Abstract
DNA polymerase β (Pol β) is essential for maintaining genomic integrity. During short-patch base excision repair (BER), Pol β incorporates a nucleotide into a single-gapped DNA substrate. Pol β may also function in long-patch BER, where the DNA substrate consists of larger gap sizes or 5′-modified downstream DNA. We have recently shown that Pol β fills small gaps in DNA during microhomology-mediated end-joining as part of a process that increases genomic diversity. Our previous results with single-nucleotide gapped DNA show that Pol β undergoes two pre-catalytic conformational changes upon binding to the correct nucleotide substrate. Here we use FRET to investigate nucleotide incorporation of Pol β with various DNA substrates. The results show that increasing the gap size influences the fingers closing step by increasing its reverse rate. However, the 5′-phosphate group has a more significant effect. The absence of the 5′-phosphate decreases the DNA binding affinity of Pol β and results in a conformationally more open binary complex. Moreover, upon addition of the correct nucleotide in the absence of 5′-phosphate, a slow fingers closing step is observed. Interestingly, either increasing the gap size or removing the 5′-phosphate group results in loss of the noncovalent step. Together, these results suggest that the character of the DNA substrate impacts the nature and rates of pre-catalytic conformational changes of Pol β. Our results also indicate that conformational changes are important for the fidelity of DNA synthesis by Pol β.
Keywords: FRET, kinetics, conformational change, DNA polymerase, crystal structure, 5′-phosphate, gap size, polymerase β
Introduction
Genomic DNA is continuously exposed to endogenous and exogenous reactive oxygen species (1, 2). The base excision repair (BER)3 pathway removes ∼20,000 lesions per human cell per day (3). DNA polymerase β (Pol β) is a small family X DNA polymerase with a molecular mass of 39 kDa. It plays an essential role in maintaining the fidelity of DNA synthesis during BER. Pol β is composed of two functional domains. The 8-kDa lyase domain binds downstream DNA and removes the 5′-deoxyribose phosphate group, generating a 5′-phosphate to permit ligation (4). The polymerase domain catalyzes the incorporation of the incoming nucleotide via a magnesium ion–dependent mechanism (5). The incorporation reaction is coordinated through Pol β's three subdomains; i.e. the thumb, palm, and fingers subdomains, which are responsible for DNA binding, catalysis, and nucleotide binding, respectively (6–8).
Early studies showed that Pol β is nonprocessive on 3′-recessed DNA templates but can fill short gapped DNA substrates to completion (9, 10). Work from our laboratory and others demonstrated that Pol β exhibits the highest catalytic efficiency and fidelity on single-nucleotide gapped DNA (11, 12). Therefore, it is suggested that the single-nucleotide gap is a physiologically relevant DNA substrate for Pol β. Additionally, the phosphate group on the 5′ end of the gap is important for both the DNA binding and nucleotide incorporation of Pol β (13, 14). Prasad et al. (14) showed that the binding affinity of Pol β to a 5-nt gapped DNA is strongly enhanced when the 5′-phosphate group is present on the downstream DNA, which allows a processive mechanism for nucleotide incorporation. The crystal structure of the binary Pol β–DNA complex revealed a lysine-rich 5′-phosphate binding pocket in the lyase domain, with Lys-35 and Lys-68 hydrogen-bonded with the 5′-phosphate on downstream DNA. Interestingly, it is thought that the lyase domain tethers the polymerase domain during a multinucleotide gap (less than 6 nt) filling step (15, 16).
Several studies have been performed to characterize the effect of gap size on the kinetics of nucleotide incorporation by Pol β (11, 17, 18). In general, increasing the gap size has a dramatic effect on the apparent equilibrium dissociation constant of dNTP (Kd (dNTP)), with the single-gapped DNA showing the lowest Kd (dNTP). In addition, increasing gap size was associated with decreased fidelity of DNA synthesis (17). However, there was little effect on the rate of polymerization (kpol) (17, 18). Under steady-state conditions, a significantly decreased affinity is observed for nucleotide binding when the 5′-phosphate group is removed, but the overall catalytic rate was less affected (11).
DNA polymerases are known to undergo conformational changes when incorporating a correct nucleotide (19–24). Structural analyses of the Klenow fragment, T7 polymerase, HIV reverse transcriptase, and Pol β show a major change of the Pol–DNA binary complex upon dNTP binding (6, 15, 25–27). Specifically, the fingers domain of the polymerase closes onto the DNA substrate, aligning the terminal 3′-OH for the nucleophilic attack on the Pα of the incoming nucleotide to form a phosphodiester bond, which is coordinated through a two- or perhaps three-metal ion-dependent mechanism (5, 28). These conformational changes have been linked to polymerase fidelity (20, 22, 29). After the chemistry step, at least one additional conformational step is likely to facilitate the release of pyrophosphate (30, 31). Several studies have characterized conformational changes during Pol β catalysis and provide evidence that the fingers closing is rapid (19, 32–34), whereas chemistry is the rate-limiting step (35–37). More recently, our laboratory developed a FRET system to monitor the movements of Pol β during nucleotide incorporation and found that Pol β undergoes two conformational changes. Upon binding to the correct nucleotide, the fingers domain of Pol β closes onto the palm domain, followed by a second pre-catalytic noncovalent step (NCS) right before nucleotidyl transfer (or chemistry) (38). We proposed that the second NCS may be involved in the binding of catalytic metal into the Pol β active site (the NCS throughout the text refers in particular to the second NCS step) (38). Using this system, we have also discovered that nucleotide release before chemistry plays an important role in the fidelity of Pol β, similar to what has been found for HIV reverse transcriptase (22, 24, 29). To characterize the nature of pre-catalytic conformational changes of Pol β with a variety of DNA substrates, we performed FRET analysis of Pol β on various DNA substrates that vary in both the gap size and the modification of 5′ termini on the downstream DNA. Our results suggest that both the gap size and the 5′-phosphate group influence the pre-catalytic conformational steps.
Results
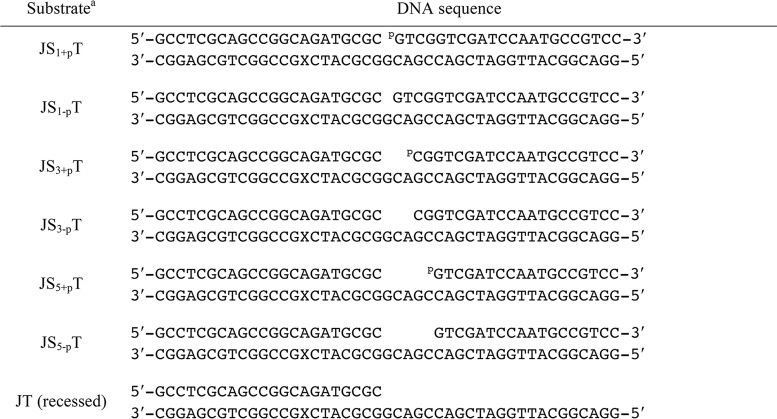
In this study, we created a series of DNA substrates to gain a deeper understanding of the catalytic mechanism of Pol β. These DNA substrates vary in gap sizes and include 1-, 3-, and 5-nt gaps with and without the 5′-phosphate group on the downstream DNA so that we could evaluate the effects of gap size and the 5′-phosphate group on catalysis. Detailed information for all DNA substrates employed in our experiments is shown in Table 1.
Table 1.
DNA substrates used in this study
a The DNA substrates used in this study. The primer, downstream, and template DNA are defined as J, S, and T, respectively. Systematic modifications were introduced to the downstream DNA (S), as indicated by the subscript, where 1, 3, and 5 represent the gap size and +p and −p represent the presence or absence of the 5′-phosphate group on the downstream DNA (S). The X on the template DNA denotes dT with a Dabcyl modification and is located at the T(−8) position of the template DNA.
Pol β binds better to DNA with a 5′-phosphate group
We first examined the ability of Pol β to bind to the various DNA substrates. Pol β binds to the canonical single-gapped DNA (JS1+pT) tightly, with a KD (DNA) of 0.5 nm (Fig. 1). When the 5′-phosphate group on the downstream single-nucleotide gapped DNA is not present (JS1-pT), the binding affinity is significantly reduced to 20 nm. Increasing the gap size of DNA with a 5′-phosphate group to 3 nt and 5 nt only slightly reduces the binding affinity to 2.0 and 1.0 nm, respectively (Fig. 1). Interestingly, the KD (DNA) does not appear to correlate with an increase in the size of the gap in these experiments, as Pol β interacts with the 3-nt gap with the lowest binding affinity for substrates with a 5′-phosphate. When the 5′-phosphate groups are not present in the 3- and 5-nt gapped DNA substrates, the binding affinity dramatically drops to 12 and 18 nm (Fig. 1). Last, when the downstream DNA is not present at all, the binding affinity is similar to that of DNA substrates without the 5′-phosphate group. In summary, our results suggest that the 5′-phosphate group has a more significant effect than gap size on the binding affinity of Pol β for DNA.
Figure 1.
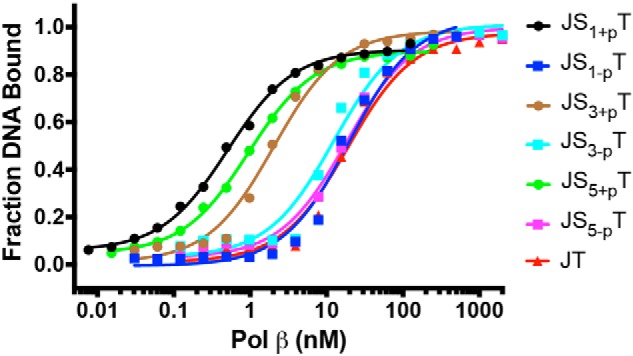
The 5′-phosphate group has a dominant effect on the binding of Pol β. DNA (0.1 nm) was incubated with varying amounts of Pol β (0.0076–2000 nm) in binding buffer for 15 min at 23 °C. Sigmoidal plots of the fraction of DNA bound versus the log of protein concentration show the differences for all seven DNA substrates. The binding affinities of Pol β to various DNA substrates are 0.5 ± 0.0 nm (JS1+pT), 20 ± 3 nm (JS1-pT), 2.0 ± 0.3 nm (JS3+pT), 12 ± 3 nm (JS3-pT), 1.0 ± 0.1 nm (JS5+pT), 18 ± 3 nm (JS5-pT), and 19 ± 3 nm (JT).
The effects of DNA gap size and 5′-phosphate on nucleotide incorporation
Single turnover kinetics measurements were performed to determine the maximum rate of nucleotidyl transfer, kpol, and apparent nucleotide dissociation constant, Kd (dNTP) for single-nucleotide incorporation for all DNA substrates (Figs. S1 and S2). The kpol and Kd (dNTP) for the single-gapped DNA substrate (JS1+pT) are 3.0 s−1 and 1.5 μm, respectively (Table 2). We observed a slight decrease of kpol and increase of Kd (dNTP), 2.5 s−1 and 4.6 μm, respectively, for single-nucleotide gapped DNA substrate without the 5′-phosphate group (JS1-pT). Increasing the gap size to 3 nt and 5 nt has a profound effect on Kd (dNTP) but does not have a strong effect on kpol. In addition, Pol β exhibits a significantly higher Kd (dNTP) with DNA substrates that do not contain a 5′-phosphate group compared with those that contain the 5′-phosphate group (Table 2). Overall, among the DNA substrates studied here, we demonstrate that the dNTP binding affinity decreases as the DNA gap size increases and that the absence of a 5′-phosphate on the downstream DNA also results in decreased affinity of Pol β for dNTP. However, the rates of incorporation have a much weaker dependence on both the gap size and 5′-phosphate groups.
Table 2.
DNA gap size and 5′-phosphate impact nucleotide binding
| Substrate | Gap size | 5′-Pa | kpol | Kd (dNTP) | kpol/Kd (dNTP) |
|---|---|---|---|---|---|
| nt | s−1 | μm | s−1 μm−1 | ||
| JS1+pT | 1 | Yes | 3.0 ± 0.1 | 1.5 ± 0.2 | 2.0 ± 0.3 |
| JS1−pT | 1 | No | 2.5 ± 0.1 | 4.6 ± 0.4 | 0.5 ± 0.1 |
| JS3+pT | 3 | Yes | 1.7 ± 0.0 | 3.9 ± 0.2 | 0.4 ± 0.0 |
| JS3−pT | 3 | No | 1.6 ± 0.0 | 43.8 ± 4.8 | 0.04 ± 0.00 |
| JS5+pT | 5 | Yes | 1.2 ± 0.0 | 12.1 ± 1.7 | 0.1 ± 0.0 |
| JS5−pT | 5 | No | 1.4 ± 0.1 | 73.0 ± 9.9 | 0.02 ± 0.00 |
| JT | Recessed | Nob | 1.8 ± 0.1 | 32.5 ± 4.0 | 0.06 ± 0.01 |
a 5′-P is the 5′-phosphate group on the downstream DNA.
b This DNA substrate does not contain the downstream DNA.
We also calculated the incorporation efficiency for the correct nucleotide dCTP for each DNA substrate (Table 2 and Fig. 2). It is clear that the 5′-phosphate groups separated the results into two groups, where DNA substrates with the 5′-phosphate have a higher incorporation efficiency than the DNA substrates of the same gap size but without the 5′-phosphate. These differences mainly result from the decreased affinity of dNTP binding rather than the rate of incorporation. For both groups, the incorporation efficiency also decreases with increasing gap size as a result of the weaker dNTP binding to Pol β binary complexes with DNA substrates containing larger gap sizes. The recessed DNA, which is a mimic of infinite gap size without the 5′-phosphate, has a better incorporation efficiency than the 3-nt and 5-nt DNA substrates without the 5′-phosphate.
Figure 2.
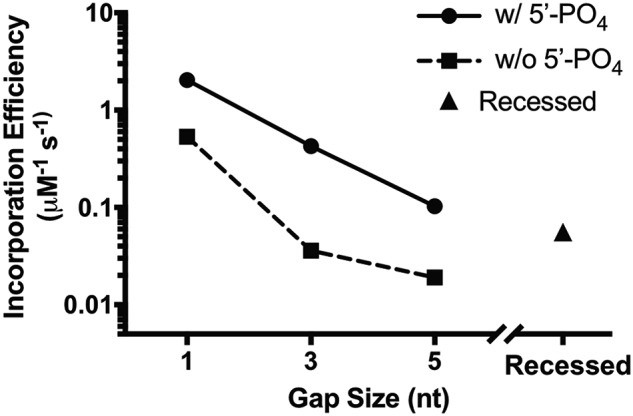
Effect of gap size and 5′-phosphate group on the correct nucleotide incorporation efficiency by Pol β.
Formation of binary complexes depends on the DNA substrates
Our FRET system is constructed to measure the fluorescence signal change that corresponds to the distance of Val-303 on the Pol β fingers domain to location X (position of the Dabcyl) on the template DNA (Table 1 and Fig. 3). Therefore, the distance (r) between the Pol β fingers domain and the DNA template can be calculated for each DNA substrate in the presence and absence of dNTP using the FRET efficiency (Table 3). Among all DNA substrates investigated, we observed the shortest distance between the dye on V303C and the DNA template upon Pol β binding to the JS1+pT DNA to form the binary complex, with a distance r at 44.8 Å (Table 3). Addition of the correct nucleotide further reduces the distance r to 40.6 Å (Table 3), suggesting the formation of a ternary complex. Interestingly, increasing the DNA gap size to 3 nt or 5 nt slightly increases the r to 47.7 Å and 47.3 Å for JS3+pT and JS5+pT, respectively (Table 3), in the Pol β binary complex. However, when the 5′-phosphate group is removed from the downstream DNA, the binary complexes appear to assume a dramatically more open form with a distance of ∼51–53 Å (Table 3), regardless of the gap size (JS1-pT, JS3-pT, and JS5-pT), including the DNA substrate that does not have the downstream DNA (JT). Although the r for binary complexes exhibit variations among different DNA substrates depending on the gap size and the 5′-phosphate group, the r for ternary structures shows little dependence on the properties of the DNA substrates, with a final distance r of around 40.3 Å to 41.9 Å for all DNA substrates (Table 3).
Figure 3.
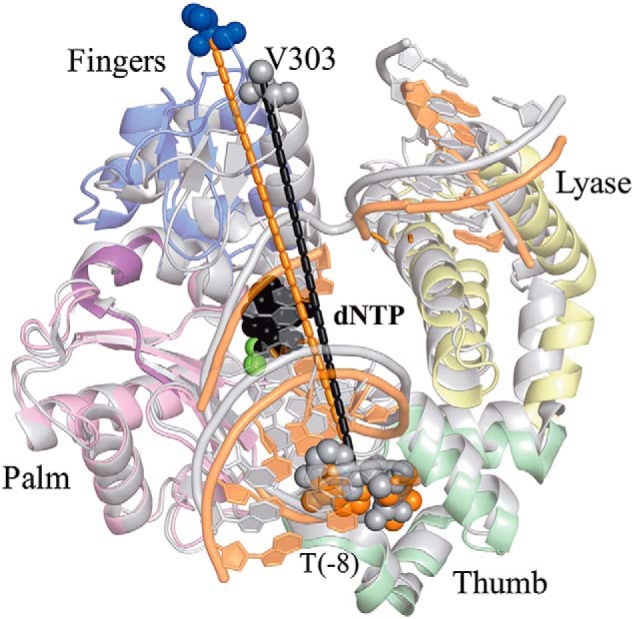
The FRET system monitors the distance (r) change between the IAEDANS label on V303C of Pol β and the Dabcyl label on template DNA at the −8 position. Addition of the correct nucleotide induces a movement of the fingers domain, resulting in shortening of the distance (r) between the IAEDANS label on V303C (blue to gray) and the Dabcyl label on the template DNA at the −8 position. The PDB codes for ternary (gray tones) and binary (colored by domain) are 2FMS and 3ISB, respectively. Structures are superimposed via the palm domain.
Table 3.
Distance (r) of Val-303 on the Pol β fingers domain to the Dabcyl quencher on the template DNA, calculated based on FRET efficiencies
| JS1+pT | JS1−pT | JS3+pT | JS3−pT | JS5+pT | JS5−pT | JT | |
|---|---|---|---|---|---|---|---|
| Binary (Å) | 44.8 ± 0.6 | 51.4 ± 1.2 | 47.7 ± 0.6 | 53.5 ± 0.6 | 47.3 ± 0.7 | 51.7 ± 1.3 | 52.7 ± 0.8 |
| Ternary (Å) | 40.6 ± 0.7 | 41.9 ± 1.0 | 40.7 ± 0.8 | 41.8 ± 1.0 | 41.8 ± 0.2 | 40.7 ± 0.1 | 40.3 ± 0.4 |
Pre-catalytic conformational changes depend on the gap size and 5′-phosphate
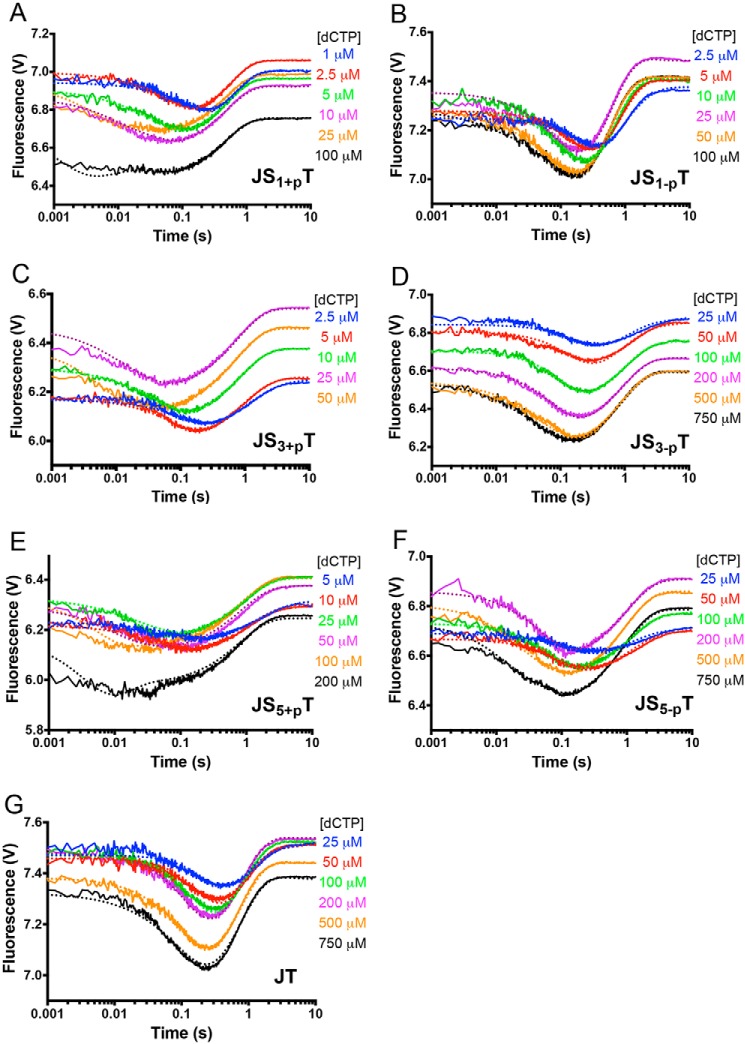
Our laboratory previously established a FRET system to investigate the kinetic parameters associated with the pre-catalytic steps of Pol β during nucleotide incorporation. We found that, after correct nucleotide binding, Pol β undergoes fingers closing, followed by a noncovalent step prior to nucleotidyl transfer (or chemistry) (38). Here we employ the same FRET system to characterize the pre-catalytic steps among different DNA substrates. For these measurements, DNA substrates of various gap sizes with or without the 5′-phosphate are pre-incubated with an excess of Pol β under single turnover conditions. Then each of the DNA–Pol β binary complexes is mixed with various concentrations of dCTP, and the fluorescence signal is recorded as a function of time (Fig. 4). Pre-catalytic conformational changes are observed upon mixing each of the Pol β-DNA binary complexes with dCTP in a manner that depends on the concentration of nucleotide for each DNA substrate (Fig. 4). First, at high dCTP concentrations, the time to reach the fluorescence signal minimum decreases compared with the low dNTP concentrations. As the fluorescent signal is a measurement of distance between the Pol β fingers domain and template DNA, the shorter time it takes to reach the signal minimum, the faster Pol β undergoes the conformational changes to its closed state. This observation applies to all DNA sequences.
Figure 4.
FRET demonstrates that the binary complex of Pol β and DNA substrates with 5′-phosphate groups undergo faster fingers closing upon binding to the correct nucleotide. A–G, the fingers domain of Pol β closes in the presence of increasing dCTP concentrations for JS1+pT (A), JS1-pT (B), JS3+pT (C), JS3-pT (D), JS5+pT (E), JS5-pT (F), and JT (G) DNA. The nucleotide concentrations used are listed in each panel. The dotted lines from the KinTek Explorer modeling results overlay the experimentally determined FRET traces, shown as solid lines.
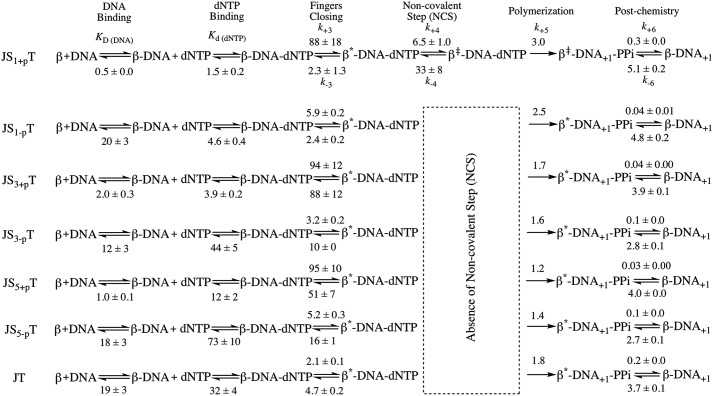
To estimate the rates of the pre-catalytic steps, we used KinTek Global Explorer and analyzed our data based on the model from our earlier publication (38), which includes a fingers closing step (step 3) and noncovalent step (step 4) prior to chemistry (step 5) (see below),
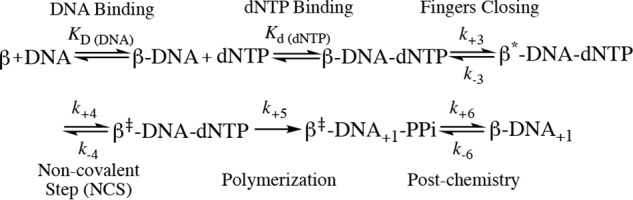
The goodness of fit of the modeling results was evaluated using FitSpace 2D, which allows us to explore the landscape over which parameters can vary while still achieving a good fit to the data (39, 40). Analysis of the single-nucleotide gapped DNA substrate (JS1+pT) gave comparable kinetic parameters compared with our previous results, with the fingers closing step proceeding at a forward rate k+3 of 88 s−1 and a reverse rate k-3 of 2.3 s−1, with an overall fingers closing forward constant K3 = k+3/k−3 of ∼ 38 (Scheme 1). The noncovalent step exhibits a forward rate k+4 of 6.5 s−1 and a reverse rate k−4 of 33 s−1 (Scheme 1). FitSpace 2D analysis of modeling the JS1+pT DNA suggests good confidence in the modeling of our data (Fig. S3). We also attempted to model the JS1+pT DNA FRET data without the noncovalent step and rejected this model upon evaluation with FitSpace 2D (Fig. S4). Using this approach, we modeled FRET data obtained with each of the DNA substrates and evaluated our models using FitSpace 2D (Scheme 1 and Figs. S5–S10). The results obtained with the rest of the DNA substrates were best modeled to a kinetic scheme that did not contain a noncovalent step between fingers closing and chemistry (Scheme 1). Interestingly, when the gap size is increased to 3 nt and 5 nt (JS3+pT and JS5+pT), the forward fingers closing rate (k+3) remains similar compared with the single-gapped DNA substrate (JS1+pT), but the reverse fingers closing rate (k−3) increases for larger gap sizes, ultimately resulting in a slow overall kinetic constant for fingers movement (K3). For DNA substrates lacking the 5′-phosphate groups (JS1-pT, JS3-pT, and JS5-pT), the fingers close at much slower forward rates (k+3) (Scheme 1) compared with what we observe with DNA substrates possessing a 5′-phosphate group. However, the reverse rate of fingers closing (k−3) is faster with DNA substrates having 3-nt and 5-nt gaps compared with a single nucleotide gap, which is similar to what we observe for DNA substrates with the 5′-phosphate groups, leading to a slow overall kinetic constant of fingers movement (K3) as well. Pol β exhibits a slow forward rate of fingers closing (k+3) with the JT DNA substrate (Scheme 1), as would be expected for DNA that does not possess a 5′-phosphate group. However, the reverse fingers closing rate (k−3) for Pol β with the JT substrate is slower compared with the DNA substrates with large gap size (JS3-pT and JS5-pT). In general, our analysis indicates that the 5′-phosphate group influences the forward rate of fingers closing and that gap size influences the rate of reverse fingers closing. In addition, K3 is extremely low for Pol β when it employs the three- and five-based gapped DNA substrates as well as the recessed DNA primer template (JT).
Scheme 1.
Kinetic scheme of Pol β with various DNA substrates. The noncovalent step is only present for JS1+pT DNA but not for other DNA substrates. The parameters are listed for each step. The overall fingers closing forward constant K3 = k+3/k−3 is calculated for each DNA substrate, and the values are 38, 1.1, 1.9, 2.4, 0.3, 0.3, and 0.4 for JS1+pT, JS3+pT, JS5+pT, JS1-pT, JS3-pT, JS5-pT, and JT, respectively. The unit for KD (DNA) is nanomolar and for Kd (dNTP) is micromolar. The units for fingers closing, the noncovalent step, and polymerization (both forward and reverse reaction) are per second. The unit for the post-chemistry step forward reaction is per second and for the reverse reaction is per micromolar per second.
Reverse fingers closing for JS1+pT and JS1-pT DNA
To provide direct measurements of the reverse rates of fingers closing, the excess nonlabeled DNA–Pol β binary complex was used to trap the dNTP from a preformed and labeled ddDNA–Pol β–dNTP ternary complex. In these experiments, the trapping of dNTP from the ternary complex will allow us to detect reverse conformational changes, which will be reflected by an increase in the fluorescence signal (Fig. 5) (22, 29). The trapping of dNTP will trigger the reverse conformational changes, as illustrated below,
Figure 5.
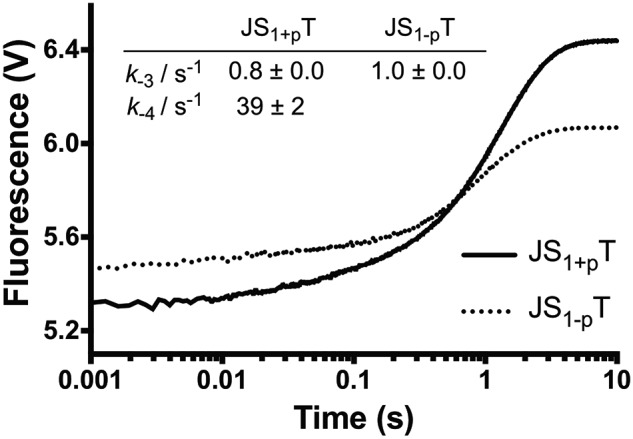
Reverse fingers closing. We measured the reverse fingers closing rates by mixing preformed ternary complex and excess of binary complex, followed by monitoring the FRET signal as described under “Experimental procedures.” The data of JS1+pT were fit to a double exponential equation to generate a k−4 at 39 ± 2 s−1 and k−3 at 0.8 ± 0.0 s−1, where the rate of k−3 corresponds to 90% of the signal. The data of JS1-pT can only be fit with a single exponential equation to generate a k−3 at 1.0 ± 0.0 s−1.
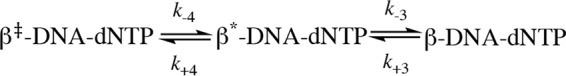
In this experiment, the trap concentrations should be higher than the dNTP concentrations to serve as an efficient trap. As a result, only 2 μm dNTP can be used in these experiments, which permits us to directly measure the reverse rates of the gapped DNA–Pol β–dCTP complex in an accurate manner. Fitting of the traces for Pol β bound to the JS1+pT DNA substrate to a double exponential equation revealed rates of 39 ± 2 and 0.8 ± 0.0 s−1 (Fig. 5), which are relatively close to our assignment for the two rates as k−4 of 33 ± 8 s−1 and k−3 at 2.3 ± 1.3 s−1 from our modeling results (Scheme 1). The contribution of the slow phase is more than 90% and possibly associated with the larger conformational changes induced by the fingers movement; the fast phase only accounts for ∼10% of the signal, suggesting a smaller conformational change, such as the noncovalent step. Importantly, the reverse reaction for JS1-pT DNA can only be fitted to a single exponential equation, with a k−3 rate of 1.0 ± 0.0 s−1 (Fig. 5), which is similar to what was obtained in our model in the absence of the noncovalent step (Scheme 1).
Discussion
In this study, we employed FRET to monitor conformational changes associated with correct nucleotide incorporation by Pol β with various DNA substrates. When the DNA substrate is a single-nucleotide gap with a 5′-phosphate, Pol β undergoes two pre-catalytic conformational changes, fingers closing and the noncovalent step. In addition, fingers closing is fast with a slow reverse rate when Pol β employs this DNA substrate. However, increasing the DNA gap size or eliminating the 5′-phosphate groups leads to loss of the pre-catalytic noncovalent step. Although fingers closing is fast, with 3- and 5-nt gapped substrates harboring a 5′-phosphate, so is the rate of the reverse fingers closing step. In addition, removal of the 5′-phosphate leads to slow fingers closing, with reverse rates similar to what is observed with the single-nucleotide gapped DNA with a 5′-phosphate. We conclude that the nature of the DNA substrate of Pol β influences its catalytic mechanism. Previous results suggest that Pol β exhibits the greatest fidelity on single-nucleotide gapped DNA with a 5′-phosphate (11, 17). These findings, in combination with the results here, show that the molecular motions are altered when Pol β employs alternative substrates other than a single nucleotide gap and suggest that these molecular motions govern the fidelity of Pol β. Our results also indicate that the pre-catalytic noncovalent step plays an important function in substrate discrimination by Pol β.
DNA substrate influences binary complex conformation
Loss of the 5′-phosphate group has a profound effect on the DNA binding affinity of Pol β with the DNA substrates examined in this study. It lowers the affinity of Pol β for the DNA by almost 40-fold compared with the single-gapped DNA with a 5′-phosphate group (Fig. 1). Interestingly, Pol β exhibits a more open conformation in a binary complex with DNA substrates lacking the 5′-phosphate, as evidenced by greater distances between FRET labels in our experimental system. Whether the binary conformations are simply more open or also have altered spatial relationships between the DNA substrate and enzyme and/or between amino acid residues within Pol β remains to be characterized. We point out that recent NMR spectroscopy analysis suggests that the conformation of the binary complex is important for substrate discrimination by Pol β using an induced fit mechanism (41, 42). Because the nature of the DNA substrate impacts the binary conformation, our results suggest that the nature of the DNA substrate is likely to influence substrate discrimination by Pol β.
The fingers and lyase domains interact upon formation of the ternary complex with single-nucleotide gapped DNA
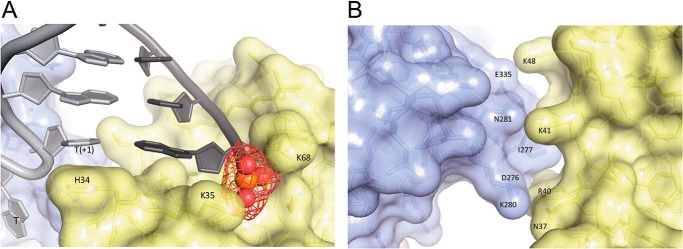
The crystal structure of Pol β with single-nucleotide gapped DNA defines several interactions. First, the lyase domain interacts with downstream DNA, and Lys-35 and Lys-68 interact with the 5′-phosphate group (Fig. 6A) (15). Second, the imidazole side chain from His-34 stacks with the nucleobase at the T+1 position of the DNA template, which further stabilizes the interactions between the lyase domain and the DNA (Fig. 6A). A similar interaction is also found in DNA polymerase l (Pol λ), another family X DNA polymerase, in which Trp-274 stacks with T+1 position, which facilitates the bending of DNA (43). Finally, several electrostatic or hydrophobic interactions are possible between the lyase and fingers domains of Pol β in the ternary structure, as illustrated in Fig. 6B. Different from the ternary structure of Pol β with a single-nucleotide gapped DNA, the ternary structure of Pol β with recessed DNA, the extreme case among the DNA substrates we studied, exhibits different structural properties, especially in the lyase domain (Fig. 7). Lack of the downstream DNA also removes the 5′-phosphate group, which eliminates the interaction with the lyase domain. The lyase domain of Pol β with a recessed DNA substrate assumes a relatively open state compared with its positioning in the presence of single-nucleotide gapped DNA (Fig. 7). Therefore, none of the interactions mentioned above exist when the downstream DNA is absent. Unfortunately, no structural information is available for Pol β with larger gap sizes, although it is likely that gap size influences the nature of the interactions between the lyase and fingers domains.
Figure 6.
A, the lyase domain of Pol β interacts with DNA in the ternary structure. Lys-35 and Lys-68 interact with the 5′-phosphate group on the downstream DNA. His-34 stacks with the nucleobase at the T+1 position of the template DNA. B, interactions between the lyase domain (yellow) and fingers domain (light blue) of Pol β in the ternary structure. The PDB code for structure is 2FMS (47).
Figure 7.
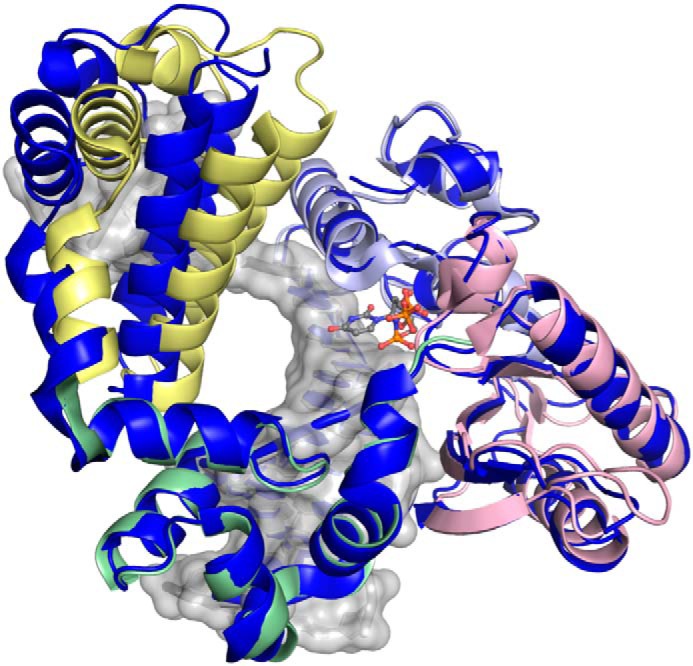
Shown is a ternary complex of Pol β containing 1-nt gapped DNA with a 5′-phosphate group (PDB code 2FMS) (47), with each domain shown by color (lyase, yellow; thumb, green; palm, pink; fingers, light blue) superimposed with a recessed ternary structure, PDB code 2BPG) (6), shown in dark blue, with the only significant deviation in global conformation observed for the lyase domain.
Gap size and the presence of 5′-phosphate influence rates of pre-catalytic conformational changes
Enlargement of the gap from one to three or five nucleotides increases the rates of reverse fingers closing compared with single-nucleotide gapped DNA. However, fast forward fingers closing remains intact for these larger gapped substrates. It is tempting to speculate that the fingers and lyase domains interact in a very stable manner in the presence of single-nucleotide gapped DNA, impeding reverse fingers closing. We speculate that the stability of the interaction between these domains with the 3- and 5-t gapped DNA substrates is compromised and is more permissive for a fast reverse fingers closing step.
DNA substrates lacking 5′-phosphate exhibit slow fingers closing, with rates similar to chemistry, even with single-nucleotide gapped DNA. This result indicates that the interaction of Lys-35 and Lys-68 with the 5′-phosphate on the DNA facilitate rapid fingers closing in some unknown manner that is not likely to be due to interactions between the lyase and fingers domains, as they occur after fingers closing. One possibility is that the more open binary complex that is observed for DNA without the 5′-phosphate group may assume a structure that is not amenable to rapid closing.
The NCS requires single-nucleotide gapped DNA
Although the function of the NCS is still vague, it is thought to be involved in metal ion coordination (38), based on the disappearance of this step when Mg2+ is replaced by Ca2+, similarly shown by Joyce et al. (20) for the Klenow fragment. For DNA substrates without the 5′-phosphate groups and gaps larger than 1 nt, FRET data cannot be modeled well when the NCS is included in the reaction scheme, suggesting that, for these substrates, only a single conformational change (namely, fingers closing) occurs prior to chemistry. Hence, removing the 5′-phosphate groups eliminates the NCS, resulting in Pol β utilizing only a single conformational change to discriminate between dNTP substrates during nucleotide incorporation. Previous results from our laboratory showed that removal of the 5′-phosphate group enhances the misincorporation frequency (11). In combination with our current results, we suggest that loss of the NCS may be a causal factor for the low fidelity exhibited by Pol β when it utilizes noncanonical DNA substrates.
Conclusion
Our pre-steady-state FRET analysis revealed significantly altered conformational change rates and the absence of the NCS with DNA substrates other than the single nucleotide gap with a 5′-phosphate, consistent with the observed differences in various crystal structures of Pol β. These results suggest that the nature of the DNA substrate influences the catalytic mechanism of Pol β. Increasing the gap size has a moderate effect by increasing the reverse rate of the fingers closing step. Removal of the 5′-phosphate group has a more profound impact along the kinetic pathway. Alteration of gap size or loss of the 5′-phosphate results in loss of the NCS. Alteration of pre-catalytic steps as a result of the nature of the DNA substrate is suggested to be responsible for the lower fidelity of Pol β when utilizing these noncanonical DNA substrates. Our findings contribute to the understanding of the mechanism of Pol β during nucleotide incorporation and the critical steps involved to govern the fidelity of polymerization.
Pol β participates in the base excision repair pathway and creates a nicked DNA, which is later sealed by DNA ligase. Our results suggest that the properties of Pol β depend on the nature of the DNA substrates so that the absence of 5′-phosphate groups dramatically reduces the conformational change rates and catalytic efficiencies. Moreover, DNA substrates lacking the 5′-phosphate groups on downstream DNA will not be substrates for DNA ligase. The poor performance of Pol β on these DNA substrates might facilitate hand-off to other BER or alternative repair pathway components, but we currently do not have data to support such a conclusion. Studies from Joyce et al. (20), Tsai and co-workers (21), Kellinger and Johnson (22), and our group (29) show that the pre-catalytic conformational step, including the reverse step, affects the fidelity of several polymerases. Therefore, the altered conformational change rates for DNA substrates other than the single-nucleotide gapped DNA might affect the fidelity of Pol β. Further studies may help to address the fidelity of DNA repair on various DNA substrates of Pol β, and we will not pursue it in this manuscript.
Experimental procedures
Generation of IAEDANS-labeled human WT Pol β
Expression and purification of human WT Pol β was conducted as described previously (38). To label the Pol β with IAEDANS, three mutations, V303C, C267S, and C239S, were introduced, and the IAEDANS was attached to the cysteine mutation at the 303 position. All following work was conducted with IAEDANS-labeled human WT Pol β except when noted otherwise.
Generation of DNA substrates
Oligonucleotides were purchased from the Keck Oligo Synthesis Resource at Yale University with the necessary modifications and purified by 20% (w/v) denaturing PAGE. Primer, downstream, and template oligonucleotides were mixed in a 1:1.5:1.2 ratio and allowed to anneal in buffer containing 50 mm Tris-HCl (pH 8.0) and 250 mm NaCl to generate DNA substrates with various gap sizes (Table 1). The annealing of the DNA was confirmed by 12% (w/v) native PAGE.
Gel mobility shift assay
A gel shift assay was performed to determine the binding affinity of Pol β to a specific DNA substrate. Various concentrations of Pol β (0.0076 to 2000 nm) were incubated with 0.1 nm each radiolabeled DNA substrate in buffer containing 10 mm Tris-Cl (pH 7.6), 100 mm NaCl, 6 mm MgCl2, 10% (v/v) glycerol, and 0.1% (v/v) IGEPAL at 23 °C for 15 min. Samples were loaded onto a 6% (w/v) native polyacrylamide gel at a voltage of 300 V during loading at 4 °C and then reduced to 150 V during electrophoresis. The samples were visualized by autoradiography. Pol β bound to DNA resulted in a shift of the DNA on the gel compared with DNA without bound Pol β. The fraction bound is the ratio of the intensity of all shifted species divided by the total. The dissociation constant for Pol β to DNA (KD (DNA)) was estimated from fitting the bound protein (Y) versus protein concentration (x) with equation 1,
| (Eq. 1) |
where m is a scaling factor and b is the apparent minimum Y value.
Single-turnover kinetics
Single-turnover conditions were determined empirically for each DNA substrate at 22 °C by titration of increasing concentrations of DNA substrate with Pol β. A minimum ratio of 4:1 Pol β to DNA was required to achieve the single-turnover conditions. Reactions were performed at 22 °C using a KinTek rapid quench flow apparatus (RQF-3). A solution consisting of 400 nm Pol β, 100 nm DNA, 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 10% (v/v) glycerol was incubated for 5 min on ice. The incorporation reaction was initiated by adding various concentrations of dCTP (listed in each figure legend) in buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 20 mm MgCl2. For each reaction in the RQF, 15 μl of DNA/Pol β and 15 μl of dCTP were loaded into the reaction loops, rapidly mixed, and incubated for the designated times before being quenched by 88 μl of 500 mm EDTA. The final reaction concentrations were 50 nm DNA, 200 nm Pol β, dCTP (various concentrations), 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10 mm MgCl2, and 10% (v/v) glycerol. The samples were resolved on a 20% (w/v) denaturing polyacrylamide gel and visualized by autoradiography. The amounts of products were quantified by ImageQuant software and plotted versus time. The data were fitted to a single exponential equation,
| (Eq. 2) |
for each nucleotide concentration, where A is the amplitude of product formation, kobs is the observed rate, and t is the time. The kobs from these fits were plotted versus [dCTP] and fitted to the hyperbolic equation 3,
| (Eq. 3) |
where kpol is the polymerization rate of the enzyme and Kd (dNTP) is the equilibrium dissociation constant of the incoming dNTP from the Pol β–DNA binary complex.
Steady-state fluorescence emission scan
The fluorescence of Pol β was measured at room temperature on a Photon Technology International spectrofluorometer in buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10% (v/v) glycerol, and 10 mm MgCl2. The sample was excited at 336 nm, and an emission scan was performed from 420 nm to 580 nm at a resolution of 1 nm. The fluorescence signal of Pol β was measured first, followed by addition of nonextendable DNA substrate and the second scan. Finally, a saturating concentration of dCTP was added to the above solution, and the fluorescence signal was recorded. Dilutions after each addition were accounted for in the final analysis.
To correlate our fluorescence data with the structural distance between our two probes, we calculated the efficiency of energy transfer (EFRET) between the IAEDANS-labeled V303C and Dabcyl-labeled DNA using equation 4 (44),
| (Eq. 4) |
where FDA is the emission of IAEDANS at 490 nm in the presence of Dabcyl and FD is the fluorescence of IAEDANS in the absence of Dabcyl-labeled DNA. The efficiency was then used in equation 5 to estimate the distance separating the two probes (r),
| (Eq. 5) |
where R is the Förster radius, defined as the distance at which energy transfer is 50%. This distance was estimated to be 37.76 Å using equation 6,
| (Eq. 6) |
where κ2 is the relative orientation of the transition dipoles of the probes and is assumed to be equal to ⅔ for a dynamic random average, and η is the refractive index, assumed to be 1.344 in a solution of Tris-HCl (45). Fd is the fluorescence quantum yield of IAEDANS in the absence of Dabcyl, assumed to be 0.7 (46), and J is the spectral overlap integral in units M−1 cm3, which was measured and calculated using equation 7,
| (Eq. 7) |
where we measured ED, the maximum normalized emission of IAEDANS and ϵA, the extinction coefficient of Dabcyl at each wavelength (λ).
Stopped-flow FRET
All experiments were conducted at 22 °C on the stopped-flow SX-20 (Applied Photophysics) instrument. Samples were excited at 336 nm, and the emission was filtered with a 400-nm filter with the voltage set at 400 V.
To monitor nucleotide-induced conformational changes, 400 nm IAEDANS-labeled Pol β was incubated with 100 nm extendable DNA or 200 nm nonextendable DNA substrate in reaction buffer (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10 mm MgCl2, and 10% glycerol (v/v)) on ice for 5 min before loading into the stopped-flow apparatus at 22 °C. The binary complex was then mixed with an equal volume of dCTP in a series of concentrations in the same reaction buffer. Data were collected using the pretrigger setting and continued for 10 s, and artifacts from the initial flow and mixing of the solutions were removed (anything prior to 0.036 s, which represents the dead time of the instrument). Each trace reported represents an average of at least four repeats.
The reverse fingers closing rate is determined by a trapping experiment (22). A solution containing a ternary complex of 400 nm IAEDANS-labeled Pol β, 200 nm nonextendable DNA, 2 μm dCTP, and 10 mm MgCl2 in reaction buffer (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10 mm MgCl2, and 10% (v/v) glycerol) was mixed with a solution containing a 10-fold excess of an unlabeled Pol β-extendable DNA binary complex. Data were collected up to 10 s.
KinTek Explorer modeling
Fluorescence data were analyzed using KinTek Explorer software to evaluate the appropriate model and the associated rate constants. FRET traces composed of at least five nucleotide concentrations were imported into the software as a concentration series, and sigma was estimated by fitting to a triple exponential (for experiments with extendable DNA) or double exponential (for experiments with nonextendable DNA) equations. A concentration series scaling factor (multiplier) was applied, and observables were entered as a*(ED+b*EDN+c*END…). The dNTP binding step was constrained such that k−/k+ reflected the Kd (dNTP) determined in single-turnover experiments. The rate of chemistry was set to the kpol obtained in our single-turnover experiments, and the reverse polymerization rate was fixed at 0. FRET data were modeled with or without the noncovalent step for extendable DNA and their χ2/degree of freedom were recorded. The modeling results were evaluated from several aspects, including visual observation, χ2/degree of freedom, and the percent of error associated with each kinetic parameter. Most importantly, FitSpace 2D calculations were performed for each modeling result. During these calculations, observable constants were fixed after modeling, and nonrate parameters are not included in the calculations.
Author contributions
J. H. and J. B. S. conceptualization; J. H. and J. B. S. resources; J. H. and J. B. S. data curation; J. H., K. S. A., M. M. M., B. E., S. D., and J. B. S. formal analysis; J. H. investigation; J. H., B. E., S. D., and J. B. S. methodology; J. H. and J. S. writing-original draft; J. H., K. S. A., M. M. M., B. E., S. D., and J. B. S. writing-review and editing; J. B. S. supervision; J. B. S. funding acquisition; J. B. S. project administration.
Supplementary Material
This work was supported by NCI, National Institutes of Health Grant R01 CA080830 (to J. B. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S10.
- BER
- base excision repair
- Pol β
- DNA polymerase β
- NCS
- non-covalent step
- IAEDANS
- ((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid
- Dabcyl
- 4-((4-(dimethylamino)phenyl)azo)benzoic acid.
References
- 1. Gates K. S. (2009) An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 22, 1747–1760 10.1021/tx900242k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cadet J., Douki T., and Ravanat J. L. (2010) Oxidatively generated base damage to cellular DNA. Free Radic. Biol. Med. 49, 9–21 10.1016/j.freeradbiomed.2010.03.025 [DOI] [PubMed] [Google Scholar]
- 3. Barnes D. E., and Lindahl T. (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38, 445–476 10.1146/annurev.genet.38.072902.092448 [DOI] [PubMed] [Google Scholar]
- 4. Matsumoto Y., and Kim K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269, 699–702 10.1126/science.7624801 [DOI] [PubMed] [Google Scholar]
- 5. Freudenthal B. D., Beard W. A., Shock D. D., and Wilson S. H. (2013) Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 10.1016/j.cell.2013.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., and Kraut J. (1994) Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science 264, 1891–1903 10.1126/science.7516580 [DOI] [PubMed] [Google Scholar]
- 7. Sawaya M. R., Pelletier H., Kumar A., Wilson S.H., and Kraut J. (1994) Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science 264, 1930–1935 10.1126/science.7516581 [DOI] [PubMed] [Google Scholar]
- 8. Pelletier H., Sawaya M. R., Wolfle W., Wilson S. H., and Kraut J. (1996) Crystal structures of human DNA polymerase β complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry 35, 12742–12761 10.1021/bi952955d [DOI] [PubMed] [Google Scholar]
- 9. Wang T. S., and Korn D. (1980) Reactivity of KB cell deoxyribonucleic acid polymerases α and β with nicked and gapped deoxyribonucleic acid. Biochemistry 19, 1782–1790 10.1021/bi00550a009 [DOI] [PubMed] [Google Scholar]
- 10. Siedlecki J. A., Szyszko J., Pietrzykowska I., and Zmudzka B. (1980) Evidence implying DNA polymerase β function in excision repair. Nucleic Acids Res. 8, 361–375 10.1093/nar/8.2.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chagovetz A. M., Sweasy J. B., and Preston B. D. (1997) Increased activity and fidelity of DNA polymerase β on single-nucleotide gapped DNA. J. Biol. Chem. 272, 27501–27504 10.1074/jbc.272.44.27501 [DOI] [PubMed] [Google Scholar]
- 12. Singhal R. K., Prasad R., and Wilson S. H. (1995) DNA polymerase conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 270, 949–957 10.1074/jbc.270.2.949 [DOI] [PubMed] [Google Scholar]
- 13. Singhal R. K., and Wilson S. H. (1993) Short gap-filling synthesis by DNA polymerase β is processive. J. Biol. Chem. 268, 15906–15911 [PubMed] [Google Scholar]
- 14. Prasad R., Beard W. A., and Wilson S. H. (1994) Studies of gapped DNA substrate binding by mammalian DNA polymerase β: dependence on 5′-phosphate group. J. Biol. Chem. 269, 18096–18101 [PubMed] [Google Scholar]
- 15. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., and Pelletier H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 10.1021/bi9703812 [DOI] [PubMed] [Google Scholar]
- 16. Beard W. A., Shock D. D., Batra V. K., Pedersen L. C., and Wilson S. H. (2009) DNA polymerase β substrate specificity: side chain modulation of the “A-rule”. J. Biol. Chem. 284, 31680–31689 10.1074/jbc.M109.029843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown J. A., Pack L. R., Sanman L. E., and Suo Z. (2011) Efficiency and fidelity of human DNA polymerases λ and β during gap-filling DNA synthesis. DNA Repair 10, 24–33 10.1016/j.dnarep.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahn J., Kraynov V. S., Zhong X., Werneburg B. G., and Tsai M. D. (1998) DNA polymerase β: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem. J. 331, 79–87 10.1042/bj3310079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong X., Patel S. S., Werneburg B. G., and Tsai M. D. (1997) DNA polymerase β: multiple conformational changes in the mechanism of catalysis. Biochemistry 36, 11891–11900 10.1021/bi963181j [DOI] [PubMed] [Google Scholar]
- 20. Joyce C. M., Potapova O., Delucia A. M., Huang X., Basu V. P., and Grindley N. D. (2008) Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry 47, 6103–6116 10.1021/bi7021848 [DOI] [PubMed] [Google Scholar]
- 21. Bakhtina M., Roettger M. P., and Tsai M. D. (2009) Contribution of the reverse rate of the conformational step to polymerase β fidelity. Biochemistry 48, 3197–3208 10.1021/bi802119f [DOI] [PubMed] [Google Scholar]
- 22. Kellinger M. W., and Johnson K. A. (2010) Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 107, 7734–7739 10.1073/pnas.0913946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Santoso Y., Joyce C. M., Potapova O., Le Reste L., Hohlbein J., Torella J. P., Grindley N. D., and Kapanidis A. N. (2010) Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 107, 715–720 10.1073/pnas.0910909107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kellinger M. W., and Johnson K. A. (2011) Role of induced fit in limiting discrimination against AZT by HIV reverse transcriptase. Biochemistry 50, 5008–5015 10.1021/bi200204m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doublié S., Tabor S., Long A. M., Richardson C. C., and Ellenberger T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 10.1038/34593 [DOI] [PubMed] [Google Scholar]
- 26. Huang H., Chopra R., Verdine G. L., and Harrison S. C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669–1675 10.1126/science.282.5394.1669 [DOI] [PubMed] [Google Scholar]
- 27. Patel P. H., Suzuki M., Adman E., Shinkai A., and Loeb L. A. (2001) Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 308, 823–837 10.1006/jmbi.2001.4619 [DOI] [PubMed] [Google Scholar]
- 28. Yang W., Weng P. J., and Gao Y. (2016) A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 6, 51 10.1186/s13578-016-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mahmoud M. M., Schechter A., Alnajjar K. S., Huang J., Towle-Weicksel J., Eckenroth B. E., Doublié S., and Sweasy J. B. (2017) Defective nucleotide release by DNA polymerase β mutator variant E288K is the basis of its low fidelity. Biochemistry 56, 5550–5559 10.1021/acs.biochem.7b00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shock D. D., Freudenthal B. D., Beard W. A., and Wilson S. H. (2017) Modulating the DNA polymerase β reaction equilibrium to dissect the reverse reaction. Nat. Chem. Biol. 13, 1074–1080 10.1038/nchembio.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reed A. J., Vyas R., Raper A. T., and Suo Z. (2017) Structural insights into the post-chemistry steps of nucleotide incorporation catalyzed by a DNA polymerase. J. Am. Chem. Soc. 139, 465–471 10.1021/jacs.6b11258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S. J., Beard W. A., Harvey J., Shock D. D., Knutson J. R., and Wilson S. H. (2003) Rapid segmental and subdomain motions of DNA polymerase β. J. Biol. Chem. 278, 5072–5081 10.1074/jbc.M208472200 [DOI] [PubMed] [Google Scholar]
- 33. Arndt J. W., Gong W., Zhong X., Showalter A. K., Liu J., Dunlap C. A., Lin Z., Paxson C., Tsai M.-D., and Chan M. K. (2001) Insight into the catalytic mechanism of DNA polymerase β: structures of intermediate complexes. Biochemistry 40, 5368–5375 10.1021/bi002176j [DOI] [PubMed] [Google Scholar]
- 34. Bose-Basu B., DeRose E. F., Kirby T. W., Mueller G. A., Beard W. A., Wilson S. H., and London R. E. (2004) Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase β. Biochemistry 43, 8911–8922 10.1021/bi049641n [DOI] [PubMed] [Google Scholar]
- 35. Alnajjar K. S., Garcia-Barboza B., Negahbani A., Nakhjiri M., Kashemirov B., McKenna C., Goodman M. F., and Sweasy J. B. (2017) A change in the rate-determining step of polymerization by the K289M DNA polymerase β cancer-associated variant. Biochemistry 56, 2096–2105 10.1021/acs.biochem.6b01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sucato C. A., Upton T. G., Kashemirov B. A., Batra V. K., Martínek V., Xiang Y., Beard W. A., Pedersen L. C., Wilson S. H., McKenna C. E., Florián J., Warshel A., and Goodman M. F. (2007) Modifying the β, γ leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase β. Biochemistry 46, 461–471 10.1021/bi061517b [DOI] [PubMed] [Google Scholar]
- 37. Sucato C. A., Upton T. G., Kashemirov B. A., Osuna J., Oertell K., Beard W. A., Wilson S. H., Florián J., Warshel A., McKenna C. E., and Goodman M. F. (2008) DNA polymerase β fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry 47, 870–879 10.1021/bi7014162 [DOI] [PubMed] [Google Scholar]
- 38. Towle-Weicksel J. B., Dalal S., Sohl C. D., Doublié S., Anderson K. S., and Sweasy J. B. (2014) Fluorescence resonance energy transfer studies of DNA polymerase β: the critical role of fingers domain movements and a novel non-covalent step during nucleotide selection. J. Biol. Chem. 289, 16541–16550 10.1074/jbc.M114.561878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson K. A., Simpson Z. B., and Blom T. (2009) FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 387, 30–41 10.1016/j.ab.2008.12.025 [DOI] [PubMed] [Google Scholar]
- 40. Johnson K. A., Simpson Z. B., and Blom T. (2009) Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 10.1016/j.ab.2008.12.024 [DOI] [PubMed] [Google Scholar]
- 41. Berlow R. B., Swain M., Dalal S., Sweasy J. B., and Loria J. P. (2012) Substrate-dependent millisecond domain motions in DNA polymerase β. J. Mol. Biol. 419, 171–182 10.1016/j.jmb.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moscato B., Swain M., and Loria J. P. (2016) Induced fit in the selection of correct versus incorrect nucleotides by DNA polymerase β. Biochemistry 55, 382–395 10.1021/acs.biochem.5b01213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Diaz M., Bebenek K., Krahn J. M., Blanco L., Kunkel T. A., and Pedersen L. C. (2004) A structural solution for the DNA polymerase λ-dependent repair of DNA gaps with minimal homology. Mol. Cell 13, 561–572 10.1016/S1097-2765(04)00061-9 [DOI] [PubMed] [Google Scholar]
- 44. Wu P., and Brand L. (1994) Resonance energy transfer: methods and applications. Anal. Biochem. 218, 1–13 10.1006/abio.1994.1134 [DOI] [PubMed] [Google Scholar]
- 45. Heiner Z., and Osvay K. (2009) Refractive index of dark-adapted bacteriorhodopsin and tris(hydroxymethyl)aminomethane buffer between 390 and 880 nm. Appl. Opt. 48, 4610–4615 10.1364/AO.48.004610 [DOI] [PubMed] [Google Scholar]
- 46. Haran G., Haas E., Szpikowska B. K., and Mas M. T. (1992) Domain motions in phosphoglycerate kinase: determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc. Natl. Acad. Sci. U.S.A. 89, 11764–11768 10.1073/pnas.89.24.11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., and Wilson S. H. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 10.1016/j.str.2006.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.