Abstract
The genomes of the malaria-causing Plasmodium parasites encode a protein fused of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK) and dihydropteroate synthase (DHPS) domains that catalyze sequential reactions in the folate biosynthetic pathway. Whereas higher organisms derive folate from their diet and lack the enzymes for its synthesis, most eubacteria and a number of lower eukaryotes including malaria parasites synthesize tetrahydrofolate via DHPS. Plasmodium falciparum (Pf) and Plasmodium vivax (Pv) HPPK–DHPSs are currently targets of drugs like sulfadoxine (SDX). The SDX effectiveness as an antimalarial drug is increasingly diminished by the rise and spread of drug-resistant mutations. Here, we present the crystal structure of PvHPPK–DHPS in complex with four substrates/analogs, revealing the bifunctional PvHPPK–DHPS architecture in an unprecedented state of enzymatic activation. SDX's effect on HPPK–DHPS is due to 4-amino benzoic acid (pABA) mimicry, and the PvHPPK–DHPS structure sheds light on the SDX-binding cavity, as well as on mutations that effect SDX potency. We mapped five dominant drug resistance mutations in PvHPPK–DHPS: S382A, A383G, K512E/D, A553G, and V585A, most of which occur individually or in clusters proximal to the pABA-binding site. We found that these resistance mutations subtly alter the intricate enzyme/pABA/SDX interactions such that DHPS affinity for pABA is diminished only moderately, but its affinity for SDX is changed substantially. In conclusion, the PvHPPK–DHPS structure rationalizes and unravels the structural bases for SDX resistance mutations and highlights architectural features in HPPK–DHPSs from malaria parasites that can form the basis for developing next-generation anti-folate agents to combat malaria parasites.
Keywords: drug resistance, malaria, plasmodium, crystal structure, infectious disease, dihydropteroate synthase, HPPK–DHPS, p-aminobenzoic acid, Plasmodium vivax, sulfadoxine
Introduction
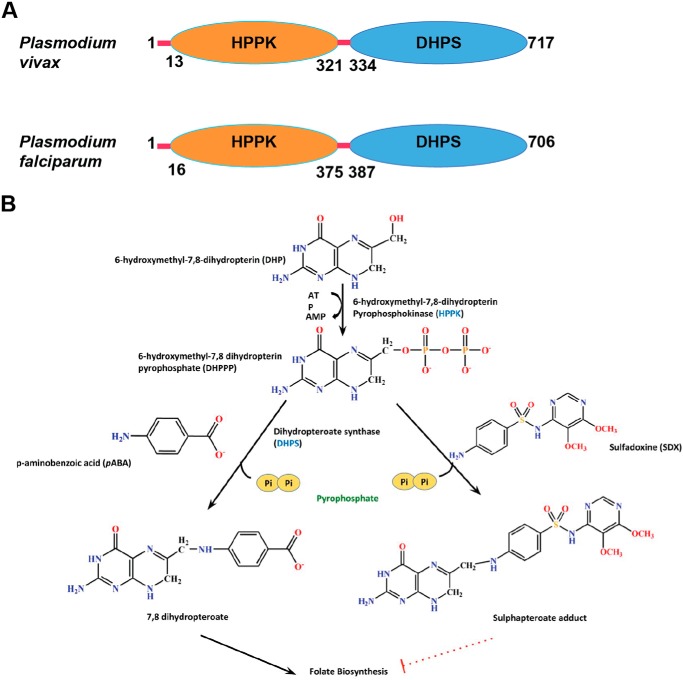
Malaria remains a central cause of morbidity and mortality in humans. The malaria parasites Plasmodium falciparum (Pf)2 and Plasmodium vivax (Pv) infect >400 million people and result in ∼0.3 to ∼0.4 million annual deaths worldwide (1). Malaria remains a constant public health threat because of the emergence of drug-resistant strains across endemic regions (2). Despite increased drug resistance, several anti-malarial drugs are still used clinically for the treatment of malaria infection (3). Therefore, the world health community needs to continually discover both new drug targets and novel chemical scaffolds. Tetrahydrofolate is an essential cofactor that is vital for metabolic reactions involving one-carbon transfer (4). Most notably, it is required for the synthesis of nucleic acid precursors like purines and thymidine and for methionine, glycine, and pantothenate (5). Higher organisms derive folate from their diet and lack the necessary enzymes for folate synthesis, but almost all eubacteria and a number of lower eukaryotes including malaria parasites synthesize tetrahydrofolate (5–7). The malaria parasite genomes encode fused 6-hydroxymethyl-7, 8-dihydropterin pyrophosphokinase (HPPK) and dihydropteroate synthase (DHPS) domains (Fig. 1A) that perform sequential reactions wherein HPPK catalyzes transfer of pyrophosphate from ATP to 6-hydroxymethyl-7,8-dihydropterin (DHP) resulting in 6-hydroxymethyl-7,8-dihydropterin-pyrophosphate (DHPPP) (8, 9). Subsequently, DHPS acts as a crucial convergence point in the folate pathway and catalyzes the condensation of 4-aminobenzoic acid (pABA) and DHPPP to form the intermediate 7,8-dihydropteroate (Fig. 1B) (10). 7,8-Dihydropteroate is then fed into the folate synthesis pathway where it is converted to 7,8-dihydrofolate by the enzyme dihydrofolate synthase and subsequently to tetrahydrofolate by the enzyme dihydrofolate reductase (Fig. 1B). The folate pathway is therefore an ideal target for anti-infectives and has been utilized for many decades (11).
Figure 1.
Structure and function of PvHPPK–DHPS. A, the domain architecture of Pv and Pf HPPK–DHPSs (717 and 706 residues, respectively) with marked domain boundaries of HPPK and DHPS in orange and blue, respectively. The red lines are regions that fall outside annotated domain boundaries. B, HPPK domain catalyzes the transfer of pyrophosphate from ATP to DHP, resulting in the product DHPPP. DHPPP is then converted to 7,8-dihydropteroate by DHPS using pABA and DHPPP. SDX inhibits DHPS via mimicry of pABA where it results in formation of an adduct that is deleterious to the downstream folate synthesis pathway. The dotted red line indicates termination of the pathway.
Sulfonamides target a key enzyme in folate biosynthesis pathway viz. DHPS (12). The aryl amine moiety of sulfa drugs forms a dihydropteroate-like product with DHPPP that is impotent toward undergoing subsequent dihydrofolate synthesis (13, 14). Sulfa drugs have remained important clinical agents since they were first discovered in the 1930s (15), but their efficacy has been severely impacted by drug resistance that began to emerge shortly after they were first introduced (16, 17). Despite increased resistance, sulfodaxine (SDX) is still used in combination with pyrimethamine (SP) to treat malaria (18, 19). Pyrimethamine inhibits the enzyme dihydrofolate reductase in the folate biosynthesis pathway, whereas SDX stalls DHPS activity; because of their synergistic effect, these two drugs (SP) are more effective in combination than either drug used alone (18, 19). SP is used to treat chloroquine-resistant Pf malaria and is the only drug combination recommended by World Health Organization for intermittent preventive treatment of infants as an additional malaria control in high transmission areas of sub-Saharan Africa (19–22). Increasing resistance toward SDX is therefore alarming and calls for the development of a new generation of anti-folates that are less susceptible to resistance generation but that retain the enzymatic target of DHPS.
Here, we present the crystal structure of PvHPPK–DHPS in complex with its four substrates/analogs. The structure contains residues from 1 to 717, is dimeric, and reveals the juxtaposition of both HPPK and DHPS domains that are connected by a linker segment. We have mapped the conserved pABA-binding residues and sites of vital mutations that tune the specificity and affinity of pABA, as well as the drug SDX. This work will enhance our understanding of molecular mechanisms used by PvHPPK–DHPS to develop anti-folate drug resistance and provides a new focus for development of novel anti-malarial agents.
Results
Crystal structure of PvHPPK–DHPS
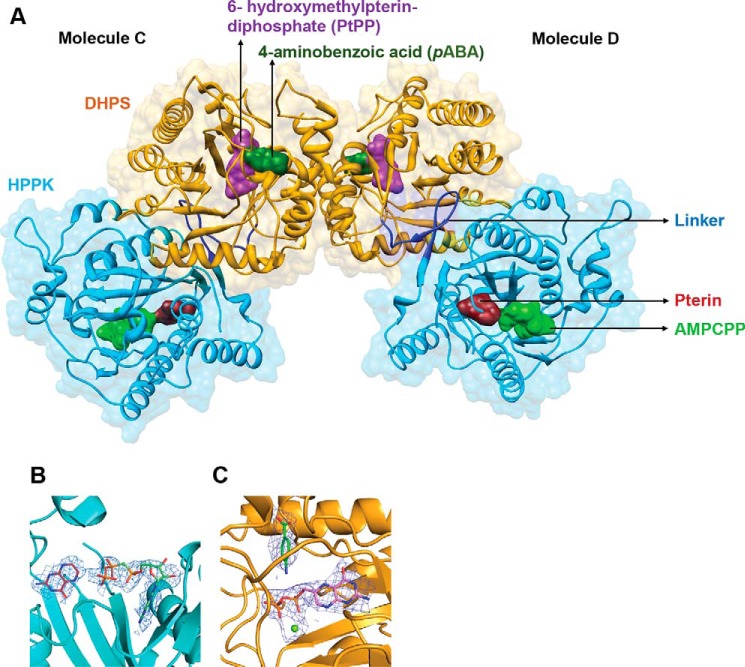
Plasmodium parasite genomes encode fused genes for HPPK and DHPS. We expressed recombinant full-length PvHPPK–DHPS consisting of 717 residues and crystallized it (P21 system with six molecules in asymmetric unit (ASU)). Each PvHPPK–DHPS chain that was traced has 600 of 717 residues along with disordered and missing loops. The six molecules of HPPK–DHPS in the ASU are designated A to F that form three biological dimers called AB, CD, and EF. The three dimers are similar, and their root mean squared deviations (RMSDs) for Cα atoms are generally <0.5 Å; they differ mostly in their long flexible loop regions. The N-terminal 10 residues and loop residues 55–80, 145–160, 189–202, 320–435, and 588–660 are disordered in all six molecules of HPPK–DHPS. The overall fold of the PvHPPK–DHPS domains is similar to their known homologs. The average B-factor for molecules A, C, and D is ∼37 Å2, whereas for the other three molecules B, E, and F it is ∼51Å2. Therefore, from hereon the structural analyses discussed are based on the CD dimer (Fig. 2A). The RMSD between the PvHPPK–DHPS and known HPPK and DHPS structures is <1 Å for the overlapping ferredoxin fold (Cα atoms 65–95) and the triosephosphate isomerase TIM barrel core (Cα atoms 165–195). The PvHPPK–DHPS was crystallized in the presence of 6-hydroxymethylpterin-diphosphate (PtPP), pterin, the ATP analog AMPCPP, and pABA (Fig. 2A). The electron densities for bound ligands are clear for the monomeric chains of PvHPPK–DHPS (Fig. 2, B and C). The electron densities for whole AMPCPP or terminal phosphate groups of the AMPCPP are weak in monomers B, D, E, and F, possibly because of their poor occupancies. The present crystal structure therefore represents a holo form of the PvHPPK–DHPS enzyme and will be discussed below in this light.
Figure 2.
Crystal structure of PvHPPK–DHPS. A, dimeric PvHPPK–DHPS where bound substrate/analogs in both HPPK and DHPS domains are shown as molecular surfaces. HPPK (cyan), DHPS (yellow), and the linker regions (blue) are marked. The bound substrates/analogs of pterin (maroon), AMPCPP (light green), PtPP (purple), pABA (green), and Mg2+ ion (lime green) are shown as sticks and spheres. B and C, the simulated annealing composite omit map contoured at 2σ levels for the bound ligands where ligands and bound Mg2+ ion are shown as sticks and spheres, respectively.
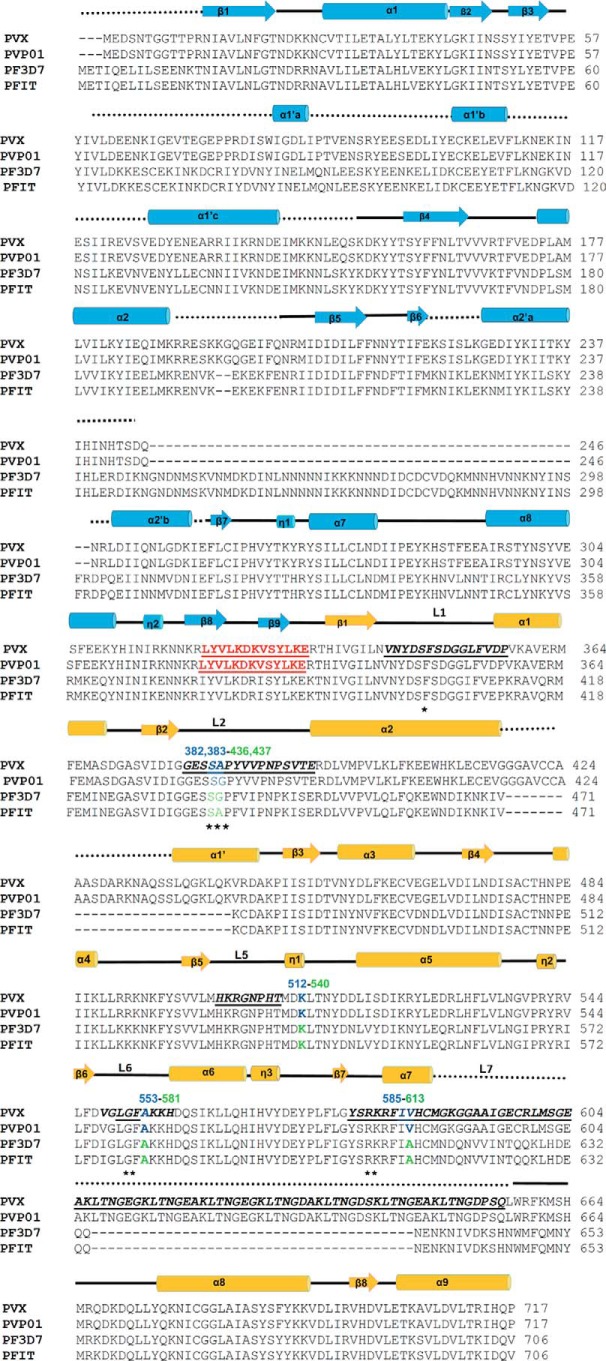
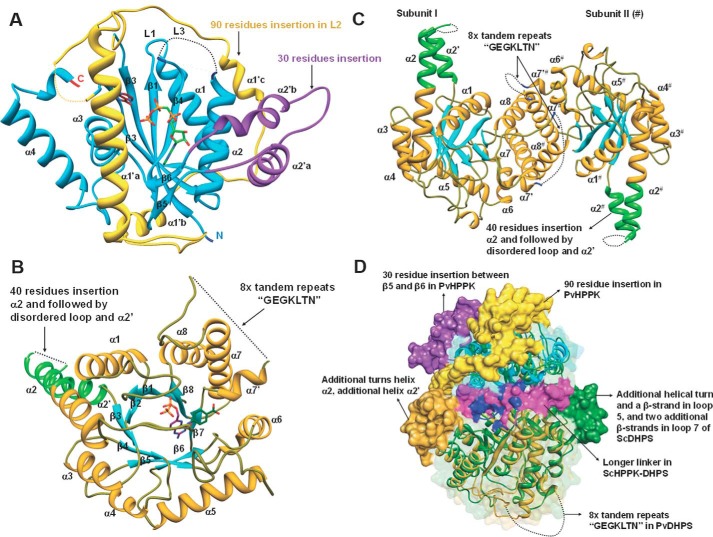
PvHPPK has a 90-residue insertion (residues 60–150) between β-strands 2 and 3 when compared with the known homologs of HPPK structures from Escherichia coli, Streptococcus pneumoniae, Yersinia pestis (Yp), Saccharomyces cerevisiae, and Francisella tularensis (Figs. 3 and 4A) (23–28). This insertion wraps around the ferredoxin fold of PvHPPK domain, and it has three α-helices: α1′a, α1′b, and α1′c. α1′a partially masks the active site of PvHPPK (Fig. 4A). The loop (L3) between β-strands β5 and β6 are important for ATP binding in HPPK (Fig. 4A). In PvHPPK, a 48-residue-long insertion (residues 222–260) is present in ATP-binding loop, and this insert has two α-helices (α2′a and α2′b) when compared with the known HPPK structures (Fig. 4A).
Figure 3.
Structure-based sequence alignment of malaria parasite HPPK–DHPSs. The secondary structure elements of PvHPPK–DHPS (blue) and PfHPPK–DHPS (yellow) are marked, with α- and 310-helices (cylinders) and β-strands (arrows). The pABA-binding loops (L1, L2, L5, L6, and L7) residues are in bold, underlined, and labeled. The disordered regions in the crystal structure of PvHPPK–DHPS are marked by black dots. The SDX resistance mutations in Pf (green) and Pv (blue) are numbered above the sequences. The linker region between HPPK and DHPS is highlighted in red. The pABA-binding residues are marked with stars.
Figure 4.
PvHPPK–DHPS domains and structural comparisons. A and B, the domains of PvHPPK–DHPS are shown with secondary structural elements. The unique features/larger insertions are also labeled. C, a view of dimer interface and the participating helices α6, α7′, α7, and α8. A possible tandem repeat motif is shown. D, the superposition of DHPS domain of PvHPPK–DHPS on ScHPPK–DHPS where their unique features are highlighted.
Clear electron densities in PvDHPS are evident for pABA and the substrate analog PtPP (Fig. 2C). In addition, well defined electron densities were observed for all the active-site loops in PvDHPS because they are highly ordered (Fig. 4B). Long insertions are not found in the core-TIM barrel fold of PvDHPS domain. However, a 40-residue insertion is present between strand β3 and helix α2, and this insertion has a 10-residue helix α2′ (Fig. 4B). The helix α2 of PvDHPS has 26 residues in 7 turns and is up to 9 residues longer than the other known DHPS structures that have between 14 and 17 residues (Fig. 4B) (23–28). A Mg2+ cofactor is known to coordinate the diphosphate group within DHPPP and involves the conserved Asn342 in PvHPPK–DHPS. In PvDHPS, another 80-residue-long insertion is present between α-helix 7′ and α7, and this insertion contains the 8× “tandem repeat-like” sequence motif of GEGKLTN (Fig. 4B).
PvHPPK–DHPS forms a dimeric assembly via C-terminal α-helices α6, α7′, α7, and α8 (Fig. 4C). In its dimeric form, the solvent-accessible surface of PvHPPK–DHPS is ∼51,800 Å2, and the buried surface is ∼3300 Å2. The association of helices α6, α7′, α7, and α8 is similar to that observed in all dimeric DHPS structures, as also in the bifunctional ScHPPK–DHPS structure (23–28). The dimer interface involves only the DHPS domain structural elements. In PvDHPS, the N-terminal end of the TIM barrel is capped by antiparallel β-strands, and they are conserved among E. coli and Yp), Thermus thermophilus and Bacillus anthracis DHPSs (23–28). The capped anti-parallel β-sheet serves as a linker region between PvHPPK and PvDHPS domains. The linker β-strand (L1) forms a short anti-parallel association with the β2-strand of PvHPPK domain (Figs. 3 and 4D). This is very similar to ScHPPK–DHPS structure (25). The orientation of HPPK domains does not overlap on the F. tularensis HPPK–DHPS (FtHPPK–DHPS) structure (27), and large displacement in HPPK is observed in context of ScHPPK–DHPS (Fig. 4D) (25). This is due to the longer β-strands in linker region of ScHPPK–DHPS and shorter linker in FtHPPK–DHPS when compared with the PvHPPK–DHPS.
The molecular mechanism of sulfadoxine resistance
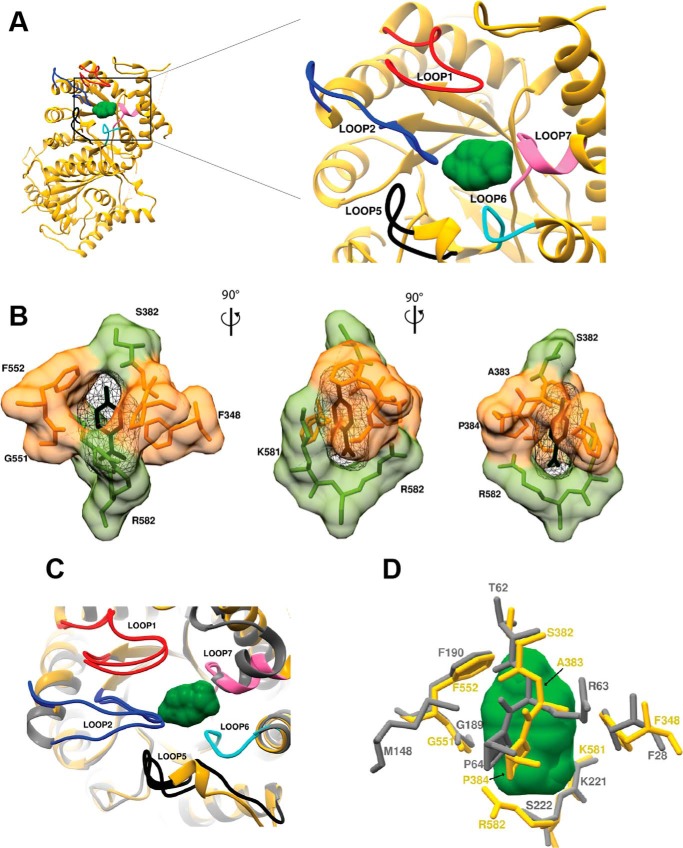
Our crystal structure of PvDHPS in complex with pABA shows a striking state of enzymatic activation in which five PvDHPS loops bind pABA onto the protein surface (Fig. 5A). The substrate pABA is ensconced by loops 1, 2, 5, and 6 and the 7′ helix within loop 7. The eight key residues envelope pABA in PvDHPS and thus form its binding site that contains Phe348, Ser382, Ala383, Pro384, Gly551, Phe552, Lys581, and Arg582 (Fig. 5B). These residues are highly conserved among Pv and PfHPPK–DHPSs (Fig. 3, highlighted in the box). In addition to the PvDHPS crystal structure reported here, the crystal structure of YpDHPS bound to pABA is known (28). Therefore, we compared the pABA-binding residues and their conformational states in PvDHPS and YpDHPS. Loops 1 and 2 encapsulating pABA in Pv and Yp are shown in (Fig. 5C), and they show no significant differences. In PvDHPS, key residues caging pABA in loop 2 are 381SSAPY385, whereas in Yp the residues are 61STRPG65 (Fig. 5, C and D). Among these, it is evident that the proline in fourth position is conserved; it nestles the benzene ring of pABA via a hydrophobic interaction (Figs. 5D and 6C). The two flexible loops 1 and 2 that cover pABA-binding site are highly ordered in the present PvHPPK–DHPS. In case of bacterial DHPS, the majority of the mutations known to confer resistance to sulfonamides are found in these two loops. However, in the case of PvHPPK–DHPS, SDX point mutations do not occur in loop 1 that contributes the single pABA-binding residue Phe348.
Figure 5.
The pABA-binding site in PvDHPS. A, overall view of the pABA-binding region with key loops labeled. B, key residues that cage pABA in PvDHPS. C, structural comparisons between loops that surround pABA in DHPSs from Y. pestis and P. vivax. D, critical pABA-binding residues in Pv (yellow) and Yp (gray) are shown. The pABA molecule is shown as molecular surface (green) and protein residues shown as sticks (gray and yellow).
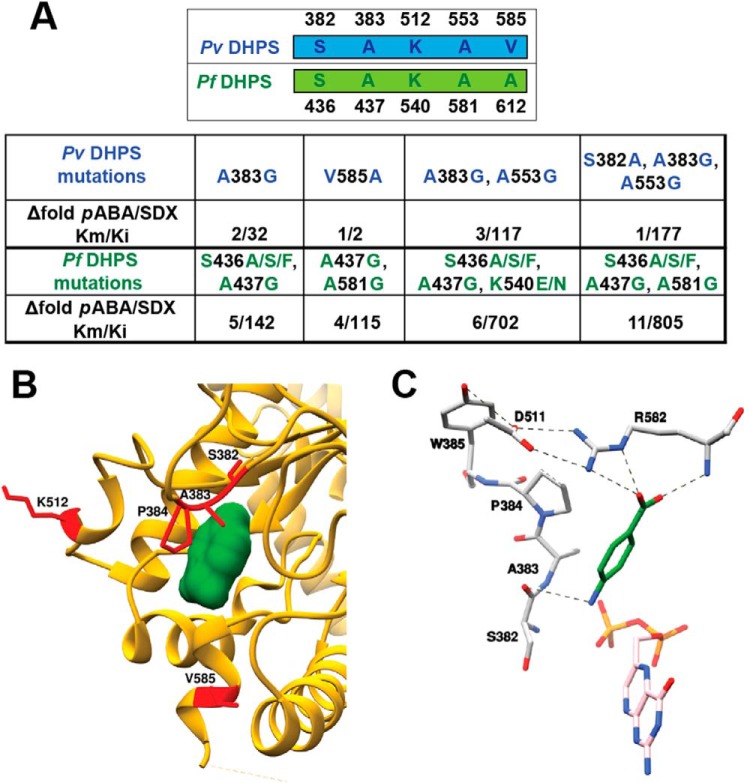
Figure 6.
Enzymatic data. A, the kinetic parameters Km and Ki values for SDX resistance mutations in Pf and Pv based on previous studies (33, 34). The fold change in substrate affinity and drug potency were calculated using published data (41, 42). B, the pABA-bound PvDHPS domain with the mutant residues is highlighted in red. C, the important salt-bridge interactions between Arg582 and Asp511 are shown with dotted lines along with hydrogen-bonding networks around pABA binding site in PvDHPS.
The pABA-binding residues Phe348, Gly551, Phe552, Ser382, Ala383, Lys581, Arg582, and Pro384, as well as the SDX resistance mutations are conserved among human malaria parasites Pv and Pf (Fig. 6A). Five SDX point mutations at positions S382F/A/C, A383G, K512E/M/T, A553G, and V585A have been reported in PvDHPS (29–34) (Fig. 6B). These amino acid changes occur as singles (V585A or A383G), in doubles (A383G and A553G), or triply (S382A, A383G and A553G) and together account for DHPS mutations in 90% of the clinical isolates from malaria endemic regions (29–34). Our PvHPPK–DHPS crystal structure shows that these resistance mutations surrounding the pABA-binding site are present within loop 2 (S382/F/A/C and A383G), loop 5 (Lys512), loop 6 (Ala553), and 7′ helix in loop 7 (Val585) (Figs. 3 and 6B).
We collated SDX resistance data from clinical isolates of Pf and Pv and analyzed available enzyme kinetic data from two elegant published studies in context of pABA and SDX affinities (33, 34). We assessed the fold difference of the substrate-binding constant (Km) of pABA with the inhibitory constant (Ki) of SDX as a function of the DHPS mutations from both studies (33, 34) (Fig. 6A). It is documented that single amino acid mutations of A383G and V585A do not seem to confer very high levels of SDX resistance, as shown in Fig. 6A (33, 34). We also observed that the five key mutations responsible for SDX drug resistance mostly precede or succeed the vital atomic interactions that fall within 4 Å of the pABA binding pocket (Fig. 6B). The residues Ser382 and Ala383 are present within loop 2 of PvDHPS, and from analyses of deposited DHPS structures, it is evident that loop 2 is highly flexible and that its sequence is conserved across DHPSs. Further, in the case of PvDHPS, loop 2 clearly stabilizes pABA binding by contributing (Ser382, Ala383, and Pro384) residues for pABA recognition. Based on analysis of our PvHPPK–DHPS crystal structure, it is evident that Ser382 and Ala383 residues make intimate interactions with pABA (Fig. 5B). Further, Ser382 and Ala383 precede the critical Pro384 residue that stacks with the benzene ring of pABA (Figs. 5D and 6C). These structural constraints within the pABA-binding site likely explain the mutational effects of residues 382 and 383, because their mutation may disturb the positioning of Pro384 that is critical for stacking with the benzene ring of pABA. Intriguingly, resistance mutation residue Lys512 lies distal to the pABA-binding site, and its role in resistance generation cannot be reliably accessed via the present HPPK–DHPS structure (Fig. 6B). The A553G mutation will likely create alteration in the presentation of loop 6 that interacts with pABA. The SDX mutant residue V585A shows a 2-fold increase in the level of resistance compared with the WT PvDHPS (Fig. 6A); this residue is located at dimerization interface and does not seem to play a role in the binding of either the substrate or SDX (not shown). Interestingly, our PvHPPK–DHPS structure reveals that residues Arg582 and Asp511 make salt-bridge interactions and are conserved in Pv and Pf HPPK–DHPSs. It is likely that the V585A mutation results in steric hindrance with Arg582. This may translate into diminished interactions with Asp511 and structural perturbation of pABA recognition (Fig. 6C).
Discussion
The X-ray structure of PvHPPK–DHPS presented here rationalizes the known sulfa drug resistance mutations that have arisen over the past four decades as a result of drug usage. The PvHPPK–DHPS structural analyses reveal an intricate dimeric assembly via a C-terminal region in the PvDHPS sequence. This long-awaited crystal structure of the malaria parasite's unique bifunctional HPPK–DHPS enzyme reveals the exquisite structural subtleties involved in SDX resistance generation. It is evident that most SDX resistance mutations map proximal to the pABA-binding site, where they are likely to subtly alter the intricate atomic interactions such that enzyme affinity for pABA is only diminished moderately (up to 11-fold), but Ki for SDX is altered substantially (more than 100-fold for double/triple mutants in both Pv/Pf DHPSs (Fig. 6A). Thus, a structural compromise is reached in the drug resistant enzyme wherein marginal lowering in the substrate (pABA) affinity (Km = ∼11-fold) is traded for substantial reductions (Ki = ∼800-fold; Fig. 6A) in SDX drug potency. This trick, although highly successful, also opens the enzyme for targeting via novel inhibitors that are designed based on the exact substrate envelope such that new inhibitors fit and colonize the substrate-binding site fully and that too in at least four different druggable pockets that are evident in the PvHPPK–DHPS structure. Indeed, inhibitors like SDX that protrude beyond the substrate envelope may encourage development of mutations that confer drug resistance. In this light, an interesting drug design approach has been applied toward potentially overcoming drug resistance in HIV by focusing on inhibitors that fit snugly in the small substrate-binding cavity of HIV protease (35, 36).
It is noteworthy that the PvHPPK–DHPS residues involved in recognition of pterin moiety in both domains are highly conserved. This presents yet another opportunity to target conserved motifs within PvHPPK–DHPS now that the crystal structure is available (37). The presented PvHPPK–DHPS structure indicates that sulfa-drug resistance mutations emanate from a structural compromise in the mutant drug resistance enzyme that enables rejection of the drug while minimally altering affinity for its substrate pABA. Although our pABA-bound PvHPPK–DHPS crystal structure can explain SDX resistance for most mutations, we feel that SDX-bound crystal structures of mutant and WT PvDHPSs are required for a deeper understanding of this enzyme/drug system (37). From our structural analysis of PvHPPK–DHPS and its mutations in context of sulfadoxine resistance, we have generated several insights including (a) the presented structure should be exploited to identify nonsulfa drugs that do not mimic pABA and thus inhibit the enzyme irreversibly, (b) the Plasmodium HPPK domain can now be utilized for focusing on pterin-based inhibitors (38), (c) designing drugs that target the triple mutant in PvHPPK–DHPS will be valuable because they can be selectively administered in regions of prevalent SDX resistance, and (d) twin targeting of Plasmodium HPPK and the DHPS subdomains within PvHPPK–DHPS may provide more potent inhibition of the enzyme. This work therefore provides exceptional opportunities to exploit the structure of PvHPPK–DHPS for screening of drug-like libraries to identify drug scaffolds that can occupy any one or more of its substrate pockets in addition to the pABA-binding site.
Experimental procedures
Gene cloning and protein production
Full-length PvHPPK–DHPS (1–717 amino acids) was purchased as a gBlock (Integrated DNA Technologies, Leuven, Belgium). The ORF of full-length PvHPPK–DHPS (residues 1–717) was optimized for expression in the E. coli strain Rosetta-pLysS and cloned into the pOPINF vector that was linearized using NcoI and SalI restriction sites. Transformed E. coli strain Rosetta-pLysS was grown in LB medium containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol to an A600 of 0.6–0.8 at 37 °C. Expression of the His6-tagged recombinant PvHPPK–DHPS was induced by the addition of 0.5 mm isopropyl β-d-galactoside, and culture was further incubated at 18 °C for 20 h. Bacterial cells were lysed by a combination of lysozyme treatment and sonication in buffer with 50 mm Tris, pH 8, 500 mm NaCl, 10% glycerol, 10 mm imidazole, 1 mm phenylmethylsulfonyl fluoride, and 1 mm benzamidine HCl. Recombinant protein was affinity captured using nickel–nitrilotriacetic acid–agarose beads (Qiagen) followed by cleavage at 20 °C with 3C-protease for removal of the His6 tag. Protein was subsequently applied to Q-Sepharose (GE Healthcare) column in buffer with 50 mm Tris, pH 8.0, 250 mm NaCl, 5 mm β-mercaptoethanol. Protein from the flow through fraction was then processed using hydrophobic interaction chromatography on a Phenyl FF 16/10 column (GE Healthcare). Pure fractions obtained from hydrophobic interaction chromatography were pooled and concentrated with 30-kDa cutoff centrifugal devices (Millipore) followed by gel permeation chromatography on a S-200–16/60 column (GE Healthcare) in a buffer containing 50 mm HEPES, pH 6.8, 200 mm NaCl, 5 mm β-mercaptoethanol. A single peak corresponding to dimeric PvHPPK–DHPS was collected from gel permeation chromatography.
Crystallization and data collection
The purified full-length PvHPPK–DHPS (∼10 mg ml−1) was used for crystallization screening in the presence of PtPP (0.5 mm), pterin (0.5 mm), AMPCPP (1 mm), and pABA (3 mm) using the hanging-drop vapor-diffusion method at 20 °C with commercially available crystallization screens (Hampton and Molecular Dimensions). The initial screening was carried out in 96-well plates using nano-drop dispensing mosquito robot (TTP Lab Tech). Diffraction quality crystals were obtained using 20% PEG 3350, 0.2 m potassium citrate tribasic monohydrate as mother liquor. The crystals were harvested using corresponding crystallization solution supplemented with 20% (v/v) ethylene glycol as a cryo-protectant and were flash-frozen into liquid nitrogen. Preliminary data collection screening was conducted at PROXIMA 1 Beamline (Soleil, France), and a high-resolution data set was collected at 100 K using Pilatus3 6 m detector (Dectris) and wavelength (λ) of 0.9763 Å at I03 Beamline, Diamond Light Source in the United Kingdom. The data were processed and scaled with XIA2 (39) using DIALS (40), and data processing statistics are shown in Table 1.
Table 1.
X-ray data collection and refinement statistics
The numbers in the parentheses are for the highest resolution bin.
| Source | I03, DLS |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9763 |
| Oscillation width (°) | 0.1 |
| Exposure time (s) | 0.02 |
| Cell parameters (Å, °) | 136.05, 113.89, 172.39; 94.24 |
| Space group | P 21 |
| Resolution | 113.89–2.79 (2.79–2.84) |
| Number of unique reflections | 129475 (5799) |
| I/σ(I) | 4.5 (0.7) |
| CC½ | 0.9 (0.2) |
| Completeness | 99.4 (90.4) |
| Redundancy | 3.5 (3.4) |
| Molecules/ASU | 6 |
| Solvent content (%) | 56 |
| Matthews coefficient Vm (Å Da−1) | 2.75 |
| Refinement | |
| Refinement resolution (Å) | 50.0–2.90 (2.95–2.90) |
| Reflections used in refinement | 114185 |
| Reflections in working set | 112179 (8347) |
| Test set | 2006 (144) |
| Rfactor/Rfree (%) | 24.2/28.5 |
| Number of atoms | |
| Protein | 28365 |
| Ligands | 342 |
| Water | 217 |
| B-factors (Å2) | |
| Protein | 43.8 |
| Ligands | 46.3 |
| Waters | 18.2 |
| Ramachandran plot (%) | |
| Favored regions | 94 |
| Allowed regions | 6 |
| RMSD | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.643 |
Structure determination
Preliminary X-ray data analysis indicated that the PvHPPK–DHPS crystals contain ∼56% solvent with Matthews coefficient of 2.75 Å3 Da−1 for six full-length PvHPPK–DHPS protomers in the ASU. Attempts to solve the phase problem using molecular replacement (MR) techniques with PHASER (41) as implemented in PHENIX (42) and coordinates of fused bifunctional HPPK–DHPS enzyme of S. cerevisiae (ScHPPK–DHPS; PDB code 2BMB) and F. tularensis (FtHPPK–DHPS; PDB code 4PZV) as template were unsuccessful. In both ScHPPK–DHPS and FtHPPK–DHPS, the orientation of the HPPK domains did not overlap while superposing the DHPS domains. Therefore, the available dimeric DHPS domain structures were fed as template, and most of the MR runs placed three dimers with log-likelihood gain value in the range of 300–600 and with translation function Z score of 5.5–6.8. The Rfree for these models were >50% for most of the templates except with Mycobacterium tuberculosis dimeric DHPS (PDB code 1EYE) model (43), which gave a starting Rfree of 49%. The initial Fo − Fc map revealed significant unbiased regions of positive connected electron density that did not form part of the DHPS probe; this thus indicated a correct MR solution. The initial atomic model was subjected to AutoBuild in PHENIX (42) that provided a partial model with Rfree of 46% for ∼1500 residues in several chains with three dimeric cores of DHPS. Subsequently, the model was manually built, extended and completed by several cycles of iterative building using COOT (44) and REFMAC (45). Map interpretation and model building was based on electron densities in difference Fourier (Fo − Fc), 2Fo − Fc and composite omit maps. In all stages, model building was guided by manual inspection of the model and Rfree. The substrate/analogs and water molecules were added into the difference Fourier maps (Fo − Fc). The modeled ligands and protein residues were validated using simulated annealing composite omit maps. The occupancies of the ligand molecules were refined and weakly bound ligands, highly disordered loops (residues numbered 1–10, 55–80, 189–202, and 420–434) and low complexity regions (residues 588–660) were not included in the final model. The final refinement statistics are shown in Table 1. The coordinates and structure factors for PvHPPK–DHPS have been deposited in the PDB under accession code 5Z79. The figures were generated using CHIMERA (46) and PyMOL (47).
Author contributions
M. Y. and Am. S. were responsible for conceptualization, formal analysis, supervision, funding acquisition, and investigation; M. Y. and Am. S. solved the structure of PvHPPK–DHPS; J. E. N. and K. H. provided the recombinant clones; K. H. collected diffraction data.; Ak.S. and A.J. performed expression, purification, and crystallization using a construct screened and developed by J. E. N; R. C., M. S., and J. C. G. maintained the clones; M. Y., Am. S., and J. C. G. analyzed the data and wrote the manuscript.
Acknowledgments
We thank the beam line staff at PROXIMA 1 and PROXIMA 2A for assistance during data collection at SOLEIL. We also thank staff at Diamond for access to Beamline I03 (MX14744).
This work was supported by grant (EMR/2016/004898) from Department of Science and Technology (DST), Ministry of Science and Technology, Government of India. This work was also supported in part by a travel grant from the Indo-French Centre for the Promotion of Advanced Research (IFCPAR/CEFIPRA) (to M. Y. and M. S.), Core Grant Award 090532/Z/09/Z from the Wellcome Centre for Human Genetics (to K. H.), and a J.C. Bose fellowship from DST, Ministry of Science and Technology, Government of India (to Am. S.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5Z79) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Pf
- P. falciparum
- Pv
- P. vivax
- HPPK
- 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase
- DHPS
- dihydropteroate synthase
- RMSD
- root mean square deviation
- SDX
- sulfadoxine
- pABA
- 4-amino benzoic acid
- DHP
- 6-hydroxymethyl-7,8-dihydropterin
- DHPPP
- 6-hydroxymethyl-7,8-dihydropterin-pyrophosphate
- SP
- SDX with pyrimethamine
- ASU
- asymmetric unit
- PtPP
- 6-hydroxymethyl pterindiphosphate
- AMPCPP
- adenosine 5′-(α,β-methylene)-diphosphate
- MR
- molecular replacement
- TIM
- triosephosphate isomerase
- PDB
- Protein Data Bank.
References
- 1. World Health Organization (2017) World Malaria Report, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Fidock D. A., Eastman R. T., Ward S. A., and Meshnick S. R. (2008) Recent highlights in antimalarial drug resistance and chemotherapy research. Trends Parasitol. 24, 537–544 10.1016/j.pt.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fidock D. A. (2010) Drug discovery: priming the antimalarial pipeline. Nature 465, 297–298 10.1038/465297a [DOI] [PubMed] [Google Scholar]
- 4. Ragsdale S. W. (2008) Catalysis of methyl group transfers involving tetrahydrofolate and B12. Vitam. Horm. 79, 293–324 10.1016/S0083-6729(08)00410-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Müller I. B., and Hyde J. E. (2013) Folate metabolism in human malaria parasites: 75 years on. Mol. Biochem. Parasitol. 188, 63–77 10.1016/j.molbiopara.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 6. Duffy S., and Avery V. M. (2013) Identification of inhibitors of Plasmodium falciparum gametocyte development. Malar. J. 12, 408 10.1186/1475-2875-12-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nzila A., Ward S. A., Marsh K., Sims P. F., and Hyde J. E. (2005) Comparative folate metabolism in humans and malaria parasites: I. pointers for malaria treatment from cancer chemotherapy. Trends Parasitol. 21, 292–298 10.1016/j.pt.2005.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shiota T. (1984) Biosynthesis of folate from pterin precursors. In Chemistry and Biochemistry of Folates (Blakley R., and Benkovic S., eds) pp. 121–134, John Wiley & Sons, New York [Google Scholar]
- 9. Blaszczyk J., Shi G., Li Y., Yan H., and Ji X. (2004) Reaction trajectory of pyrophosphoryl transfer catalyzed by 6-hydroxymethyl-7,8-dihydropterinpyrophosphokinase. Structure 12, 467–475 10.1016/j.str.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 10. Chotpatiwetchkul W., Boonyarattanakalin K., Gleeson D., and Gleeson M. P. (2017) Exploring the catalytic mechanism of dihydropteroate synthase: elucidating the differences between the substrate and inhibitor. Org. Biomol. Chem. 15, 5593–5601 10.1039/C7OB01272A [DOI] [PubMed] [Google Scholar]
- 11. Wright D. L., and Anderson A. C. (2011) Antifolate agents: a patent review (2006–2010). Expert Opin. Ther. Pat. 21, 1293–1308 10.1517/13543776.2011.587804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammoudeh D. I., Zhao Y., White S. W., and Lee R. E. (2013) Replacing sulfa drugs with novel DHPS inhibitors. Future Med. Chem. 5, 1331–1340 10.4155/fmc.13.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roland S., Ferone R., Harvey R. J., Styles V. L., and Morrison R. W. (1979) The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J. Biol. Chem. 254, 10337–10345 [PubMed] [Google Scholar]
- 14. Achari A., Somers D. O., Champness J. N., Bryant P. K., Rosemond J., and Stammers D. K. (1997) Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4, 490–497 10.1038/nsb0697-490 [DOI] [PubMed] [Google Scholar]
- 15. Gaudillière J.-P. (2009) The first miracle drugs: how the sulfa drugs transformed medicine. Bull. Hist. Med. 83, 218–220 10.1353/bhm.0.0197 [DOI] [Google Scholar]
- 16. Thaithong S., Chan S. W., Songsomboon S., Wilairat P., Seesod N., Sueblinwong T., Goman M., Ridley R., and Beale G. (1992) Pyrimethamine resistant mutations in Plasmodium falciparum. Mol. Biochem. Parasitol. 52, 149–157 10.1016/0166-6851(92)90047-N [DOI] [PubMed] [Google Scholar]
- 17. Pinichpongse S., Doberstyn E. B., Cullen J. R., Yisunsri L., Thongsombun Y., and Thimasarn K. (1982) An evaluation of five regimens for the outpatient therapy of falciparum malaria in Thailand 1980–81. Bull. World Health Organ. 60, 907–912 [PMC free article] [PubMed] [Google Scholar]
- 18. Ferone R. (1970) Dihydrofolate reductase from pyrimethamine-resistant Plasmodium berghei. J. Biol. Chem. 245, 850–854 [PubMed] [Google Scholar]
- 19. Chulay J. D., Watkins W. M., and Sixsmith D. G. (1984) Synergistic antimalarial activity of pyrimethamine and sulfadoxine against Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 33, 325–330 10.4269/ajtmh.1984.33.325 [DOI] [PubMed] [Google Scholar]
- 20. Hurly M. G. (1959) Potentiation of pyrimethamine by sulphadiazine in human malaria. Trans. R. Soc. Trop. Med. Hyg. 53, 412–413 10.1016/0035-9203(59)90044-6 [DOI] [PubMed] [Google Scholar]
- 21. World Health Organization (2010) WHO policy recommendation on intermittent preventive treatment during infancy for Plasmodium falciparum malaria control in Africa contra-indications, pp. 4–6, World Health Organization, Geneva, Switzerland [Google Scholar]
- 22. Conteh L., Sicuri E., Manzi F., Hutton G., Obonyo B., Tediosi F., Biao P., Masika P., Matovu F., Otieno P., Gosling R. D., Hamel M., Odhiambo F. O., Grobusch M. P., Kremsner P. G., et al. (2010) The cost-effectiveness of intermittent preventive treatment for malaria in infants in sub-Saharan Africa. PLoS One 5, e10313 10.1371/journal.pone.0010313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blaszczyk J., Li Y., Cherry S., Alexandratos J., Wu Y., Shaw G., Tropea J. E., Waugh D. S., Yan H., and Ji X. (2007) Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics. Acta Crystallogr. D Biol. Crystallogr. 63, 1169–1177 10.1107/S0907444907047452 [DOI] [PubMed] [Google Scholar]
- 24. Garçon A., Levy C., and Derrick J. P. (2006) Crystal structure of the bifunctional dihydroneopterin aldolase/6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Streptococcus pneumoniae. J. Mol. Biol. 360, 644–653 10.1016/j.jmb.2006.05.038 [DOI] [PubMed] [Google Scholar]
- 25. Lawrence M. C., Iliades P., Fernley R. T., Berglez J., Pilling P. A., and Macreadie I. G. (2005) The three dimensional structure of the bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/dihydropteroate synthase of Saccharomyces cerevisiae. J. Mol. Biol. 348, 655–670 10.1016/j.jmb.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 26. Chhabra S., Dolezal O., Collins B. M., Newman J., Simpson J. S., Macreadie I. G., Fernley R., Peat T. S., and Swarbrick J. D. (2012) Structure of S. aureus HPPK and the discovery of a new substrate site inhibitor. PLoS One 7, e29444 10.1371/journal.pone.0029444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pemble C. W. 4th, Mehta P. K., Mehra S., Li Z., Nourse A., Lee R. E., and White S. W. (2010) Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase·dihydropteroate synthase bifunctional enzyme from Francisella tularensis. PLoS One 5, e14165 10.1371/journal.pone.0014165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yun M. K., Wu Y., Li Z., Zhao Y., Waddell M. B., Ferreira A. M., Lee R. E., Bashford D., and White S. W. (2012) Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 335, 1110–1114 10.1126/science.1214641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snounou G., and White N. J. (2004) The co-existence of Plasmodium: sidelights from falciparum and vivax malaria in Thailand. Trends Parasitol. 20, 333–339 10.1016/j.pt.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 30. Auliff A., Wilson D. W., Russell B., Gao Q., Chen N., Anh le N., Maguire J., Bell D., O'Neil M. T., and Cheng Q. (2006) Amino acid mutations in Plasmodium vivax DHFR and DHPS from several geographical regions and susceptibility to antifolate drugs. Am. J. Trop. Med. Hyg. 75, 617–621 [PubMed] [Google Scholar]
- 31. Hawkins V. N., Suzuki S. M., Rungsihirunrat K., Hapuarachchi H. C., Maestre A., Na-Bangchang K., and Sibley C. H. (2009) Assessment of the origins and spread of putative resistance-conferring mutations in Plasmodium vivax dihydropteroate synthase. Am. J. Trop. Med. Hyg. 81, 348–355 [PubMed] [Google Scholar]
- 32. Rungsihirunrat K., Sibley C. H., Mungthin M., and Na-Bangchang K. (2008) Geographical distribution of amino acid mutations in Plasmodium vivax DHFR and DHPS from malaria endemic areas of Thailand. Am. J. Trop. Med. Hyg. 78, 462–467 [PubMed] [Google Scholar]
- 33. Pornthanakasem W., Riangrungroj P., Chitnumsub P., Ittarat W., Kongkasuriyachai D., Uthaipibull C., Yuthavong Y., and Leartsakulpanich U. (2016) Role of Plasmodium vivax dihydropteroate synthase polymorphisms in sulfa drug resistance. Antimicrob. Agents Chemother. 60, 4453–4463 10.1128/AAC.01835-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Triglia T., Menting J. G., Wilson C., and Cowman A. F. (1997) Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 94, 13944–13949 10.1073/pnas.94.25.13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nalam M. N., Ali A., Altman M. D., Reddy G. S., Chellappan S., Kairys V., Ozen A., Cao H., Gilson M. K., Tidor B., Rana T. M., and Schiffer C. A. (2010) Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 84, 5368–5378 10.1128/JVI.02531-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altman M. D., Ali A., Reddy G. S., Nalam M. N., Anjum S. G., Cao H., Chellappan S., Kairys V., Fernandes M. X., Gilson M. K., Schiffer C. A., Rana T. M., and Tidor B. (2008) HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J. Am. Chem. Soc. 130, 6099–6113 10.1021/ja076558p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bertacine Dias M. V., Santos J. C., Libreros-Zúñiga G. A., Ribeiro J. A., and Chavez-Pacheco S. M. (2018) Folate biosynthesis pathway: mechanisms and insights into drug design for infectious diseases. Future Med. Chem. 10, 935–959 10.4155/fmc-2017-0168 [DOI] [PubMed] [Google Scholar]
- 38. Yun M. K., Hoagland D., Kumar G., Waddell M. B., Rock C. O., Lee R. E., and White S. W. (2014) The identification, analysis and structure-based development of novel inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase. Bioorg. Med. Chem. 22, 2157–2165 10.1016/j.bmc.2014.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winter G., Lobley C. M., and Prince S. M. (2013) Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69, 1260–1273 10.1107/S0907444913015308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waterman D. G., Winter G., Gildea R. J., Parkhurst J. M., Brewster A. S., Sauter N. K., and Evans G. (2016) Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr. D Struct. Biol. 72, 558–575 10.1107/S2053230X16009237,10.1107/S2059798316002187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., and Richardson D. C. (2000) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baca A. M., Sirawaraporn R., Turley S., Sirawaraporn W., and Hol W. G. (2000) Crystal structure of Mycobacterium tuberculosis 6-hydroxymethyl-7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J. Mol. Biol. 302, 1193–1212 10.1006/jmbi.2000.4094 [DOI] [PubMed] [Google Scholar]
- 44. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 47. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]