Supplemental Digital Content is available in the text.
Keywords: animals, atherosclerosis, choline, gastrointestinal microbiome, mice
Abstract
Objective—
To investigate the effect of gut microbiota and diet on atherogenesis.
Approach and Results—
Here, we investigated the interaction between the gut microbiota and diet on atherosclerosis by feeding germ-free or conventionally raised Apoe−/− mice chow or Western diet alone or supplemented with choline (which is metabolized by the gut microbiota and host enzymes to trimethylamine N-oxide) for 12 weeks. We observed smaller aortic lesions and lower plasma cholesterol levels in conventionally raised mice compared with germ-free mice on a chow diet; these differences were not observed in mice on a Western diet. Choline supplementation increased plasma trimethylamine N-oxide levels in conventionally raised mice but not in germ-free mice. However, this treatment did not affect the size of aortic lesions or plasma cholesterol levels. Gut microbiota composition was analyzed by sequencing of 16S rRNA genes. As expected, the global community structure and relative abundance of many taxa differed between mice fed chow or a Western diet. Choline supplementation had minor effects on the community structure although the relative abundance of some taxa belonging to Clostridiales was altered.
Conclusions—
In conclusion, the impact of the gut microbiota on atherosclerosis is dietary dependent and is associated with plasma cholesterol levels. Furthermore, the microbiota was required for trimethylamine N-oxide production from dietary choline, but this process could not be linked to increased atherosclerosis in this model.
The gut microbiota has been implicated in cardiovascular disease (CVD) and atherogenesis.1–3 Accumulation of cholesterol in the artery wall and immune mechanisms are involved in lesion formation, and atherosclerosis is characterized by chronic inflammation that develops over decades.4 Accordingly, reducing plasma cholesterol levels and inflammation markers by statin or other cholesterol-lowering therapies are associated with a lower risk of CVD.5–7
See accompanying editorial on page 2269
The human gut is inhabited by trillions of bacteria (microbiota). Patients with CVD are associated with an altered microbiota,8 but whether the commensal microbiota is causally linked to atherogenesis remains unresolved. Diet has been described as one of the key factors affecting gut microbiota composition.9,10 Earlier studies in Apoe−/− mice have demonstrated that the effect of microbiota on the development of atherosclerosis is dependent on the macronutrient composition of the diet, showing that the gut microbiota was protective in mice fed a low cholesterol but not in mice fed a high cholesterol diet.11
Recent evidence suggests that bacteria-derived metabolites, such as trimethylamine N-oxide (TMAO), may influence the disease. TMAO is associated with CVD in humans and with atherosclerosis in chow-fed mice.9,12 However, even though the association between circulating TMAO and the risk of cardiovascular events has been demonstrated, reports are inconsistent,13 and the role of TMAO in metabolic disease is not fully elucidated.14
Materials and Methods
Data available on request from the authors.
Mice and Diet
The mice were maintained under standard specific pathogen-free (conventionally raised [CONV-R]) or germ-free (GF) conditions with a 12:12-hour light:dark cycle. Starting at the age of 8 weeks, male and female apolipoprotein E knockout mice (C57BL/6J Apoe−/−) were fed either chow (Envigo TD.130104), chow supplemented with 1.2% choline (Envigo TD.09041), Western diet (D11042101; Research Diets), or Western diet supplemented with 1% choline (D11042102; Research Diets) for 12 weeks before anaesthetization with isoflurane and collection of blood and tissues. All procedures were approved by the Ethics Committee on Animal Care and Use in Gothenburg, Sweden.
Blood was collected through vena cava, centrifuged in EDTA-containing tubes, and plasma was stored in −80°C. The hearts were perfused with phosphate buffered saline, put in OCT Cryomount (Histolab Products AB, Gothenburg, Sweden) and slowly frozen in liquid nitrogen, other tissues were snap-frozen in liquid nitrogen. Tissues were stored in −80°C until further analyses.
Atherosclerosis Assessment and Macrophage Staining
Aortic roots were sectioned in 8-µm sections using a cryostat and stained with Oil Red O and hematoxylin as described previously.15 The lesion and vessel areas were defined manually and calculated by an image program. Atherosclerosis was expressed as the percentage of the vessel area covered by lesion quantified as the mean value of 2 sections per mouse.
Macrophages were stained in aortic root sections with an Alexa Fluor 594 anti-mouse CD68 antibody (Biolegend, San Diego, CA) diluted 1:300, and images were taken in an Axio Imager Z2 microscope (Zeiss). Stained area representing macrophages was quantified in Adobe Photshop CS5 and related to total vessel area, the mean of 2 sections is presented.
Plasma Analyses
TMAO and bile acids were extracted from plasma samples and measured by ultraperformance liquid chromatography–tandem mass spectrometry as described previously.16 Low abundant bile acids, <0.1%, were excluded from further analyses. Plasma cholesterol and lipids were analyzed as described previously.17
Plasma cytokines were analyzed in duplicate according to the manufacturer’s instructions using a V-PLEX Plus Proinflammatory Panel 1 (mouse) kit (Meso Scale Discovery), which measures IFN (interferon)-γ, IL (interleukin)-1β, IL-2, IL-4, IL-5, IL-6, KC/GRO (keratinocyte chemoattractant/growth-regulated oncogene), IL-10, IL-12p70, and TNF-α (tumor necrosis factor-α). Meso Scale Discovery plates were analyzed in a MESO QuickPlex SQ120 using the software Discovery Workbench, version 4.0.12 (Meso Scale Discovery).
Profiling of Cecal Microbiota Composition
Total genomic DNA was isolated from 100-mg cecal content using a repeated bead beating method as described previously.18 Briefly, samples were placed in Lysing Matrix E tubes (MP Biomedicals) and extracted twice in lysis buffer (4% wt/vol SDS; 500 mmol/L NaCl; 50 mmol/L EDTA; 50 mmol/L Tris·HCl; pH 8) with bead beating at 5.5 m/s for 45 s in a FastPrep-24 instrument (MP Biomedicals). After each bead beating cycle, samples were heated at 85°C for 15 minutes and then centrifuged at full speed for 5 minutes at 4°C. Supernatants from the 2 extractions were pooled and purified as described previously.18 Total genomic DNA was eluted in DNA elution buffer (10 mmol/L Tris·Cl; 0.5 mmol/L EDTA; pH 9.0).
The cecal microbiota composition was profiled by sequencing the V4 region of the 16S rRNA gene on an Illumina MiiSeq instrument (llumina RTA v1.17.28; MCS v2.5) with the V2 Illumina kit (2×250 bp paired-end reads), as described previously.19 The V4 region was amplified using 515F and 806R primers designed for dual indexing.20 Two 25-µL polymerase chain reactions were pooled, each containing 1× Five Prime Hot Master Mix (5 PRIME GmbH), 200 nmol/L of each primer, 0.4 mg/mL BSA, 5% DMSO (dimethyl sulfoxide), and 20 ng of genomic DNA. Polymerase chain reaction was performed under the following conditions: initial denaturation for 3 minutes at 94°C, followed by 25 cycles of denaturation for 45 s at 94°C, annealing for 60 s at 52°C and elongation for 90 s at 72°C, and a final elongation step for 10 minutes at 72°C. Duplicates were combined, purified with the NucleoSpin Gel and Polymerase Chain Reaction Clean-up kit (Macherey-Nagel), and quantified using the Quant-iT PicoGreen dsDNA kit (Invitrogen). Purified polymerase chain reaction products were diluted to 5 ng/μL and pooled in equal amounts. The pooled amplicons were purified again using Ampure magnetic purification beads (Agencourt) to remove short amplification products.
Illumina paired-end reads were merged using PEAR (pair-end read merger)21 and quality filtered with FASTX (Phred score ≥20 for 100% of the bases in a sequence). Quality-filtered Illumina reads were analyzed with the software package QIIME22 (version 1.9.0). Sequences were clustered into operational taxonomic units (OTUs) at a 97% identity threshold using an open-reference OTU picking approach with UCLUST23 against the Greengenes reference database24 (13_8 release). All sequences that failed to cluster when tested against the Greengenes database were used as input for picking OTUs de novo. Representative sequences for the OTUs were Greengenes reference sequences or cluster seeds and were taxonomically assigned using the Greengenes taxonomy and the Ribosomal Database Project Classifier.25 Representative OTUs were aligned using PyNAST26 and used to build a phylogenetic tree with FastTree,27 which was used to estimate α- and β-diversity of samples using phylogenetic diversity28 and UniFrac.29 Three-dimensional principal coordinates analysis plots were visualized using Emperor.30 Chimeric sequences were identified with ChimeraSlayer31 and excluded from all downstream analyses. Similarly, sequences that could not be aligned with PyNAST, singletons, and low abundant sequences (relative abundance <0.002%) were also excluded. To correct for differences in sequencing depth, the same number of sequences was randomly subsampled from each sample (14 060 sequences per sample), which excluded 3 samples because of low sequencing depth, and a total of 1 758 521 sequences and 571 OTUs were included in diversity analyses.
Statistical Analysis
Statistical analyses were done using GraphPad Prism 6 software and presented as mean±SEM if not otherwise stated. Two-way ANOVA followed by Tukey post hoc test was performed on log transformed data to examine the influence of 2 variables. Pearson correlation coefficient was calculated to test correlation of TMAO, IL-6, lesion size, taxa, and bile acids. Differences in microbiota composition based on phylogenetic distance were tested with a nonparametric test using Monte Carlo permutations, and results are presented as mean±SD. Statistical significance of sample grouping based on unweighted UniFrac was tested with a multivariate nonparametric analysis of variance (adonis with 999 permutations) in R. Differences in OTU abundance was determined with Mann-Whitney U test with Bonferroni correction. Differences in bile acid proportions were analyzed with Mann-Whitney U test. P<0.05 was considered as statistically significant.
Results
The effect of microbiota on the development of atherosclerosis is dietary dependent
To address how the microbiota and diet interact to affect TMAO levels and atherogenesis, we fed GF and CONV-R male Apoe−/− mice chow or Western diet alone or supplemented with choline for 12 weeks. TMAO levels were barely detectable in GF mice on either diet and were not increased by choline supplementation (Figure 1A), consistent with the fact that the gut microbiota is required for production of the TMAO precursor trimethylamine from choline.32 In contrast, TMAO levels were significantly increased by choline supplementation in CONV-R mice on either diet (Figure 1A). Neither the microbiota nor choline supplementation had any effect on body weight in chow-fed mice. Epididymal fat weight was higher in CONV-R than GF mice but was independent of choline supplementation (Figure I in the online-only Data Supplement). However, choline supplementation reduced body weight and epididymal fat weight in CONV-R but not GF mice when fed Western diet (Figure I in the.
Figure 1.
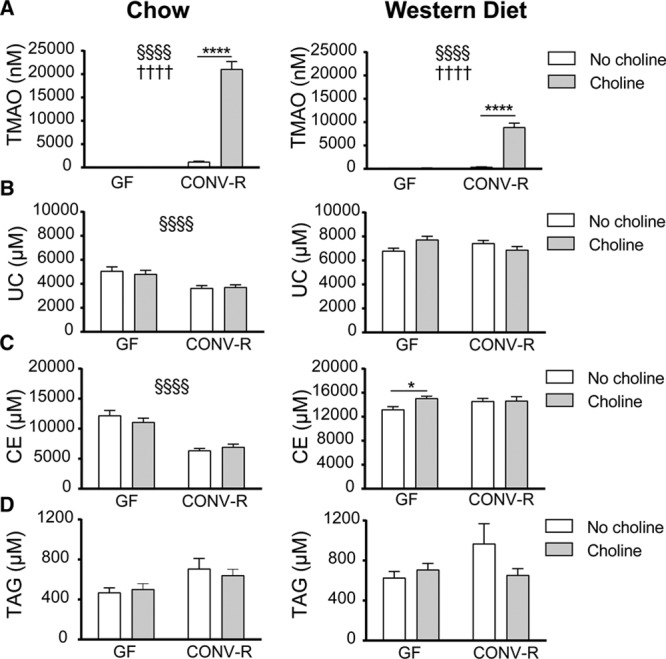
Choline and cholesterol metabolism is regulated by microbiota and diet. Plasma concentrations of trimethylamine N-oxide (TMAO; A), unesterified cholesterol (UC; B), cholesteryl esters (CE; C), and triacylglycerols (TAGs; D) were analyzed in Apoe−/− mice fed chow (n=10–18, left) or Western diet (n=15–20, right) with or without supplementation of choline for 12 wk. Data are presented as mean±SEM. CONV-R indicates conventionally raised; and GF, germ free. Variation induced by the gut microbiota: §P<0.05, §§§§P<0.0001. Variation induced by choline: ††††P<0.0001. Post hoc multiple comparison analysis: *P<0.05, ****P<0.0001.
We also observed lower plasma levels of unesterified cholesterol and cholesteryl esters and a trend toward higher levels of triacylglycerols in chow-fed CONV-R compared with GF Apoe−/− mice; choline supplementation did not have any additional effect (Figure 1B through 1D). In mice fed Western diet, neither the presence of a microbiota nor choline supplementation had any effect on plasma triacylglycerol or cholesterol levels (Figure 1B and 1D). Choline supplementation to a Western diet increased plasma levels of cholesteryl esters in GF but not CONV-R mice (Figure 1C).
Aortic lesions were observed in both GF and CONV-R Apoe−/− mice after 12 weeks on chow (range, 9728–144 033 µm2) or Western diet (range, 51 927–165 848 µm2; Figure 2A; Figure II in the online-only Data Supplement). Aortic lesions were smaller in CONV-R compared with GF mice on chow, whereas there was no difference in lesion size between CONV-R and GF mice on a Western diet (Figure 2B). In contrast to previous findings,9 we did not observe any effect of choline supplementation on lesion size in chow-fed mice (Figure 2B; Figure II in the online-only Data Supplement). In mice on Western diet, choline supplementation did not affect lesion size in GF mice but resulted in a greater absolute aortic lesion size in CONV-R mice (Figure II in the online-only Data Supplement); however, the lesion size normalized to vessel area was similar compared with GF mice and chow diet (Figure 2B). There was no correlation of plasma TMAO concentration with lesion size (Figure 2C). In contract, lesion size correlated with cholesteryl esters in mice fed chow but not in mice fed Western diet (Figure III in the online-only Data Supplement).
Figure 2.
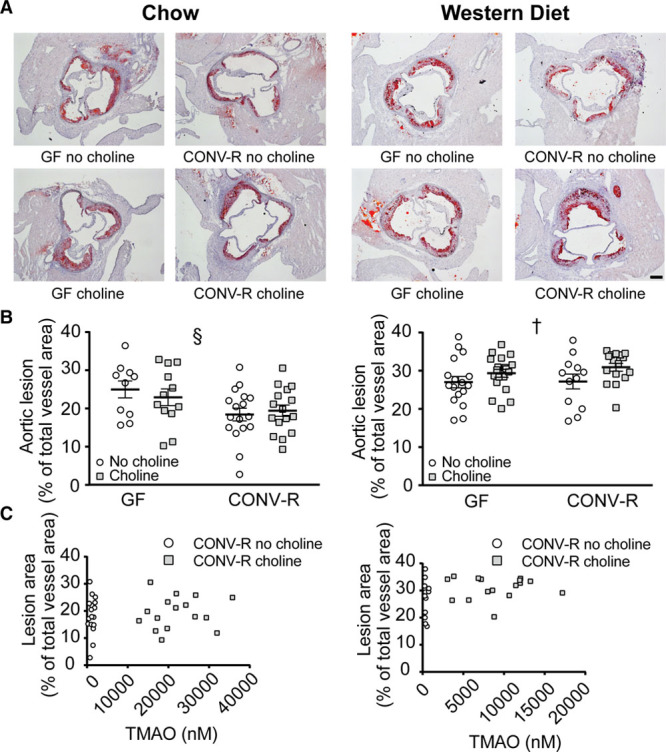
Atherosclerotic lesion size is dependent on microbiota and diet. Germ-free (GF; n=10–19) and conventionally raised (CONV-R; n=12–17) Apoe−/− mice were fed chow (left) or Western diet (right) with or without supplementation of choline for 12 wk. Representative Oil Red O-stained aortic root sections are shown in A (scale bar=200 µm). The size of the aortic lesions normalized to vessel area is shown in B. Trimethylamine N-oxide (TMAO) concentrations in plasma were tested for correlation with aortic lesion area (C). Data are presented as mean±SEM. Variation induced by the gut microbiota: §§P<0.01. Variation induced by choline: †P<0.05.
To investigate how sex affects the impact of gut microbiota and dietary choline on aortic lesions, we fed female GF and CONV-R Apoe−/− mice chow or Western diet alone or supplemented with choline for 12 weeks. Female mice developed similarly sized aortic lesions on both chow (30 923–197 076 µm2) and Western diet (100 630–220 967 µm2; Figure IV in the online-only Data Supplement) as male mice. The effects of microbiota and choline on atherosclerosis were comparable with those in males, and, therefore, further analyses were only done in males.
In summary, lesions were smaller in CONV-R compared with GF Apoe−/− mice on a chow diet but not on a Western diet. Furthermore, the increased plasma TMAO levels observed in CONV-R Apoe−/− mice fed a choline-supplemented diet did not correlate with atherosclerotic lesion size.
Microbial and Dietary Effects on Inflammatory Processes
To assess whether the microbiome modulates atherosclerosis-associated inflammation, we quantified macrophages in the aortic root lesions using antibodies against the glycoprotein CD68, which is expressed in macrophages in inflamed tissues (Figure 3A). The CD68-stained area was similar in GF and CONV-R mice on a chow diet and was not affected by choline supplementation (Figure 3B). In mice on a Western diet, the CD68-stained area was smaller in CONV-R compared with GF mice (P<0.01), and again, choline supplementation had no effect (Figure 3B).
Figure 3.
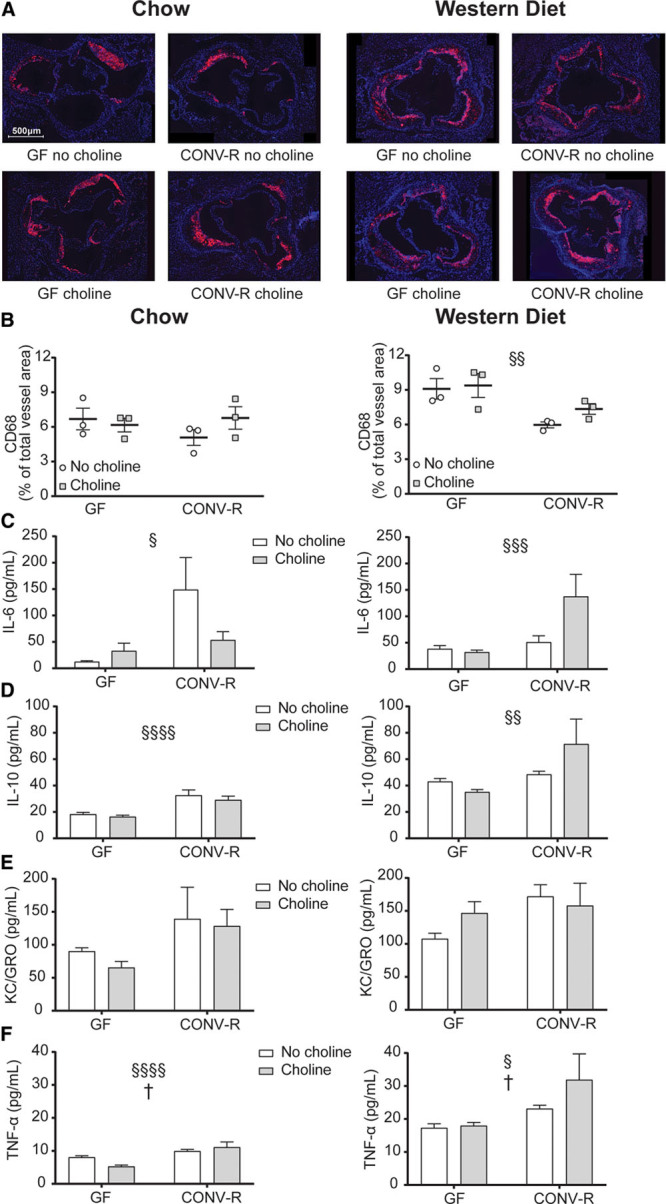
Microbial regulation of inflammation. Representative CD68-stained aortic root sections from Apoe−/− mice fed chow (left) or Western diet (right) with or without supplementation of choline are shown in A (scale bar=500 μm). CD68-stained area quantified and normalized to total vessel area is shown in B (n=3). Plasma cytokine concentrations of IL (interleukin)-6 (C), IL-10 (D), KC/GRO (keratinocyte chemoattractant/growth-regulated oncogene; E), and TNF-α (tumor necrosis factor-α; F) determined by using a multiplex assay from Meso scale (n=11–19). Data are presented as mean±SEM. CONV-R indicates conventionally raised; and GF, germ free. Variation induced by the gut microbiota: §P<0.05, §§P<0.01, §§§§P<0.0001. Post hoc multiple comparison analysis: *P<0.05. Variation induced by choline: †P<0.05.
To further study the inflammation-related response of microbiota and choline, 10 cytokines and chemokines were measured in plasma from Apoe−/− mice using a multiplex assay. IL-6, IL-10, KC/GRO, and TNF-α were higher in CONV-R compared with GF mice fed chow (Figure 3C through 3F, left). Higher concentrations of these cytokines, except KC/GRO, were also observed in Western diet-fed CONV-R compared with GF mice (Figure 3C through 3F, right). Note that several cytokines were only detected in low concentrations, including IFN-γ, IL-1β, IL-2, IL-4, IL-5, and IL-12p70 (Table I in the online-only Data Supplement).
To conclude, the microbiota affected the plasma concentration of several cytokines in both chow- and Western diet-fed mice. CD68-stained area did not associate with aortic lesion size.
Diet Alters the Gut Microbiota Composition
To investigate how diet and choline supplementation affected the microbiota composition, we sequenced and analyzed the 16S rRNA gene from cecal content of CONV-R Apoe−/− mice. Western diet-fed mice had reduced phylogenetic diversity compared with chow-fed mice (P=0.001), but choline supplementation did not affect this parameter (Figure 4A). Principal coordinate analysis of the unweighted UniFrac showed that microbiota clustered according to chow or Western diet: R2=0.46 (P<0.001; Figure 4B). Supplementation with choline had a much smaller effect on the microbial community compared with switching from chow to Western diet: chow, R2=0.07 (P=0.003); Western diet, R2=0.11 (P=0.001; Figure 4B). We identified OTUs from 8 different phyla; the relative abundance of these phyla (with the exception of TM7) differed in mice on a Western diet compared with those on a chow diet (P<0.01; Figure 4C). Choline supplementation increased the relative abundance of Actinobacteria in chow-fed mice (no choline, 0.34%; choline, 1.6%; P<0.01) but not in Western diet-fed mice (no choline, 0.18%; choline, 0.25%; nonsignificant) and did not alter the relative abundance of any other phyla.
Figure 4.
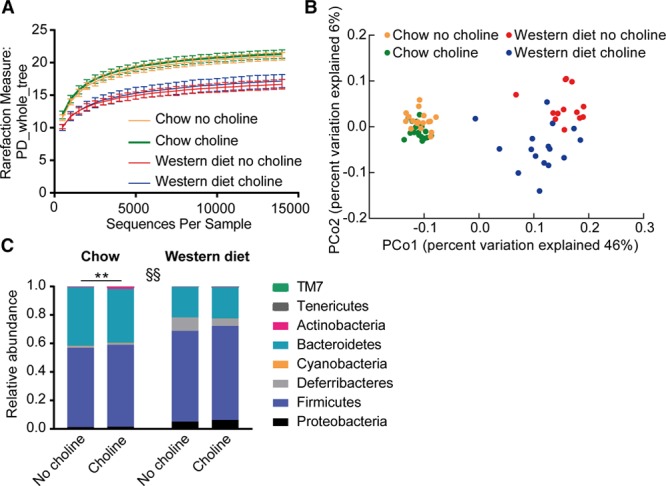
Global community structure of microbiota in cecal content from chow and Western diet-fed mice. Microbial composition was determined in conventionally raised Apoe−/− mice fed chow or Western diet with or without choline for 12 wk (n=13–18) by sequencing 16S rRNA genes. Alpha diversity measurements with rarefaction curves using phylogenetic distance as rarefaction measure showing mean±SD in mice fed chow: no choline (orange) and choline (green); or Western diet: no choline (red) and choline (blue; A). Principal coordinate analysis plot with the 2 major components determining the microbial gene profile based on unweighted UniFrac (B). Each dot represents a cecal content sample from 1 mouse fed chow with no choline (orange) or choline (green) and Western diet with no choline (red) or choline (blue). Mean relative abundance of the 8 different phyla present in the cecal content (C). **P<0.01 indicates difference induced by choline determined by Mann-Whitney U test with Bonferroni correction. Variation induced by the diet for all phyla except TM7: §§P<0.01.
To address whether the altered microbiota composition also translated to altered microbial metabolism, we assessed the plasma bile acid profile. Bile acids are produced from cholesterol in the liver and deconjugated and further metabolized to secondary bile acids by the gut microbiota.33 Supplementation with choline increased the proportion of deconjugated bile acids in plasma from CONV-R Apoe−/− mice on a chow diet (choline, 50.9±3.5%; no choline, 34.1±4.2%; P=0.0046) and on a Western diet (choline, 36.5±4.1%; no choline, 16.6±2.4%; P=0.0003; Figure V in the online-only Data Supplement). In contrast, supplementation with choline only increased the proportion of secondary bile acids in plasma from CONV-R Apoe−/− mice on a Western diet (choline, 34.0±2.5%; no choline, 19.2±1.9%; P=0.0002; Figure V in the online-only Data Supplement). As expected, we only observed primary and conjugated bile acids in plasma from GF Apoe−/− mice (Figure V in the online-only Data Supplement).
Choline supplementation also altered the abundance of 4 and 6 OTUs, all belonging to the order Clostridiales, in chow- and Western diet-fed mice, respectively (Figure VI in the online-only Data Supplement; Table II in the online-only Data Supplement). These OTUs were also found to be significantly correlated, both positively and negatively, with plasma TMAO concentrations and some of the choline-regulated bile acids (Table II in the online-only Data Supplement) but not with aortic lesion size (nonsignificant, data not shown).
To conclude, the macronutrient composition of the diet had large effects on the overall microbial composition, confirming previous data. In addition, choline supplementation also affected the microbial composition but to a much lower extent than switching between chow and Western diet.
Discussion
In this study, we demonstrated that the impact of the gut microbiota on the development of atherosclerosis is dietary dependent. Atherosclerotic lesion size and plasma cholesterol levels were lower in CONV-R compared with GF Apoe−/− mice on a chow diet but not on a Western diet. Furthermore, we showed that choline supplementation, which promoted increased TMAO levels in CONV-R but not GF mice, did not affect lesion size in chow-fed mice. In mice on a Western diet, choline supplementation did not alter the normalized aortic lesion size but decreased body weight and body fat in CONV-R but not GF mice. Thus suggesting that microbial metabolism of choline may have additional effects on host metabolism in agreement with earlier studies.34
In agreement with previous findings, we showed that the effect of gut microbiota on atherosclerosis is dietary dependent,11 and atherosclerotic lesions develop regardless of microbial status.35 We found that lesion size in the Apoe−/− mouse model paralleled plasma cholesterol levels in a microbiota-dependent fashion, that is, chow-fed CONV-R mice had both lower plasma cholesterol levels and smaller aortic lesions than GF mice. We, therefore, hypothesize that the microbiota is protective on fiber-rich chow diet through production of beneficial metabolites, such as short-chain fatty acids, that may reduce cholesterol levels.8
Dietary choline and plasma TMAO levels have previously been shown to associate with an increased risk for CVD.9,12,36 A recent study showed that inhibition of trimethylamine lyases—enzymes expressed by gut microbes that convert choline to trimethylamine—reduces atherosclerosis in mice,37 and thus, microbial regulation of choline has been proposed to be important in the development of atherosclerosis. We showed that choline supplementation promoted an increase in plasma TMAO levels in chow-fed CONV-R mice but did not affect aortic lesion size or macrophage content, plasma cholesterol, or cytokine levels. In Western diet-fed mice, choline supplementation had no effect on normalized aortic lesion size and did not affect plasma cholesterol.38
Our results contrast with previous publications demonstrating a clear association between choline/carnitine diets and atherosclerosis.9,12 This discrepancy may be because of different experimental setups. In previous studies, antibiotics were administered to deplete the gut microbiota, and choline supplementation was started at weaning (aged around 4 weeks).9,12 Here, we used GF mice, which are born and raised under sterile conditions, and we supplemented choline at 8 weeks of age when atherosclerotic disease already started to develop.39 Therefore, we cannot exclude that dietary choline may be an important factor at an early-stage atherosclerosis development. Consistent with this possibility, a recent study showed that female Apoe−/− mice with hypercholesterolemic mothers develop larger aortic root lesions compared with mice with normocholesterolemic mothers—an effect that was associated with TMAO levels in the offspring.40
Interestingly, CD68-stained macrophage content in aortic lesions was not associated with lesion size when mice were fed chow diet, and thus, we did not observe increased abundance of CD68-positive macrophages in GF mice, despite larger lesions compared with CONV-R mice. In contrast, GF mice had increased CD68 content compared with CONV-R mice when fed Western diet, despite similar plaque size. These findings are in agreement with our observation that GF mice had elevated cholesterol levels and that cholesterol crystals may form sterile inflammation by activating inflammasomes.41
We did not observe any effects of choline on accumulation of CD68-positive macrophages in the plaque, which is in contrast to a previous study, demonstrating that inflammation markers and CD68 mRNA were increased in aorta in low-density lipoprotein receptor knockout mice receiving choline in the drinking water.42 These differences could reflect different time points in atherosclerosis development because Seldin et al42 studied mice treated with choline for 3 weeks, whereas we studied mice treated with choline for 12 weeks or that we used different animal models. However, we found that several cytokines were microbially regulated, which may reflect an immune response mediated by colonization of the gut but is not sufficient to affect atherosclerosis. Rather, atherosclerosis development in our model was more associated with cholesterol levels.
The diet has a large impact on the gut microbiota composition and physiology, and shifting the diet from a low-fat, fiber-rich to a high-fat, high-sugar Western diet rapidly changes the microbiome, even within a day.43,44 In this study, the macronutrient composition of the diet was the most important factor influencing the overall composition of the gut microbiota, and we observed distinct clustering of chow- and Western diet-fed mice. Dietary choline had smaller effects on global community structure although some taxa belonging to Clostridiales, which have previously been shown to correlate with TMAO and aortic lesion size,37 were either downregulated or upregulated and associated with TMAO levels but not aortic lesion size. Interestingly, we observed that choline supplementation affected bile acid profile, suggesting that despite minor taxonomic effects, choline may affect the functionality of the microbiome.
In conclusion, we demonstrate that the development of atherosclerosis in Apoe−/− mice is associated with the macronutrient composition of the diet and cholesterol levels, and there is no evidence that interaction between the gut microbiota and dietary choline further promotes atherogenesis. Importantly, in this atherosclerosis model, the gut microbiota reduces atherogenesis, but interaction with the diet can influence the disease process.
Acknowledgments
We thank the following people at the Wallenberg Laboratory in Gothenburg: Anna Hallén for technical assistance, Marcus Ståhlman and Per-Olof Bergh for plasma analyses, Valentina Tremaroli for assisting in data analyses, Liliana Håversen and Mikael Rutberg for advice in staining procedures, and Rosie Perkins for editing the manuscript.
Sources of Funding
This work was supported by grants from the Swedish Heart-Lung Foundation, AFA Insurance, Swedish Research Council, Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, Novo Nordisk Foundation, the FP7 sponsored program METACARDIS (Metagenomics in Cardiometabolic Diseases), and LUA-ALF grants from Västra Götalandsregionen. F. Bäckhed is Torsten Söderberg Professor in Medicine and recipient of European Research Council consolidator grant 2013 (615362-METABASE).
Disclosures
F. Bäckhed is a shareholder and founder of Metabogen AB. The other authors report no conflicts.
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CONV-R
- conventionally raised
- CVD
- cardiovascular disease
- GF
- germ free
- IFN-γ
- interferon
- IL
- interleukin
- KC/GRO
- keratinocyte chemoattractant/growth-regulated oncogene
- OTU
- operational taxonomic unit
- TMAO
- trimethylamine N-oxide
- TNF-α
- tumor necrosis factor-α
Current address for C. Reinhardt: Center for Thrombosis and Hemostasis, University Medical Center Mainz, Germany.
Current address for F. Fåk Hållenius: Food for Health Science Centre, Lund University, Sweden.
The online-only Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.118.311233.
Highlights.
The interaction between gut microbiota and diet affects atherogenesis, and conventionally raised mice develop smaller lesions compared with germ-free mice on chow diet but not Western diet.
Plasma cholesterol levels, but not trimethylamine N-oxide, are associated with aortic lesion sizes.
Inflammation markers are influenced by the interaction of the macronutrient composition of the dietand gut microbiota.
The macronutrient composition of the diet determines the gut microbiota composition, and choline alters the abundance of several operational taxonomic units belonging to the order Clostridiales.
References
- 1.Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jie Z, Xia H, Zhong SL, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 5.Nissen SE, Tuzcu EM, Schoenhagen P, Crowe T, Sasiela WJ, Tsai J, Orazem J, Magorien RD, O’Shaughnessy C, Ganz P Reversal of Atherosclerosis With Aggressive Lipid Lowering (REVERSAL) Investigators. Statin therapy, LDL cholesterol, C-reactive protein, and coronary artery disease. N Engl J Med. 2005;352:29–38. doi: 10.1056/NEJMoa042000. doi: 10.1056/NEJMoa042000. [DOI] [PubMed] [Google Scholar]
- 6.Ridker PM, Cannon CP, Morrow D, Rifai N, Rose LM, McCabe CH, Pfeffer MA, Braunwald E Pravastatin or Atorvastatin Evaluation and Infection Therapy-Thrombolysis in Myocardial Infarction 22 (PROVE IT-TIMI 22) Investigators. C-reactive protein levels and outcomes after statin therapy. N Engl J Med. 2005;352:20–28. doi: 10.1056/NEJMoa042378. doi: 10.1056/NEJMoa042378. [DOI] [PubMed] [Google Scholar]
- 7.Bohula EA, Giugliano RP, Cannon CP, Zhou J, Murphy SA, White JA, Tershakovec AM, Blazing MA, Braunwald E. Achievement of dual low-density lipoprotein cholesterol and high-sensitivity C-reactive protein targets more frequent with the addition of ezetimibe to simvastatin and associated with better outcomes in IMPROVE-IT. Circulation. 2015;132:1224–1233. doi: 10.1161/CIRCULATIONAHA.115.018381. doi: 10.1161/CIRCULATIONAHA.115.018381. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson AL, Bäckhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14:79–87. doi: 10.1038/nrcardio.2016.183. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stepankova R, Tonar Z, Bartova J, Nedorost L, Rossman P, Poledne R, Schwarzer M, Tlaskalova-Hogenova H. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. J Atheroscler Thromb. 2010;17:796–804. doi: 10.5551/jat.3285. [DOI] [PubMed] [Google Scholar]
- 12.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi J, You T, Li J, Pan T, Xiang L, Han Y, Zhu L. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J Cell Mol Med. 2018;22:185–194. doi: 10.1111/jcmm.13307. doi: 10.1111/jcmm.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, Fändriks L, le Roux CW, Nielsen J, Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fåk F, Bäckhed F. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe-/- mice. PLoS One. 2012;7:e46837. doi: 10.1371/journal.pone.0046837. doi: 10.1371/journal.pone.0046837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caesar R, Nygren H, Orešič M, Bäckhed F. Interaction between dietary lipids and gut microbiota regulates hepatic cholesterol metabolism. J Lipid Res. 2016;57:474–481. doi: 10.1194/jlr.M065847. doi: 10.1194/jlr.M065847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Mobini R, Tremaroli V, Ståhlman M, Karlsson F, Levin M, Ljungberg M, Sohlin M, Bertéus Forslund H, Perkins R, Bäckhed F, Jansson PA. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: a randomized controlled trial. Diabetes Obes Metab. 2017;19:579–589. doi: 10.1111/dom.12861. doi: 10.1111/dom.12861. [DOI] [PubMed] [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina paired-end read merger. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 29.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW Human Microbiome Consortium. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.al-Waiz M, Mikov M, Mitchell SC, Smith RL. The exogenous origin of trimethylamine in the mouse. Metabolism. 1992;41:135–136. doi: 10.1016/0026-0495(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 33.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright SD, Burton C, Hernandez M, Hassing H, Montenegro J, Mundt S, Patel S, Card DJ, Hermanowski-Vosatka A, Bergstrom JD, Sparrow CP, Detmers PA, Chao YS. Infectious agents are not necessary for murine atherogenesis. J Exp Med. 2000;191:1437–1442. doi: 10.1084/jem.191.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartman J, Frishman WH. Inflammation and atherosclerosis: a review of the role of interleukin-6 in the development of atherosclerosis and the potential for targeted drug therapy. Cardiol Rev. 2014;22:147–151. doi: 10.1097/CRD.0000000000000021. doi: 10.1097/CRD.0000000000000021. [DOI] [PubMed] [Google Scholar]
- 39.Coleman R, Hayek T, Keidar S, Aviram M. A mouse model for human atherosclerosis: long-term histopathological study of lesion development in the aortic arch of apolipoprotein E-deficient (E0) mice. Acta Histochem. 2006;108:415–424. doi: 10.1016/j.acthis.2006.07.002. doi: 10.1016/j.acthis.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Trenteseaux C, Gaston AT, Aguesse A, Poupeau G, de Coppet P, Andriantsitohaina R, Laschet J, Amarger V, Krempf M, Nobecourt-Dupuy E, Ouguerram K. Perinatal hypercholesterolemia exacerbates atherosclerosis lesions in offspring by altering metabolism of trimethylamine-N-oxide and bile acids. Arterioscler Thromb Vasc Biol. 2017;37:2053–2063. doi: 10.1161/ATVBAHA.117.309923. doi: 10.1161/ATVBAHA.117.309923. [DOI] [PubMed] [Google Scholar]
- 41.Duewell P, Kono H, Rayner KJ, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N-oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor- B. J Am Heart Assoc. 2016;5:e002767. doi: 10.1161/JAHA.115.002767. doi: 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]


