Abstract
Glucose is a critical nutrient for cell proliferation. However, the molecular pathways that regulate glucose metabolism are still elusive. We discovered that co-activator-associated arginine methyltransferase 1 (CARM1) suppresses glucose metabolism toward serine biosynthesis. By tracing the 13C-labeled glucose, we found that Carm1 knockout mouse embryonic fibroblasts exhibit significantly increased de novo serine synthesis than WT cells. This is caused, at least in part, by the reduced pyruvate kinase (PK) activity in these cells. The M2 isoform of PK (PKM2) is arginine-methylated by CARM1, and methylation enhances its activity. Mechanistically, CARM1 methylates PKM2 at arginines 445 and 447, which enhances PKM2 tetramer formation. Consequently, Carm1 knockout cells exhibit significant survival advantages over WT cells when extracellular serine is limited, likely due to their enhanced de novo serine synthesis capacity. Altogether, we identified CARM1 as an important regulator of glucose metabolism and serine synthesis.
Keywords: protein methylation, pyruvate kinase, serine, metabolic regulation, proliferation, CARM1
Introduction
Glucose is metabolized via glycolysis to produce pyruvate, which is catalyzed through oxidative phosphorylation in the tricarboxylic acid (TCA)2 cycle to generate ATP (1). When completely oxidized, one glucose molecule can generate 36 ATP molecules, providing a major source of energy for cellular activity. However, fast-growing cells exhibit enhanced aerobic glycolysis, which converts glucose to lactate even in the presence of oxygen. This phenomenon, initially observed in cancer cells by Otto Warburg, was named the Warburg effect (2–4). We now know that enhanced aerobic glycolysis is a hallmark of metabolic reprogramming in most proliferating cells (5, 6). Although inefficient in terms of ATP production (two ATP molecules per molecule of glucose), aerobic glycolysis maintains high levels of glycolytic intermediates to support macromolecular synthesis and biomass accumulation, providing the building blocks necessary for cell division (5). The de novo serine biosynthesis pathway (SSP), which uses the glycolytic intermediate 3-phosphoglycerate (3PG) to produce serine, is one such side branch of glucose metabolism (7, 8). Serine is both a proteinogenic amino acid and an important source of one-carbon units. One-carbon units from serine are required for several key metabolic processes, including nucleotide synthesis for DNA replication, the generation of NADPH for antioxidant defense, and the production of S-adenosylmethionine (SAM), a methyl donor for protein, nucleic acids, and lipid methylation (9, 10). Enhanced de novo serine biosynthesis has been linked to enhanced cell proliferation and survival, particularly under nutrient-deprived conditions (11–13). However, the molecular pathways that regulate de novo serine biosynthesis remain elusive.
Protein arginine methylation is an important post-translational modification (PTM) that modulates protein functions in various cellular processes, such as transcriptional regulation, DNA damage repair, mRNA processing, and signaling transduction (14, 15). In mammalian cells, three types of arginine methylation have been identified, including mono-methylated arginine, asymmetrical dimethylated arginine (ADMA), and symmetrical dimethylated arginine. The methylation of arginine is catalyzed by a family of enzymes called protein arginine methyltransferases (PRMTs) (14, 15). The human genome encodes nine PRMTs (PRMT1–9). PRMT4, also called the co-activator-associated arginine methyltransferase 1 (CARM1), specifically catalyzes ADMA modifications (14, 15). Genetic studies in mice revealed that complete loss of Carm1 is lethal: Carm1 knockout mice fail to breathe and die shortly after birth (16). This lethality is caused by the overproliferation of immature alveolar type II (AT2) cells in the lung during embryonic development, which reduces the airspace and blocks the oxygen exchange (16). CARM1 has also been found to be overexpressed in human malignancies, including breast, prostate, and ovarian cancers (15, 17, 18), suggesting its role in tumorigenesis. Characterization of CARM1-catalyzed arginine-methylated protein substrates, including histones, transcription factors, transcription co-regulators, and mRNA-binding proteins, revealed its convergent function in transcriptional activation (19–23) and mRNA processing (24–26). As enthusiasm for developing small molecule inhibitors to target CARM1 in human malignancies is rising (27–30), further elucidation of CARM1 function in normal and cancer cell biology is in great need.
Recent work by Liu et al. (31) demonstrated that CARM1 methylates the M2 isoform of pyruvate kinase (PKM2) and shifts the balance of glucose metabolism from oxidative phosphorylation to aerobic glycolysis. Although pyruvate kinase catalyzes the final step of glycolysis (32–35), whether PKM2 serves as a rate-limiting step of glycolysis and the extent to which PKM2 contributes to the Warburg effect remain controversial (36–38).
In this study, we revealed a novel function of CARM1 in the regulation of de novo serine synthesis in mouse embryonic fibroblasts (MEFs) and human breast cancer MCF7 cells. By tracing the 13C-labeled glucose in cells, we observed significantly increased glucose flux toward serine synthesis in MEFs and MCF7 cells after CARM1 knockout. This altered flux is mainly caused by lower pyruvate kinase (PK) activity in CARM1 knockout cells. Mechanistically, CARM1 methylates PKM2 at arginines 445 and 447, leading to enhanced intramolecular interactions that promote PKM2 tetramerization and PK activity in vitro and in cells. Reduced PK activity has been reported to enhance glucose flux toward the de novo serine biosynthesis pathway to favor cell proliferation in nutrient-limited conditions (11–13). Consistent with the role of enhanced de novo serine synthesis in promoting cell proliferation and survival, CARM1 knockout MEFs and MCF7 cells exhibit significant proliferation and survival advantages over their respective WT cells when extracellular serine is limited. Altogether, we have uncovered a novel function of CARM1 in regulating cell metabolism and identified a metabolic vulnerability of CARM1-overexpressing cells.
Results
CARM1-loss promotes de novo serine synthesis
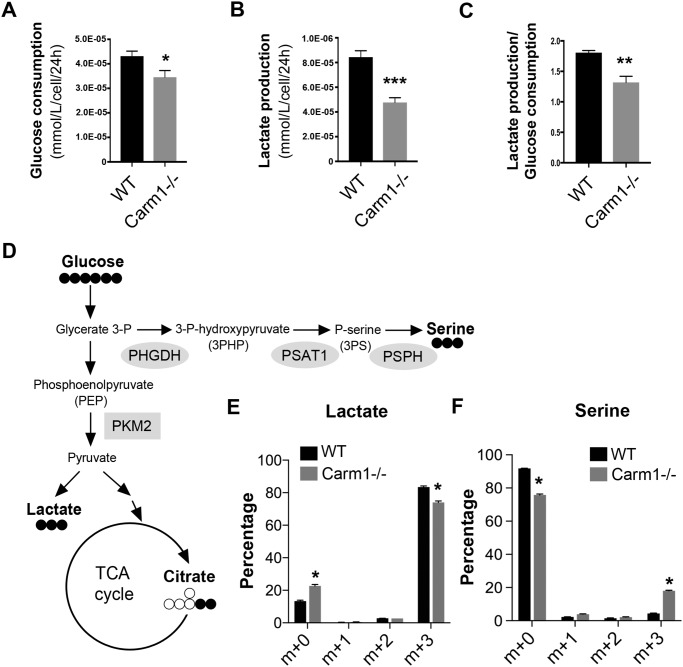
Whereas CARM1 has been increasingly recognized as an important regulator of lipid and amino acid metabolism (19, 39), its role in glucose metabolism remains largely unexplored. During daily cell culture maintenance, we observed that the cell culture medium in which Carm1 knockout (−/−) MEFs were grown turned yellow/orange much slower than the medium in which WT MEFs were grown, even at 100% confluence. This observation suggested a slower buildup of acidic metabolites, such as lactate, in the Carm1 knockout cells. To confirm this hypothesis, we used a bioanalyzer to determine the glucose consumption and lactate production of the WT and Carm1 knockout MEFs. Cellular glucose consumption (Fig. 1A) and lactate production (Fig. 1B) were significantly lower in the Carm1 knockout cells. Importantly, the Carm1 knockout MEFs exhibited a significantly lower lactate production/glucose consumption ratio compared with the WT cells (Fig. 1C), indicating that reduced lactate is not due to decreased glucose uptake but reduced glucose flux toward lactate production. We also detected similar glucose metabolism shift in CARM1 knockout breast cancer MCF7 cells (data not shown).
Figure 1.
CARM1 suppresses de novo serine synthesis in MEFs. A, glucose consumption, normalized to cell number, in wildtype (WT) and Carm1 knockout (−/−) MEFs over 24 h. Data represent mean ± S.E., *, p < 0.05. B, lactate production, normalized to cell number, in WT and Carm1−/− MEFs over 24 h. C, lactate production/glucose consumption ratio is significantly lower in Carm1−/− MEFs than in WT MEFs. D, simplified schematic representing glucose flux in cells. E and F, mass isotopomer distributions representative of the relative abundances of different 13C-labeled (E) lactate and serine (F) isomers in WT and Carm1−/− MEFs, measured using GC-MS. Data represent mean ± S.E., n = 3.
To further define the role of Carm1 in regulating cellular glucose flux, we performed a 13C-labeled glucose tracing experiment in WT and Carm1 knockout MEFs. A simplified schematic of the glycolysis pathway, focusing on several significantly altered glucose-derived metabolites related to this study, is shown in Fig. 1D. Consistent with a reduced lactate production in the cell culture medium (Fig. 1B), intracellular lactate enrichment was also significantly reduced in Carm1 knockout MEFs (Fig. 1E). We also observed significant increases in the enrichment of several key TCA cycle metabolites, such as citrate and α-ketoglutarate (αKG) (Fig. S1), indicating an increased mitochondrial oxidative phosphorylation in Carm1 knockout MEFs. Surprisingly, the enrichment of serine was significantly increased in Carm1 knockout cells (Fig. 1F), demonstrating that lack of Carm1 leads to increased de novo serine synthesis.
Reduced pyruvate kinase activity enhances serine synthesis in Carm1 knockout cells
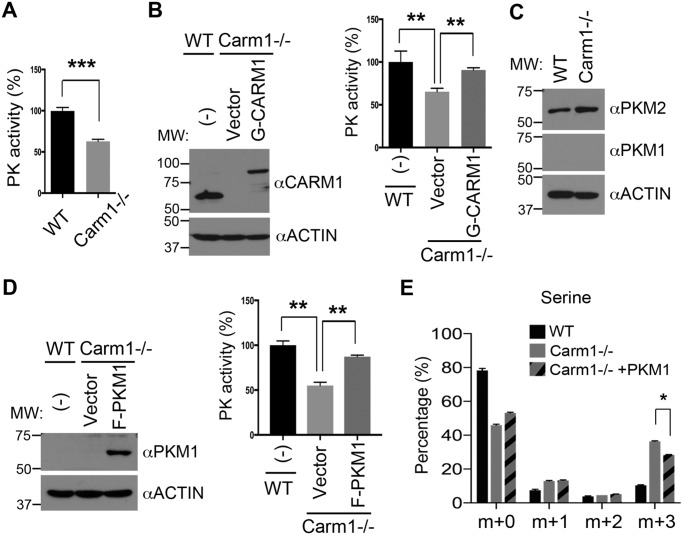
Enhanced de novo serine synthesis might result from the overexpression of enzymes involved in the SSP. However, we examined the published gene expression (RNA-Seq) data from WT and Carm1 knockout MEFs (GSE72901) (40) and did not identify significant differences between groups in the expression of SSP enzymes, such as phosphoglycerate dehydrogenase (Phgdh), phosphoserine aminotransferase (Psat1), and phosphoserine phosphatase (Psph) (Fig. 1D). Recent studies revealed that the flux of glucose to de novo serine synthesis is also determined by another important pathway that involves the regulation of cellular PK activity. For example, the less active M2 form of PK (Pkm2) promotes the channeling of the glycolytic intermediate 3-phosphoglycerate (3-PG) to the SSP (11), whereas small molecule activators of PKM2 or re-expression of the highly active M1 isoform of PK (Pkm1) channels the glucose flux away from the SSP (12, 13, 41, 42). To test the hypothesis that enhanced de novo serine synthesis in Carm1 knockout cells is caused by reduced PK activity, we examined the PK activity of WT and Carm1 knockout MEFs. Knockout of Carm1 in MEFs significantly reduced cellular PK activity (Fig. 2A and Fig. S2A). This reduction is caused by loss of Carm1, because when we restored Carm1 expression via transient transfection with GFP-tagged CARM1 cDNA (G-CARM1), the reduced PK activity was rescued (Fig. 2B and Fig. S2B). MEFs predominantly express the less active Pkm2 instead of Pkm1 (Fig. 2C). To test whether Carm1 regulates Pkm2 gene expression, we examined both mRNA and protein levels of Pkm2 in WT and Carm1 knockout MEFs. Loss of Carm1 does not impact Pkm2 mRNA levels (data not shown). However, Pkm2 protein levels were higher in the Carm1 knockout than in WT cells, indicating that the Pkm2 protein may be stabilized to compensate for reduced PK activity (Fig. 2C).
Figure 2.
CARM1 promotes pyruvate kinase activity and suppresses serine synthesis. A, Carm1 knockout (−/−) MEFs exhibit lower PK activity than WT MEFs. In vitro PK assays were performed using total cell lysates of WT and Carm1−/− MEFs. B, re-expression of GFP-CARM1 in Carm1−/− MEFs restores PK activity. The rescued expression of CARM1 was confirmed by Western blot analysis (left panel). PK activity assays (right panel) were performed using total cell lysates from untransfected (−) and vector- or G-CARM1–transfected Carm1−/− MEFs. Actin was used as a loading control. C, expression of PKM1 and PKM2 was examined in WT and Carm1−/− MEFs by Western blot analysis. D, re-expression of FLAG-PKM1 (F-PKM1) in Carm1−/− MEFs restores PK activity. The expression of PKM1 was confirmed by Western blot analysis (left panel). PK activity assays (right panel) were performed using total cell lysates from the indicated cells. E, overexpression of PKM1 in Carm1−/− MEFs suppresses de novo serine synthesis. The level of 13C-labeled serine in the indicated cells was measured using GC-MS. Data represent mean ± S.E., n = 3.
To determine whether enhanced serine synthesis in Carm1 knockout cells is caused by reduced PK activity, we introduced the constitutively active FLAG-tagged PKM1 (F-PKM1) to Carm1 knockout MEFs and restored cellular PK activity (Fig. 2D and Fig. S2C). We repeated 13C-labeled glucose tracing in these cells and found that expression of highly active PKM1 significantly reduced the flux of glucose to the SSP, attributing the augmented serine synthesis in Carm1 knockout cells, at least in part, to the reduced PKM2 activity (Fig. 2E). Interestingly, the increased TCA cycle metabolites in Carm1 knockout MEFs were also reversed by PKM1 overexpression (Fig. S2D), indicating that alteration of PK activity might be a major mechanism by which Carm1 regulates glucose metabolism.
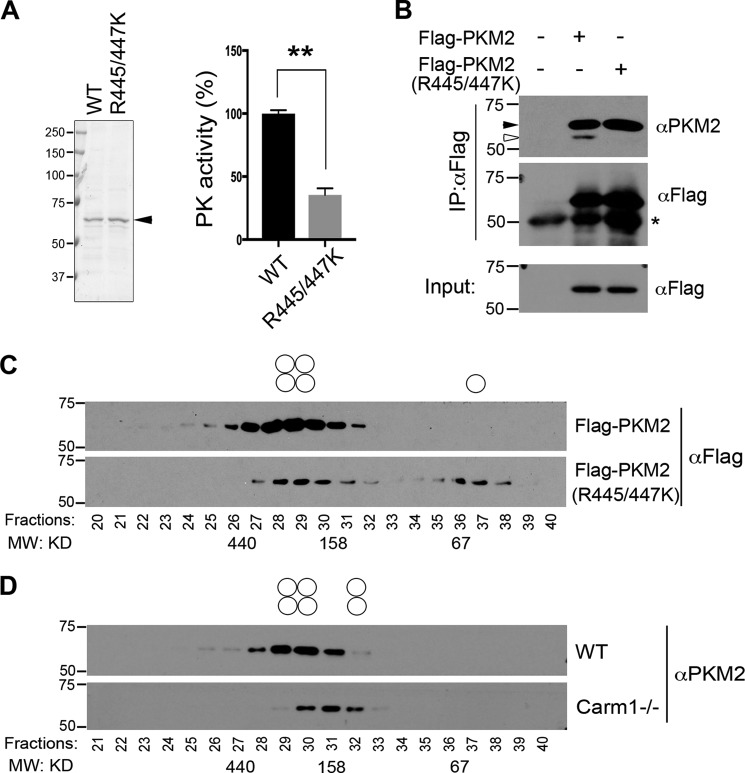
PKM2 is arginine-methylated by CARM1 at arginines 445 and 447
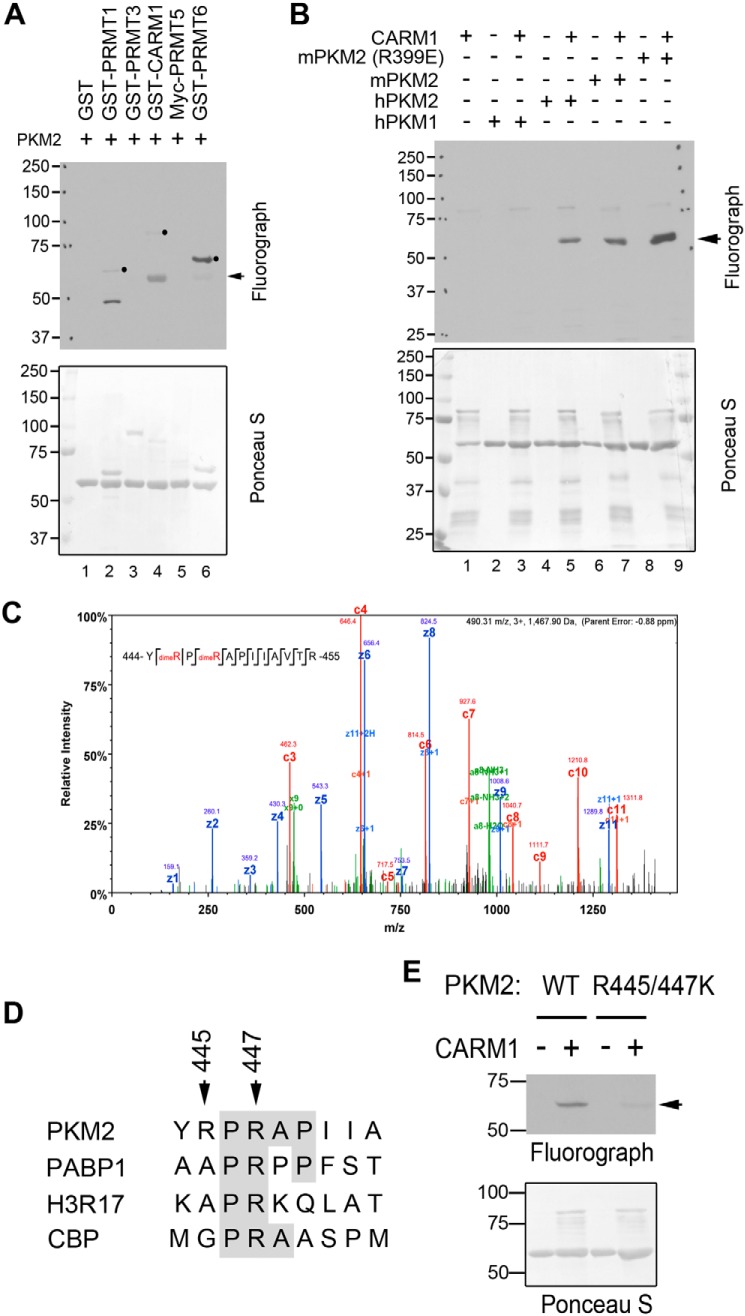
To determine how CARM1 regulates PKM2 activity, we tested whether PKM2 is arginine-methylated by CARM1. In vitro methylation assays were performed by incubating recombinant mouse PKM2 proteins with a panel of PRMTs, including PRMT1, PRMT3, CARM1, PRMT5, and PRMT6, in the presence of 3H-labeled SAM. PKM2 is robustly methylated by CARM1 and to a lesser extent by PRMT6 (Fig. 3A). Both mouse and human PKM2 can be methylated by CARM1, but surprisingly, PKM1 cannot (Fig. 3B, lanes 2–7). There are 22 amino acid differences between PKM1 and PKM2 due to alternative splicing (32), only one of which is an arginine, Arg-399. A previous study has shown that mutation of positively charged Arg-399 to negatively charged glutamic acid disrupts PKM2 tetramer formation at one of the dimer interfaces, thereby producing an excess of dimeric PKM2 (43). When mutant PKM2 (R399E) was examined for methylation by CARM1, the methylation signal was much stronger than that of WT PKM2 (Fig. 3B, lanes 8 and 9). Further quantitative analysis of in vitro methylation products also confirmed that PKM2 R399E is a better CARM1 substrate than the WT protein (Fig. S3, A and B). These results suggested that the arginine methylation sites of PKM2 are shared with PKM1, but they are only accessible for methylation when the protein is in its nontetrameric form. In support of this, GSH S-transferase (GST) pulldown experiments examining the interactions of PKM2 with CARM1 revealed that CARM1 preferentially interacts with the dimeric PKM2 (R399E) compared with the WT protein (Fig. S3C). To further confirm PKM2 methylation by CARM1 and to identify the methylation sites, we performed LC-tandem MS (LC-MS/MS) analysis of in vitro-methylated PKM2, and we detected mono- and di-methylation of Arg-445 and Arg-447 (Fig. 3C and Fig. S3D). Notably, these two sites are located within a proline-rich motif, a consensus sequence found in several other CARM1 substrates (Fig. 3D). Mutation of both Arg-445 and Arg-447 to lysine (R445K/R447K) dramatically reduced CARM1-mediated PKM2 methylation (Fig. 3E), confirming that Arg-445 and Arg-447 are the major sites methylated by CARM1.
Figure 3.
CARM1 methylates PKM2 at Arg-445 and Arg-447 sites. A, PKM2 is methylated by CARM1. In vitro methylation assays were performed by incubating GST-tagged recombinant PRMTs (PRMT1, PRMT3, CARM1, Myc-PRMT5, and PRMT6) with purified His-tagged PKM2 proteins. A, B, and E, arrows indicate PKM2 methylation, and solid dots indicate PRMT automethylation. Membranes were stained with Ponceau S to ensure equal protein loading. B, recombinant human PKM1 (hPKM1) and PKM2 (hPKM2), mouse PKM2 (mPKM2), and tetramerization-deficient mutant mPKM2 (R399E) were subjected to in vitro methylation assays with CARM1. C, LC-MS/MS was performed to identify the arginine methylation sites of in vitro methylated His–mPKM2. Arginines 445 and 447 were found to be both mono- and dimethylated. D, PKM2 arginine methylation sites, Arg-445 and Arg-447, share sequence consensus with several known CARM1 substrates, including poly(A)-binding protein 1 (PABP1), histone H3R17, and cAMP-response element-binding protein-binding protein (CBP). E, in vitro methylation assays were performed using WT and R445K/R447K mutant PKM2, confirming that Arg-445 and Arg-447 are the major sites of methylation by CARM1.
Arginine methylation promotes PKM2 pyruvate kinase activity
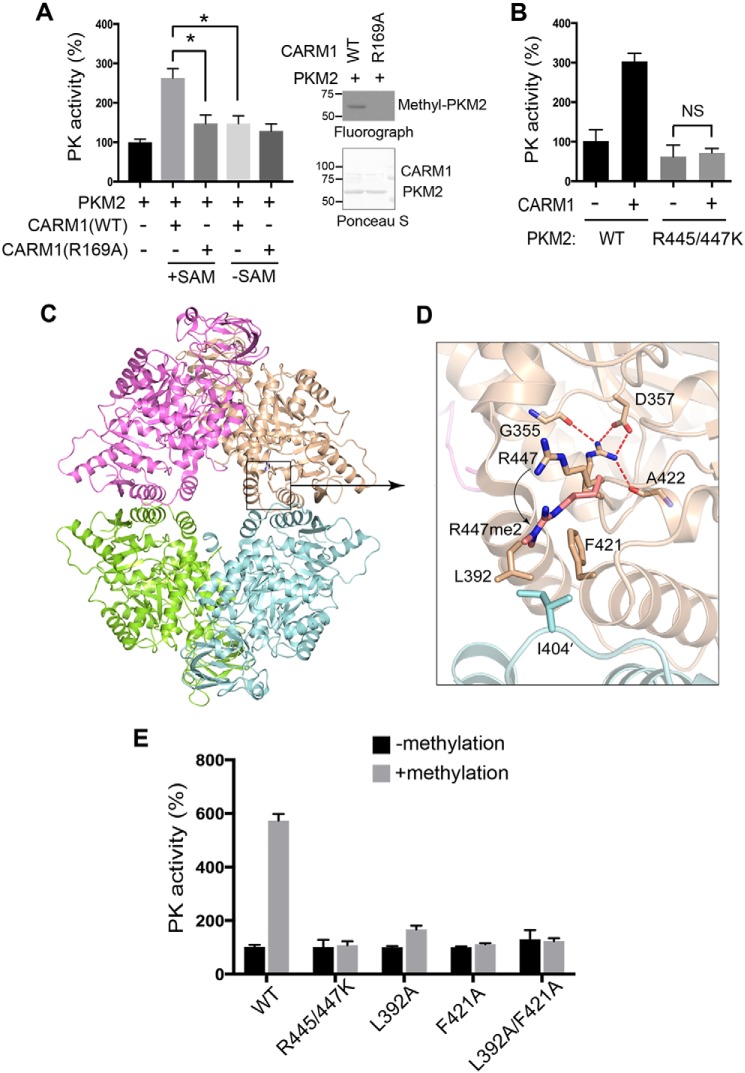
Various PTMs, such as phosphorylation and acetylation, have been reported to negatively regulate PKM2 activity (44). The reduced PK activity observed in Carm1 knockout MEFs (Fig. 2, A and B) prompted us to test whether CARM1-mediated arginine methylation regulates PKM2 activity. Bacteria-expressed recombinant PKM2 was subjected to in vitro methylation by CARM1 in the presence or absence of the methyl donor SAM, and the methylation reaction products were dialyzed into PK activity assay buffer. Arginine methylation by CARM1 enhanced PK activity of PKM2 to about 3-fold, compared with that of unmethylated PKM2 (Fig. 4A and Fig. S4A). This increase was indeed caused by arginine methylation, because incubation with catalytically-deficient CARM1 (R169A) and the removal of the methyl donor SAM abolished this enhanced PK effect (Fig. 4A). Consistent with the observation that PKM1 is not methylated by CARM1 (Fig. 3B), incubating PKM1 with CARM1 and SAM did not impact its PK activity (Fig. S4B). Additionally, although PKM2 (R399E) is strongly methylated by CARM1, its activity was not promoted by CARM1-mediated arginine methylation (Fig. S4C). This is likely due to the constitutively dimeric nature of the PKM2 (R399E) mutant. To further confirm that CARM1 promotes PKM2 activity through methylation of Arg-445 and Arg-447, we compared the PK activity of WT and R445K/R447K mutant PKM2 with and without methylation by CARM1. The mutant PKM2 (R445K/R447K) exhibited slightly decreased PK activity, but more importantly, incubation with CARM1 in the methylation reaction did not promote greater PK activity (Fig. 4B). These results demonstrated that CARM1-mediated arginine methylation of PKM2 Arg-445 and Arg-447 enhances its PK activity.
Figure 4.
Arginine methylation PKM2 promotes its PK activity in vitro. A, arginine-methylated PKM2 exhibits significantly greater PK activity than unmethylated PKM2. Recombinant PKM2 was subjected to in vitro methylation by either WT or catalytic-deficient CARM1 (R169A) in the presence or absence of SAM. The methylation products were dialyzed and subjected to in vitro PK activity assays (left panel). In vitro methylation assays were performed to confirm that CARM1 (R169A) is catalytically deficient in methylating PKM2 (right panel). *, p < 0.05. B, CARM1 promotes PKM2 PK activity through arginine methylation. Recombinant WT and methylation-defective PKM2 (R445/447K) were subjected to in vitro methylation by CARM1. The reaction products were dialyzed and subjected to in vitro PK activity assays. NS, not significant. C, structural analysis of PKM2 reveals that methylation of Arg-447 (R447me2a) potentiates intramolecular interactions with distal amino acids, Leu-392 and Phe-421. D, mutations of amino acids involved in the intramolecular interactions of PKM2 abolish methylation-mediated regulation of PKM2 activity. E, recombinant WT, methylation-deficient (R445/447K), and intramolecular interaction-deficient (L392A, F421A, and L392A/F421A) PKM2 were subjected to in vitro methylation by CARM1. The reaction products were dialyzed and subjected to in vitro PK activity assays.
To determine the molecular mechanism by which Arg-445/447 methylation promotes PKM2 PK activity, we analyzed the arginine methylation sites on the crystal structure of PKM2 (PDB code 1ZJH). Both Arg-445 and Arg-447 reside in the C domain of PKM2 that fosters tetramer formation; however, neither of the sites is directly involved in intermolecular interactions. Interestingly, both sites can only be accessed by CARM1 for methylation when PKM2 is in its monomeric or dimeric state, consistent with our in vitro methylation results showing that tetrameric PKM1 cannot be methylated by CARM1 and that dimeric PKM2 (R399E) is a better substrate than WT PKM2 (Fig. 3B). Structural modeling analysis further suggested that methylation of Arg-445 and Arg-447 might on the one hand disrupt Arg-445–mediated intramolecular interactions and on the other hand promote nonpolar contacts between Arg-447 and residues located at the tetrameric interface, including leucine 392 and phenylalanine 421 from the same monomer and isoleucine 404 from the tetramerization-related monomer (Fig. 4, C and D). To test this model and determine the importance of the methylation-mediated interactions in enhancing PKM2 activity, we increased the distance between Arg-447 and Leu-392 or Phe-421 by replacing leucine or phenylalanine with alanine. Both L392A and F421A mutants of PKM2 exhibited baseline levels of PK activity comparable with that of the WT enzyme (Fig. S4D) and were able to be methylated by CARM1 (data not shown). Surprisingly, CARM1-mediated arginine methylation failed to enhance their activities (Fig. 4E), suggesting that Phe-392 and Leu-421 are crucial for mediating methylation-promoted PKM2 activity. Altogether, these results demonstrated that arginine methylation enhances PKM2 activity, likely through affecting its intramolecular and/or intermolecular interactions.
Arginine methylation enhances PKM2 tetramerization
We next determined the impact of arginine methylation on PKM2 PK activity using WT and arginine methylation-deficient (R445K/R447K) PKM2 purified from MCF7 cells (Fig. 5A, left panel). Consistent with the results obtained using bacteria-expressed in vitro-methylated PKM2 (Fig. 4, A and B), depletion of arginine methylation significantly reduces PKM2 activity in cells (Fig. 5A, right panel). The activity of PKM2 is determined by its oligomerization, with its tetrameric form being the most active. To test whether methylation-mediated intramolecular interactions enhance PKM2 tetramerization in cells, we performed two independent experiments. First, co-immunoprecipitation assays were performed to examine the interactions of FLAG-tagged WT and arginine methylation-deficient (R445K/R447K) PKM2 with endogenous PKM2. Unlike WT PKM2, which efficiently co-immunoprecipitates with the endogenous protein, PKM2 (R445K/R447K) failed to interact with endogenous PKM2 (Fig. 5B), indicating impaired PKM2 oligomerization in the absence of methylation. Next, to further determine the impact of arginine methylation on PKM2 tetramerization, we performed gel-filtration chromatography on cell lysates from MCF7 cells overexpressing either FLAG-tagged WT or arginine methylation-deficient (R445K/R447K) PKM2. In contrast to the WT protein, which mainly exists as tetramers, the methylation-deficient PKM2 (R445K/R447K) exists in both tetrameric and monomeric forms (Fig. 5C), suggesting reduced oligomerization. We also examined PKM2 oligomerization in WT and Carm1 knockout MEFs. Although we did not observe as dramatic a shift in PKM2 oligomerization, possibly because other PRMTs (such as PRMT6; Fig. 3A) may compensate for CARM1 loss, we did observe a greater proportion of dimeric PKM2 in Carm1 knockout compared with WT MEFs (Fig. 5D). Therefore, our data demonstrated that CARM1-mediated methylation of PKM2 enhances its intramolecular interactions, which might indirectly promote PKM2 tetramerization or the stability of its tetramer states.
Figure 5.
Arginine methylation enhances PKM2 tetramerization. A, arginine methylation-deficient PKM2 (R445/447K) expressed in mammalian cells is less active than the WT enzyme. FLAG-tagged WT and R445K/R447K mutant PKM2 were expressed and purified from MCF7 cells (left panel). Eluted proteins were subjected to in vitro PK activity assays (right panel). B, loss of arginine methylation reduces PKM2 intermolecular interactions. Co-immunoprecipitation assays were performed to examine the interactions of endogenous PKM2 with FLAG-tagged WT or arginine methylation-deficient (R445/447K) PKM2 in MCF7 cells. * indicates the IgG heavy chain. Black arrow indicates transfected FLAG-tagged constructs, and white arrow indicates the endogenous PKM2. C, arginine methylation promotes PKM2 tetramerization. MCF7 cells were transfected with FLAG-tagged WT and arginine methylation-deficient PKM2 (R445/447K). Total cell lysates from transfected cells were separated by gel filtration, followed by Western blot analysis using an anti-FLAG antibody to determine the proportion of mono-, di-, and tetramers (indicated with open circles above the gels). D, reduced PKM2 tetramerization in Carm1−/− MEF cells. Total cell lysates from WT and Carm1 knockout (−/−) MEFs were separated by gel filtration, followed by Western blot analysis using an anti-PKM2 antibody.
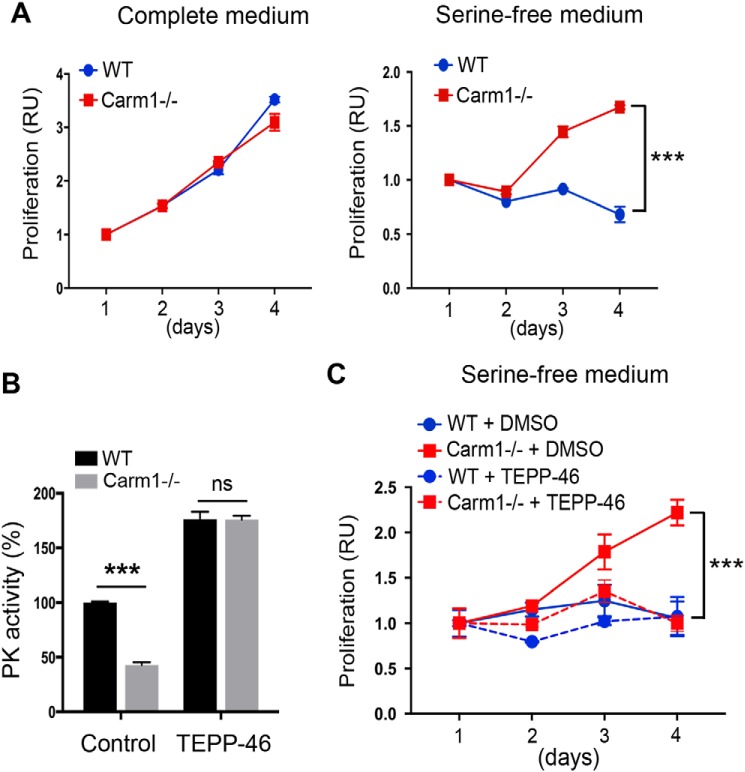
Knockout of Carm1 confers a growth advantage upon depletion of extracellular serine
The nonessential amino acid serine supports several aspects of cell metabolism that are crucial for the growth and survival of proliferating cells, including protein, GSH, and nucleotide synthesis, methylation reactions, and the generation of NADPH for antioxidant defense (7, 8). We reason that enhanced de novo serine synthesis in Carm1 knockout MEFs (Figs. 1 and 2) renders them more resistant to deprivation of exogenous serine. Indeed, cell proliferation assays revealed that although WT and Carm1 knockout MEFs proliferate at a comparable rate in complete medium (Fig. 6A, left panel), Carm1 knockout cells exhibit superior growth compared with WT cells when the medium is depleted of serine (Fig. 6A, right panel). To further determine whether the resistance to serine deprivation observed in Carm1 knockout MEFs was due to reduced PK activity, we treated WT and Carm1 knockout MEFs with a small molecule PKM2 activator, TEPP-46 (45). TEPP-46 elevated the PK activity of both WT and Carm1 knockout cells to a comparable level (Fig. 6B). Surprisingly, activating PKM2 using TEPP-46 completely abolished the resistance of serine depletion in Carm1 knockout MEFs (Fig. 6C), supporting the argument that reduced PK activity is the primary mechanism driving this resistance. This result further confirmed that reduced PK activity in Carm1 knockout cells channels glucose flux to de novo serine synthesis. In support of our results, others have reported that enhanced PKM2 activity can lead to serine auxotrophy and proliferation arrest when extracellular serine is limited (12, 13, 42). Similarly, CARM1 knockout MCF7 cells exhibited reduced PK activity and were also resistant to depletion of extracellular serine (Fig. S5), suggesting that the mechanism identified in MEFs also applies to some cancer cell lines.
Figure 6.
CARM1 knockout cells are more resistant to serine deprivation than WT cells, likely due to reduced PK activity. A, CARM1 knockout (−/−) MEFs are more resistant to serine deprivation. The proliferation of WT and Carm1−/− MEFs in complete (left panel) and serine-free (right panel) medium was assayed over a 4-day period. B, WT and CARM1−/− MEFs respond similarly to the PKM2 small molecule activator TEPP-46. To activate PK activity, WT and Carm1−/− MEFs were either untreated or treated with TEPP-46 (25 μm) for 24 h. The total cell lysates were subjected to in vitro PK activity assays. C, activating PK activity using PKM2 activator TEPP-46 abolishes the survival advantage of CARM1−/− MEFs in serine-free medium. WT and Carm1−/− MEFs were treated with DMSO or TEPP-46 (25 μm) and subjected to culture in serine-free medium over a 4-day period. The cell proliferation was monitored as described in A.
Discussion
The arginine methyltransferase CARM1 has emerged as an important regulator of normal and cancer cell biology, particularly through regulation of cell proliferation and differentiation (15, 16, 19, 20, 22, 46). Adding to the compelling evidence defining the role of CARM1 in transcription regulation and mRNA splicing, our study revealed a novel aspect of CARM1 function in regulating cell metabolism, which may shed light on the role of this important enzyme in development and disease.
CARM1 and the regulation of cell metabolism
Genetic studies on Carm1 knockout mice have revealed the blockade of adipocyte differentiation (19) and hyperproliferation of immature AT2 cells in the embryonic lung (16). In contrast, overexpression of CARM1 is observed in many human cancers and is associated with cancer cell proliferation and metastasis (15). Interestingly, however, studies of CARM1 function in human cancer revealed that overexpression of CARM1 inhibits the proliferation and induces differentiation of breast cancer MCF7 cells (47) and that CARM1-overexpressing breast cancer cells are more sensitive to chemotherapy drug-induced cell death (46). We reason that these seemingly contradictory functions of CARM1 in development and human malignancies could be due to the context-dependent nature of CARM1 regulation of cell metabolism in normal and cancerous cells. Knockout of Carm1 in immortalized MEFs and breast cancer MCF7 cells increases glucose flux to de novo serine synthesis (Fig. 1), thus allowing Carm1 knockout cells to be more resistant to serine deprivation (Fig. 6A). Because serine is an important source of one-carbon units for nucleotide synthesis, the generation of NADPH, and the production of SAM (9, 10), enhanced de novo serine synthesis has been characterized as a critical molecular pathway to promote cancer cell proliferation and survival, particularly when exogenous serine is limited (7, 8, 48, 49). It is reasonable to hypothesize that the hyperproliferative phenotype of lung AT2 cells in Carm1 knockout mice is due to enhanced de novo serine synthesis. However, whether serine availability becomes limited during any period of mouse embryonic lung development has not been tested.
Considering the vulnerabilities of CARM1-overexpressing cells, what would be the advantages of CARM1 overexpression for cancer cell metabolism? Our results showed that CARM1-expressing cells exhibited greater lactate production than knockout cells (Fig. 1, A–C). Once considered a waste product of glycolysis, lactate has emerged as a critical regulator of cancer development, maintenance, and metastasis (50, 51). Tumor cells can metabolize lactate as an energy source or shuttle lactate to adjacent stromal and vascular endothelial cells to alter the tumor microenvironment to promote angiogenesis and metastasis (52–54). It is likely that increased lactate production contributes, at least in part, to the increased metastatic potential of CARM1 overexpressing breast cancer cells (17, 22).
Regulation of PKM2 activity by arginine methylation
PK catalyzes the final step in glycolysis (44, 55). Unlike PKM1, which is mainly expressed in differentiated tissues and exists in a constitutively active form, PKM2 is selectively expressed in actively proliferating cells, such as lymphocytes and tumor cells (44, 55). The activity of PKM2 is allosterically regulated by the glycolytic intermediate fructose 1,6-bisphosphate (33), serine (56), and a variety of PTMs (44). Although cancer cells revert to using the highly regulated PKM2 (rather than PKM1), whether it serves as a rate-limiting step of glycolysis and the extent to which PKM2 contributes to the Warburg effect remain controversial (36–38). Recently, the glyceraldehyde 3-phosphate dehydrogenase, once considered as a “housekeeping gene” and devoid of any significant research impact, has emerged as a key regulator of aerobic glycolysis, particularly in highly glycolytic cancer (57, 58) and activated immune cells (59).
Nevertheless, reduced PK activity has been consistently linked to enhanced de novo serine synthesis. For example, expression of PKM2 promotes de novo serine synthesis and sustains the activity of mechanistic target of rapamycin complex 1 (mTORC1), thus allowing cells to proliferate in serine-depleted medium (11). Increasing PKM2 activity in cancer cells using small molecules reduces de novo serine biosynthesis activity and suppresses tumorigenesis (12, 13). In this study, we determined that PKM2 is arginine-methylated by CARM1 (Fig. 3). Through structural analysis, we defined the molecular basis by which arginine methylation, particularly at Arg-447, enhances the intramolecular interactions of PKM2 and stabilizes its tetramer state (Figs. 4 and 5). Although the stoichiometry of PKM2 arginine methylation was not determined in this study, methylated PKM2 does exhibit significantly increased PK activity compared with the unmethylated PKM2 (Figs. 4 and 5). This is a unique mechanism of PKM2 regulation, because all other previously identified PTMs suppress PKM2 activity (44, 55). However, the extent to which this altered PK activity contributes to enhanced glucose to serine flux in CARM1 knockout cells remains to be determined. It is expected that reduced PKM2 activity, due to loss of CARM1-mediated methylation, would cause buildup of glycolytic intermediates, in particular 3PG, to feed into the SSP (8, 49). However, MS analysis of 13C-labeled glycolytic metabolites failed to identify an increase of 3PG enrichment in CARM1 knockout cells (data not shown). This could be caused by compensatory alterations in the activity of other glycolytic enzymes that may neutralize the accumulation of intermediate metabolites, because CARM1 has been reported to regulate the activity as well as the expression of several metabolic enzymes (19, 39, 47).
Liu et al. (31) recently reported that inhibiting PKM2 arginine methylation increases mitochondrial oxidative phosphorylation by increasing Ca2+ uptake and mitochondrial membrane potential. Specifically, PKM2 arginine methylation is inversely correlated with the expression of the endoplasmic reticulum calcium-releasing channel protein InsP3R, which is responsible for Ca2+ transfer to mitochondria to promote oxidative phosphorylation (31). Consistent with this finding, our [13C]glucose tracing experiments revealed increased enrichment of TCA cycle metabolites, such as citrate and αKG, in CARM1 knockout cells (Fig. S1). However, this increase is caused, at least in part, by reduced PK activity in these cells, because enhancing PK activity by expressing more active PKM1 largely restores these metabolite levels (Fig. S2). Interestingly, direct cross-talk between altered PK activity and the level of mitochondrial oxidative phosphorylation has been reported; enhanced PK activity suppresses mitochondrial oxidative phosphorylation due to their competition for the available ADP (60, 61).
Although arginine methylation was once considered a stable PTM, evidence suggesting the active removal of arginine methylation is emerging (15, 62, 63). It is intriguing to consider the mechanism by which PKM2 arginine methylation might be dynamically regulated to enable cells to adapt to various metabolic conditions. Because the PKM2 arginine methylation sites (Arg-445 and Arg-447) are completely buried within the tetramer (Fig. 4, C and D), the demethylation reaction would occur only in its monomeric or dimeric states. Interestingly, the Jumonji domain containing protein 5 (JMJD5), which belongs to the αKG-dependent demethylase family, has been reported to only interact with dimeric PKM2 to promote its nuclear translocation (64). Future work is needed to determine whether JMJD5 functions as an arginine demethylase for PKM2 methylation.
Enhanced serine biosynthesis promotes cell proliferation and survival
The nonessential amino acid serine has been increasingly recognized as an important fuel for cell proliferation and survival (7–10, 49). Cancer cells meet their high demand for serine through a combination of exogenous serine uptake and de novo synthesis from glucose (7, 8). Rapidly proliferating cancer cells avidly consume exogenous serine and show a precipitous drop in intracellular serine levels following transfer to serine-free culture conditions. Under such conditions, cancer cells respond by slowing proliferation as their metabolism adjusts to enable an increase in de novo serine synthesis. Two critical molecular processes that enhance de novo serine synthesis in various cancers are the amplification of the rate-limiting SSP enzyme PHGDH (49) and the reduction of PKM2 activity (11, 48, 65). Consistent with these adaptive changes, we found that Carm1 knockout cells exhibit increased glucose flux to the SSP (Fig. 1F) and are more resistant to the deprivation of exogenous serine (Fig. 6A). This is likely due to reduced PKM2 activity, because treating Carm1 knockout cells with the PKM2 small molecule activator TEPP-46 restores their sensitivity to serine depletion (Fig. 6, B and C), consistent with previous reports demonstrating that lower PK activity promotes cancer cell survival and that enhancing PK activity leads to proliferation arrest, when extracellular serine is limited (12, 13, 42). Our studies provide further evidence to support efforts to develop PKM2 activators for cancer therapy (12, 13, 41, 45).
Experimental procedures
Chemicals and antibodies
Lactate dehydrogenase (LDH; L1254), NADH (N8129), ADP (A5285), and BAY-876 (SML1774) were purchased from Sigma. PEP (21459) was purchased from Chem-Impex International. NCT-503 (catalog no. 19718) and TEPP-46 (catalog no. 13942) were purchased from Cayman Chemical. Adenosyl-l-methionine, S-methyl-3H (NET155250UC), and EN3HANCE 1× (6NE9701) were purchased from PerkinElmer Life Sciences. Anti-β-actin (A5316) and anti-FLAG M2 (F3165) antibodies were purchased from Sigma. Anti-PKM2 (4053) and anti-PKM1 (7067) antibodies were purchased from Cell Signaling Technology. Anti-CARM1 (A300–421A) antibody was purchased from Bethyl Laboratories.
Plasmids and siRNA
Human PKM1 (44241)- and PKM2 (44242)-expressing constructs for His-tagged recombinant protein purification were purchased from Addgene. The His-tagged mouse PKM2 construct was a gift from Dr. Matthew Pratt (University of Southern California). Both PKM1 and PKM2 were subcloned into a p3×FLAG-CMV-7.1 vector for expression in mammalian cells. GST-tagged PRMT1, PRMT3, CARM1, PRMT6, and Myc-PRMT5 used for in vitro methylation have been described before (66). All mutant constructs, including CARM1 (R169A), PKM2 (R399E), PKM2 (R445K/R447K), PKM2 (L392A), PKM2 (F421A), and PKM2 (L392A/F421A), were generated using the QuickChange II XL site-directed mutagenesis kit (Agilent Technologies). The PKM2 siRNA (J-006781-07-0002) was purchased from Dharmacon.
Cell culture and transfection
MEF and breast cancer MCF7 cells were cultured at 37 °C with 5% CO2 in high-glucose DMEM (11965092, Life Technologies, Inc.) supplemented with 10% FBS and 1% penicillin/streptomycin solution. CARM1 knockout MEFs have been described before (19). CARM1 knockout MCF-7 cells were kindly provided by Dr. Wei Xu (University of Wisconsin). Plasmid and siRNA transfection were performed using Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer's protocol.
Glucose consumption and lactate production analysis
Glucose uptake and lactate production were measured using the YSI 7100 MBS Multiparameter Bioanalytical System (YSI Inc.). After a 24-h cell culture, glucose and lactate concentrations in the medium were determined. The amount consumed or produced by cells was determined by subtracting the concentrations in the sample medium from the concentrations in medium incubated without cells and then normalized to cell number.
Metabolic stable isotope tracing
[U-13C]Glucose was purchased from Sigma. Stable isotope tracing experiments to determine isotopologue distributions in soluble metabolites were performed as described previously (67). WT and Carm1 knockout MEFs, as well as control and CARM1 knockout MCF7 cells, were incubated for 4 h in [U-13C]glucose-containing medium (glucose-free DMEM supplemented with 10% dialyzed FBS). Intracellular metabolites were harvested, derivatized, and measured with GC-MS.
Purification of recombinant proteins
To express His-tagged proteins, plasmids were transformed into Escherichia coli BL21 (DE3) cells, and protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 30 °C. Cells were lysed by sonication in lysis buffer: 50 mm Tris, pH 8.0, 10 mm MgCl2, 200 mm NaCl, 100 mm KCl, 10% glycerol, 10 mm imidazole containing protease inhibitor (ThermoFisher Scientific). After centrifugation, the supernatant was bound to HisLinkTM nickel-nitrilotriacetic acid resin (Promega) for 4 h at 4 °C. Proteins were eluted with lysis buffer containing 250 mm imidazole and dialyzed into 100 mm Tris, pH 8.0, 120 mm NaCl buffer for the in vitro methyltransferase assay or into PKM2 reaction buffer (30 mm Tris, pH 7.5, 25 mm KCl, 5 mm MgCl2, 10% glycerol, 1 mm DTT) for the PK activity assay. The expression and purification of GST-tagged recombinant proteins for in vitro methylation have been described previously (66, 68).
In vitro methylation assay
In vitro methylation reactions were carried out in 30 μl of 1× PBS, pH 7.4, containing 0.5–1.0 μg of substrate, 3 μg of recombinant enzymes, and 0.42 μm S-adenosyl-l-[methyl-3H]methionine (79 Ci mmol−1 from a 7.5 μm stock solution). The reaction was incubated at 30 °C for 1 h and then separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, treated with EN3HANCE, and exposed to a film for 1–3 days at −80 °C.
PK activity assay
PK activity was measured using the NADH/LDH-coupled assay according to published protocols (33). Reactions for each assay contained recombinant PK (50–200 ng) or cell lysate (2–4 μg), 50 mm Tris, pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.6 mm ADP, 0.5 mm PEP, 180 μm NADH, and 8 units LDH. The change in absorbance at 340 nm due to oxidation of NADH by LDH was measured using a Synergy H4 hybrid multimode microplate reader (BioTek). In addition to the normalized PK activity presented in Figs. 1–6, the PK activity reflected by the turnover of NADH calculated as the unit of nanomoles/min/μg cell lysate or nanomoles/min/μg recombinant enzyme was also presented in Figs. S1–S5.
Identification of methylation sites by LC-MS/MS
In vitro-methylated PKM2 samples were resolved on 8% SDS-polyacrylamide gel and stained with SimplyBlueTM SafeStain (InvitrogenTM, catalog no. LC6065). The protein bands of interest were excised and destained, followed by trypsin/Lys-C digestion and MS.
Immunoprecipitation
Cells were lysed in lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA) supplemented with protease inhibitor mixture. Supernatants were clarified by centrifugation at top speed (17,000 × g) for 15 min at 4 °C, and the supernatants (500 μl) were incubated with 40 μl of anti-FLAG M2 affinity agarose beads (Sigma) at 4 °C overnight. Beads were washed three times with lysis buffer, and elution was performed by incubating the washed beads in 40 μl of 2× SDS sample buffer at 98 °C for 5 min. The input lysates and immunoprecipitates were separated by SDS-PAGE and analyzed by Western blot analysis. When eluting the proteins for kinetic assays, washed beads were incubated with 200 ng/μl 3× FLAG peptide in TBS, pH 7.4, for 4–6 h at 4 °C.
Size-exclusion chromatography
Size-exclusion chromatography (gel filtration) was performed with a Superose 6 10/300GL (Tricorn) column. Cells were lysed in lysis buffer (50 mm Tris·Cl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, and 1× protease inhibitors (ThermoFisher Scientific)), and 4–7 mg/ml total protein (500 μl) was loaded into the column and eluted with elution buffer (50 mm phosphate, 0.15 m NaCl, pH 7.0). Every 500-μl fraction was collected, and 20 μl of each fraction was analyzed by Western blot analysis.
Cell proliferation assay
To measure cell proliferation, cells were plated at a density of 500 cells per well in 96-well plates, using four replicates for each time point. Cell growth was measured using the Cell Counting Kit-8 (Dojindo Molecular Technologies) according to the manufacturer's instructions. The day after initial seeding was considered day 1, and the data for all other time points were normalized to day 1 readings. For serine-dependence analysis, cells were cultured in serine-free minimal essential medium (M0446, Sigma).
Author contributions
T. A. and Yanzhong Yang conceptualization; T. A., M. O., L. J., and J. S. data curation; T. A., L. J., J. S., and Yanzhong Yang formal analysis; T. A. and Yanzhong Yang investigation; T. A. and Yanzhong Yang writing-original draft; M. O., Ying Yang, and M. K. methodology; M. K. resources; Yanzhong Yang supervision; Yanzhong Yang funding acquisition; Yanzhong Yang project administration; Yanzhong Yang writing-review and editing.
Supplementary Material
Acknowledgments
We thank Lei Zhang and Yunan Miao from TBDC for their assistance with LC-MS/MS. We thank Kerin Higa for providing helpful comments on scientific writing. Research reported in this publication includes work performed in the Translational Biomarker Discovery Core supported by NCI, National Institutes of Health, under Award P30CA33572.
This work was funded by The V Foundation for Cancer Research and CONquer canCER Now Award from Concern Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- TCA
- tricarboxylic acid
- MEF
- mouse embryonic fibroblast
- CARM1
- co-activator-associated arginine methyltransferase 1
- PK
- pyruvate kinase
- SSP
- serine synthesis pathway
- 3PG
- 3-phosphoglycerate
- αKG
- α-ketoglutarate
- PTM
- post-translational modification
- PEP
- phosphoenolpyruvate
- DMEM
- Dulbecco's modified Eagle's medium
- LDH
- Lactate dehydrogenase
- SAM
- S-adenosyl-l-methionine
- PRMT
- protein arginine methyltransferase
- ADMA
- asymmetrical dimethylated arginine
- AT2
- alveolar type II
- PHGDH
- phosphoglycerate dehydrogenase.
References
- 1. Mulukutla B. C., Yongky A., Le T., Mashek D. G., and Hu W. S. (2016) Regulation of glucose metabolism–a perspective from cell bioprocessing. Trends Biotechnol. 34, 638–651 10.1016/j.tibtech.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 2. Warburg O., Wind F., and Negelein E. (1927) The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530 10.1085/jgp.8.6.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liberti M. V., and Locasale J. W. (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218 10.1016/j.tibs.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lunt S. Y., and Vander Heiden M. G. (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464 10.1146/annurev-cellbio-092910-154237 [DOI] [PubMed] [Google Scholar]
- 6. Vander Heiden M. G., Locasale J. W., Swanson K. D., Sharfi H., Heffron G. J., Amador-Noguez D., Christofk H. R., Wagner G., Rabinowitz J. D., Asara J. M., and Cantley L. C. (2010) Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499 10.1126/science.1188015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Labuschagne C. F., van den Broek N. J., Mackay G. M., Vousden K. H., and Maddocks O. D. (2014) Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258 10.1016/j.celrep.2014.04.045 [DOI] [PubMed] [Google Scholar]
- 8. Yang M., and Vousden K. H. (2016) Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- 9. Mehrmohamadi M., Liu X., Shestov A. A., and Locasale J. W. (2014) Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 9, 1507–1519 10.1016/j.celrep.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao X., and Locasale J. W. (2016) Serine metabolism links tumor suppression to the epigenetic landscape. Cell Metab. 24, 777–779 10.1016/j.cmet.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 11. Ye J., Mancuso A., Tong X., Ward P. S., Fan J., Rabinowitz J. D., and Thompson C. B. (2012) Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 109, 6904–6909 10.1073/pnas.1204176109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kung C., Hixon J., Choe S., Marks K., Gross S., Murphy E., DeLaBarre B., Cianchetta G., Sethumadhavan S., Wang X., Yan S., Gao Y., Fang C., Wei W., Jiang F., et al. (2012) Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem. Biol. 19, 1187–1198 10.1016/j.chembiol.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parnell K. M., Foulks J. M., Nix R. N., Clifford A., Bullough J., Luo B., Senina A., Vollmer D., Liu J., McCarthy V., Xu Y., Saunders M., Liu X. H., Pearce S., Wright K., O'Reilly M., et al. (2013) Pharmacologic activation of PKM2 slows lung tumor xenograft growth. Mol. Cancer Ther. 12, 1453–1460 10.1158/1535-7163.MCT-13-0026 [DOI] [PubMed] [Google Scholar]
- 14. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 16. O'Brien K. B., Alberich-Jordà M., Yadav N., Kocher O., Diruscio A., Ebralidze A., Levantini E., Sng N. J., Bhasin M., Caron T., Kim D., Steidl U., Huang G., Halmos B., Rodig S. J., et al. (2010) CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 137, 2147–2156 10.1242/dev.037150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng H., Qin Y., Fan H., Su P., Zhang X., Zhang H., and Zhou G. (2013) Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn. Pathol. 8, 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim Y. R., Lee B. K., Park R. Y., Nguyen N. T., Bae J. A., Kwon D. D., and Jung C. (2010) Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer 10, 197 10.1186/1471-2407-10-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yadav N., Cheng D., Richard S., Morel M., Iyer V. R., Aldaz C. M., and Bedford M. T. (2008) CARM1 promotes adipocyte differentiation by coactivating PPARγ. EMBO Rep. 9, 193–198 10.1038/sj.embor.7401151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. El Messaoudi S., Fabbrizio E., Rodriguez C., Chuchana P., Fauquier L., Cheng D., Theillet C., Vandel L., Bedford M. T., and Sardet C. (2006) Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the cyclin E1 gene. Proc. Natl. Acad. Sci. U.S.A. 103, 13351–13356 10.1073/pnas.0605692103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., and Stallcup M. R. (1999) Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 10.1126/science.284.5423.2174 [DOI] [PubMed] [Google Scholar]
- 22. Wang L., Zhao Z., Meyer M. B., Saha S., Yu M., Guo A., Wisinski K. B., Huang W., Cai W., Pike J. W., Yuan M., Ahlquist P., and Xu W. (2014) CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36 10.1016/j.ccr.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Y., Lu Y., Espejo A., Wu J., Xu W., Liang S., and Bedford M. T. (2010) TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023 10.1016/j.molcel.2010.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng D., Côté J., Shaaban S., and Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 10.1016/j.molcel.2006.11.019 [DOI] [PubMed] [Google Scholar]
- 25. Ohkura N., Takahashi M., Yaguchi H., Nagamura Y., and Tsukada T. (2005) Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 280, 28927–28935 10.1074/jbc.M502173200 [DOI] [PubMed] [Google Scholar]
- 26. Sanchez G., Bondy-Chorney E., Laframboise J., Paris G., Didillon A., Jasmin B. J., and Côté J. (2016) A novel role for CARM1 in promoting nonsense-mediated mRNA decay: potential implications for spinal muscular atrophy. Nucleic Acids Res. 44, 2661–2676 10.1093/nar/gkv1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaniskan H. Ü., Eram M. S., Liu J., Smil D., Martini M. L., Shen Y., Santhakumar V., Brown P. J., Arrowsmith C., Vedadi M., and Jin J. (2016) Design and synthesis of selective, small molecule inhibitors of coactivator-associated arginine methyltransferase 1 (CARM1). Medchemcomm. 7, 1793–1796 10.1039/C6MD00342G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen Y., Szewczyk M. M., Eram M. S., Smil D., Kaniskan H. Ü., de Freitas R. F., Senisterra G., Li F., Schapira M., Brown P. J., Arrowsmith C. H., Barsyte-Lovejoy D., Liu J., Vedadi M., and Jin J. (2016) Discovery of a potent, selective, and cell-active dual inhibitor of protein arginine methyltransferase 4 and protein arginine methyltransferase 6. J. Med. Chem. 59, 9124–9139 10.1021/acs.jmedchem.6b01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferreira de Freitas R., Eram M. S., Smil D., Szewczyk M. M., Kennedy S., Brown P. J., Santhakumar V., Barsyte-Lovejoy D., Arrowsmith C. H., Vedadi M., and Schapira M. (2016) Discovery of a potent and selective coactivator associated arginine methyltransferase 1 (CARM1) inhibitor by virtual screening. J. Med. Chem. 59, 6838–6847 10.1021/acs.jmedchem.6b00668 [DOI] [PubMed] [Google Scholar]
- 30. Drew A. E., Moradei O., Jacques S. L., Rioux N., Boriack-Sjodin A. P., Allain C., Scott M. P., Jin L., Raimondi A., Handler J. L., Ott H. M., Kruger R. G., McCabe M. T., Sneeringer C., Riera T., et al. (2017) Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci. Rep. 7, 17993 10.1038/s41598-017-18446-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu F., Ma F., Wang Y., Hao L., Zeng H., Jia C., Wang Y., Liu P., Ong I. M., Li B., Chen G., Jiang J., Gong S., Li L., and Xu W. (2017) PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat. Cell Biol. 19, 1358–1370 10.1038/ncb3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., and Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
- 33. Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., and Cantley L. C. (2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 10.1038/nature06667 [DOI] [PubMed] [Google Scholar]
- 34. Luo W., and Semenza G. L. (2012) Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol. Metab. 23, 560–566 10.1016/j.tem.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang W., and Lu Z. (2015) Pyruvate kinase M2 at a glance. J. Cell Sci. 128, 1655–1660 10.1242/jcs.166629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Israelsen W. J., Dayton T. L., Davidson S. M., Fiske B. P., Hosios A. M., Bellinger G., Li J., Yu Y., Sasaki M., Horner J. W., Burga L. N., Xie J., Jurczak M. J., DePinho R. A., Clish C. B., et al. (2013) PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155, 397–409 10.1016/j.cell.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morita M., Sato T., Nomura M., Sakamoto Y., Inoue Y., Tanaka R., Ito S., Kurosawa K., Yamaguchi K., Sugiura Y., Takizaki H., Yamashita Y., Katakura R., Sato I., Kawai M., et al. (2018) PKM1 confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell 33, 355–367.e7 10.1016/j.ccell.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 38. Xie J., Dai C., and Hu X. (2016) Evidence that does not support pyruvate kinase M2 (PKM2)-catalyzed reaction as a rate-limiting step in cancer cell glycolysis. J. Biol. Chem. 291, 8987–8999 10.1074/jbc.M115.704825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y. P., Zhou W., Wang J., Huang X., Zuo Y., Wang T. S., Gao X., Xu Y. Y., Zou S. W., Liu Y. B., Cheng J. K., and Lei Q. Y. (2016) Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol. Cell 64, 673–687 10.1016/j.molcel.2016.09.028 [DOI] [PubMed] [Google Scholar]
- 40. Shin H. J., Kim H., Oh S., Lee J. G., Kee M., Ko H. J., Kweon M. N., Won K. J., and Baek S. H. (2016) AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534, 553–557 10.1038/nature18014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walsh M. J., Brimacombe K. R., Anastasiou D., Yu Y., Israelsen W. J., Hong B. S., Tempel W., Dimov S., Veith H., Yang H., Kung C., Yen K. E., Dang L., Salituro F., Auld D. S., et al. (2010) ML265: a potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information, Bethesda, MD: [PubMed] [Google Scholar]
- 42. Lunt S. Y., Muralidhar V., Hosios A. M., Israelsen W. J., Gui D. Y., Newhouse L., Ogrodzinski M., Hecht V., Xu K., Acevedo P. N., Hollern D. P., Bellinger G., Dayton T. L., Christen S., Elia I., et al. (2015) Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57, 95–107 10.1016/j.molcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao X., Wang H., Yang J. J., Liu X., and Liu Z. R. (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 10.1016/j.molcel.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dayton T. L., Jacks T., and Vander Heiden M. G. (2016) PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730 10.15252/embr.201643300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anastasiou D., Yu Y., Israelsen W. J., Jiang J. K., Boxer M. B., Hong B. S., Tempel W., Dimov S., Shen M., Jha A., Yang H., Mattaini K. R., Metallo C. M., Fiske B. P., Courtney K. D., et al. (2012) Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 8, 839–847 10.1038/nchembio.1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang L., Zeng H., Wang Q., Zhao Z., Boyer T. G., Bian X., and Xu W. (2015) MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci. Adv. 1, e1500463 10.1126/sciadv.1500463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Al-Dhaheri M., Wu J., Skliris G. P., Li J., Higashimato K., Wang Y., White K. P., Lambert P., Zhu Y., Murphy L., and Xu W. (2011) CARM1 is an important determinant of ERα-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res. 71, 2118–2128 10.1158/0008-5472.CAN-10-2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mattaini K. R., Sullivan M. R., and Vander Heiden M. G. (2016) The importance of serine metabolism in cancer. J. Cell Biol. 214, 249–257 10.1083/jcb.201604085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doherty J. R., and Cleveland J. L. (2013) Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692 10.1172/JCI69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Faubert B., Li K. Y., Cai L., Hensley C. T., Kim J., Zacharias L. G., Yang C., Do Q. N., Doucette S., Burguete D., Li H., Huet G., Yuan Q., Wigal T., Butt Y., et al. (2017) Lactate metabolism in human lung tumors. Cell 171, 358–371 e359 10.1016/j.cell.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie H., Hanai J., Ren J. G., Kats L., Burgess K., Bhargava P., Signoretti S., Billiard J., Duffy K. J., Grant A., Wang X., Lorkiewicz P. K., Schatzman S., Bousamra M. 2nd, Lane A. N., et al. (2014) Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 19, 795–809 10.1016/j.cmet.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. San-Millán I., and Brooks G. A. (2017) Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg effect. Carcinogenesis 38, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Romero-Garcia S., Moreno-Altamirano M. M., Prado-Garcia H., and Sánchez-García F. J. (2016) Lactate contribution to the tumor microenvironment: mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 7, 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Israelsen W. J., and Vander Heiden M. G. (2015) Pyruvate kinase: function, regulation and role in cancer. Semin. Cell Dev. Biol. 43, 43–51 10.1016/j.semcdb.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chaneton B., Hillmann P., Zheng L., Martin A. C. L., Maddocks O. D. K., Chokkathukalam A., Coyle J. E., Jankevics A., Holding F. P., Vousden K. H., Frezza C., O'Reilly M., and Gottlieb E. (2012) Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462 10.1038/nature11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shestov A. A., Liu X., Ser Z., Cluntun A. A., Hung Y. P., Huang L., Kim D., Le A., Yellen G., Albeck J. G., and Locasale J. W. (2014) Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 3, e03342 10.7554/eLife.03342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liberti M. V., Dai Z., Wardell S. E., Baccile J. A., Liu X., Gao X., Baldi R., Mehrmohamadi M., Johnson M. O., Madhukar N. S., Shestov A. A., Chio I. I. C., Elemento O., Rathmell J. C., Schroeder F. C., et al. (2017) A predictive model for selective targeting of the Warburg effect through GAPDH inhibition with a natural product. Cell Metab. 26, 648–659.e8 10.1016/j.cmet.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kornberg M. D., Bhargava P., Kim P. M., Putluri V., Snowman A. M., Putluri N., Calabresi P. A., and Snyder S. H. (2018) Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 10.1126/science.aan4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gosalvez M., Pérez-García J., and Weinhouse S. (1974) Competition for ADP between pyruvate kinase and mitochondrial oxidative phosphorylation as a control mechanism in glycolysis. Eur. J. Biochem. 46, 133–140 10.1111/j.1432-1033.1974.tb03605.x [DOI] [PubMed] [Google Scholar]
- 61. Gosalvez M., López-Alarcón L., García-Suarez S., Montalvo A., and Weinhouse S. (1975) Stimulation of tumor-cell respiration by inhibitors of pyruvate kinase. Eur. J. Biochem. 55, 315–321 10.1111/j.1432-1033.1975.tb02165.x [DOI] [PubMed] [Google Scholar]
- 62. Walport L. J., Hopkinson R. J., Chowdhury R., Schiller R., Ge W., Kawamura A., and Schofield C. J. (2016) Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 10.1038/ncomms11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li S., Ali S., Duan X., Liu S., Du J., Liu C., Dai H., Zhou M., Zhou L., Yang L., Chu P., Li L., Bhatia R., Schones D. E., Wu X., et al. (2018) JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403 10.1016/j.celrep.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang H. J., Hsieh Y. J., Cheng W. C., Lin C. P., Lin Y. S., Yang S. F., Chen C. C., Izumiya Y., Yu J. S., Kung H. J., and Wang W. C. (2014) JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 111, 279–284 10.1073/pnas.1311249111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pacold M. E., Brimacombe K. R., Chan S. H., Rohde J. M., Lewis C. A., Swier L. J., Possemato R., Chen W. W., Sullivan L. B., Fiske B. P., Cho S., Freinkman E., Birsoy K., Abu-Remaileh M., et al. (2016) A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12, 452–458 10.1038/nchembio.2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang L., Wang Z., Narayanan N., and Yang Y. (2018) Arginine methylation of the C-terminal RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 46, 3061–3074 10.1093/nar/gky103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jiang L., Boufersaoui A., Yang C., Ko B., Rakheja D., Guevara G., Hu Z., and DeBerardinis R. J. (2017) Quantitative metabolic flux analysis reveals an unconventional pathway of fatty acid synthesis in cancer cells deficient for the mitochondrial citrate transport protein. Metab. Eng. 43, 198–207 10.1016/j.ymben.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Narayanan N., Wang Z., Li L., and Yang Y. (2017) Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells. Cell Discov. 3, 16048 10.1038/celldisc.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.