen in MiCS and topoisomerase II was also present in NuMat. Because ribonucleoproteins(RNPs) and RNA is known to be an integral part of NuMat, we estimated the amount of RNA that is retained in NuMat and MiCS and expressed it as percentage of that in nuclei and mitotic chromosomes respectively (supplemental Fig. S2). We find that ∼11% RNA is retained in both the structures.
Fig. 1.
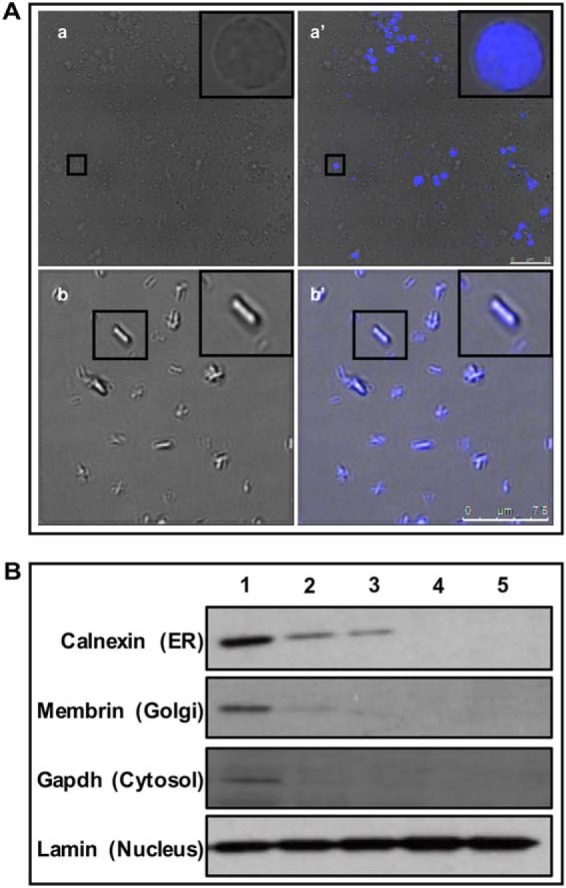
Isolation of S2 cells nuclei and mitotic chromosomes. A, Transmission images of (a) nuclear and (b) mitotic chromosomal preparations. Scale bar - 10 μm for (a), 25 μm for (b). Insets show enlarged pictures of single nuclei/mitotic chromosome merged with DAPI to show that purified nuclei/mitotic chromosomes were free of adhering membranous contamination. B, Western blotting hybridization with nuclei at different stages of purification. Antibodies to marker proteins from various cellular organelles were used. Lane 1 - Embryonic homogenate; Lane 2 - Crude nuclear pellet; Lane 3 - Nuclear pellet after sucrose gradient centrifugation; 4 - Nuclear pellet after 1st wash; 5 - Final nuclear pellet. The blots show that the final nuclear pellet was free of ER, golgi and cytosolic contamination.
Fig. 2.
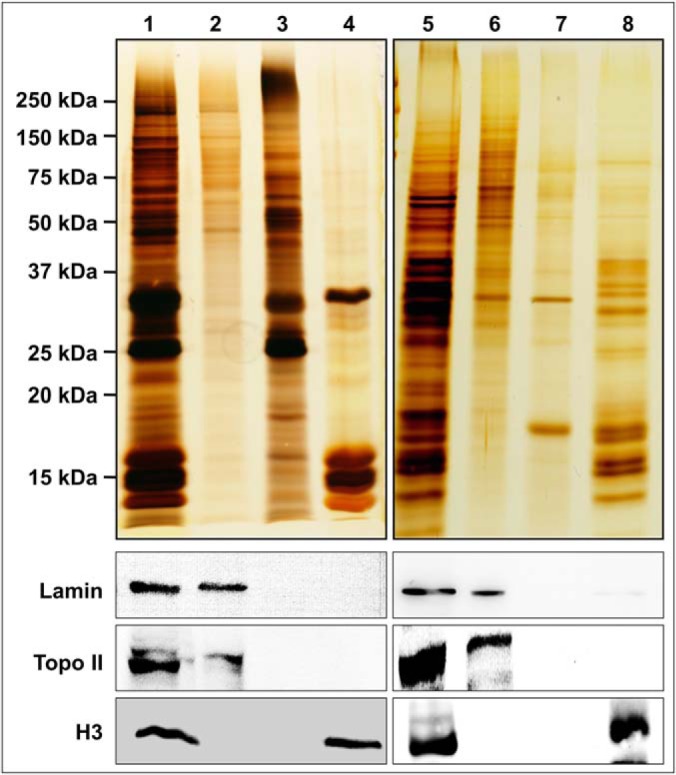
Protein profile of NuMat and MiCS. The upper panel shows silver stained gel profile of NuMat/MiCS proteins. Lane 1 - nuclear proteins, Lane 2 - NuMat proteins, Lane 3 - nuclear proteins extracted with DNaseI, Lane 4 - nuclear protein extracted with 2 m NaCl, Lane 5 - mitotic chromosome proteins, Lane 6 - MiCS proteins, Lane 7 - chromosomal proteins extracted with DNaseI + MNase, Lane 8 - chromosomal proteins extracted with 2 m NaCl. Lower panels show immuno-blots corresponding to similarly loaded protein gels, probed with Lamin Dm0, Topoisomerase II and Histone H3 antibodies. Silver stained profile shows enrichment of high molecular weight proteins in NuMat/MiCS fractions. Western blots show that Lamin Dm0 and Topoisomerase II, were retained in NuMat/MiCS respectively, whereas histone H3 was extracted with 2 m NaCl.
DIGE Analysis of NuMat and MiCS Proteome
To get an estimate of total number of protein in NuMat and MiCS and to get an idea of overlap among the two proteomes, DIGE analysis was carried out (supplemental Fig. S1A). NuMat proteins were labeled with Cy5 (supplemental Fig. S1A, a), MiCS proteins were labeled with Cy2 (supplemental Fig. S1A, b) and an internal control comprising of pooled proteins from both the fractions was labeled with Cy3 (supplemental Fig. S1A, d). The spots were detected and analyzed by Image Master 2D Platinum Version 7.0, DIGE analysis software (GE Health Care) keeping threshold values at default level. Spot volume was chosen after manual assessment and exclusion of artifacts. The software identified a total of 1479 proteins spots of which 793 were common to NuMat and MiCS (supplemental Fig. S1B). The experiment was performed twice and each time the numbers of unique and overlapping spot was nearly the same. This gave us a rough estimate that more than half of NuMat and MiCS proteome overlap with each other.
LC-MS/MS Profiling of NuMat and MiCS Proteome
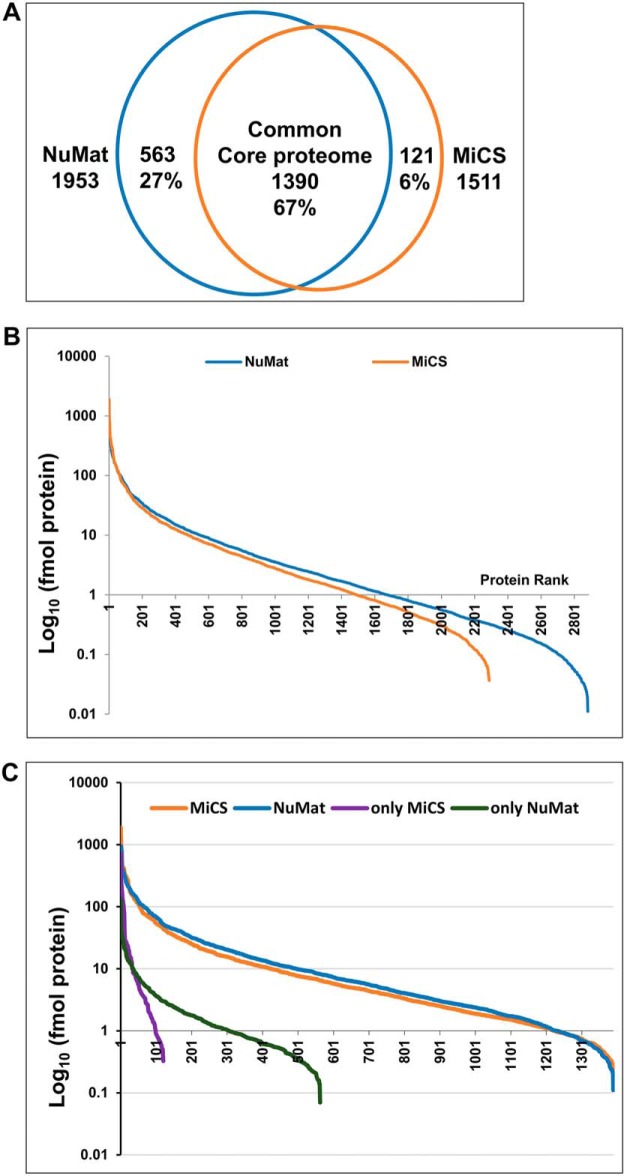
LC-MS/MS analysis was performed for NuMat and MiCS from three biological replicates, using 2 μg of proteins per run. The FT-MS raw spectra were analyzed using MaxQuant with integrated Andromeda search engine. A total of 2892 and 2300 proteins were identified with a minimum of one unique peptide in NuMat and MiCS respectively (supplemental Tables S1, S2, and S5). Although, proteins identified by single unique peptide may also be NuMat/MiCS components, to reduce the possibility of false positive identifications, we used very stringent criteria for data analysis and considered only the proteins, that occur in all three biological replicates and were identified with two or more unique peptides, as true NuMat and MiCS components. Hence, 1953 proteins in NuMat and 1511 proteins in MiCS were considered for further analysis. Comparing NuMat and MiCS proteome we find that 67% (1390) of proteins were common to both, 27% (563) proteins were unique to NuMat and 6% (121) were unique to MiCS (Fig. 3A and supplemental Table S3).
Fig. 3.
LC/MSMS analysis of NuMat and MiCS proteins. A, Venn diagram to show percentage overlap among NuMat and MiCS proteomes (based on LC/MSMS analysis). B, Dynamic range of NuMat and MiCS proteins based on single run analysis. C, Dynamic range of proteins unique to, and common between NuMat and MiCS based on single run analysis.
The proteins were assigned an abundance value that was calculated using MS signal of peptides of each protein, adjusted for protein length and compared with total MS signal. These quantifications give an approximate idea of abundance of individual proteins in a sample. Quantification was carried out based on added peptide intensities for each protein obtained for each of three biological replicate single runs. It has been reported earlier that there is a good correlation between added peptide intensity of each protein and total amount of proteins (19, 21–23). Spearmen rank correlation of protein quantification between biological replicates was greater than 0.78 for NuMat (p < 0.0001) and greater than 0.77 for MiCS (p < 0.0001) indicating that the quantification measurements were reliable among biological replicates. Protein abundance ranged over five orders of magnitude, with sub-femtomole amounts. The dynamic range of NuMat proteome was higher than that of the MiCS proteome, with about 300 additional proteins detected at sub-femtomole level (Fig. 3B). Although the experiment does not provide accurate copy numbers for each of the proteins measured, as this would require isotope labeled standards, the method has been shown to grossly estimate protein abundance in a sample (19). Even though the quantification may not be very accurate for individual proteins there is no deviation in global ranks from these values. However, at very low abundance level of proteins, their abundance measurement may be unreliable because the relationship between absolute amount of protein and MS signal may be less accurate. Interestingly, the 1390 proteins common to NuMat and MiCS show similar abundance in the respective proteomes with a wide range of dynamics of MS signal and sensitivity. (Fig. 3C).
General Features of the NuMat and MiCS Proteome
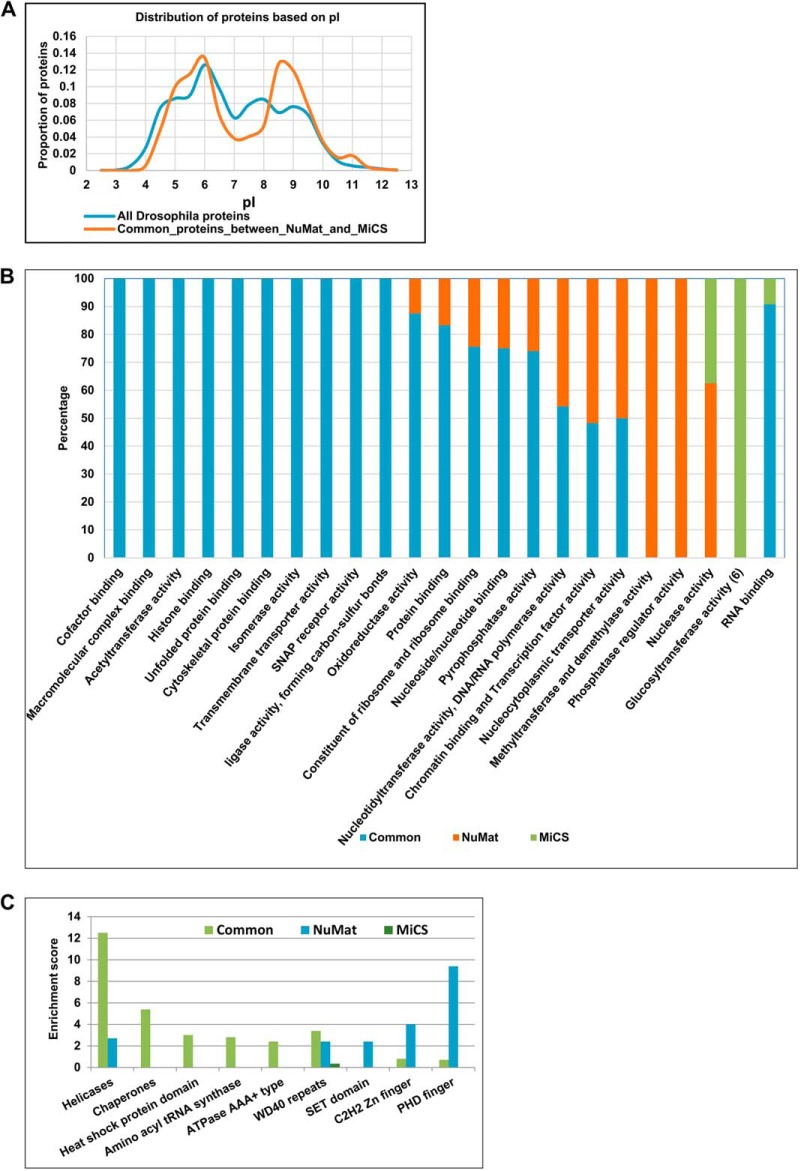
To elucidate the biochemical nature of common proteins of NuMat and MiCS we plotted the distribution of pI values of these proteins over the range of 2–13 with a class interval of 0.5 (Fig. 4A). The plot shows that the proteins common to NuMat and MiCS have a bimodal distribution as compared with the whole Drosophila proteome, which shows a trimodal distribution. Although the proteins common to NuMat and MiCS and the background Drosophila proteome show an abundance of proteins of pI value of ∼6, the former is enriched in proteins with pI value of ∼8.5 as opposed to the latter which shows slight abundance of proteins with pI values of ∼7.5 and ∼9.
Fig. 4.
General properties of NuMat and MiCS proteome. A, Distribution of NuMat and MiCS proteins based on pI value. B, Dynamics of NuMat and MiCS proteins based on molecular function of proteins where annotation was derived using clusterProfiler, an R package. C, Bar chart to show protein domains enriched in NuMat and MiCS proteome based on InterPro database where analysis was done using DAVID annotation tool (v6.7 (http://david.abcc.ncifcrf.gov).
Gene ontology classification was done using clusterProfiler, an R package, to identify the molecular function of the proteins present in NuMat and MiCS (supplemental Table S4) (24). Based on the molecular function ontology, proteins that hydrolyze nucleoside triphosphates to release energy are highly enriched in common NuMat/MiCS proteome. Further analysis of the enrichment Table indicates that DNA/RNA binding proteins that have hydrolase as well as helicase activity are found abundantly in common proteome of NuMat and MiCS. Many chromatin binding transcription factors were uniquely enriched in NuMat whereas several proteins that bind to and hydrolyze RNA were uniquely associated with MiCS.
Depending on their molecular function in NuMat or MiCS, we observe that certain classes of proteins like RNA binding proteins, cytoskeletal proteins and unfolded protein binding chaperones were the least dynamic class, with almost all such proteins being common to NuMat/MiCS. Proteins with acetyltransferase, ligase and isomerase activity were also commonly associated with NuMat as well as MiCS. On the other hand, structural constituents of ribosome, nucleotide/nucleoside binding proteins and proteins with pyrophosphatase activity were moderately dynamic as almost 75% of them were present in NuMat as well as MiCS whereas the rest of them were present only in the NuMat. Proteins involved in DNA/RNA polymerization, nucleocytoplasmic transport and transcription factors were highly dynamic with half of them (∼50%) common to NuMat as well as MiCS and the other half present only in NuMat. Interestingly, enzymes with methyltransferase/demethylase and phosphatase activity were exclusively present in NuMat. Similarly, ribonucleases were only present in MiCS (Fig. 4B).
Using DAVID v6.7, we looked into the enrichment of functional domain among NuMat/MiCS proteins (Fig. 4C) (25, 26). The tool provides an enrichment score using the whole Drosophila proteome as background. This score is a modified Fisher Exact p value score. The results obtained were in parallel to the enrichment results obtained based on molecular function gene ontology classification. Based on the enrichment score for different categories, we find that proteins that carry a helicase domain are enriched in both NuMat and MiCS proteomes. This was also evident in the molecular function categorization where pyrophosphatases with helicase activity were found to be significantly enriched in common NuMat/MiCS proteome. Proteins with WD40 repeat, SET domain, C2H2 zinc finger and PHD domain that are mostly found in DNA binding transcription factors, were uniquely enriched in NuMat. Four proteins with WD40 repeats were found uniquely associated with MiCS.
Validation of NuMat/MiCS Proteins by Western Blotting
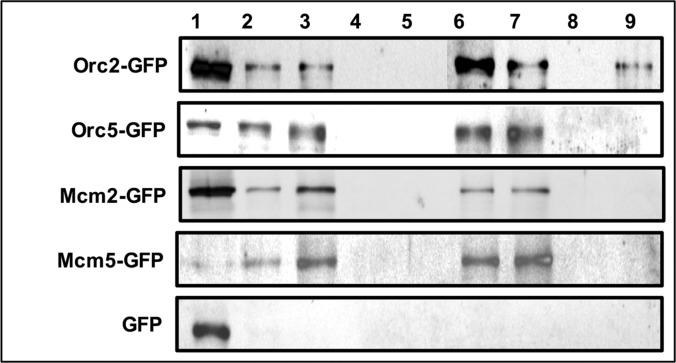
We observed that many proteins involved in specific functions in interphase nucleus such as DNA replication were also present in MiCS. As DNA replication occurs in association with the NuMat during S-phase of cell cycle, presence of these proteins in NuMat is understandable, but the relevance of these proteins in MiCS seemed confounding. We thus chose four proteins, namely ORC2, ORC5, MCM2 and MCM5, involved in replication licensing for validation. We tagged these proteins with eGFP and expressed them in S2 cells. Stably transfected cell lines were obtained by growing the transfected cells in the presence of antibiotic (hygromycin) for several passages. NuMat and MiCS were prepared from these cells and Western blotting was carried out using anti GFP antibody (Fig. 5). The results confirmed that the queried proteins were present in NuMat as well as MiCS, thus validating the result obtained by LC-MS/MS.
Fig. 5.
Western blot analysis to validate the presence of DNA replication associated proteins in NuMat as well as MiCS. S2 cell were transfected and selected to derive stable cell lines expressing eGFP-tagged Ocr2, Orc5, Mcm2 and Mcm5 proteins. NuMat and MiCS proteins were prepared from these cells, western blotted and probed with GFP antibody. Lane 1 - Whole cell extract, Lane 2 - nuclear proteins, Lane 3 - NuMat proteins, Lane 4 - nuclear proteins extracted with DNaseI, Lane 5 - nuclear protein extracted with 2 m NaCl, Lane 6 - mitotic chromosome proteins, Lane 7 - MiCS proteins, Lane 8 - chromosomal proteins extracted with DNaseI + MNase, Lane 9 - chromosomal proteins extracted with 2 m NaCl.
Visualization of NuMat/MiCS Proteins by Immunostaining
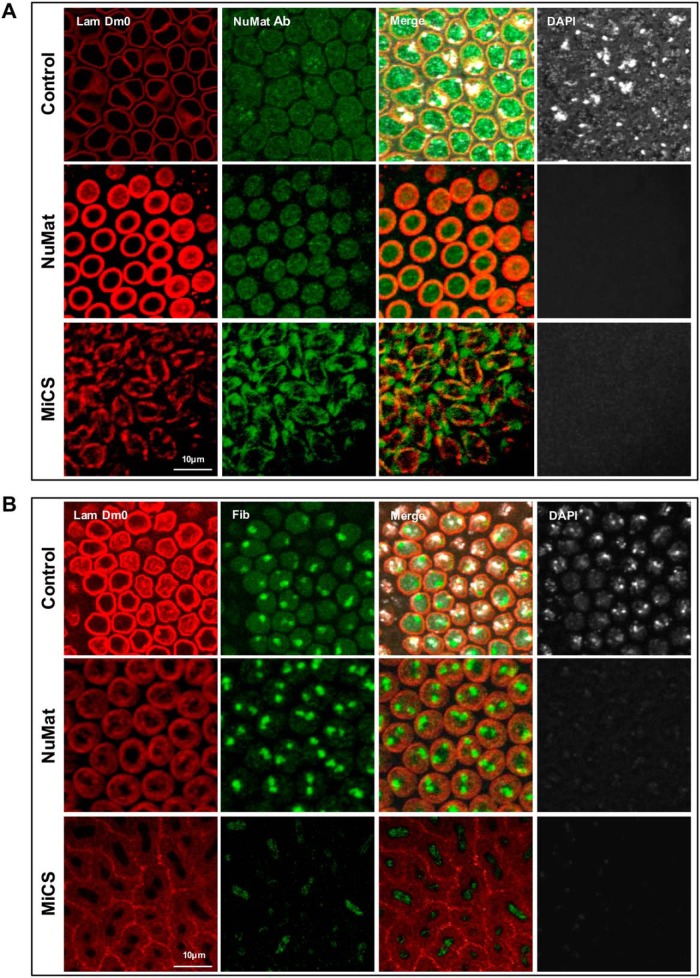
To directly visualize the reorganization of NuMat proteins during mitosis we used an antibody that was raised in-house, using the whole NuMat proteins from S2 cell nuclei as antigen. The antibody was used for immuno-staining Drosophila early (60–90 min) embryo that is an excellent model to visualize dynamics of mitotic events because the nuclear divisions are synchronous and it is relatively easy to spot an embryo with a mitotic wave sweeping through. In intact nuclei (control), the NuMat antibody gives a punctate staining confined to the nuclear space defined by the lamin rim (Fig. 6A, nuclei panel). After in vivo NuMat/MiCS preparation, when bulk of chromatin and soluble proteins had been removed, the antibody continues to stain the NuMat proteins that undergo a striking reorganization into mitotic chromosomes and spindle like structure, which colocalizes with Lamin Dm0, during mitosis (Fig. 6A, NuMat, MiCS panels). Quality of NuMat/MiCS preparation was judged by the absence of DAPI signal that indicates complete digestion and removal of chromatin. Similar results were obtained when we carried out the immuno-staining with a known NuMat protein, fibrillarin. In intact nuclei (control), the protein remains associated with the nucleolus, remains distinctly associated with nucleolar remnant in NuMat preparation and were seen to distribute in the chromosomal space during mitosis (Fig. 6B). For this experiment, slightly older embryos (100–140 min) were used, because nucleolus forms only after the thirteenth division when the nuclei have been separated into separate cells because of cellularization.
Fig. 6.
Visualization of NuMat/MiCS proteins in situ. Drosophila embryos were immuno-stained with antibodies against A. Lamin Dm0 and an in-house raised antibody against total NuMat proteins, B, Lamin Dm0 and fibrillarin. DNA was stained with DAPI. The upper panel shows untreated embryos with intact nuclei whereas the middle and lower panel shows embryos where in situ NuMat/MiCS had been prepared. Scale bar - 10 μm.
Distribution of Proteins Involved in Specific Functions in NuMat/MiCS
We then analyzed the distribution of few proteins with known cellular functions in NuMat/MiCS. We observe that many proteins show interesting and differential distribution in the two structures (Summarized in Table I). Whereas epigenetic modifiers such as histone methyltransferases and demethylases were mostly present in NuMat, histone acetyl transferases and deacetylases were found to associate with both NuMat and MiCS. Mediator complex proteins and most Polycomb group (PcG) of proteins were present only in NuMat, whereas RISC complex proteins, Trithorax group (trxG) of proteins and insulator binding proteins were present in NuMat as well as MiCS. Cohesin and Condensin complex proteins were not only present on MiCS but were also associated with NuMat. Further, we investigated the role of NuMat/MiCS in organizing the various nuclear sub-compartments, which lack a delimiting membrane and are known to be assembled by the hierarchical recruitment of component proteins from the surrounding nucleoplasm. The protein components responsible for biogenesis of several nuclear sub-compartments were found in NuMat as well as MiCS (Table II).
Table I. Distribution of a few nuclear proteins in NuMat and MiCS.
| Nuclear proteins | Proteins common to NuMat/MiCS | Proteins present only in NuMat | Proteins present only in MiCS | Function | Comments |
|---|---|---|---|---|---|
| Histone methyl transferases (HMTs) | Art1, Su(var)3–9 | Ash1, Caf1, esc, egg, gpp, Mes-4, Set1, Set2, Su(z)12 | – | To create active/repressive chromatin marks | Most HMTs are NuMat associated |
| Histone demethylases | – | Su(var)3–3, Kdm2, JHDM2, Kdm4A, Jarid2 | – | To create active chromatin marks | Most Histone demethylases are NuMat associated |
| Histone acetyl transferases (HATs) | lid, mof, Hcf, wds, Sgf29, enok | Acf1, Ada2b, D12, dik | – | To create active chromatin marks | HATs are NuMat associated and a subset of HATs persist in MiCS |
| Histone deacetylases (HDACs) | Sin3A, lid, Sap130, Bin1, Mi-2, Rpd3, simj, MEP-1, Caf1 | – | HDAC4 | To create repressive chromatin marks | HDACs are NuMat associated and persist in MiCS |
| Mediator complex | – | MED1, MED8, MED10, MED14, MED15, MED17, MED30 | – | To link transcription factors to RNA Pol II | Transcription is NuMat associated activity |
| Dosage compensation complex (DCC) | mle, mof, JIL-1 | MSL-1, MSL-2, MSL-3 | – | To assemble activation complex on male X-chr | Enzymatic components of DCC persists in MiCS while DCC complex assembly occurs in interphase |
| RISC complex | AGO2, bel, Dcr2, Fmr1, Tudor-SN, vig, Rm62, blanks | – | – | Establishment of heterochromatin | RNAi machinery remains associated with heterochromatic loci through mitosis |
| PcG proteins | Pc, RAF2, rept | Sce, ph-d, esc, Su(z)12, Pcl, lolal, Jarid2, Kdm2, calypso, sxc | – | Establishment of H3K27me associated silenced domains | PcG proteins are enriched in NuMat |
| trxG proteins | brm, mor, osa, Snr1, Bap55, Bap60, dalao,Iswi, E(bx), Nurf-55, Nurf-38, pont, lid, kis | ash1, ash2, dom | – | Establishment of active chromatin domains | trxG proteins associate with NuMat and persist in MiCS |
| Insulator binding proteins | BEAF, GAF, Su(Hw), Mod(mdg4), Cp190 | – | – | Establishment of chromatin domain boundaries | Chromatin domain boundary proteins persist in MiCS |
| Condensin complex | Barr, Cap-D2, Glu, SMC2, Smc5 | Cap-G | – | Mitotic chromosome condensation and segregation | Condensin complex proteins are present in NuMat as well as MiCS |
| Cohesin complex | Cap, Nipped-B, SA, san, SMC1, vtd | pds5 | – | Cohesion of sister chromosomes | Cohesin complex proteins are present in NuMat as well as MiCS |
Table II. Presence of protein components suspected to be responsible for post-mitotic assembly of nuclear bodies in NuMat and MiCS.
| Nuclear bodies | Putative function | Protein component(s) present in NuMat | Protein component(s) present in MiCS | Mode of inheritance | References |
|---|---|---|---|---|---|
| Nucleolus | Ribosome biogenesis | Nopp140 | Nopp140 | Disassembly, persists as mitotic nucleoli | (63) |
| Nuclear speckles | Storage and recycling of splicing factors | SF2 | SF2 | Disassembly/Reassembly | (64) |
| Nuclear stress bodies (Omega speckles) | Response to stress | Hsf, HRB87F, hnRNPs | Hsf, HRB87F, hnRNPs | Unknown | (65, 66) |
| Histone locus bodies | Transcription and processing of histone mRNAs | Mxc, Spt6 | Mxc | Disassembly/Reassembly | (60) |
| Cajal bodies | Biogenesis, maturation and recycling of snRNPs and snoRNPs | Coillin, snRNPs, snoRNPs, Fib, Nopp140 | Coillin, snRNPs, snoRNPs, Fib, Nopp140 | Disassembly, persists as mitotic Cajal bodies | (61) |
| Polycomb bodies | Polycomb proteins-mediated gene pairing and silencing | Pc, sxc, Su(z)12, esc, ph-d, Sce, Kdm2 | Pc | Disassembly/Reassembly | (67) |
DISCUSSION
In an earlier study, NuMat and MiCS proteomes were shown to be largely similar in HeLa cells, however the exact protein constituents were not known till now (17). This extensive proteomics approach not only uncovers the fact that the MiCS proteome is a subset of NuMat proteome but also gives a glimpse into the complexity of both the structures indicated by the wide variety of proteins. We for the first time report an in depth quantitative proteomics of Drosophila NuMat and MiCS exemplified by detection of proteins present in as low as sub-femtomole amount. Although the common proteins of NuMat and MiCS were detected with comparable intensities, the higher number of unique proteins in NuMat also included a significant proportion of low abundance proteins compared with MiCS. This is expected because the interphase nucleus is functionally more complex than mitotic chromosomes.
Proteome analysis studies have elucidated that bacterial and archaeal proteomes exhibit a bimodal distribution of pI values, whereas eukaryotic proteomes show a trimodal distribution (27, 28). Nuclear proteins are almost evenly distributed throughout the pI range of 4.5–10 in contrast to the bimodal distribution around pI values ∼6 and ∼8.5 observed in our study. This observation is interesting as proteins are generally least soluble at their isoelectric point (29). As intracellular pH is close to neutrality, the pI values of greater than or less than pH 7 of NuMat/MiCS proteins suggests their inherent insoluble nature. This also points to potential role of NuMat/MiCS proteins as structural units in nondiffusible nuclear architecture.
We observed many heat shock proteins/chaperones that are mostly common between NuMat/MiCS were also one of the most abundant proteins as observed based on protein intensities. Presence of molecular chaperones and protein folding catalysts such as heat shock proteins are known to associate with nuclear multiprotein complexes and their presence in NuMat is well documented (30–32). Because, chaperones protect nonspecific aggregation of their substrates in a crowded micro-environment, perhaps they assist in the proper folding and ordered assembly of the proteinaceous network in NuMat and MiCS and maintain them under changing conditions of the cell cycle.
Several studies in past have shown that RNA is an essential component of NuMat. Considering the similarity between the NuMat and MiCS proteome, including number of RNA binding proteins, RNA components in the later one is also likely to be important. Although we maintain strict RNase free conditions during our preparation process, that we used both MNase and DNase I for digestion of metaphase chromosomes for preparation of MiCS, whereas only DNase I was used for digestion of nuclear chromatin. However, it is known that the specific activity of MNase for DNA is ∼3-fold greater than that for RNA (33). This and the short digestion time for MNase used in this study, is unlikely to significantly affect the RNP content in MiCS. This was also confirmed by the fact that both NuMat and MiCS retain ∼11% of nuclear and chromosomal RNA respectively. Therefore, the effect of MNase digestion on the structural integrity and protein composition of MiCS is expected to be minor, if any. This is further supported by the observation that most of the RNPs, proteins which are expected to be associated in an RNA dependent manner, detected in this study are common between both NuMat and MiCS.
One of the limitations of the NuMat field has been the suspicion that because of high salt concentration (2 m NaCl) various components may acquire interactions that are irrelevant to their normal context (34, 35). These concerns are at least partially addressed by stepwise elution (0.4 m followed by 2 m Nacl), used in our study, thereby reducing the complexity by removing bulk of extracted proteins in the first step. As discussed above, high degree of consistency among biological replicates further supports the relevant and non-random nature of components identified in NuMat. Finally, remarkable similarity in the constituents of NuMat and MiCS also indicates the reliability of these constituents and argues against any significant artifactual/random aggregation.
One of the interesting findings of this study is the fact that cytoskeletal proteins like cheerio and zipper (fly homologs of filamin and myosin II, respectively) were present in both NuMat and MiCS, whereas abundant proteins of the cytoskeleton were found only in NuMat (e.g. actin) or were present in very low amounts (e.g. tubulin) in NuMat/MiCS, also strengthens the idea that cytoskeletal proteins in NuMat/MiCS (filamin and myosin) are associated with each of these structures as important structural/functional components. It has been shown earlier that actin is associated with regulation of gene expression in the nucleus, by interacting with transcription factors and chromatin remodeling complexes, which are associated only with interphase nucleus, and hence, is presence only in NuMat (36). Cheerio has been reported to be involved in repression of rRNA transcription whereas zipper is known to help attach chromatin to the nucleoskeleton, functions which are essential in both nucleus and mitotic chromosomes (37, 38). The role of several cytosolic proteins in nuclear architecture is also supported by our finding that ∼22% of the NuMat/MiCS proteins are known to localize to different cytosolic organelles like mitochondria, golgi, endoplasmic reticulum, etc. Association of such proteins with the NuMat as well as MiCS raises the, possibility of these proteins having a role in maintaining cellular memory through mitosis to re-establish similar cellular state in daughter cells. In recent years, many proteins have been found to behave as moonlighting proteins (39, 40).
The role of MiCS in mitotic cellular memory would also suggest several nuclear function proteins might be retained at the appropriate loci in mitotic chromosomes. For example, the replication licensing proteins (Orc2, Orc5, Mcm2 and Mcm5) that we chose for validation of our results, are also known to show chromosome condensation defects indicating that these proteins may have functions other than replication (41, 42). The replication machinery itself is correlated with differential organization of the genome in the interphase nuclei in the form of early and late replication associated domains (RADs), which overlap with actively transcribing and inactive regions, respectively (43). Replicating units are also known to be differentially distributed between euchromatin and heterochromatin. Their presence in both NuMat and MiCS suggests that replication licensing proteins might be the information carriers of RADs from mother to daughter cell during mitosis. On the other hand, several proteins known to have a defined function in mitotic chromosomes are also found in NuMat. For example, cohesins involved in cohesion and segregation of sister chromatids are found in NuMat. Recently, they have been shown to be involved in mediating enhancer promoter interactions and in organizing chromatin loops during replication (44, 45). Interestingly, these proteins have also been implicated in re-establishment of transcription foci after cell division suggesting their role in transmission of transcriptional memory to daughter nuclei (46–48).
One of the known mechanisms by which cellular memory is transmitted from mother to daughter cells during mitosis is by bookmarking of genes in the form of locus specific histone modifications and the recruitment of complexes which maintain open/closed chromatin state thus enabling/repressing transcription (49). Our results provide a glimpse into the distribution/localization of histone modifiers, mediator complex proteins, RISC complex proteins, insulator body proteins and proteins of trxG and PcG proteins in NuMat and MiCS, all of which have plausible roles in cellular memory. We observe that mediator complex proteins that regulate the process of active transcription are present only in NuMat, but trxG proteins that mark the active genes, are present in NuMat as well as MiCS (50, 51). On the other hand, PcG proteins that maintain silenced chromatin are uniquely present in NuMat, indicating that the marks identifying active genes are transmitted through mitosis whereas those identifying repressed genes are probably deposited and maintained during interphase. Our data also suggests that though active transcription complexes are assembled in interphase, some seed components of these complexes persist through mitosis by associating with mitotic chromosomes. These complexes might also be responsible for the low levels of transcription that has recently been shown to occur during mitosis (52).
At the syncytial blastoderm stage in Drosophila, nuclear division is characterized by semi-closed mitosis where the nuclear membrane ruptures only partially near the spindle poles (53–55). The localization of Lamin Dm0 (Figure 6A), where it forms a spindle like structure between the spindle poles but is discontinuous at the spindle poles, also reflects the partially closed mitosis in early embryos. On the other hand, in post syncytial stage embryos (Figure 6B), when mitosis is completely open, Lamin Dm0 localizes to the cell periphery. Interestingly, Lamin Dm0, which is one of the most abundant proteins in NuMat, is also present in MiCS, which is intriguing as nuclear membrane and lamina disintegrate during mitosis. Interaction between chromatin and nuclear lamina is essential for higher order organization of chromatin because lamin associated domains comprise 30–40% of whole genome in mammals (56). Several nuclear pore proteins, in addition to their role as transport channel have been implicated in chromatin organization and gene regulation and are shown to bind at distinct genomic loci that are not nuclear envelope contact sites (57, 58). Retention of lamina components and nuclear pore complex proteins in MiCS hints toward the structural basis for cellular memory during cell division. The relevance of such protein in MiCS needs to be studied further.
Nuclear bodies that lack a delimiting lipid membrane represent yet another aspect of nuclear organization because they are the sites of many important nuclear functions. Most nuclear bodies, just like the rest of interphase nucleus, disassemble during mitosis but are efficiently and rapidly reassembled after mitosis from components present in the surrounding nucleoplasm (59). We observed that protein components responsible for the formation of several nuclear bodies were present in NuMat as well as MiCS indicating that the information for facilitating the formation of these nuclear bodies is carried over from the mother to daughter nuclei in association with MiCS. For example, Drosophila histone locus bodies form when transcription of histone genes commences in interphase by hierarchical recruitment of components, first of which is Mxc (60). Mxc persists as a MiCS protein probably to aid the re-assembly of histone locus body after mitosis. Similarly, coillin, which is considered as the marker protein for Cajal bodies that are the site for snRNP biogenesis, is abundant in both NuMat and MiCS (61). Thus, the presence of key components of nuclear bodies in MiCS supports the model in which nuclear body formation is driven by initial seeding present in the mitotic chromosome followed by self-assembly as cell enters interphase (62). Taken together, based on these observations, we suggest that mitotic chromosome may represent a miniaturized version of interphase chromosome territory, which retains key protein components as seed elements to re-establish functionally equivalent organization of the nucleus in daughter cells. MiCS constituents, therefore, play a role in mitotic cellular memory and provide a structural basis for the same.
In conclusion, the retention of majority of NuMat proteins in MiCS might be instrumental in transmitting structural and functional information of nuclear architecture to daughter cells. NuMat proteins retained in MiCS may also provide structural basis, to re-establish the nuclear architecture and gene expression pattern specific to the cell type. This study gives a flavor of the rich complexity involved in the multifaceted process of maintaining cellular memory and serves as an useful resource for future candidate-based studies. Functional studies on the constituents of nuclear architecture will be needed to establish the precise molecular basis of cellular memory.
DATA AVAILABILITY
The Proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (20) partner repository with the data set identifier PXD008404; PubMed ID: 29991507; Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD008404. The MS/MS spectra can be viewed at the MS-Viewer website with the search key 5upcevyfxh or by clicking on the following link: http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MSViewer&search_key=5upcevyfxh&search_name=msviewer.
Supplementary Material
Acknowledgments
We thank RKM lab members for helpful discussions. We thank Dr. Nandini Rangaraj and N. R. Chakravarthi for helping us in confocal microscopy. We thank the proteomics facility staff of CCMB for maintaining the facility and helping in proteomics analysis.
Footnotes
* Funding work in RKM lab was supported by the Department of Biotechnology (DBT) and Council for Scientific and Industrial research (CSIR), Govt. of India. RS thanks CSIR for fellowship.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- NuMat
- nuclear matrix
- MiCS
- mitotic chromosome scaffold
- ACN
- acetonitrile
- FBS
- fetal bovine serum
- ODU
- optical density units
- RT
- room temperature
- DIGE
- difference gel electrophoresis
- pI
- isoelectric point.
REFERENCES
- 1. Cremer T., and Cremer C. (2001) Chromosome Territories, Nuclear Architecture and Gene Regulation in Mammalian Cells. Nat. Rev. Genet. 2, 292–301 [DOI] [PubMed] [Google Scholar]
- 2. Schneider R., and Grosschedl R. (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 21, 3027–3043 [DOI] [PubMed] [Google Scholar]
- 3. Misteli T. (2007) Beyond the sequence: Cellular organization of genome function. Cell 128, 787–800 [DOI] [PubMed] [Google Scholar]
- 4. Lanctôt C., Cheutin T., Cremer M., Cavalli G., and Cremer T. (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8, 104–115 [DOI] [PubMed] [Google Scholar]
- 5. Mirkovitch J., and Laemmli U. K. (1984) Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 39, 223–232 [DOI] [PubMed] [Google Scholar]
- 6. Jackson D., Dolle A., Robertson G., and, Cook. P. (1992) The attachments of chromatin loops to the nucleoskeleton. Cell Biol. Int. Rep. 16, 687–696 [DOI] [PubMed] [Google Scholar]
- 7. Prescott D. M., and Bender M. A. (1962) Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260–268 [DOI] [PubMed] [Google Scholar]
- 8. Liang K., Woodfin A. R., Slaughter B. D., Unruh J. R., Box A. C., Rickels R. A., Gao X., Haug J. S., Jaspersen S. L., and Shilatifard A. (2015) Mitotic transcriptional activation: clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol. Cell 60, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Probst A. V., Dunleavy E., and Almouzni G. (2009) Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 [DOI] [PubMed] [Google Scholar]
- 10. Zaidi S. K., Young D. W., Montecino M., Van Wijnen A. J., Stein J. L., Lian J. B., and Stein G. S. (2011) Bookmarking the genome: Maintenance of epigenetic information. J. Biol. Chem. 286, 18355–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadauke S., and Blobel G. A. (2013) Mitotic bookmarking by transcription factors. Epigenetics Chromatin 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerlich D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., and Ellenberg J. (2003) Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112, 751–764 [DOI] [PubMed] [Google Scholar]
- 13. Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., and Joffe B. (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 [DOI] [PubMed] [Google Scholar]
- 14. Lewis C., and Laemmli U. (1982) Higher order metaphase chromosome structure: Evidence for metalloprotein interaction. Cell 29, 171–181 [DOI] [PubMed] [Google Scholar]
- 15. Adolphs K. W., Cheng S. M., Paulson J. R., and Laemmli U. K. (1977) Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc. Natl. Acad. Sci. U.S.A. 74, 4937–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keller J., and Riley D. (1976) Nuclear ghosts: a nonmembranous structural component of mammalian cell nuclei. Science 193, 399–401 [DOI] [PubMed] [Google Scholar]
- 17. Detke S., and Keller J. M. (1982) Comparison of the proteins present in HeLa cell interphase nucleoskeletons and metaphase chromosome scaffolds. J. Biol. Chem. 257, 3905–3911 [PubMed] [Google Scholar]
- 18. Wray W., and Stubblefield E. (1970) A new method for the rapid isolation of chromosomes, mitotic apparatus, or nuclei from mammalian fibroblast at near neutral pH. Exp. Cell Res. 59, 469–478 [DOI] [PubMed] [Google Scholar]
- 19. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Fröhlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Godoy L. M. F., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., and Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 22. Malmström J., Beck M., Schmidt A., Lange V. Deutsch E. W., and Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., and Mann M. (2014) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu G., Wang L. G., Han Y., and He Q. Y. (2012) clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omi. A J. Integr. Biol. 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang D. W., Sherman B. T., and Lempicki R. A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27. Kozlowski L. P. (2017) Proteome- pI : proteome isoelectric point database. Nucleic Acids Res. 45, D1112–D1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwartz R., Ting C. S., and King J. (2001) Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 11, 703–709 [DOI] [PubMed] [Google Scholar]
- 29. Arakawa T., and Timasheff S. N. (1985) Theory of protein solubility. Methods Enzymol. 114, 49. [DOI] [PubMed] [Google Scholar]
- 30. Saurin A. J., Shao Z., Erdjument-Bromage H., Tempst P., and Kingston R. E. (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412, 655–660 [DOI] [PubMed] [Google Scholar]
- 31. Gerner C., Holzmann K., Meissner M., Gotzmann J., Grimm R., and Sauermann G. (1999) Reassembling proteins and chaperones in human nuclear matrix protein fractions. J. Cell. Biochem. 74, 145–151 [PubMed] [Google Scholar]
- 32. Willsie J. K., and Clegg J. S. (2001) Small heat shock protein p26 associates with nuclear lamins and HSP70 in nuclei and nuclear matrix fractions from stressed cells. J. Cell. Biochem. 84, 601–614 [PubMed] [Google Scholar]
- 33. Heins J. N., Suriano J. R., Taniuchi H., and Anfinsen C. B. (1967) Characterization of a nuclease produced by Staphylococcus aureus. J. Biol. Chem. 242, 1016–1020 [PubMed] [Google Scholar]
- 34. Pederson T. (2000) Half a century of 'the nuclear matrix'. Mol. Biol. Cell 11, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pederson T. (1998) Thinking about a nuclear matrix. J. Mol. Biol. 277, 147–159 [DOI] [PubMed] [Google Scholar]
- 36. Miyamoto K., and Gurdon J. B. (2013) Transcriptional regulation and nuclear reprogramming: Roles of nuclear actin and actin-binding proteins. Cell. Mol. Life Sci. 70, 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Lanerolle P., and Serebryannyy L. (2011) Nuclear actin and myosins: Life without filaments. Nat. Cell Biol. 13, 1282–1288 [DOI] [PubMed] [Google Scholar]
- 38. Deng W., Lopez-Camacho C., Tang J. Y. J., Mendoza-Villanueva D., Maya-Mendoza A., Jackson D. A., and Shore P. (2012) Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc. Natl. Acad. Sci. 109, 1524–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeffery C. J. (2003) Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19, 415–417 [DOI] [PubMed] [Google Scholar]
- 40. Huberts D. H., and van der Klei I. J. (2010) Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta - Mol. Cell Res. 1803, 520–525 [DOI] [PubMed] [Google Scholar]
- 41. Pflumm M. F., and Botchan M. R. (2001) Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 128, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 42. Christensen T. W., and Bik K. T. (2003) Drosophila Mcm10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 14, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rivera-Mulia J. C., and Gilbert D. M. (2016) Replication timing and transcriptional control: Beyond cause and effect - part III. Curr. Opin. Cell Biol. 40, 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guillou E., Ibarra A., Coulon V., Casado-Vela J., Rico D., Casal I., Schwob E., Losada A., and Méndez J. (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 24, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo Y., Monahan K., Wu H, Gertz J., Varley K. E., Li W., Myers R. M., Maniatis T., and Wu Q. (2012) CTCF/cohesin-mediated DNA looping is required for protocadherin promoter choice. Proc. Natl. Acad. Sci. 109, 21081–21086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan J., Enge M., Whitington T., Dave K., Liu J., Sur I., Schmierer B., Jolma A., Kivioja T., Taipale M., and Taipale J. (2013) Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813 [DOI] [PubMed] [Google Scholar]
- 47. Hirano T. (2005) Condensins: Organizing and segregating the genome. Curr. Biol. 15, 265–275 [DOI] [PubMed] [Google Scholar]
- 48. Kong X., Stephens J., Ball A. R., Heale J. T., Newkirk D. A., Berns M. W., and Yokomori K. (2011) Condensin I recruitment to base damage-enriched DNA lesions is modulated by PARP1. PLoS ONE 6, e23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao R., Nakamura T., Fu Y., Lazar Z., and Spector D. L. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carlsten J. O. P., Zhu X., and Gustafsson C. M. (2013) The multitalented Mediator complex. Trends Biochem. Sci. 38, 531–537 [DOI] [PubMed] [Google Scholar]
- 51. Schuettengruber B., Martinez A.-M., Iovino N., and Cavalli G. (2011) Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 [DOI] [PubMed] [Google Scholar]
- 52. Palozola K. C., Donahue G., Liu H., Grant G. R., Becker J. S., Cote A., Yu H., Raj A., and Zaret K. S. (2017) Mitotic transcription and waves of gene reactivation during mitotic exit. Science 122, eaal4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arnone J. T., Walters A. D., and Cohen-Fix O. (2013) The dynamic nature of the nuclear envelope. Nucleus 4, 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kiseleva E., Rutherford S., Cotter L. M., Allen T. D., and Goldberg M. W. (2001) Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J. Cell Sci. 114, 3607–3618 [DOI] [PubMed] [Google Scholar]
- 55. Harel a. Zlotkin E., Nainudel-Epszteyn S., Feinstein N., Fisher P. A., and Gruenbaum Y. (1989) Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J. Cell Sci. 94, 463–470 [DOI] [PubMed] [Google Scholar]
- 56. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., and van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 57. Capelson M., Liang Y., Schulte R., Mair W., Wagner U., and Hetzer M. W. (2010) Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Light W. H., and Brickner J. H. (2013) Nuclear pore proteins regulate chromatin structure and transcriptional memory by a conserved mechanism. Nucleus 4, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dundr M., and Misteli T. (2010) Biogenesis of Nuclear Bodies. Cold Spring Harb. Perspect. Biol. 2, a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. White A. E., Burch B. D., Yang X. C., Gasdaska P. Y., Dominski Z., Marzluff W. F., and Duronio R. J. (2011) Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193, 677–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hearst S. M., Gilder A. S., Negi S. S., Davis M. D., George E. M., Whittom A. A., Toyota C. G., Husedzinovic A., Gruss O. J., and Hebert M. D. (2009) Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J. Cell Sci. 122, 1872–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mao Y. S., Sunwoo H., Zhang B., and Spector D. L. (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. James A., Cindass R. J., Mayer D., Terhoeve S., Mumphrey C., and DiMario P. (2013) Nucleolar stress in Drosophila melanogaster: RNAi-mediated depletion of Nopp140. Nucleus 4, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tripathi V., Song D. Y., Zong X., Shevtsov S. P., Hearn S., Fu X. D., Dundr M., Prasanth K. V. (2012) SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell 23, 3694–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prasanth K. V., Rajendra T. K., Lal A. K., and Lakhotia S. C. (2000) Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 113 Pt 19, 3485–3497 [DOI] [PubMed] [Google Scholar]
- 66. Biamonti G., and Vourc'h C. (2010) Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Müller J., and Verrijzer P. (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (20) partner repository with the data set identifier PXD008404; PubMed ID: 29991507; Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD008404. The MS/MS spectra can be viewed at the MS-Viewer website with the search key 5upcevyfxh or by clicking on the following link: http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MSViewer&search_key=5upcevyfxh&search_name=msviewer.