Abstract
Cells organize their actions partly through tightly controlled protein-protein interactions—collectively termed the interactome. Here we use crosslinking mass spectrometry (XL-MS) to chart the protein-protein interactions in intact human nuclei. Overall, we identified ∼8,700 crosslinks, of which 2/3 represent links connecting distinct proteins. From these data, we gain insights on interactions involving histone proteins. We observed that core histones on the nucleosomes expose well-defined interaction hot spots. For several nucleosome-interacting proteins, such as USF3 and Ran GTPase, the data allowed us to build low-resolution models of their binding mode to the nucleosome. For HMGN2, the data guided the construction of a refined model of the interaction with the nucleosome, based on complementary NMR, XL-MS, and modeling. Excitingly, the analysis of crosslinks carrying posttranslational modifications allowed us to extract how specific modifications influence nucleosome interactions. Overall, our data depository will support future structural and functional analysis of cell nuclei, including the nucleoprotein assemblies they harbor.
Keywords: Protein Cross-linking, Protein-Protein Interactions, Histones, Chromatin function or biology, Mass Spectrometry, Histone interaction network – PTM interplay, Histone variant specific interactions, Nuclear interactome
Many biological processes in cells involve tightly regulated noncovalent interactions between proteins. Highly stable interactions functionalize molecular machines, such as the ribosome or proteasome, and provide more complex and efficient behavior than the sum of the individual parts (1, 2). Protein kinase A on the other hand, which interacts with its different substrates through signal transduction cascades, represents a prime example of more transient protein-protein interaction. Both stable and transient interactions can be regulated by additional co-factors and/or posttranslational modifications (PTMs)1, providing an additional level of complexity to the cellular interactome. The sheer number of proteins involved, and the complexity introduced by PTMs, makes interactome studies challenging. Nevertheless, these studies are important as the fundamental understanding of the principles that regulate protein-protein interactions (PPIs) can open the way to elucidate biochemical mechanisms that govern basic cellular functions. Different approaches have been developed to chart the cellular protein interaction network. Notably, affinity purification of tagged proteins coupled with mass spectrometry (AP-MS) for the identification of the bait protein and its interactors has successfully been used to dissect the composition of various protein assemblies present in cellular lysates (3–5). This method allows sampling the interaction profile of soluble protein complexes while preserving the PTMs that could be required to maintain the complex integrity. Recent approaches rely on the proximity-dependent labeling of proteins localized in the vicinity of a specific bait protein fused to enzymes able to generate a reactive biotin protein label (6–8). The biotinylation allows for efficient isolation and identification by mass spectrometry of the potential bait interactors. While these methods can be applied to insoluble proteins, they still rely on the engineering and exogenous expression of the fusion protein that may not entirely preserve the function and/or the endogenous interaction profile of the bait protein.
An alternative approach to map PPIs, which in recent years has seen more traction, is crosslinking mass spectrometry (9–12). For this methodology homo bifunctional chemicals are mostly used that integrate two amine reactive groups (e.g. NHS-esters) separated by a spacer arm of a specific length. The spacer arm together with the flexibility of the amino acids captured by the amine reactive groups imposes a distance constraint between those amino acids, providing useful information about protein structure and the model of protein complexes. As such, chemical crosslinking coupled with mass spectrometry (XL-MS) has been used extensively to gain structural insights on reconstituted protein complexes. However, recently, the methodology has expanded its reach to dissect PPIs at a broader proteome-wide level due to advances in crosslinking reagents, mass spectrometry, and data analysis. To illustrate, technological advances in chemistry, for instance, with the introduction of MS cleavable and/or affinity-tagged crosslinkers (13–16), dedicated search algorithms (17–21), and chromatography (22, 23) have led to increased efficiency in the identification of crosslinked peptides from complex mixtures. Fueled by these advances, XL-MS has recently been used to study the architecture and composition of very large and complex purified or reconstituted protein complexes (24–30) as well as to characterize PPIs at the proteome level (17–19, 31–35). Despite these recent accomplishments, proteome-wide XL-MS experiments are still challenging and have so far reached the most abundant proteins and protein complexes. The high complexity of the samples in combination with the relatively low abundance of crosslinked peptides makes the identification of these latter peptides difficult (9).
Here we set out to analyze proteome-wide PPIs in the human cellular nucleus by XL-MS, aiming to enhance the crosslinking efficiency while preserving the cell nuclear environment. Our strategy started with the treatment of isolated nuclei with the MS-cleavable crosslinker disuccinimidyl sulfoxide (DSSO), followed by several sample fractionation steps both at the protein and at the peptide level and analyzed by mass spectrometry. Using this approach, we identified overall 8,710 unique crosslinks with a false discovery rate (FDR) of 1%. Between two independent biological replicates we find ∼58% overlap. To illustrate the potential and novelty of our resource, we zoom in to various parts, revealing among others structural information on endogenous PPIs, including on the binding mode of many known and novel nucleosome-interacting proteins. Moreover, our approach allowed us to dissect PPIs of histone H1. Finally, the identification of crosslinks between different proteins carrying PTMs sets the basis to investigate PTM-dependent PPIs in the cell nucleus to shed light on the regulation of nuclear activities.
EXPERIMENTAL PROCEDURES
Cell Culture and Intact Nuclei Isolation
U2OS cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (Lonza) and 1% penicillin/streptomycin (Lonza). For both the biological independent unfractionated and fractionated nuclear preparations, the cells collected at different times were washed twice with ice cold PBS, scraped in PBS, and counted, then resuspended at a concentration of 2 million cells/ml in the following buffer: 25 mm HEPES, pH 7.5; 50 mm NaCl; 5 mm KCl; 10 mm iodoacetamide; protease inhibitors (Roche, Basel, Switzerland); and 10 μm MG132. The cells were incubated for 20 min on ice. Then digitonin was added at a final concentration of 40 μg/ml, and the suspension passed 80 times through a dounce tissue grinder (pestle B) (Sigma) and subsequently centrifuged 10 min at 400 g. The pelleted nuclei were washed once in resuspension buffer and then crosslinked.
Recombinant Protein Expression, Native Gel and Western Blotting
HeLa mononucleosomes were purchased from Epicypher (Durham, NC) (#16-0002). Recombinant RCC1 and Ran-E70A were purchased from Abcam (Cambridge, United Kingdom) (#ab90625 and #ab90203).
For the native electrophoresis analysis of the nucleosome-Ran-RCC1 complex, 1.5 μg of mono-nucleosomes were incubated for 15 min at room temperature with 4 molar excess of recombinant Ran and/or RCC1 and then mixed 1:1 in Native Gel sample buffer. Each sample was loaded and run onto a Criterion TGX 4–15% gel (Bio-Rad) in the absence of SDS. For Western blotting, each sample was loaded and run onto a Criterion XT 4–12% gel (Bio-Rad). The proteins were then transferred onto nitrocellulose, the membrane blocked for 1 h with 5% BSA in TBS supplemented with 0.1% Tween 20, and the antibody staining performed according to the manufacturer instructions. The following primary antibodies were used in this study: anti- heterogeneous nuclear ribonucleoprotein K (hn-RNPK) (3C2) (Abcam), anti-Histone H3 (Cell Signaling Technology, Danvers, MA), anti-Lamin A/C (Clone 14) (BD biosciences, Franklin Lakes, NJ), anti-Calnexin (C5C9) (Cell Signaling Technology), and anti-Ubiquitin (clone FK2) (Enzo Life Sciences, Zandhoven, Belgium).
Crosslinking and Detergent Fractionation
The nuclei samples were crosslinked in resuspension buffer with DSSO obtained from Thermo Fisher Scientific (Waltham, MA). The crosslinker solution was prepared fresh in DMSO to obtain a stock concentration of 10 mm and applied to the resuspended nuclei (4 million nuclei/ml) at a final concentration of 250 μm unless otherwise stated. The estimated amount of protein crosslinked each time was 12 mg. The nuclei were crosslinked for 10 min at room temperature and the reaction stopped by adding 1 m Tris, pH 8, to a final concentration of 50 mm; then the nuclei were pelleted. For the unfractionated nuclei, the total nuclear fraction was obtained, collecting the supernatant after the nuclei were lysed in 50 mm Tris, pH 8, 2% SDS, then heated at 95 °C for 3 min, sonicated, and centrifuged for 30 min at 20,000 g. For the fractionated nuclei, the Triton X-100 nuclear soluble and insoluble fractions were obtained by resuspending the crosslinked nuclei first in 50 mm Tris, pH 8; 150 mm NaCl; 5 mm EDTA; 50 mm iodoacetamide; 1% TX100; protease inhibitors (Roche); phospho-stop (Roche); and 10 μm MG132. The solution was incubated on ice 15 min and centrifuged for 30 min at 20,000 g. The supernatant was used as TX100 soluble fraction, while the pellet was resuspended in 50 mm Tris, pH 8, 2% SDS; heated at 95 °C for 3 min; sonicated; and centrifuged for 30 min at 20,000 g. The supernatant represented the TX100 insoluble fraction. All the protein samples were quantified using the BCA protein Assay Kit (Thermo Fisher Scientific). All the in vitro crosslinking experiments were performed three times with 2 mm final concentration of DSSO for 15 min at room temperature.
Proteolytic Digestion
To remove the detergents, all the protein samples were subjected to acetone precipitation. Then the pellets were resuspended in 50 mm AMBIC, 8 m urea, reduced by addition of DTT at a final concentration of 15 mm for 1 h at room temperature, and alkylated for 2 h at room temperature in the dark by addition of iodoacetamide at a final concentration of 50 mm. Alkylation was then stopped by addition of thiourea at a final concentration of 150 mm. The samples were digested in two rounds. In the first round, the samples were digested with Lys-C at an enzyme-to-protein ratio of 1:50 (w/w) at 37 °C for 4 h. In the final round, the samples were diluted four times in 50 mm AmBic and further digested with trypsin at an enzyme-to-protein ratio of 1:100 (w/w) at 37 °C for 16 h. The digested samples were desalted using Sep-Pak C18 cartridges (Waters, Milford, MA), dried, and stored at −80 °C for further use.
The samples deriving from the in vitro crosslinking reaction were digested following the same urea workflow, but desalted with the Oasis HLB 96-well μElution Plate (Waters) before MS analysis.
Strong Cation Exchange (SCX) Chromatography
The SCX separation was performed as previously described (19, 22). Briefly, the desalted digests were resuspended in 10% formic acid and loaded onto a Zorbax BioSCX-Series II column (0.8-mm inner diameter, 50-mm length, 3.5 μm). SCX solvent A consisted of 0.05% formic acid in 20% acetonitrile, and solvent B consisted of 0.05% formic acid, 0.5 m NaCl in 20% acetonitrile. The SCX gradient was as follows: 0–0.01 min (0–2% B); 0.01–8.01 min (2–3% B); 8.01–14.01 min (3–8% B); 14.01–28 min (8–20% B); 28–38 min (20–40% B); 38–48 min (40–90% B); 48–54 min (90% B); and 54–60 min (0% B). We collected 50 fractions for each sample that were desalted with an Oasis HLB 96-well μElution Plate (Waters), dried, and stored at −20 °C before further use.
Mass Spectrometry
The late desalted SCX fractions from the crosslinked nuclear samples and the digested in vitro crosslinked complexes were analyzed by LC-MS/MS using an Agilent 1290 Infinity System (Agilent Technologies, Santa Clara, CA) in combination with an Orbitrap Fusion (Thermo Fisher Scientific). Reverse phase chromatography was carried out using a 100-μm inner diameter 2-cm trap column (packed in-house with ReproSil-Pur C18-AQ, 3 μm) coupled to a 75-μm inner diameter 50-cm analytical column (packed in-house with Poroshell 120 EC-C18, 2.7 μm) (Agilent Technologies). Mobile-phase solvent A consisted of 0.1% formic acid in water, and mobile-phase solvent B consisted of 0.1% formic acid in 80% acetonitrile. A 120-min gradient was used and start and end percentage buffer B adjusted accordingly for each SCX fraction to maximize the samples separation. For the MS/MS experiment, we selected the ten most abundant precursors and subjected them to sequential CID-MS/MS and ETD-MS/MS acquisitions. All spectral data were acquired in the Orbitrap mass analyzer. For the MS scans, the scan range was set to 300–1,500 m/z at a resolution of 60,000, and the automatic gain control target was set to 1e6. For the MS/MS scans, the resolution was set to 30,000, the automatic gain control target was set to 1e5, the precursor isolation width was 1.6 Da, and the maximum injection time was set to 120 ms. The CID normalized collision energy was 30%; the charge-dependent ETD reaction time was enabled; and the ETD automatic gain control target was set to 1e5.
For bottom-up analysis of the noncrosslinked nuclear samples or the in vitro reconstituted protein complexes, the isolated nuclei or recombinant proteins were processed as described above and injected for a single-shot LC-MS/MS analysis using the same LC-MS setup.
Data Analysis
Proteome Discoverer 2.2 (beta, version 2.2.0.196) was used for data analysis with the XlinkX (beta, version 0.1.3) nodes integrated (36). The processing workflow was set up with the following nodes. The built-in nodes 'Spectrum Files' and 'Spectrum Selector' were used to extract the MS2 scans, together with a precise precursor m/z and charge. To extract precursor intensity information, we added the built-in node “Minora feature detection.” The following crosslinking workflow consists of the following nodes. The “XlinkX Detect” node performs diagnostic peak detection specific for the used labile crosslinker DSSO (13, 36). The following “XlinkX Filter” nodes only filters out all MS2 scans for which no diagnostic peak set was detected. The remaining MS2 scans were identified with the dedicated crosslink peptide search engine “XlinkX Search” node, for which the following settings were used: Uniprot human protein database from January 2016 containing 42,150 proteins, protease trypsin (full), two allowed missed cleavages, precursor mass tolerance of 10 ppm, fragment mass tolerance of 20 ppm, carbamidomethyl on C as static modification, oxidation on M as variable modification, and where appropriate acetylation or ubiquitination on K were also set as variable modification. The results from the search were FDRs corrected to 1% using the “XlinkX Validator” node, which utilizes a specific set of crosslink peptide spectral features and machine learning to define the cutoff as developed for peptide spectral matches in Percolator (37). We used Percolator version 1.3 as provided with the release of Proteome Discoverer 2.2 and included a set of 47 values calculated individually for each fragmentation spectrum as features (described in supplemental Table S6). The features themselves were inspired by the standard set of features used by Percolator but elaborated on for XL-MS data. Alterations for this more complex environment included: splitting of features for the individual peptides (e.g. the standard feature “number matches” is calculated for each peptide individually), inclusion of cleavable crosslinking specific features (e.g. likelihood score for detecting the reporter ions as “reporter score”), inclusion of new features (e.g. “PTM score A,” denoting the accuracy of pinpointing the lysine where the crosslink is detected), and retention of a number of features (e.g. “Delta mass”). This minimal set for XL-MS data was derived from a much larger set of features that was culled by recursive feature elimination (38), giving insight into the collection of features providing the best separation of false and true positives and consequently the best possible means of obtaining results at 1% FDR. The final node is the “Crosslink Consensus” node where individual crosslink spectral matches were grouped in those cases where they represent the same peptide sequence and modification state. The workflow described above was used also for the analysis of the in vitro crosslinked samples using databases generated from the protein sequences obtained from the bottom-up analysis of the recombinant protein samples. The bottom-up proteomics files to calculate the proteins copy numbers were analyzed with MaxQuant (version 1.5.5.0) (39) using the database described above and with Perseus (version 1.5.5.0) using the proteomic ruler plugin (40). The standard searching parameters were used: protease trypsin, two allowed missed cleavages, precursor mass tolerance of 4.5 ppm, fragment mass tolerance of 20 ppm, and carbamidomethyl on C was set as static modification, oxidation on M, acetylation on K, methylation on K and R, and N terminus acetylation were set as variable modifications. FDR was set to 0.01 for PSM FDR, protein FDR, and site decoy fraction.
For 3D protein structure prediction, we utilized I-Tasser (41, 42). The structure of the bHLH domain of USF3 (amino acids 18–69) was generated through I-Tasser providing PDB: 1AN4 as template. To predict the structure hn-RNPK KH2 domain, we uploaded the protein sequence corresponding to amino acids 144 to 209 (Uniprot ID: P61978) and selected the models with the best score. To sample the interaction space of nucleosome-interacting proteins through DisVis (61), we defined the nucleosome (PDB: 1EQZ) as fixed chain and USF3-bHLH (generated through I-Tasser), the hn-RNPK KH2 domain (generated through I-Tasser). As input for the calculation, we provided five interlinks between USF3-bHLH and the nucleosome and two interlinks between the hn-RNPK KH2 and the nucleosome. The density maps obtained represent the position of the scanning chain that satisfies the highest number or restraints provided. The models of the HMGN2-nucleosome complex were constructed by docking an ensemble of 1,000 HMGN2 conformations (either the extended nucleosomal-binding domain (NBD), residues 9–51, or full-length) to the nucleosome (PDB: 2PYO) using HADDOCK (43). The HMGN2 conformations were randomly selected from the 50% most expanded structures from a pool of 10,000 conformations, generated according to a statistical coil model using FlexibleMeccano (44). The nucleosome structure was modified in case of full-length HMGN2 by extending the DNA by one turn on each end and adding the missing histone tails according to their conformation in PDB 1KX5. Docking was guided by: (i) ambiguous interaction restraints between HMGN2 R22, S24, R26, and L27 and the acidic patch, based on NMR and mutagenesis data; (ii) ambiguous interaction restraints between HMGN2 K35, K39, K41, and K42 and the DNA close to the H2A C terminus, based on NMR and mutagenesis data; (iii) unambiguous interaction restraints according to the XL-MS data (11 for the extended NBD and 26 for full-length HMGN2). For the full-length model, crosslinks to H2A K5, H2B K3/K7, and H3 K4 (the most N-terminal positions) as well as crosslinks between HMGN2 K81 and H2B K11/K117 (incompatible with the majority of K81 crosslinks) were excluded. In the rigid body stage of HADDOCK, 10,000 solutions were calculated, of which 1,000 out of the best 2,000 structures, according to the HADDOCK score, were further refined by semi-flexible annealing and subsequent refinement in explicit solvent. The final best 200 solutions, according to HADDOCK score, were used for analysis and display. All the crosslinking maps were generated with xiNET (45). All the interaction networks were generated with Cytoscape (46).
Experimental Design and Statistical Rationale
The XL-MS analysis was performed on two independent biological replicates (the detergent-fractionated and the unfractionated nuclear sample). The two nuclei preparations were obtained at different times. The following SCX desalted fractions from the two biological replicates were analyzed with the described XL-MS workflow:
- Detergent fractionated nuclear sample (Insol and Sol): fractions Insol- 17 to 38 and reinjected Insolbis/or Th- 17 to 38 from the TX100 insoluble material and fractions Sol- 17 to 37 and reinjected Solbis- 17 to 37 from the TX100 soluble material.
- Unfractionated nuclear sample (Tot): fractions Tot- 25 to 38 and reinjected Totbis- 17 to 38.
Crosslinks identified in both the unfractionated and fractionated nuclear datasets and crosslinks-defining interactions observed in both the unfractionated and fractionated nuclear dataset were used for the analysis unless otherwise stated. Novel PPIs defined by crosslinks observed in one nuclear dataset were not considered in the analysis.
Each in vitro complex reconstitution and in vitro crosslinking experiment was performed three times. All the blots and native gel scans displayed in the study are representative of three independent experiments.
RESULTS
A Crosslinking Strategy Preserving Endogenous Nuclear Protein-Protein Interactions
We hypothesized that the large transporters, such as the nuclear pore complexes, embedded in the nuclear envelope could expedite diffusion of the crosslinking reagent DSSO into the nucleus, enabling crosslinking of the nuclear proteins in their natural environment (Fig. 1A). To test whether DSSO indeed diffuses into the nucleus, we isolated intact nuclei from human U2OS cells by mechanical rupture, coupled with soft centrifugation (supplemental Fig. S1) and treated them with the crosslinking reagent. The strongly shifted band on the SDS-Page gel toward the high molecular weight region demonstrates that this is indeed the case (supplemental Fig. S2A). Importantly, DSSO was effective in crosslinking proteins inside the intact nuclei already at low concentrations (supplemental Figs. S2B and S3A), highlighting the efficiency of the diffusion and reaction. By using our earlier introduced XlinkX-based workflow (36), in the LC-MS/MS data acquired from these samples, 3,936 unique crosslinks could be identified at an FDR of 1% (supplemental Fig. S2C; supplemental Table S1). More than 2,300 of these crosslinks connected peptides originating from distinct proteins (interprotein crosslinks or interlinks), while the remaining crosslinks mapped different regions of the same protein or protein oligomers (intraprotein crosslinks or intralinks). The higher ratio of interlinks over intralinks detected compared with previous studies reflects the way the crosslinking reaction is carried out. The intact nuclei display organized structures like chromatin and high internal protein concentration, thus the crosslinking reagent can more effectively react with the side chains of lysines from distinct polypeptide chains. However, ∼40% of the detected proteins were not clearly assigned as nuclear (supplemental Fig. S2D). This is expected given the simple strategy adopted to isolate the nuclei, which leaves the endoplasmic reticulum, and its associated ribosomes, largely connected to the nucleus and could result in the co-isolation of other cellular organelles like mitochondria. To further increase the depth of our XL-MS analysis and to obtain a better separation between the nuclear chromatin fraction from the nuclear soluble and other subcellular fractions, we next performed detergent fractionation of the crosslinked nuclei. After the DSSO reaction, we lysed the nuclei in a buffer containing 1% Triton X-100 to release the nucleoplasma and to solubilize the proteins associated with the endoplasmic reticulum and the nuclear membrane. The Triton X-100 insoluble fraction containing chromatin was further solubilized in a buffer containing sodium dodecyl sulfate (SDS) to maximize protein extraction (Fig. 1B). As expected, histone proteins were largely detected in the Triton X-100 insoluble fraction (supplemental Fig. S3A). From the acquired mass spectrometry data of both the soluble as well as the insoluble fractions, we identified 7,095 unique crosslinks at an FDR of 1% (Fig. 1C; supplemental Table S2). Of these, 4,606 connected peptides originated from distinct proteins. As anticipated, the fraction of identifiable nuclear crosslinks increased to 85% in the insoluble fraction. To uncover the depth our crosslinking methodology is reaching, we calculated the protein copy numbers from the nuclear proteome from the combined fractions using the proteomic ruler approach (40) and transferred the copy numbers from the second set to the detected crosslinks. As expected, the crosslinked dataset does not completely reach the full depth of the nuclear proteome but, excitingly, is delving already close to halfway in the dynamic range of the proteins copy number down to ∼1.5e4 proteins per cell (supplemental Fig. S3B). At this level, we are starting to observe crosslinks between components of the spliceosome (estimated copy numbers 6e5–2e6, seven crosslinks), the Ku heterodimer (estimated copy numbers ∼ 2.5e5–5e5, nine crosslinks), the NPC (estimated copy numbers ∼ 9e3–1.6e5; 22 crosslinks), down to the condensins complex (estimated copy numbers ∼ 1.2e4–1.8e4; eight crosslinks) (supplemental Fig. S4). We next evaluated the overlap between the fractionated and unfractionated nuclear samples. Both samples derive from two independent nuclear preparations but differ in terms of the analytical workflow only in the application of the detergent fractionation. As mentioned, the detergent fractionation achieves a far greater depth of analysis; however, the two datasets also provide insight into the variability of the crosslinking data in terms of uncovering PPIs. The unfractionated dataset consists of 3,936 crosslinks, of which a remarkable 58% (or >2,300) are also in the fractionated dataset (Fig. 1D). A subset of crosslinks identified in only one of the two samples was still defining PPIs shared between the two datasets and thus included in the subsequent analysis (supplemental Table S3). The merged crosslinking dataset defines over 850 PPIs, of which 778 have not been previously reported, while the others have been previously annotated in the IntAct (47) or in the CORUM (48) databases (Fig. 1D).
Fig. 1.
Crosslinking mass spectrometry (XL-MS) based strategy to investigate assemblies and interactions of nuclear proteins. (A) The MS-cleavable crosslinker DSSO diffuses inside the cell nucleus, possibly through the nuclear pore complex, facilitating efficient crosslinking of the nuclear proteins. (B) U2OS nuclei were isolated and crosslinked. After detergent fractionation, the proteome is digested and the crosslinked peptides are enriched by using strong cation exchange chromatography. The crosslinked peptides are analyzed by LC MS/MS, and by making use of the cleavable crosslinker efficiently identified using the XlinkX PD nodes. (C) Pie chart indicating the total number of unique crosslinks at an FDR of 1%. Unique intralinks are unique crosslinks between peptides derived from the same protein/gene. Unique interlinks connect peptides from distinguishable proteins/genes. The bar charts indicate the nuclear crosslinks identified in the TX100 insoluble and soluble fraction. (D) The top panel displays a Venn diagram indicating the overlap between unique crosslinks identified in the independent biological replicates of the fractionated and unfractionated nuclei samples. The bar chart below indicates the PPIs reproducibly identified in both these XL-MS datasets. In dark brown are indicated the PPIs annotated in the IntAct (47) and/or in the CORUM (48) databases.
Validation on Available High-Resolution Structures
We validated our results mapping a subset of crosslinks onto available high-resolution structures of nuclear complexes (Fig. 2 and supplemental Fig. S4). As the maturation of the 60S and 40S ribosomal particles occurs in the nucleus and our datasets include crosslinks involving ribosomal proteins and ribosome maturation factors, we evaluated the agreement to the DSSO-imposed distance constraint for these crosslinks. As the data analysis software has no a priori knowledge that this complex will be used for validation and the detected crosslinks span the full dynamic range in terms of precursor intensities, this represents an excellent validation model. For the pre-60S ribosomal particle, a well-resolved cryo-EM structure is available that we here used for validation. Of the 87 crosslinks identified from both datasets, 35 (3 interlinks and 32 intralinks) could be mapped onto the cryo-EM structure of the yeast pre-60S ribosome; PDB: 3JCT (Figs. 2A and 2B). The other 52 crosslinks lacked the homologous sequence on the yeast complex or were within regions of the complex where the structural details are not sufficiently resolved in the available PDB structure. Out of 35 crosslinks, we could map on the structure, 34 satisfied the 28 Å Cα-Cα distance constraint of DSSO. Thus, of all crosslinks mapped on the structure, 97% were within the expected distance constraints, providing validation for our XL-MS approach. We also validated a subset of crosslinks between the components of the histone octamer. From both datasets, we identified 243 unique crosslinks (200 interlinks and 43 intralinks) involving the different histone variants, of which 177 connected regions of the histone tails. The crosslinks that could be included in the analysis involved residues in the folded region of the histone octamer and residues of the histone tails closest to the folded regions. Since the nucleosome contains two copies of each histone protein and this could lead to ambiguities for the crosslinks assignment, we mapped all the possible Lys-Lys linkages and displayed the crosslinks defining the shortest distance between all the possible combinations. From the 21 crosslinks that could be mapped on the histone octamer structure, 76% fall within the set constraint (Figs. 2C and 2D). The crosslinks exceeding the maximal DSSO distance involved residues of the histone tail regions.
Fig. 2.
Validation of the identified crosslinks onto available high-resolution structures. Crosslinks satisfying the DSSO distance constraint (28 Å) were indicated in red (interlinks) and blue (intralinks), while crosslinks exceeding the maximum DSSO-imposed distance constraints are in cyan. (A) Crosslinks mapped onto the cryo-EM structure of the structurally homologue yeast pre-60S ribosome (PDB: 3JCT). In yellow are indicated the ribosomal proteins and light blue the ribosome maturation factors. (B) Cα-Cα distance distribution for the crosslinks mapped onto the pre-60S complex. A total of 97% crosslinks fall within the set constraint. (C) Crosslinks mapped on the X-ray structure of the homologue chicken histone octamer (PDB: 1EQZ). Histone H2A is indicated in yellow, H2B in pink, H3 in light blue, and H4 in green. The histone tails are indicated in purple. (D) Cα- Cα distance distribution for the crosslinks mapping on the nucleosome. The crosslinks violating the DSSO distance constraint map to the histone tails regions and can also be explained by higher order structures.
To illustrate the potential of our XL-MS data resource, we will zoom in in the next section on interactions involving histone-binding proteins with the aim to gain structural insights on the interaction with the nucleosome.
Interaction Hotspots on the Nucleosome
The PPIs identified involving histones are defined by 1,017 interlinks. To extract detailed insights in the nucleosome PPIs, we generated a Lys-reactivity map to highlight the residues on the core histones that were involved in interprotein crosslinks (Fig. 3A). The resulting heat maps display both the frequency of interlinks normalized for the number of lysine present in each histone, as well as the median intensity of the crosslinked peptides. This provides insight into the region of the protein most accessible to interact and can help guide the identification of specific domains engaged in nucleosome PPIs. We noticed that, as expected, a relevant fraction of the identified interlinks mapped at the flexible and more exposed (49) histone tails. In particular, Lys5 of histone H2B and Lys4, 18, 23, and 27 of histone H3 are the residues on the histone N-terminal tails found to be engaged in the highest number of interprotein crosslinks (Fig. 3A). Importantly, the histone H3 N-terminal tail was extensively crosslinked to the linker histone H1 in agreement with the proximity of these proteins in the context of the chromatin fiber (50). More remarkable, about 25% of the identified interprotein crosslinks occurring between the nucleosome and its binding proteins were located at the α-helical C-terminal region of histone H2B (Figs. 3A and 3B). This H2B C-terminal α-helix is in close proximity to the nucleosome acidic patch (Fig. 3B), a negatively charged region involved in nucleosome-nucleosome interactions (49) and required to establish interactions between the nucleosome and chromatin-binding proteins (49, 51–55). Importantly, many of the proteins found crosslinked to lysines adjacent to this region were previously reported to engage the acidic patch, including the HMGN family proteins and components of chromatin-remodeling complexes CHD4 and BAZ1B. The heat maps based on the crosslinks median precursor intensity values reveal a slightly different lysine reactivity profile on the nucleosome. Of note, Lys85 of histone H2B and Lys8, 12, 16, 20, 31, and 77 of histone H4 are engaged in a few albeit abundant interlinks. This may hint at a specific stronger binding mode on the nucleosome for this subset of proteins, although we cannot exclude that the high abundance for this subset is a consequence of the possibly better ionization efficiency of these peptides. H2B Lys85 is located close to a DNA contact point (Fig. 3B), suggesting that this residue can react only with the crosslinker when it is not in close contact with the DNA (e.g. during histone deposition or after the action of chromatin remodelers that promote histone variant exchange (56, 57)). On histone H4 Lys 8, 12, 16, and 20 are located at the N-terminal tail, and thus exposed. Lys31 and 77 together with Lys79 of histone H3 are located at the lateral surface of the nucleosome (Fig. 3B). Interestingly, we found that both the transcription factors USF3 and YY1 are engaged in interprotein crosslinks with some of these residues. Both proteins were found crosslinked with H4 Lys20, 31, and 77, with H3 Lys79, and with Lys5, 108, and 121 of histone H2B. USF3 belongs to the basic loop-helix-loop (bHLH) superfamily of transcription factors, a group of proteins conserved from yeast to humans and involved in critical developmental processes (58). Members of this family (e.g. c-Myc, Max, MyoD) contain the bHLH domain responsible of the recognition of consensus DNA sequences throughout the genome. While previous studies have shown that members of this family can interact with nucleosomal DNA (59), structural information on how this interaction takes place is lacking. Modeling of the structure of the USF3 bHLH domain shows it resembles the previously described structure of the closely related USF1 bHLH domain (60) (Fig. 3D). We next applied the DisVis tool (61) to map the interaction surface between the bHLH domain of USF3 and the nucleosome providing as restraints the crosslinks that mapped on the structured regions of the nucleosome (Figs. 3E and 3F). The resulting model demonstrates that USF3 interacts with the peripheral surface of the nucleosome (Figs. 3E and 3F), thus establishing contact with the DNA with the H3-H4 tetramer and likely with the histone H2B N-terminal tail.
Fig. 3.
Defining interaction hot spots on the nucleosome. (A) Heat maps indicating the residues in each core histone engaged in interlinks with proteins other than the core histones. The red heat maps displays the frequency of interlinks normalized for the number of lysines present in the corresponding histone protein occurring between the specified lysine and proteins other than the core histones, including histone H1. The blue heat maps are generated calculating the median crosslink precursor intensity for the interlinks occurring between the specified lysine and proteins other than the core histones, including histone H1. The tails are indicated in red, and the α-helices indicated by the blue rods. (B) X-ray structure of the nucleosome (PDB: 1EQZ) highlighting the crosslinking hot spots. The red dots indicate the position of lysines engaged in a high number of interlinks. The purple dots indicate the position of lysines engaged in interlinks with high intensity. Histone H2A is indicated in yellow, H2B in pink, H3 in light blue, and H4 in green. (C) X-ray structure of the nucleosome (PDB: 1EQZ) indicating the region of the nucleosome involved in the establishment of interactions with protein partners. In yellow is highlighted the position of the nucleosome acidic patch, in light blue the region around the dyad, and in light green the lateral surface close to the H4 N-terminal tail. (D) Overlay of the model of the dimeric bHLH domain of USF3 (yellow), with the X-ray structure of the bHLH domain of USF1 (light blue) (PDB: 1AN4). (E) Crosslinking map defining the interaction between USF3 (amino acids 1–700) and the nucleosome. The blue box indicates the bHLH domain. (F) Visualization of the accessible interaction space of the bHLH domain of USF3, with the nucleosome (PDB: 1EQZ). The interaction space is defined by five interprotein crosslinks mapping between the structured regions of the nucleosome and the bHLH domain of USF3.
Nucleosome Interactions Regulated by Posttranslational Modifications
Histone proteins are known to be densely decorated with functionally relevant PTMs. These PTMs can alter the chromatin structure and regulate gene expression (62, 63). When we searched for crosslinks carrying PTMs, one of the most abundant modifications observed was acetylation of histone H3 on Lys23 (H3-K23Ac) (supplemental Table S4). Thus, we analyzed this subset of crosslinks to dissect the chromatin interaction landscape of H3-K23Ac. The involved peptides carrying the acetylation on Lys23 were crosslinked to other peptides through Lys18 of histone H3. Unfortunately, but not surprisingly, we did not identify some of the known readers of histone tail acetylated lysines (Fig. 4A), such as the bromo-domain containing proteins BRD2 and BRD4, as these are low abundant and below the detection limit of our approach (supplemental Fig. S3B). Still, our XL-MS data did reveal new potential interactors contacting the modified histone tail.
Fig. 4.
Characterization of PTM-regulated protein-protein interactions. (A) Overview of the proteins found crosslinked to histone H3 carrying acetylation at Lys23. (B) Crosslinking maps of hn-RNPK representing the interactions identified with and without ubiquitylation of Lys168. The interlinks (in red), the intralink (in blue), and the ubiquitylation site (in orange) are indicated. (C) Western blotting of the TX100 nuclear soluble or insoluble (chromatin) fraction obtained from nuclei purified in the presence of the cysteine alkylator iodoacetamide. The arrow indicates modified hn-RNPK. (D) Western blotting of the chromatin fraction obtained from U2OS nuclei purified with or without the cysteine alkylator iodoacetamide. The arrow indicates the band corresponding to modified hn-RNPK. (E) The left panel displays the model of the KH2 domain of hn-RNPK generated with I-Tasser indicating the ubiquitylated Lys168 (orange). The right panel displays the accessible interaction space of the KH2 domain of hn-RNPK with the nucleosome (PDB: 1EQZ) generated with the DisVis tool.
We next searched our datasets for crosslinks carrying the Gly-Gly remnant of ubiquitin modification on the Lys side chain. From the potential protein-protein interaction identified when the ubiquitin modification was included (supplemental Table S4), we highlight the interactions found for the hn-RNPK, which contains three K-homology domains responsible for the interaction with RNA and single strand DNA (64, 65). This protein was exclusively found to be crosslinked to the nucleosome when the Gly-Gly remnant was included in the search (Fig. 4B, lower panel). Hn-RNPK can act both as transcriptional co-activator or repressor and plays a role in the transcriptional regulation of p53 after DNA damage (66, 67). Remarkably, Lys166 localized on the KH2 domain of hn-RNPK was found engaged on an intralink with Lys34 (Fig. 4B, top panel). However, when we searched the crosslinked peptides allowing the Gly-Gly ubiquitin remnant as variable modification the same residue was found crosslinked with the core histone proteins (Fig. 4B, lower panel). The residue modified by ubiquitin was Lys168, which suggests that preferentially Lys168 ubiquitin-modified hn-RNPK is localized onto the chromatin. Since this interaction is detected in the SDS soluble fraction, it cannot be validated with co-immunoprecipitation as the components cannot be brought into solution while preserving the interaction. Consequently, to validate this finding we analyzed through Western blotting the hn-RNPK levels in the TX100 soluble and insoluble fractions derived from U2OS nuclei and could detect a slower migrating form of hn-RNPK in the chromatin fraction (Fig. 4C). Since the nuclei were prepared in the presence of the cysteine alkylation reagent iodoacetamide to prevent the ubiquitin removal by the deubiquitinating enzymes (supplemental Fig. S5), we next compared the hn-RNPK levels in the chromatin fraction obtained from nuclei isolated with or without iodoacetamide. Distinctively, the hn-RNPK band profile changed in the chromatin fraction prepared without iodoacetamide, suggesting that the slow migrating band detected is ubiquitin-modified hn-RNPK (Fig. 4D). To gain structural insights on the interaction between hn-RNPK and the nucleosome, we generated a model of the KH2 domain and located the lysines mapped with XL-MS (Fig. 4E, left panel). Both lysines appeared solvent accessible, and Lys168 was located on a predicted loop region. The scanning of the nucleosome surface to map the interaction with the KH2 domain of hn-RNPK with the DisVis tool providing two interlinks as restraints revealed that the KH2 domain of hn-RNPK engages the nucleosome in a region close to the acidic patch (Fig. 4E, right panel).
Locating the Binding Regions of Nucleosome-Interacting Proteins
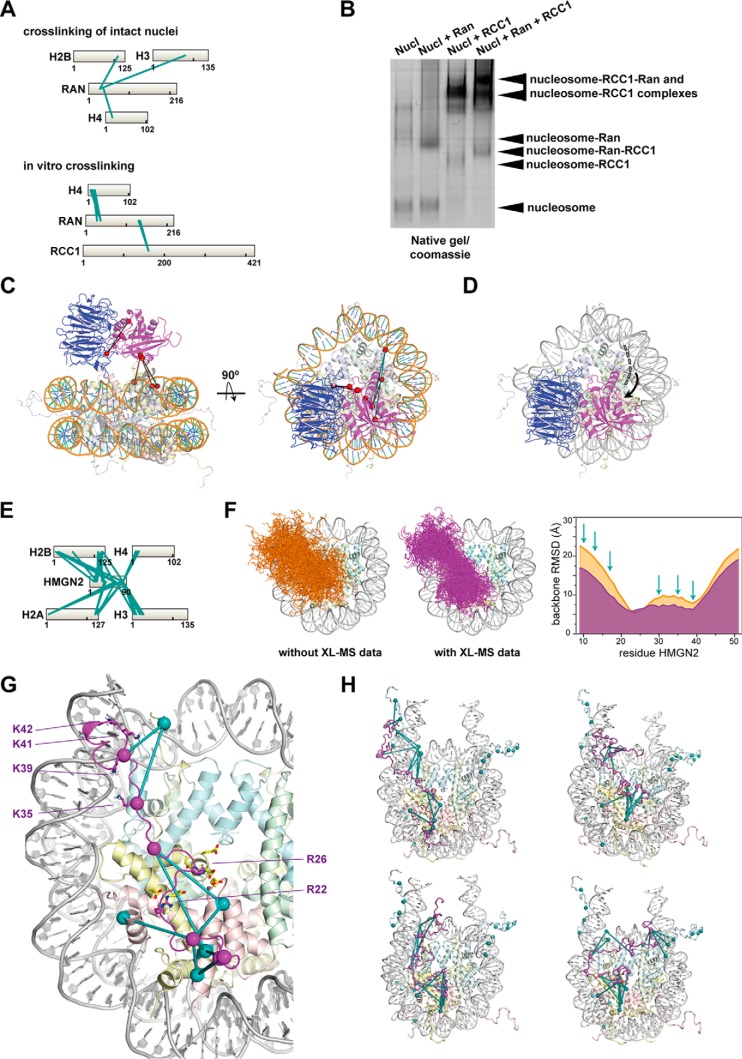
Furthermore, the distance constraints provided by our XL-MS data, yield topological information on how proteins like Ran GTPase and HMGN2 engage with the nucleosome. Ran is a GTPase responsible for nucleocytoplasmic transport that also binds chromatin to regulate spindle formation during mitosis (68). RCC1 is the nucleotide exchange factor of Ran, and both proteins can interact with chromatin in a nonmutually exclusive manner (68). A high-resolution structure of RCC1 interacting with the nucleosome is available, and it allowed proposing a model of the interaction of Ran with the RCC1-nucleosome complex (69). However, this model does not explain how Ran establishes contacts with the nucleosome. Since in our crosslinking experiments on intact nuclei we found Ran crosslinked the nucleosome (Fig. 5A, top), we next reconstituted the interaction to gain structural insights on this complex. The native gel electrophoresis coupled with MS analysis of the recombinant proteins confirmed that both Ran and, to a higher extent RCC1, interact with the nucleosome (Fig. 5B). However, when we performed in vitro crosslinking experiments to map the interaction space, we could detect crosslinks between Ran and the nucleosome only in the presence of RCC1 (Fig. 5A, bottom). The histone H4 N-terminal tail was extensively crosslinked to Ran, and two crosslinks were detected between Ran and RCC1 (Fig. 5A, bottom). Most of the crosslinks satisfied the DSSO-imposed distance constraint when mapped onto a model of the complex while the crosslinks mapping on the histone H4 satisfied the distance constraint only if we would allow the tail to extend toward Ran (Fig. 5C). This finding together with the previous observation that Ran interacts with the H3-H4 tetramer (68) demonstrates that the protein interacts with the lateral surface of the nucleosome, i.e. away from the acidic patch that RCC1 is already occupying. In addition, our result does not exclude the possibility that the N-terminal tail of histone H4 contribute to this interaction establishing contacts with Ran (Fig. 5D).
Fig. 5.
Defining structural models of nucleosome-interacting proteins using XL-MS complemented by other structural biology data and modeling. Binding mode of Ran and HMGN2 to the nucleosome. (A) Crosslinking maps of the identified unique interlinks between Ran and the core histone proteins from intact nuclei (top) and in vitro (bottom) crosslinking experiments. (B) Native electrophoresis analysis of mono-nucleosomes incubated with or without 4 molar excess of recombinant Ran E70A GDP and RCC1. The bands indicated by the arrow were excised and analyzed through bottom-up MS. (C) Crosslinks form panel A mapped onto the model of the Ran-RCC1-nucleosome complex. The model was generated superimposing the structure of the nucleosome (PDB: 1EQZ) and the structure of the Ran-RCC1 complex (PDB: 3GJ0 overlaid over PDB: 1I2M) onto the structure of the RCC1-nucleosome complex (PDB: 3MVD). In cyan are indicated the interlinks that map on the histone H4 N-terminal tail. RCC1 is in blue, and Ran is in magenta. (D) Model of the Ran-RCC1-nucleosome complex. The dotted green line indicates the histone H4 N-terminal tail. RCC1 is in blue, and Ran is in magenta. (E) Crosslinking map of the identified unique interlinks between HMGN2 and the core histone proteins from crosslinking of intact nuclei. (F) Ensembles of 200 models of the extended NBD of HMGN2 bound to the nucleosome without (left) and with (right) XL-MS obtained crosslinks imposed during docking. The heavy atom backbone RMSD over the ensemble of 200 solutions for each HMGN2 residue is plotted on the right. Position of crosslinked lysines indicated by the arrows. (G) Detailed view on the extended NBD on the nucleosome surface, taken from a structure close to the ensemble average. Important side chains for the acidic patch or DNA interaction are shown in sticks, spheres indicate the Cα positions of crosslinked lysines, crosslinks are indicated as teal lines. (H) Four models of the full-length HMGN2 bound to the nucleosome. Crosslinks with violations < 3 Å are indicated as in panel G. Crosslinks from HMGN2 K81 either place the this region close to H3 tail (top left), the proximal linker DNA (top right), the dyad (bottom left), or the distal linker DNA (bottom right).
We also probed the interaction surface between HMGN2 and the nucleosome. This intrinsically disordered protein belongs to the High Mobility Group N (HMGN) protein family and binds the nucleosome through a conserved NBD (70) and regulates nucleosome dynamics and affects chromatin organization (70). The complementation of available NMR and mutagenesis data defining the HMGN2-nucleosome interaction (71) with the crosslinks identified allowed us to obtain a better defined and more comprehensive model of the interaction between an extended NBD of HMGN2 and the nucleosome compared with using NMR and mutagenesis data only (Figs. 5F and 5G). We next attempted to apply the distance constraints provided by the crosslinks to guide the construction of a model of the full-length HMGN2 engaging the nucleosome. In the wide range of HMGN2 conformations, that are equally compatible with the crosslinking data, the C-terminal regulatory domain of HMGN2 is close to the linker DNA and dyad (Fig. 5H). This reinforces and validates the suggestion by Kato et al. that nucleosome binding positions the C-terminal domain correctly for interference with histone H1 (72, 73). In addition, proximity to the H3-tail may explain the modulation of H3 PTMs by HMGN2 (73). The dynamic binding mode of HMGN2 may reflect the ability of the protein to engage the nucleosome in the context of the different chromatin states co-existing in the cell nucleus. Importantly, the above examples demonstrated that such hybrid approaches (e.g. NMR complemented by XL-MS) can aid the development of atomistic structural models of interactions.
The Crosslinking Observed Nuclear Interaction Network
The XL-MS dataset included 646 unique intralinks and 1,493 unique interlinks involving nuclear proteins (supplemental Table S5; Fig. S6A). We considered “nuclear” links if the interlinks contained at least one peptide deriving from proteins annotated as nuclear based on the recently described Cell atlas database (74). These interprotein crosslinks defined ∼630 potential PPIs. We constructed a protein interaction network with the unique PPIs identified (supplemental Fig. S6A; Table S5). The defined 715 nodes represent the different proteins containing inter- and intralinks, and the 636 edges correspond to interactions mapped by the interprotein crosslinks. As expected, the major hubs of the network, or highly connected nodes, are represented by the nucleosome and by the linker histone H1 (including the variants H1.1, H1.2, H1.3, H1.4, H1.5), displaying the highest number of interactions identified (supplemental Fig. S6A). Other hubs were formed by the HMGNs, the heterogeneous ribonucleoproteins and NPM1 all known to be quite abundant in the nucleus and involved in many PPIs (supplemental Fig. S6A). Interestingly, a fraction of the interactions displayed in the network occurred across subcellular compartments (supplemental Fig. S6B). Notably, 20% of the interactions were nucleus-cytosol/plasma membrane, 6% nucleus-mitochondria and 4% nucleus-endoplasmic reticulum. This result is in line with the multiple subcellular localization reported for many proteins but could also be partly influenced by contamination with other subcellular compartments during our nuclei isolation.
Digging down into the crosslinking observed nucleosome interactome, we classified the proteins found crosslinked to core histone proteins or the linker histone H1 (Figs. 6A and 6B). As expected, the most highly represented groups were formed by nuclear proteins annotated with the terms “nucleic acid binding” and “transcription factor” (defining more than half of the proteins) (Fig. 6B). We built a protein interaction network with the subset of interlinks involving individual histone proteins (Fig. 6A). More than 130 proteins were found crosslinked with the core histone proteins (i.e. the nucleosome hub), albeit that also more than 100 proteins were connected with histone H1 variants (Fig. 6A). The majority of the nucleosome-interacting proteins were found crosslinked to histone H2B (Fig. 6A). Despite the important role of the nucleosome acidic patch in establishing contacts with nucleosome interactors, this unbalance in interlinks can readily be explained by the position of the H2B N terminus in relation to the chromatin fiber (75), providing a higher degree of accessibility for the reactive groups of DSSO. In contrast, the N-terminal tail of histone H4 displays substantially less crosslinks with nonnucleosomal proteins, potentially due to involvement of this tail in internucleosomal interactions with neighboring H2A-H2B dimers to maintain compact chromatin (49, 54). This notion is confirmed by the higher number of interlinks detected between the N-terminal tail of histone H4 and histones H2A and H2B (supplemental Table S3). Histone H3, the core histone with the longest N-terminal tail is found crosslinked to several nonnucleosome proteins (Fig. 6A). As this histone interacts primarily with the linker DNA and histone H1 through its N-terminal tail (54) (supplemental Figs. S7A and S7B), interlinks with other proteins are preferentially detected on the lateral surface of the nucleosome. Notably, a subset of proteins was found crosslinked exclusively to the histone H2A.Z variant and histone H1.0 variant (Fig. 6A). While the presence of shared interactors between the different histone variants cannot be excluded, this finding still demonstrates that XL-MS can aid the identification of histone-variant-specific interactions.
Fig. 6.
Snapshot of the histone interaction network. (A) Interaction network of the proteins found crosslinked to the nucleosome and histone H1 variants. Different color of each edge connecting the nucleosome hub indicates which histone variant is found crosslinked to each protein: H2B (blue), H3 (orange), H2A (purple), H4, (green), more than one histone (red). (B) Bar chart representing the functional analysis of the proteins from panel A crosslinked to the core histones and to histone H1 variants. (C) Venn diagram displaying the limited overlap between histone H2B interactors (other than core histone proteins) identified through XL-MS with the interactors reported in studies applying AP-MS (76) and BioID (77). Clearly all three methods show very limited overlap, whereby the BioID and AP-MS studies were even done on the same cells and by the same group.
Since a comparison between XL-MS and other approaches to map PPIs has not been attempted before, we next focused on histone H2B interactors and evaluated the overlap of the histone interaction profile obtained with XL-MS with AP-MS (76) and BioID (77). Both the AP-MS and the BioID studies employed the expression of exogenous tagged histone H2B in HEK293 cells on top of the endogenous histone background. While a comparable number of interactors was observed between XL-MS and AP-MS, more interactors were reported in the BioID study (Fig. 6C), possibly due to the fact that BioID can identify both direct interactors and the surrounding environment of the bait protein (77). The overlap between each of these three methods is marginal. We were not surprised to observe little overlap between the XL-MS and the AP-MS (Fig. 6C) as the latter is mostly suited for the detection of soluble PPIs and consequently samples a more restricted interaction space (76). The small overlap between XL-MS and BioID/AP-MS can also originate from the different cell lines used for the studies (U2OS for XL-MS and HEK293 for BioID/AP-MS) together with differences in sample preparation, with a SDS solubilization step, and protein precipitation in the XL-MS workflow may have led to such unexpected differences in the interaction profiles. Moreover, the cell-engineering step required for BioID could have introduced a bias on the interactors detected, influencing the fraction of nucleosome free or engaged in high-order chromatin structures present in the cell. Despite these differences, the ability of all these methods to identify different subset of previously reported histone interactors let us conclude that XL-MS is successful in the identification of histone interactors and provides a complementary readout to other technologies in the exploration of the histone interaction space. But more generally, all three methods still require further improvements to come to a much needed higher overlap in charting protein-protein interaction landscapes.
DISCUSSION
XL-MS experiments have so far been mostly applied to characterize the architecture of purified complexes or reconstituted recombinant complexes (24, 28, 29). More proteome-wide XL-MS studies have been challenging due to the inability to obtain a deep sampling of the cellular PPIs. Here we describe a strategy to apply XL-MS to the cell nucleus that maximizes crosslinking efficiency, while preserving the cellular environment. The crosslinking of intact nuclei coupled with sequential detergent fractionation and the crosslinks search with the Proteome Discoverer integrated XlinkX software (19, 36) allowed the overall unambiguous identification of over ∼8,700 crosslinks at an FDR of 1% and down to a reliable depth of ∼1.5e4 copies per cell (defined here as the first quartile of the detected copy numbers). The validation of our XL-MS data was carried out through mapping of a subset of crosslinks onto available high-resolution structures of nuclear complexes and through the validation of the interaction of several nucleosome-interacting proteins. In addition, the XL-MS data revealed an overview of chromatin associated PPIs and for the proteins engaged in a high number of crosslinks also provided structural details of specific protein-protein interactions. Notably, we provide/refine structural models of the interaction of USF3, HMGN2 and Ran to the nucleosome.
The ability to search through XlinkX for crosslinks carrying PTMs allowed the identification of additional crosslinked peptides harboring at least one PTM. While the road to characterize PTM-dependent PPIs is still very challenging due to the transient nature of such interactions, further often hampered by the low stoichiometry of PTMs, through the identification of the interaction between hn-RNPK and the nucleosome potentially regulated by ubiquitin, we demonstrate that XL-MS can be harnessed to investigate how PTMs regulate PPIs.
Despite the potential of XL-MS studies in the characterization of endogenous PPIs, there are still limitations that complicate the application of this technology for proteome-wide interactome studies. The major challenge is represented by the ability to identify and filter out nonspecific interactions and by challenges in validating novel observed PPIs. Performing the analysis on crosslinks defining PPIs observed across biological replicates can help in the identification of genuine specific interactions. However, if crosslinking is performed after cell lysis even reproducible crosslinks could reveal nonspecific interactions due to protein mislocalization occurring upon cell disruption. To take this into account, our approach relied on a gentle lysis protocol to keep the cell nuclei intact while preserving as much as possible the native nuclear environment. This approach aimed to reduce contamination across cellular compartments to decrease the number of nonspecific interactions identified compared with other nuclear lysate preparations. Regarding the validation of XL-MS data, while the mapping of crosslinks onto available resolution structure represent a reliable test to validate crosslinks, the orthogonal validation of novel PPIs especially if detected in fractions that are not soluble in physiological buffer conditions like chromatin remains problematic. These interactions are difficult to validate endogenously with classic biochemical methods like co-immunoprecipitation. A solution around this problem is the attempt to in vitro reconstitute the interaction under study; however, this is labor intensive and can be limited by the ability to recombinantly express the proteins involved in the interaction or by the absence of co-factors or adapter proteins needed to stabilize the interaction. While there is a long way to go to establish a method for the statistical validation of interactions from proteome-wide XL-MS studies, the integration with other quantitative mass spectrometry approaches has been shown to be beneficial (78) if the analysis is focused on specific protein targets obtained with similar experimental setup.
Despite these challenges, our findings demonstrate that XL-MS can contribute to the investigation of endogenous PPIs and at the same time, for the proteins displaying an extensive crosslinking profile, provide complementary insight into their regulation complete with distance constraints allowing for mapping of the subunits into models as has been demonstrated previously in many crosslinking MS publications. The application of more targeted approaches for the identification of interactors of endogenous proteins (i.e. through specific enrichment) will help to overcome the limitations of XL-MS for the investigation of cellular PPIs. Moreover, further increases in data acquisition rates and proteome depth are to be expected as newer generations of MS platforms with higher sequencing rates and novel hybrid fragmentation techniques (e.g. EThcD and UV-PD) (79–81) become more routinely available. As such, our work sets the basis to tackle the next challenges in the XL-MS field: the characterization of transient PPIs and their regulation by the different PTMs.
It has been clearly stated that the future in structural biology is hybrid (82), whereby the structural models are defined by combination of methods, such as NMR, X-ray crystallography, and electron microscopy. Here, we show by using our nuclear XL-MS dataset in conjunction with NMR, X-ray crystallography and electron microscopy data that XL-MS represents a clear valuable pillar to this hybrid future in structural biology.
DATA AVAILABILITY
The raw data, all the associated output and databases used in this study have been deposited to the ProteomeXchange Consortium (83) via the PRIDE partner repository with the dataset identifier PXD007513.
Supplementary Material
Acknowledgments
We thank all Heck-group members for their helpful contributions and, in particular, Fan Liu for her initial development of the XlinkX workflow. We thank Harm Post and Mirjam Damen for the SCX support. This work was supported by the Proteins@Work, a program of the Netherlands Proteomics Centre financed by the Netherlands Organisation for Scientific Research as part of the National Roadmap Large-Scale Research Facilities of the Netherlands (project number 184.032.201). This project also received funding from the European Union's Horizon 2020 research and innovation program (grant agreement MSMed No. 686547).
Footnotes
 This article contains supplemental material Tables S1–S6 and Figs. S1–S7.
This article contains supplemental material Tables S1–S6 and Figs. S1–S7.
Conflict of interest: The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- PTMs
- post-translational modifications
- AP-MS
- affinity purification coupled with mass spectrometry
- bHLH
- basic loop-helix-loop
- BioID
- proximity-dependent biotin identification
- CID
- collision-induced dissociation
- cryo-EM
- cryo electron microscopy
- DSSO
- disuccinimidyl sulfoxide
- ETD
- electron transfer dissociation
- EThcD
- electron-transfer/higher-energy collision dissociation
- FDR
- false discovery rate
- NBD
- nucleosomal-binding domain
- NHS
- N-hydroxysuccinimide
- NMR
- nuclear magnetic resonance
- NPC
- nuclear pore complex
- PPIs
- protein-protein interactions
- PRIDE
- proteomics identifications database
- UV-PD
- ultraviolet photodissociation
- RMSD
- root-mean-square deviation
- XL-MS
- crosslinking mass spectrometry.
REFERENCES
- 1. Alberts B. (1998) The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 2. Robinson C. V., Sali A., and Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982 [DOI] [PubMed] [Google Scholar]
- 3. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., and Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 4. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., Erickson B. K., Wühr M., Chick J., Zhai B., Kolippakkam D., Mintseris J., Obar R. A., Harris T., Artavanis-Tsakonas S., Sowa M. E., De Camilli P., Paulo J. A., Harper J. W., and Gygi S. P. (2015) The BioPlex Network: A systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hein M. Y., Hubner N. C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I. A., Weisswange I., Mansfeld J., Buchholz F., Hyman A. A., and Mann M. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 [DOI] [PubMed] [Google Scholar]
- 6. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roux K. J., Kim D. I., Burke B., and May D. G. (2018) BioID: A screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.1–19.23.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhee H. W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., Carr S. A., and Ting A. Y. (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu F., and Heck A. J. (2015) Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr. Opin. Struct. Biol. 35, 100–108 [DOI] [PubMed] [Google Scholar]
- 10. Rappsilber J. (2011) The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinz A. (2017) Divide and conquer: Cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal. Bioanal. Chem. 409, 33–44 [DOI] [PubMed] [Google Scholar]
- 12. Leitner A., Faini M., Stengel F., and Aebersold R. (2016) Crosslinking and mass spectrometry: An integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 [DOI] [PubMed] [Google Scholar]
- 13. Kao A., Chiu C. L., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., and Huang L. (2011) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang X., and Bruce J. E. (2010) A new cross-linking strategy: Protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol. Biosyst. 6, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaake R. M., Wang X., Burke A., Yu C., Kandur W., Yang Y., Novtisky E. J., Second T., Duan J., Kao A., Guan S., Vellucci D., Rychnovsky S. D., and Huang L. (2014) A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell. Proteomics 13, 3533–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Müller M. Q., Dreiocker F., Ihling C. H., Schäfer M., and Sinz A. (2010) Cleavable cross-linker for protein structure analysis: Reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 [DOI] [PubMed] [Google Scholar]
- 17. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., and Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 18. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., and Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F., Rijkers D. T., Post H., and Heck A. J. (2015) Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Anania V. G., Knott J., Rush J., Lill J. R., Bourne P. E., and Bandeira N. (2014) Combinatorial approach for large-scale identification of linked peptides from tandem mass spectrometry spectra. Mol. Cell. Proteomics 13, 1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benda C., Ebert J., Scheltema R. A., Schiller H. B., Baumgärtner M., Bonneau F., Mann M., and Conti E. (2014) Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell 56, 43–54 [DOI] [PubMed] [Google Scholar]
- 22. Fritzsche R., Ihling C. H., Götze M., and Sinz A. (2012) Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun. Mass Spectrom. 26, 653–658 [DOI] [PubMed] [Google Scholar]
- 23. Leitner A., Reischl R., Walzthoeni T., Herzog F., Bohn S., Förster F., and Aebersold R. (2012) Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmström L., and Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 [DOI] [PubMed] [Google Scholar]
- 25. Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C. B., Lakomek K., Aebersold R., Beckmann R., and Hopfner K. P. (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219 [DOI] [PubMed] [Google Scholar]
- 26. Erzberger J. P., Stengel F., Pellarin R., Zhang S., Schaefer T., Aylett C. H., Cimermančič P., Boehringer D., Sali A., Aebersold R., and Ban N. (2014) Molecular architecture of the 40S·eIF1·eIF3 translation initiation complex. Cell 158, 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E. V., Borchers C. H., Baumeister W., Herzog F., Villa E., and Cramer P. (2015) Architecture of the RNA polymerase II-mediator core initiation complex. Nature 518, 376–380 [DOI] [PubMed] [Google Scholar]
- 28. Bui K. H., von Appen A., DiGuilio A. L., Ori A., Sparks L., Mackmull M. T., Bock T., Hagen W., Andrés-Pons A., Glavy J. S., and Beck M. (2013) Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., and Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi Y., Fernandez-Martinez J., Tjioe E., Pellarin R., Kim S. J., Williams R., Schneidman-Duhovny D., Sali A., Rout M. P., and Chait B. T. (2014) Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol. Cell. Proteomics 13, 2927–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Navare A. T., Chavez J. D., Zheng C., Weisbrod C. R., Eng J. K., Siehnel R., Singh P. K., Manoil C., and Bruce J. E. (2015) Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 23, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schweppe D. K., Chavez J. D., Lee C. F., Caudal A., Kruse S. E., Stuppard R., Marcinek D. J., Shadel G. S., Tian R., and Bruce J. E. (2017) Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, 1732–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., and Bruce J. E. (2009) Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chavez J. D., Weisbrod C. R., Zheng C., Eng J. K., and Bruce J. E. (2013) Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteomics 12, 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larance M., Kirkwood K. J., Tinti M., Brenes Murillo A., Ferguson M. A., and Lamond A. I. (2016) Global membrane protein interactome analysis using in vivo crosslinking and mass spectrometry-based protein correlation profiling. Mol. Cell. Proteomics 15, 2476–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu F., Lössl P., Scheltema R., Viner R., and Heck A. J. R. (2017) Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 38. Guyon I., Weston J., Barnhill S., and Vapnik V. (2002) Gene selection for cancer classification using support vector machines. Machine Learning 46, 389 10.1023/A:1012487302797 [DOI] [Google Scholar]
- 39. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 40. Wiśniewski J. R., Hein M. Y., Cox J., and Mann M. (2014) A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics 13, 3497–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Zundert G. C. P., Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., and Bonvin A. M. J. J. (2016) The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 [DOI] [PubMed] [Google Scholar]
- 44. Ozenne V., Bauer F., Salmon L., Huang J. R., Jensen M. R., Segard S., Bernadó P., Charavay C., and Blackledge M. (2012) Flexible-Meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 28, 1463–1470 [DOI] [PubMed] [Google Scholar]
- 45. Combe C. W., Fischer L., and Rappsilber J. (2015) xiNET: Cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N. H., Chavali G., Chen C., del-Toro N., Duesbury M., Dumousseau M., Galeota E., Hinz U., Iannuccelli M., Jagannathan S., Jimenez R., Khadake J., Lagreid A., Licata L., Lovering R. C., Meldal B., Melidoni A. N., Milagros M., Peluso D., Perfetto L., Porras P., Raghunath A., Ricard-Blum S., Roechert B., Stutz A., Tognolli M., van Roey K., Cesareni G., and Hermjakob H. (2014) The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., and Mewes H. W. (2010) CORUM: The comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 38, D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 50. Zhou B. R., Jiang J., Feng H., Ghirlando R., Xiao T. S., and Bai Y. (2015) Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell 59, 628–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barbera A. J., Chodaparambil J. V., Kelley-Clarke B., Joukov V., Walter J. C., Luger K., and Kaye K. M. (2006) The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 [DOI] [PubMed] [Google Scholar]
- 52. Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., and Kingston R. E. (2011) Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334, 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McGinty R. K., Henrici R. C., and Tan S. (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R. M., Zhu P., and Li G. (2014) Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 [DOI] [PubMed] [Google Scholar]
- 55. Dann G. P., Liszczak G. P., Bagert J. D., Müller M. M., Nguyen U. T. T., Wojcik F., Brown Z. Z., Bos J., Panchenko T., Pihl R., Pollock S. B., Diehl K. L., Allis C. D., and Muir T. W. (2017) ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clapier C. R., and Cairns B. R. (2009) The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 57. Venkatesh S., and Workman J. L. (2015) Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 [DOI] [PubMed] [Google Scholar]
- 58. Jones S. (2004) An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soufi A., Garcia M. F., Jaroszewicz A., Osman N., Pellegrini M., and Zaret K. S. (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., and Burley S. K. (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Zundert G. C., and Bonvin A. M. (2015) DisVis: Quantifying and visualizing accessible interaction space of distance-restrained biomolecular complexes. Bioinformatics 31, 3222–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jenuwein T., and Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 63. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 64. Matunis M. J., Michael W. M., and Dreyfuss G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tomonaga T., and Levens D. (1995) Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J. Biol. Chem. 270, 4875–4881 [DOI] [PubMed] [Google Scholar]
- 66. Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., Kenzelmann-Broz D., Khalil A. M., Zuk O., Amit I., Rabani M., Attardi L. D., Regev A., Lander E. S., Jacks T., and Rinn J. L. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moumen A., Masterson P., O'Connor M. J., and Jackson S. P. (2005) hnRNP K: An HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 68. Bilbao-Cortés D., Hetzer M., Längst G., Becker P. B., and Mattaj I. W. (2002) Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 12, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 69. Makde R. D., England J. R., Yennawar H. P., and Tan S. (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kugler J. E., Deng T., and Bustin M. (2012) The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochim. Biophys. Acta 1819, 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kato H., van Ingen H., Zhou B. R., Feng H., Bustin M., Kay L. E., and Bai Y. (2011) Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 108, 12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Catez F., Brown D. T., Misteli T., and Bustin M. (2002) Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3, 760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ueda T., Postnikov Y. V., and Bustin M. (2006) Distinct domains in high mobility group N variants modulate specific chromatin modifications. J. Biol. Chem. 281, 10182–10187 [DOI] [PubMed] [Google Scholar]
- 74. Thul P. J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L. M., Bäckström A., Danielsson F., Fagerberg L., Fall J., Gatto L., Gnann C., Hober S., Hjelmare M., Johansson F., Lee S., Lindskog C., Mulder J., Mulvey C. M., Nilsson P., Oksvold P., Rockberg J., Schutten R., Schwenk J. M., Sivertsson Å Sjöstedt E, Skogs M., Stadler C., Sullivan D. P., Tegel H., Winsnes C., Zhang C., Zwahlen M., Mardinoglu A., Pontén F., von Feilitzen K., Lilley K. S., Uhlén M., and Lundberg E. (2017) A subcellular map of the human proteome. Science 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- 75. Arya G., and Schlick T. (2006) Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. U.S.A. 103, 16236–16241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lambert J. P., Tucholska M., Pawson T., and Gingras A. C. (2014) Incorporating DNA shearing in standard affinity purification allows simultaneous identification of both soluble and chromatin-bound interaction partners. J. Proteomics 100, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lambert J. P., Tucholska M., Go C., Knight J. D., and Gingras A. C. (2015) Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics 118, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Solis-Mezarino V., and Herzog F. (2017) compleXView: A server for the interpretation of protein abundance and connectivity information to identify protein complexes. Nucleic Acids Res. 45, W276–W284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fort K. L., Dyachenko A., Potel C. M., Corradini E., Marino F., Barendregt A., Makarov A. A., Scheltema R. A., and Heck A. J. (2016) Implementation of ultraviolet photodissociation on a benchtop Q Exactive Mass Spectrometer and its application to phosphoproteomics. Anal. Chem. 88, 2303–2310 [DOI] [PubMed] [Google Scholar]
- 80. Frese C. K., Altelaar A. F., van den Toorn H., Nolting D., Griep-Raming J., Heck A. J., and Mohammed S. (2012) Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673 [DOI] [PubMed] [Google Scholar]
- 81. Brodbelt J. S. (2016) Ion activation methods for peptides and proteins. Anal. Chem. 88, 30–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Steven A. C., and Baumeister W. (2008) The future is hybrid. J. Struct. Biol. 163, 186–195 [DOI] [PubMed] [Google Scholar]
- 83. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data, all the associated output and databases used in this study have been deposited to the ProteomeXchange Consortium (83) via the PRIDE partner repository with the dataset identifier PXD007513.