Abstract
Sulfated glycans are known to be involved in several glycan-mediated cell adhesion and recognition pathways. Our mRNA transcript analyses on the genes involved in synthesizing GlcNAc-6-O–sulfated glycans in human colon cancer tissues indicated that GlcNAc6ST-2 (CHST4) is preferentially expressed in cancer cells compared with nonmalignant epithelial cells among the three known major GlcNAc-6-O-sulfotransferases. On the contrary, GlcNAc6ST-3 (CHST5) was only expressed in nonmalignant epithelial cells, whereas GlcNAc6ST-1 (CHST2) was expressed equally in both cancerous and nonmalignant epithelial cells. These results suggest that 6-O-sulfated glycans that are synthesized only by GlcNAc6ST-2 may be highly colon cancer–specific, as supported by immunohistochemical staining of cancer cells using the MECA-79 antibody known to be relatively specific to the enzymatic reaction products of GlcNAc6ST-2. By more precise MS-based sulfoglycomic analyses, we sought to further infer the substrate specificities of GlcNAc6STs via a definitive mapping of various sulfo-glycotopes and O-glycan structures expressed in response to overexpression of transfected GlcNAc6STs in the SW480 colon cancer cell line. By detailed MS/MS sequencing, GlcNAc6ST-3 was shown to preferentially add sulfate onto core 2–based O-glycan structures, but it does not act on extended core 1 structures, whereas GlcNAc6ST-1 prefers core 2–based O-glycans to extended core 1 structures. In contrast, GlcNAc6ST-2 could efficiently add sulfate onto both extended core 1– and core 2–based O-glycans, leading to the production of unique sulfated extended core 1 structures such as R-GlcNAc(6-SO3−)β1-3Galβ1–4GlcNAc(6-SO3−)β1–3Galβ1–3GalNAcα, which are good candidates to be targeted as cancer-specific glycans.
Keywords: mass spectrometry (MS), glycobiology, glycomics, analytical chemistry, colorectal cancer, CHST, GlcNAc-6-O-sulfotransferase, MECA-79, sulfoglycomics
Introduction
Sulfation on N- and O-glycans alters their physico-chemical properties and thereby their cognate recognition by specific endogenous lectins involved in normal biological processes and disease states, including chronic inflammation, cancer cell metastasis, and hormone regulation (1–4). A well-known example is the process of lymphocyte homing initiated by interaction between L-selectin on the homing lymphocytes and its sulfated ligands expressed on the high endothelial venules (HEV)3 of lymph node, namely 6-sulfo sialyl Lewis X (5) carried on either core 2 or extended core 1 O-glycans or both (6). In fact, sulfated terminal glycotopes are increasingly implicated as the preferred high-affinity ligands for several other lectins, including members of the galectins and Siglec families through glycan array binding studies (7–12). However, the actual occurrence of sulfated glycans on physiologically relevant tissues, the precise sulfated glycotope structures involved, and their regulated biosynthesis in these cases remain mostly unknown.
6-O-Sulfation of β-GlcNAc is commonly recognized as the initial step in the biosynthesis of sulfated LacNAc, with and without subsequent β-galactosylation, polyLacNAc elongation, sialylation, and fucosylation (13). Five GlcNAc-6-O-sulfotransferases (GlcNAc6STs) have been cloned in humans, four of which have orthologs in mice (1, 14). Among these GlcNAc6STs, only the roles of GlcNAc6ST-1 (HUGO Gene Nomenclature Committee–approved name, CHST2) and GlcNAc6ST-2 (CHST4) as the major enzymes involved in synthesizing the L-selectin ligands mediating lymphocyte homing have been confirmed by studies using GlcNAc6ST-1 and GlcNAc6ST-2 double-deficient mice (15, 16). GlcNAc6ST-1 was the first identified GlcNAc6ST with rather broad expression in human and mouse, whereas GlcNAc6ST-2 exhibits limited expression in HEV among human and mouse normal tissues and is therefore also called HEC-GlcNAc6ST (3). GlcNAc6ST-3 (CHST5) is also called I-GlcNAc6ST due to its restricted expression pattern in gut-associated tissues (17). Previous studies have shown that GlcNAc6ST-3 prefers mucin-type glycoprotein substrates, whereas GlcNAc6ST-1 and GlcNAc6ST-2 exhibit strong activity on both mucins and glycoproteins with predominantly N-linked chains (17). An in vitro substrate specificity assay showed that all of these three GlcNAc6STs can add sulfate on core 2–based GlcNAcβ1-6(Galβ1–3)GalNAc-pNP structure, but only GlcNAc6ST-2 can additionally act on GlcNAcβ1–3GalNAc-pNP of core 3 structure (18). GlcNAc6ST-4 (CHST7), like GlcNAc6ST-1, shows a broad expression pattern in human tissues (19) but acts more like GalNAc-6-O-sulfotransferase (chondroitin-6-sulfotransferase) (20). GlcNAc6ST-5 (CHST6) shows a restricted expression in cornea (21) and appears to be responsible for macular corneal dystrophy (22) (see Table 1 for a summary).
Table 1.
Substrate specificities of human GlcNAc6STs
| Enzyme | HGNCa | Alternative names | Predominant tissue distribution | Preferred substrates (in vitro) | Reference(s) |
|---|---|---|---|---|---|
| GlcNAc6ST-1 | CHST2 | Ubiquitously expressed | GlcNAcβ2/6ManOMe, GlcNAcβ6 (Galβ3)GalNAc-pNP, not core 3–based structure | 18 | |
| GlcNAc6ST-2 | CHST4 | HEC-GlcNAc6ST, LSST | HEC cells | GlcNAcβ2/6ManOMe, GlcNAcβ6(Galβ3)GalNAc-pNP, GlcNAcβ3GalNAc-pNP | 17, 18, 23 |
| GlcNAc6ST-3 | CHST5 | I-GlcNAc6ST | Colon, intestine | GlcNAcβ6(Galβ3)GalNAc-pNP, not core 3–based structure, GlcNAcβ2Manα6Manβ1-octyl, GlcNAcβ2(GlcNAcβ6)-Manα6Manβ1-octyl | 17, 18, 24 |
| GlcNAc6ST-4 | CHST7 | C6ST-2 | Ubiquitously expressed | GlcNAcβ2/6ManOMe, GlcNAcβ6(Galβ3)GalNAc-pNP, GlcNAcβ3GalNAc-pNP | 19 |
| GlcNAc6ST-5 | CHST6 | C-GlcNAc6ST | Cornea | GlcNAcβ6Manα6Manβ1-octyl, GlcNAcβ3Galβ4GlcNAcβ6-Manα6Manβ1-octyl, GlcNAcβ3Galβ4GlcNAcβ3Galβ4-GlcNAcβ6Manβ6Manβ1-octyl, GlcNAcβ6(Galβ3)GalNAc-octyl | 22, 24 |
a HGNC (HUGO Gene Nomenclature Committee)-approved gene names; CHST, carbohydrate (N-acetylglucosamine-6-O) sulfotransferase.
In general, the expression of sulfated glycans is down-regulated in the course of human colonic carcinogenesis (25–27), which implies possible alteration in the expression level of galactose-3-O-sulfotransferases (Gal3STs) (28–30), GlcNAc6STs (31, 32), and/or sulfate transporters (33) in colonic cancer tissues. Differential expressions of GlcNAc6ST-2 and -3 in human colonic adenocarcinomas and the adjacent normal mucosa have been reported (31). By competitive RT-PCR, GlcNAc6ST-3 was shown to be expressed in normal colonic mucosa, whereas GlcNAc6ST-2 was ectopically expressed in colonic mucinous adenocarcinoma (32). Due to broad substrate specificity of GlcNAc6ST-2 (18, 32, 34), mucinous adenocarcinomas may thus express unique sulfated glycans, which may serve as a good clinical marker for detecting colonic adenocarcinomas. A major shortcoming in our understanding of the onco-developmental expression of sulfated glycans and the GlcNAc6STs is the uncertainty associated with their causal relationship. To date, the substrate specificities of these GlcNAc6STs were mostly determined in vitro using rather limited synthetic substrates as acceptors (31, 34) or in vivo by co-transfection with reporter glycoprotein substrates (17, 35). The actual supporting structural data are insufficient and have mostly resulted from the use of mAbs against overlapping glycotopes instead of definitive structural determination of the sulfated glycan products. Nor has the structural consequence of the elevated expression of GlcNAc6ST-2 in colon cancer cells been critically examined.
Aiming to better delineate the causal relationship, if any, between the expression level of GlcNAc6STs on one hand and the sulfoglycome on the other, we have applied our recently developed MS approach (36–41) to systematically and directly identify the sulfoglycomic changes in response to transfected overexpression of select human GlcNAc6STs in colon cancer cells of the same genetic background (SW480). The prime advantage of MS analysis resides in its ability to offer a global view of the relative abundance of all major sulfated glycans, including any unanticipated novel structures. This is in contrast to the use of mAbs, limited by availability against few known sulfated glycotopes, or an HPLC-based mapping method, limited by the need for reference glycan standards (42). In this work, we confirmed by RT-PCR and analysis of the Cancer Genome Atlas (TCGA) transcriptomic data set that the GlcNAc6ST-2 mRNA transcript indeed increases drastically in human colon cancer tissues compared with undetectable expression in normal human colon, whereas the reverse is true for GlcNAc6ST-3. By MS/MS analysis, we demonstrated that GlcNAc6ST-2 supports the synthesis of 6-sulfo LacNAc on the extended core 1 O-glycan structures and thus MECA-79 epitope, corroborated by a strong staining of colon cancer tissues by the MECA-79 mAb. GlcNAc6ST-3, on the other hand, was shown to be unable to efficiently put a sulfate on the extended core 1. The specificity of GlcNAc6ST-1 is somewhat in between that of GlcNAc6ST-2 and GlcNAc6ST-3, favoring the addition of sulfate onto the 6-arm LacNAc unit of core 2–based O-glycan structures. We further identified a series of new cancer-associated carbohydrate antigens represented by GlcNAc-6–sulfated extended core 1 structures, namely R-GlcNAc(6-SO3−)β1–3Galβ1–4GlcNAc(6-SO3−)β1–3Galβ1-3GalNAcα, which can only be synthesized by GlcNAc6ST-2 due to its unique substrate specificity.
Results
Expression of GlcNAc6ST gene and MECA-79 determinant in human colon cancer tissues and nonmalignant colonic epithelial cells
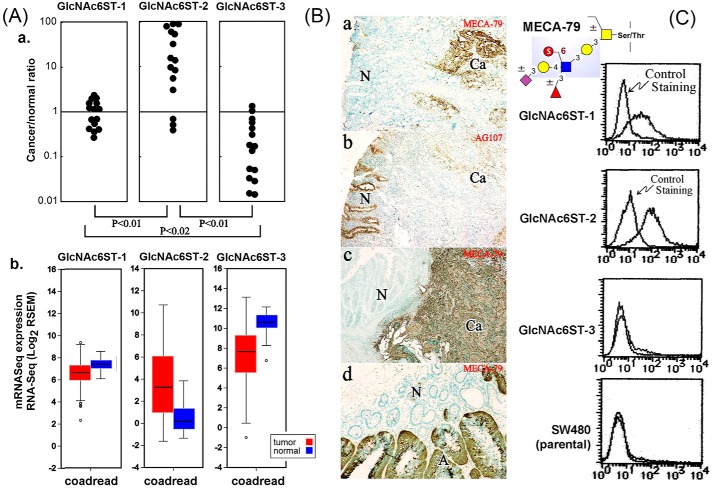
Previous reports have indicated that GlcNAc6ST-2, which has a highly restricted expression in HEC, was ectopically expressed in colonic mucinous adenocarcinoma and not in normal colonic mucosa (32). To further confirm the expression level of different GlcNAc6STs in colonic tissue, the amounts of mRNAs for GlcNAc6ST-1, -2, and -3 in colon cancer tissues and adjacent nonmalignant colonic epithelial cells from the same patient were examined. RT-PCR analyses and results summarized from 15 patients (Fig. 1A (a)) showed that a marked decrease of GlcNAc6ST-3 mRNA was registered in cancer cells compared with nonmalignant epithelial cells, whereas GlcNAc6ST-2 mRNA showed a drastic increase in colon cancer tissues. In contrast, the expression of GlcNAc6ST-1 mRNA was at a similar level in both cancer cells and nonmalignant colonic epithelial cells. These findings are consistent with what could be retrieved from the TCGA database, which contains a larger sampling of colorectal adenocarcinomas (n = 433). RNA-Seq data (Fig. 1A (b)) showed that GlcNAc6ST-2 markedly increases in cancer compared with normal tissues, whereas GlcNAc6ST-3 significantly decreases and GlcNAc6ST-1 only marginally decreases in cancers (43).
Figure 1.
Expression of GlcNAc6STs and MECA-79 epitope on colonic tissues and cells. A (a), real-time quantitative PCR analyses of the GlcNAc6ST transcripts were performed on cancer and adjacent nonmalignant mucosa tissues derived from the same patients, and a total of 15 patients were examined. The differential mRNA expression levels of these three GlcNAc6STs were also extracted from the TCGA database (RNA Seq V2, sample size 433), as shown in A (b). RSEM, RNA-Seq by expectation maximization; coadread, colon and rectal adenocarcinoma. Immunohistochemical localization of MECA-79 epitope (B (a, c, and d)) was additionally compared against that of AG107-defined determinants (B (b)) on sections taken from the same tissue specimen. N, nonmalignant; Ca, cancer; A, adenoma. C, flow cytometry analyses of different GlcNAc6ST-transfected SW480 cell lines by MECA-79 mAb. The sulfated glycotope defined by MECA-79 is represented by cartoon symbols.
Because GlcNAc6ST-2 is known to contribute to the expression of MECA-79 epitope on HEV (15), immunohistochemical staining was performed on malignant and nonmalignant colonic cells from patients using the MECA-79 mAb, which recognizes a 6-sulfo LacNAc moiety on the extended core 1 branch of O-glycans (6). This sulfated glycotope was found to be virtually absent in nonmalignant epithelial cells in all examined cases, whereas a sporadic but significant expression was observed in cancer cells (Fig. 1B (a and c)). It was expressed in 10 of 31 cancer tissue samples examined, but virtually absent in nonmalignant epithelia (statistically significant at p < 0.005, n = 31) (data not shown). Two adenomatous polyp samples showed moderate staining with MECA-79 (see an example in Fig. 1B (d), shown by A), whereas four hyperplastic polyp samples did not (data not shown). This expression pattern of the extended core 1 O-glycan–restricted, MECA-79–defined 6-sulfo LacNAc epitope was in clear contrast to that of the nonsialylated 6-sulfo determinants defined by AG107, which were expressed preferentially in nonmalignant colonic epithelial cells, consistent with that reported previously (33) (Fig. 1B (b)). It suggested that increased level of GlcNAc6ST-2 in colonic cancer cells was responsible for the expression of MECA-79 epitope, but the respective specificities of the three GlcNAc6STs remained unclear. This prompted us to focus our sulfoglycomic analysis on determining the in vivo O-glycan substrate specificities of individual GlcNAc6STs overexpressed in an otherwise homogenous genetic background, namely the SW480 colonic cancer cells. Flow cytometry analysis indicated that MECA-79 epitope was not expressed in either the parental SW480 or GlcNAc6ST-3–transfected cells, but highly expressed upon GlcNAc6ST-2 transfection and expressed at a lower level in the presence of GlcNAc6ST-1 (Fig. 1C).
Sulfoglycomic mapping of the O-glycans from different GlcNAc6ST-transfected cell lines
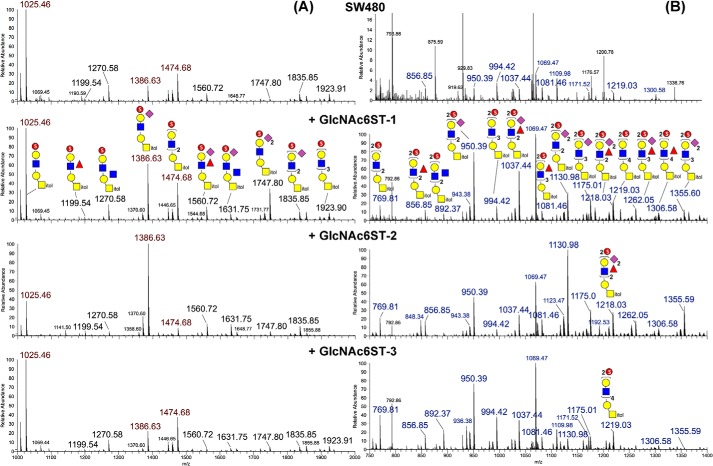
Initial MALDI-MS survey mapping of the permethylated sulfated O-glycans derived from the GlcNAc6ST-transfected cell lines in the negative ion mode afforded many ion signals at 14 mass units lower than those assigned as monosulfated permethylated O-glycans, indicative of disulfated O-glycans having lost one sulfite (36). To facilitate analysis, further amine fractionation was undertaken to separate the disulfated and multiply sulfated species away from the monosulfated ones, and the resulting two fractions were both subjected to nanoLC-nanoESI-MS/MS analyses in negative ion mode after MALDI-MS screening. Averaging the nanoLC-MS survey scans across the time span where the major O-glycans eluted showed that the parental SW480 and all three transfected cell lines afforded many common signals, albeit at different relative intensities (Fig. 2). Strikingly, for the monosulfated O-glycans, a major signal at m/z 1386.63 corresponding to monosulfated, monosialylated Hex2HexNAc2-itol dominated the mass spectrum of GlcNAc6ST-2–transfected cells, whereas the signal at m/z 1025.46 corresponding to its nonsialylated version was the more prominent base peak in the parental and other GlcNAc6ST-transfected cells. When further considered along with the nonsialylated Hex3HexNAc3-itol at m/z 1474.68, the overall pattern indicated that the relative abundance of the few simplest monosulfated O-glycans made by GlcNAcST-2 was significantly different from those produced by GlcNAc6ST-3, which were more similar to those of parental SW480 and its GlcNAc6ST-1–transfected cells (Fig. 2A). However, at higher mass range, as revealed by MALDI-MS analysis (Fig. S1), the GlcNAc6ST-3–transfected cell line alone carried a more extensive range of larger monosulfated O-glycans. On the other hand, much less of the larger disulfated O-glycans were detected for the GlcNAc6ST-3–transfected cells relative to the GlcNAc6ST-1 and -2–transfected cells, whereas only a very low level of disulfated O-glycan could be found in the parental SW480 cells (Fig. 2B). Notably, the disialylated disulfated O-glycans at m/z 1130.982−, 1218.032−, and 1355.592− were prominent and significantly more abundant in the GlcNAc6ST-2–transfected than in the GlcNAc6ST-1–transfected cells but barely detectable in the GlcNAc6ST-3–transfected cells.
Figure 2.
Negative ion mode nanoESI-MS profiles of permethylated monosulfated (A) and disulfated (B) O-glycans from parental SW480 and GlcNAc6ST-transfected cell lines. All monosulfated O-glycans were detected as [M-H]−, whereas the disulfated O-glycans were detected as [M-2H]2−. Larger singly charged monosulfated O-glycans occurring at m/z >2000 were excluded by the acquired nanoLC-MS/MS mass range but could be additionally detected by MALDI-MS, particularly for the GlcNAc6ST-3–transfected cells (Fig. S1). Only the major signals that could be assigned reasonable glycosyl composition based on their detected molecular masses and knowledge of the biosynthetic constrains were nonredundantly annotated on the spectra from the +GlcNAc6ST-1 cells using the standard Symbol Nomenclature for Glycan system (57) (except sulfate, which was represented by a red sphere enclosing an S). A complete set of MS2 spectra for the annotated monosulfated O-glycans (except the lowest abundant m/z 1119 and 1923) from all four cells is provided in Fig. S4, with those of m/z 1025, 1386, and 1474 (peaks labeled in red) further shown in Fig. 3. For the disulfated O-glycans, only that of m/z 769.812− was analyzed in detail and shown in Fig. 5. The signals at m/z 1218.032− and 1219.032− corresponding to two distinct compositions differing by 2 Da could be resolved by high-resolution and mass accuracy measurement. Both were present in the +GlcNAc6ST-1 cells. The signals at m/z 1069.472− and 1171.522− detected among the disulfated O-glycans were identified by MS/MS as corresponding to diphosphorylated, reduced N-glycans carrying two methyl-esterified phosphate groups, most likely Man-6-P–containing Man7GlcNAc2 and Man8GlcNAc2, respectively. All of the labeled m/z values for assigned peaks refer to monoisotopic accurate masses. Doubly charged peaks are labeled in blue. The disulfated O-glycan MS profile for SW480 is shown off scale to allow the view to see the signals at the less than 5% level.
Identification of monosulfated O-glycan structures
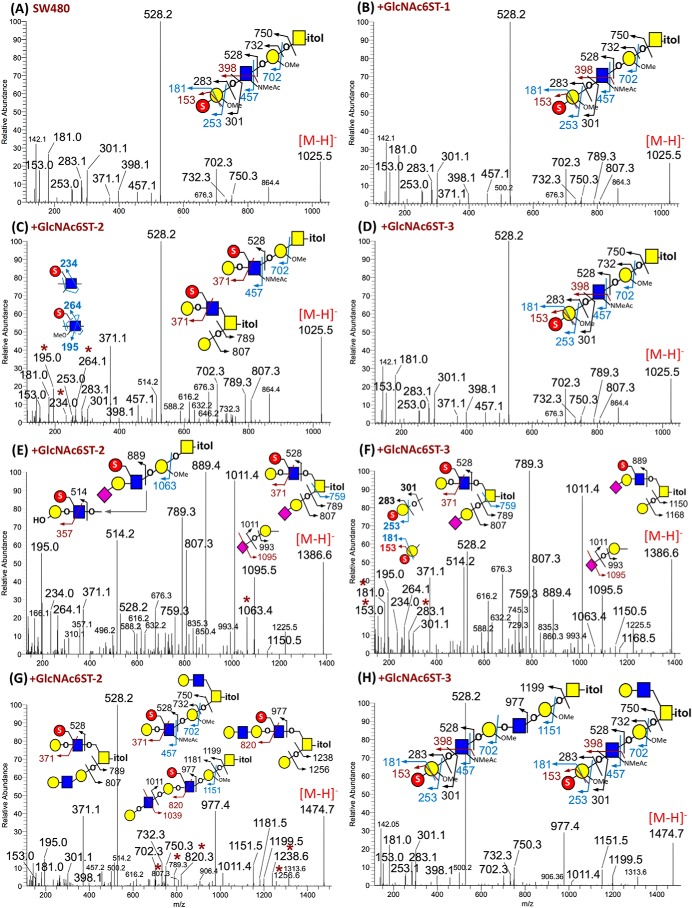
To account for the observed sulfoglycomic differences contributed by different GlcNAc6STs, the nanoLC-MS/MS spectra for the few major monosulfated O-glycans commonly found in the parental SW480 and all three transfected cell lines were extracted and manually interpreted to delineate their isomeric structures and location of sulfate based on detecting a combination of previously reported diagnostic ions (38, 40, 41). In the case of m/z 1025, 1386, and 1474, which seemingly defined the most obvious differences in the MS profiles of monosulfated O-glycans made by cells overexpressing the three GlcNAc6STs (Fig. 2A), structure assignments based on nanoESI-HCD MS2 spectra (Fig. 3) were further corroborated by the MALDI-CID MS2 data described in Figs. S2 and S3 and Table S1). Full sets of HCD MS2 spectra for the eight major O-glycans from each of the GlcNAc6ST-transfected and parental cells were further collated as Fig. S4 (complete with additional notes describing the ions and their interpretations).
Figure 3.
Negative ion mode nanoESI-MS/MS analyses of permethylated monosulfated O-glycans from nontransfected and different GlcNAc6ST-transfected SW480 cells. A complete set of HCD MS2 spectra for eight major monosulfated O-glycans detected in all four cell lines is collated as Fig. S4. Only those for the precursors at m/z 1025 (A–D), 1386 (E and F), and 1474 (G and H) are shown here. A minimum number of possible isomeric structures are provided on each spectrum as cartoon drawings, with the origins of assigned fragment ions indicated. For nonsialylated O-glycans at m/z 1025, the diagnostic ions for GlcNAc6S were only detected in the spectrum of +GlcNAc6ST-2 cells (C). The sialylated structures at m/z 1386 carried exclusively GlcNAc6S in the +GlcNAc6ST-2 cells (E), but the additional presence of Gal3S-terminated type 1 chain was evident by the diagnostic ions for the corresponding structures made by the +GlcNAc6ST-3 cells (F). Signals at m/z 789/807 and 1238/1256 in G identified the additional presence of isomeric core 2 structures carrying 6-sulfo LacNAc on the 6-arm in the +GlcNAc6ST-2 cells, which were absent from the +GlcNAc6ST-3 cells (H). In each case, the critical ions indicative of isomeric structure assignment were marked with an asterisk.
Overall, the simplest HCD MS2 spectrum for a particular precursor ion was the one produced by structure derived from SW480 parental cells. It is obvious from examining all of the relevant MS2 spectra that terminal 3-sulfo Gal (Gal3S) defined by diagnostic ions at m/z 153/181 and m/z 253/283 is the predominant endogenous form of sulfation in SW480, with very low residual endogenous GlcNAc6ST activity. Only in one case here, when both termini of a core 2 O-glycan (m/z 1747) were sialylated, could the diagnostic ions at m/z 195/234 for an internal 6-sulfo GlcNAc (GlcNAc6S) be detected for the precursor derived from the non-GlcNAc6ST–transfected parent SW480 cells. The predominance of Gal3S is consistent with previous reports indicating the dominant presence of Gal3ST2 in normal and cancer colonic cells (30, 44). Due to this lack of other sulfation possibilities, the monosulfated O-glycan structures that could be deduced for SW480 have the lowest isomeric variation, with the major ones shown by the cartoon drawings on their respective MS2 spectra (Fig. 3A and Fig. S4).
In the GlcNAc6ST-transfected cells, GlcNAc6S as a terminal unit on its own or as an internal residue of LacNAc, LeX, and sialyl LacNAc was additionally present alongside or in place of the Gal3S-terminated structures made by nontransfected cells. The possible isomeric permutation thus increased significantly. For the simplest monosulfated O-glycans represented by the precursor ion signal at m/z 1025 (Hex2HexNAc2-itol), it is clear that the predominant structure made by SW480–, GlcNAc6ST-1–, and GlcNAc6ST-3–expressing cells was an extended core 1 O-glycan with a terminal Gal3S, as indicated by (i) the trios of m/z 702/732/750 corresponding to E, B, and C ions produced at the Gal 3-linked to the GalNAcitol; (ii) the very low abundance or absence of Y1 and Z1 ion pair at m/z 807/789; and (iii) exclusive presence of m/z 153/181/253/283 and no m/z 195/234/264 (Fig. 3, A, B, and D). The single Gal3S was mostly carried on a type 1 chain as defined by m/z 301/398, but the additional presence of type 2 LacNAc chain with Gal3S was indicated by a very minor m/z 371 ion. In contrast, in GlcNAc6ST-2–transfected cells, m/z 371 was much more abundant than m/z 301/398, and m/z 195/234/264 were additionally detected, suggesting that a 6-sulfo LacNAc with an internal GlcNAc6S is preferred on either an extended core 1 or core 2 structure (Fig. 3C). The corresponding MALDI-MS/MS data for m/z 1025 derived from the GlcNAc6ST-2–transfected cells (Fig. S3B) yielded only m/z 139/234 and no m/z 153/253 at all, whereas the reverse is true for the other cells (Fig. S3, A and C). The data thus suggest that only GlcNAc6ST-2 could efficiently use the GlcNAc extending from the 3-arm of a core 1 structure as substrate, which could then be extended by β4-galactosylation to 6-sulfo LacNAc. Non-6-O-sulfated GlcNAc would instead be preferentially β3-galactosylated as type 1 chain and then 3-O-sulfated at the terminal Gal by endogenous Gal3ST activity in SW480 colonic cancer cell lines. Interestingly, a branched core 2 structure carrying a Gal3S directly 3-linked to the GalNAcitol coupled with a nonsulfated LacNAc on the 6-arm was not detected, which indicated that the endogenous Gal3ST activity acted preferentially on the Gal of extended core 1 and not core 1 itself.
In agreement with this interpretation, the 6-sulfo LacNAc–carrying extended core 1 structure found in GlcNAc6ST-2–transfected cells was commonly sialylated to yield the major monosulfated O-glycan signal at m/z 1386. MS/MS analysis showed that the sulfate was similarly carried on the 6-position of a 4-linked GlcNAc based on the diagnostic ions at m/z 195/234/264 (Fig. 3E). A sialylated sulfated type 2 LacNAc glycotope was identified by the B ion at m/z 889 and the characteristic D ion at m/z 875 produced only by MALDI-MS/MS (Fig. S3D). Importantly, a prominent E ion at m/z 1063 showed that this terminal glycotope was primarily carried on an extended core 1 structure, although an isomeric core 2–based structure in which a 6-sulfo LacNAc was carried on the 6-arm of GalNAc was also identified by the cleavage ions at m/z 759/789/807. It could, however, be inferred from the relative dominance of m/z 514 (sulfo (OH)LacNAc derived from and indicative of sulfo sialyl LacNAc) over m/z 528 (sulfo LacNAc) that the core 2 structure was much less abundant than the extended core 1 structure.
In contrast, m/z 1386 from the GlcNAc6ST-3–transfected cells (Fig. 3F) afforded more of the ions at m/z 528 relative to m/z 889 and 514, with the additional presence of m/z 153/181/253/283 to suggest that structures carrying sulfo LacNAc with either Gal3S or GlcNAc6S were more abundant than that containing 6-sulfo sialyl LacNAc. The more prominent Y1 and Z1 ion pairs detected at m/z 807/789 and 1168/1150 were indicative of core 2 structures carrying the sulfated ±sialyl LacNAc unit on the 6-arm, whereas m/z 1063 that would identify an extended core 1 structure was barely detected. The same applied to m/z 1386 from the GlcNAc6ST-1–transfected cells (Fig. S4C). The dominance of core 2 structures supports the earlier interpretation that GlcNAc6ST-1 and -3 were unable to efficiently add a sulfate to the internal GlcNAc of extended core 1 structure, unlike GlcNAc6ST-2. It follows that to accommodate for a terminal sialylation, which competes with terminal Gal3S, an extra terminus is needed. A linear sulfo sialylated extended core 1 structure would therefore be precluded. As another nonsialylated example, the major structures for m/z 1474 in the GlcNAc6ST-1– and -3–transfected cells (Fig. 3H and Fig. S4D) could be assigned as carrying a Gal3S-terminated chain extending from the 3-arm, with complete absence of diagnostic ions indicative of GlcNAc6S that were found in those from the GlcNAc6ST-2–transfected cells (Fig. 3G). In this case, many alternative isomeric structures are possible when not restricted by terminal sialylation and were indeed detected. This conclusion also explains why disulfated disialylated O-glycans were significantly much more abundant in the GlcNAc6ST-2–transfected cells (Fig. 2B). For core 2 structures where both termini are sialylated, the internal GlcNAc extending from the 3-arm must also be sulfated in addition to the one on the 6-arm, which can only be efficiently made by GlcNAc6ST-2.
Identification of MECA-79 epitope on disulfated O-glycans
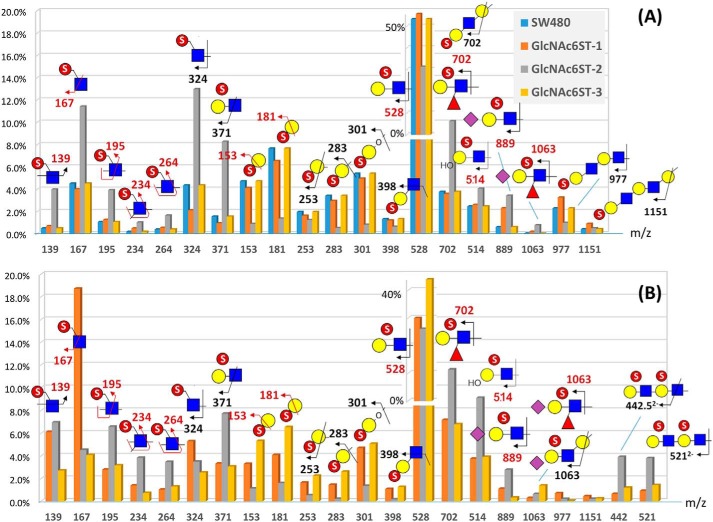
Although careful examination of the MS and MS2 data did reveal subtle differences between the very similar range of monosulfated O-glycan structures made by the GlcNAc6ST-1– and GlcNAc6ST-3–transfected cells, direct evidence for the ability of GlcNAc6ST-1 to additionally support the synthesis of MECA-79 epitope as revealed by FACS analyses (Fig. 1C) was lacking. Unbiased interpretation of the HCD MS2 data set for the eight major monosulfated O-glycans (Fig. S4) led mostly to the same isomeric constituents for any particular structure made by GlcNAc6ST-1 and -3. Structures consistent with carrying a MECA-79 epitope were convincingly found only among those made by GlcNAc6ST-2 cells. To resolve this apparent inconsistency, we used a slightly different nanoLC-MS/MS approach for the analyses of disulfated O-glycans in which the first-level data-dependent global acquisition of HCD/CID MS2 was supplemented further by targeted ion trap CID MSn analysis on a few preselected MS2 ions. Instead of manually interpreting each of the MS2 spectra acquired, the range of distinctive sulfated glycotopes expressed in each of the GlcNAc6ST-transfected cells and their relative abundance was first mapped out (Fig. 4B) based on the summed intensities of their respective diagnostic MS2 ions (45). It showed an overall pattern similar to that processed from the data set of monosulfated O-glycans (Fig. 4A). In both cases, sulfo Gal-GlcNAc (m/z 528) was the most abundant terminal unit. Internal GlcNAc6S (m/z 195/234/264) and type 2 chain defined by m/z 371 were more abundantly made by GlcNAc6ST-2–transfected cells, whereas terminal Gal3S (m/z 153/181/253/283) on type 1 chain (m/z 301/398) was the more abundant in the GlcNAc6ST-1– and -3–expressing cells and parental SW480. Importantly for the disulfated O-glycans, a disulfated diLacNAc unit was evident by the doubly charged 3,5A4 and B4 ions at m/z 442.52− and 5212−, respectively, and both ions were relatively more represented among the sulfated glycotopes on the GlcNAc6ST-2–transfected cells compared with the other two cells (Fig. 4B).
Figure 4.
Relative abundance of a full range of distinctive sulfated glycotopes expressed on the monosulfated (A) and disulfated (B) O-glycans from different GlcNAc6ST-transfected cells. The characteristic HCD/CID MS2 ions indicative of each of the annotated sulfated glycotopes were extracted from all MS2 spectra acquired within the elution time span of mono- and disulfated O-glycans identified. Their respective ion intensities were summed and then calculated as a percentage of the total for comparative analysis purposes only and not to be taken as true quantification. Most ions totaled to less than 10% of all summed ion intensities, with m/z 528 dominating at 40–50%, and were additionally plotted at a different scale to better present the differences registered for the other ions. Only a very low amount of disulfated O-glycans was detected for SW480 parental cells (Fig. 2B), which made the ion counts and summed intensity not statistically reliable and thus excluded from B. In general, the relative abundance of the sulfated ions derived from the GlcNAc6ST-1– and -3–transfected cells (and SW480 for monosulfated ones) were more similar, with ions attributed to Gal3S (m/z 153/181/253/283/301/398) contributing to a significantly higher percentage of the total than those of the same set from the GlcNAc6ST-2–expressing cells. In contrast, ions attributed to internal GlcNAc6S (m/z 195/234/264/371) and 6-sulfo LacNAc–based monosulfated (m/z 702/514/889) and disulfated (m/z 442/521) glycotopes were more represented in the GlcNAc6ST-2–expressing cells. m/z 702 and 1063 correspond to the B ion of fucosylated sulfo LacNAc and fucosylated sulfo sialyl LacNAc, respectively, but can also be assigned as the isomeric E ions. The B and E ions comprise both type 1 and 2 chains, which could only be discriminated by the presence of other ions or by further MSn. Terminal GlcNAc6S defined by m/z 167/324 was highly abundant in the GlcNAc6ST-2– and GlcNAc6ST-1–expressing cells for mono- and disulfated O-glycans, respectively, for unclear reasons.
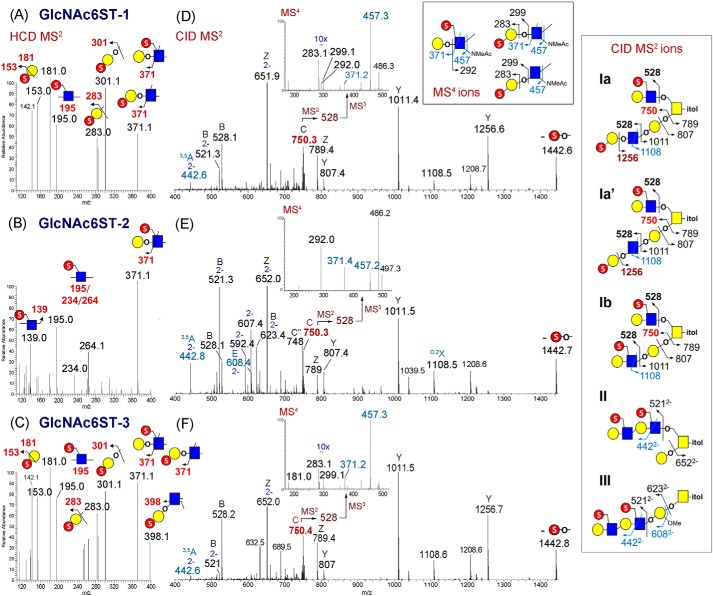
Homing in on the smallest disulfated O-glycan commonly detected in samples from all three transfected cell lines at m/z 769.81 ([M-2H]2−, corresponding to a disulfated Hex3HexNAc3-itol), the low-mass HCD MS2 ions (Fig. 5, A–C) indicated that internal GlcNAc6S was commonly present, but terminal Gal3S was only detected in samples derived from the GlcNAc6ST-1– and GlcNAc6ST-3–transfected cells. This is corroborated by the conspicuous absence of the ion at m/z 1256 derived from loss of a terminal sulfated Gal in the GlcNAc6ST-2–transfected sample (Fig. 5, D–F). To determine whether a MECA-79 epitope may be differentially expressed, the common sulfo Gal-GlcNAc–containing fragment ion derived specifically from the 3-arm of the GalNAcitol (i.e. the C3 ion at m/z 750) was targeted for further stages of MS/MS. As expected, it yielded predominantly a B2 ion at m/z 528 upon MS3, which was further subjected to MS4 to resolve its fine structure. Importantly, the location of sulfate on the internal HexNAc could be firmly established by loss of a nonsulfated terminal Hex via elimination to give the ion at m/z 292, most prominently observed in the MS4 spectrum derived from the GlcNAc6ST-2–transfected sample (Fig. 5E, inset). In the absence of an ion at m/z 283 indicative of sulfated terminal Hex, the 3,5A2 ion at m/z 371 further confirmed the identity of a 6-sulfo LacNAc epitope. In contrast, the corresponding MS4 spectrum derived from the GlcNAc6ST-3–transfected sample (Fig. 5F, inset) afforded both m/z 283 and 371 but not 292, which implicated a terminal sulfated Gal on the Gal-GlcNAc extending from core 1 instead of MECA-79. The MS4 spectrum of m/z 528 derived from the GlcNAc6ST-1–transfected sample (Fig. 5D, inset) resembled that of the GlcNAc6ST-3–transfected sample except for the additional presence of the critical ion at m/z 292. As defined by the common Y1 and Z1 ions at m/z 807 and 789, a related set of isomeric disulfated core 2 structures with a sulfated Gal-GlcNAc unit on each of the two arms could thus be established (structures I, Fig. 5). The major structural isomer in this set made by the GlcNAc6ST-2–transfected cells was the MECA-79–carrying core 2 structure Ib, with 6-sulfo LacNAc on both arms. A very low amount of this structural isomer could also be evidently synthesized by the GlcNAc6ST-1–transfected cells. However, in both the GlcNAc6ST-1– and -3–expressing cells, the Gal-GlcNAc unit extending from the 3-arm was mainly 3-O-sulfated at the terminal Gal to yield structure Ia and/or Ia′.
Figure 5.
Negative ion mode nanoESI-MS/MS analyses of permethylated disulfated O-glycans from different GlcNAc6ST-transfected cells. Only the spectra acquired on the doubly charged precursor ion at m/z 769.81 corresponding to disulfated Hex3HexNAc3itol were analyzed in detail and shown here. A–C, low-mass region of the respective HCD MS2 spectra containing the diagnostic ions indicative of the location of sulfate (41). D–F, trap CID MS2 spectra showing the presence of the C3 ion at m/z 750.3 selected for further MS3 to yield m/z 528.2 for MS4 (inset) analyses to identify the presence of MECA-79. Fragment ions were singly charged unless indicated by “2−” for doubly charged. The expected CID MS2 and MS4 ions from various isomeric structures are shown in cartoon drawings in two separate boxes. For the MS4 spectra, the ion at m/z 292 identified an internal sulfated GlcNAc, whereas ions at m/z 283/299 identified a terminal sulfated Gal. The 3,5A ion at m/z 371 would confirm the presence of a sulfated LacNAc unit on the 3-arm, but the diagnostic 1,3A ion at m/z 398 for type 1 chain was not readily afforded by trap CID to allow its positive identification. From these ions, three distinctive isomeric series could be inferred. Structure I comprised at least three possible isomers, denoted as Ia, Ia′, and Ib, which differ by the sulfated Gal-GlcNAc unit attached to the 3-arm. Isomers Ia and Ia′ could not be resolved by CID MS2 or MS4 alone. Both isomers were likely present in the GlcNAc6ST-1– and -3–expressing cells, but the latter obviously made more of the sulfated type 1 chain, as evidenced from the prominent HCD ion at m/z 398 (C). Structure Ib carrying MECA-79 was mainly synthesized by GlcNAc6ST-2 at the expense of Ia and Ia′, but a low amount of it could also be made by GlcNAc6ST-1, as corroborated by the MS4 data. Importantly, only GlcNAc6ST-2 could synthesize structure III, whereas structure II was commonly found in all three transfected cells. The ion at m/z 1442.8 corresponds to direct elimination of a sulfate moiety from the precursor.
Identification of unique disulfated O-glycan structures synthesized by GlcNAc6ST-2
In addition to a sulfo Gal-GlcNAc unit carried on each arm of core 2 O-glycans, a disulfated diLacNAc unit was evident by the aforementioned doubly charged disulfated B4 ion at m/z 521.32−, whereby sulfation on the internal LacNAc unit was identified by the 3,5A4 ion at m/z 442.82−. These two ions were significantly more abundantly produced by the GlcNAc6ST-2–expressing cells (Fig. 5E), consistent with the global mapping of sulfoglycotopes described earlier (Fig. 4B). A very strong doubly charged Z1 ion at m/z 652.02− in all three GlcNAc6ST–transfected samples implicated a common core 2 structure (structure II), which carried the disulfated diLacNAc on the 6-arm in combination with nonsulfated, nonextended 3-arm. The MS/MS data therefore not only reinforced the earlier conclusion that all three GlcNAc6STs can catalyze GlcNAc-6-O-sulfation on the 6-arm of core 2 structures but also revealed that an internal 6-sulfo LacNAc unit can be further extended by another 6-sulfo LacNAc. On the other hand, it is observed that an extended core 1 structure (structure III) carrying a disulfated diLacNAc could only be efficiently made by GlcNAc6ST-2. The existence of such a structure was supported by the doubly charged B and E ions at m/z 623.42− and 608.42−, respectively, along with a further loss of a MeOH moiety from both to give the ions at m/z 607.42− and 592.42− (Fig. 5E, inset). The fact that these ions were relatively strong suggested that structure III was a major isomeric constituent in the GlcNAc6ST-2–transfected sample, along with structures Ib and II. In contrast, both the extended core 1 structures Ib and III were not detected in the GlcNAc6ST-3–transfected cells, whereas the GlcNAc6ST-1–transfected cells did make a minor amount of MECA-79 epitope-carrying structure Ib but not any appreciable amount of structure III.
Discussion
MS-based glycomics can afford more structural information than what can be inferred from immunostaining using a panel of monoclonal antibodies, which normally precludes identification of novel epitopes and/or the underlying glycan carriers, compounded further by their often not clearly defined cross-reactivities. On the other hand, the common limitation of the MS-based approach is insufficient sensitivity, especially when applied to more problematic glycans, such as the sulfated glycans and/or to derive linkage-specific information requiring advanced MS/MS. Over the last few years, we have steadily established a reasonably robust and sensitive sulfoglycomic platform based initially on MALDI-MS/MS and then nanoLC-MS/MS analyses of permethylated sulfated glycans fractionated away from the otherwise dominant nonsulfated components (36, 40, 41, 45, 46). These technical advances now enable us to meaningfully dissect the in vivo specificities of various GlcNAc6STs on O-glycans by sulfoglycomic analysis that focused on a few representative base structures, with a good balance of precision and sensitivity. The sulfated N-glycans were also mapped at the MALDI-MS level but due to their larger size and m/z values, inefficient MS2 precluded detailed delineation of the sulfated glycotopes as can be performed on the smaller O-glycans.
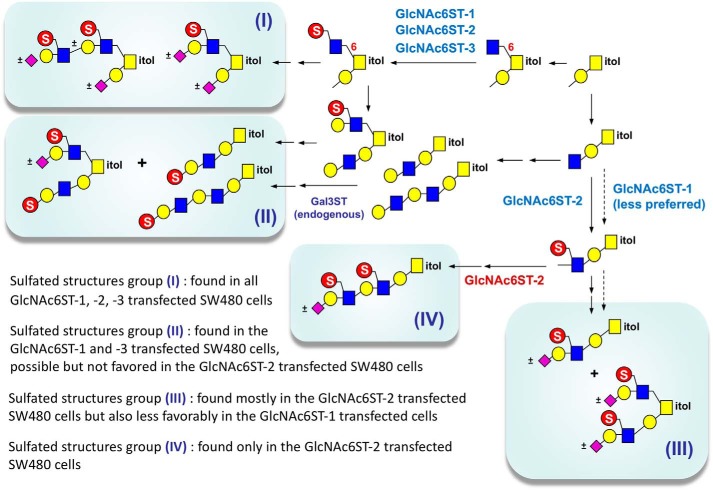
We demonstrated that the core 1 O-glycan of SW480 cancer cell line not only can branch into core 2–based structures but also has a high propensity to be further extended from the 3-arm. This core 1 extension is consistent with a previous study, which demonstrated the expression of core 1 extension β3GlcNAc transferase in human normal colon by Northern blot analysis (6). All three human GlcNAc6STs investigated appeared to be capable of adding a sulfate to the GlcNAc on the 6-arm of core 2 structures (as shown in Fig. 6, group I), but human GlcNAc6ST-2 alone is capable of efficiently utilizing the β-GlcNAc on extended core 1 as substrate. Consequently, although both GlcNAc6ST-1 and GlcNAc6ST-2 support the synthesis of MECA-79 epitope, as identified here by our CID-MSn analyses, GlcNAc6ST-2 drives the synthesis of 6-sulfo LacNAc on the 3-arm of both core 2 and extended core 1 structures (Fig. 6, group III) much more efficiently. Any failure to add sulfate on the GlcNAc appears to favor β3-galactosylation, which can then be 3-O-sulfated. Alternatively, β3- and β4-galactosylation on the nonsulfated GlcNAc can both proceed readily, but only the former is preferentially sulfated. There is a clear and strong correlation between Gal3S and diagnostic ions for type 1 chain at m/z 301/398 on one hand and the type 2 chain-specific 3,5A ion at m/z 371 for internal GlcNAc6S on the other, especially for the sulfo Gal-GlcNAc extending from the 3-arm (Fig. 3). Our data are less clear cut for the disulfated O-glycans (Fig. 5) because m/z 371 would have been contributed by the 6-sulfo LacNAc on the 6-arm. For simplicity, we nevertheless opted to depict all of the terminal 3-O-sulfated Gal as being carried on a type 1 chain in our model (Fig. 6, groups I and II), without actually ruling out the type 2 chain alternative. In contrast, 6-sulfo LacNAc is clearly preferred on the 6-arm, although LacNAc with a terminal Gal3S can also be present.
Figure 6.
The inferred in vivo substrate specificities of different GlcNAc6STs from current MS-based structural studies along with a proposed model for the most likely biosynthesis pathways, taking into consideration that GlcNAc-6-O-sulfation is known to precede its galactosylation. The 6-sulfo LacNAc could be not only sialylated but also fucosylated, as evident from several detected structures at the MS level (Fig. 2 and Fig. S1) and MS/MS analysis on the monosulfated structure at m/z 1560 from all four cells (Fig. S4E). However, because the fucosylated structures were not unambiguously identified by MS2 as sulfated LeX or sialyl LeX in most other cases and not further described in this work, they are not depicted here.
Due to this SW480 O-glycomic background and the unique capability of GlcNAc6ST-2 to catalyze 6-O-sulfation of GlcNAc on both arms of core 2 structures, the most abundant monosulfated O-glycans produced in the GlcNAc6ST-2–transfected cells were core 1 structures extended with 6-sulfo LacNAc, with additional isomers having 6-sulfo LacNAc on the 6-arm of core 2 structures. It also led to disulfated core 2 structures carrying 6-sulfo LacNAc on both arms of a Galβ3GalNAc core (Fig. 6, group III). Moreover, by detailed MS/MS sequencing of disulfated O-glycans, we could successfully identify unique structures contributed by GlcNAc6ST-2 only, namely R-GlcNAc(6-SO3−)β1–3Galβ1-4GlcNAc(6-SO3−)β1–3Galβ1-3GalNAcα (Fig. 6, group IV), which may serve as a good candidate for cancer-specific glycans. In contrast, for the GlcNAc6ST-3–transfected cells, the only sulfate that can be efficiently put onto the Gal-GlcNAc chain extending from the 3-arm of extended core 1 and core 2 structures is the 3-O-sulfate on the terminal Gal (Fig. 6, group II). Because a relatively high abundance of extended core 1 structures with terminal Gal3S on type 1 Gal-3GlcNAc chain was identified (m/z 1025, 1474; Fig. 2A), it represents a favored pathway of sulfation catalyzed by endogenous Gal3ST activity in the absence of ectopically expressed or transfected and overexpressed GlcNAc6STs. Enhanced GlcNAc6ST-3 activity would catalyze the additional synthesis of 6-sulfo LacNAc on the 6-arm of core 2 structures, resulting in a series of larger monosulfated core 2 O-glycans (Fig. S1C). Added GlcNAc6ST-2 activity will instead promote formation of disulfated core 2 O-glycans at the expense of monosulfated O-glycans because GlcNAc on both arms can be 6-O-sulfated.
This fine substrate specificity of human GlcNAc6ST-2 as deduced from MS analyses is in agreement with other independent studies using Chinese hamster ovary cells transfected with core 1 extension β3GlcNAcT and GlcNAc6ST-2, immunostained by MECA-79 antibody (6), and GlcNAc6ST-2/core 2 GlcNAcT double-null mice (47). On the other hand, our conclusion that human GlcNAc6ST-3 cannot use extended core 1 structure as substrate is not consistent with a previous study showing that mouse GlcNAc6ST-2 and GlcNAc6ST-3 can both efficiently transfer sulfate to extended core 1 structure, resulting in MECA-79 epitope (15). A likely explanation may be species-specific differences among the fine specificities of GlcNAc6STs, which is also the case for Gal3ST-2 (48).
Previous studies by cationic dye staining showed that sulfomucin tends to decrease upon malignant transformation of colonic epithelial cells (25, 49). Sulfate modification at the C3 position of terminal Gal in nonmalignant colonic epithelial cells is reported to be one of the major sulfated glycotopes (50). Expression of sialyl 6-sulfo Lewis X was also preferred in the nonmalignant colonic epithelial rather than cancer cells by immunostaining (33), and N-linked glycans having sulfation were reported to occur preferentially in the normal counterpart of the carcinoembryonic antigen (51). Our results showed that a marked decrease of GlcNAc6ST-3 mRNA was detected in cancer cells compared with nonmalignant epithelium prepared from the same patient, whereas GlcNAc6ST-2 mRNA showed a drastic increase in colonic cancer tissues, which is consistent with the previous studies (32). In addition, the MECA-79 determinant was observed in human colon cancer tissues, not nonmalignant colonic epithelial cells. These data correlated well with our MS-based biochemical analyses indicating that only GlcNAc6ST-2, not GlcNAc6ST-3, prefers to add sulfate to extended core 1 O-glycan structure in transfected SW480 cells. GlcNAc6ST-2 mRNA has also been demonstrated to increase significantly in gastric cancer tissues compared with normal mucosa (52). Indeed, immunohistochemical staining showed that gastric and colorectal cancer patients were positive for MECA-79, whereas no expression was found in normal mucosa. Although MECA-79 seems like a strongly cancer-associated antigen, FACS analysis and MS-based sulfoglycomic analysis showed that MECA-79 also could be synthesized by GlcNAc6ST-1. Here, by advanced MS/MS analyses, we detected unique disulfated O-glycan structures synthesized by GlcNAc6ST-2 only, which may be more specific than the MECA-79 glycotope as a cancer-associated antigen and therefore is worthy of further investigation in the near future.
Experimental procedures
Real-time RT-PCR analysis
Tumor specimens for RT-PCR analysis were obtained from 20 patients with primary colorectal cancer at surgical operation and processed as described previously (53). These samples were collected in accordance with the guidelines of the World Medical Association's Declaration of Helsinki. The carcinomas were classified according to the Astler–Coller modification of Dukes' classification. Malignant and nonmalignant tissues of each specimen were used for RNA extraction. Nonmalignant mucosa was scraped off using slide glasses, and tissue specimens of cancer were carefully excised so as to eliminate noncancerous tissue components. Samples were frozen rapidly and stored at −80 °C until extraction of total RNA. Specimens were powdered in liquid N2, and total cellular RNA was extracted with guanidine isothiocyanate and purified by cesium chloride gradient centrifugation. Total cellular RNA was reverse-transcribed into cDNA using SuperScript II with oligo(dT) primer as per the manufacturer's instructions (Invitrogen). Real-time RT-PCR analysis was performed using ABI prism 7000 (PerkinElmer Life Sciences) with TaqMan probes for GlcNAc6ST-1, -2, and -3 provided by the manufacturer (assay ID Hs01921028_s1, Hs00428480_m1, and Hs00375495_m1, respectively). The results were calculated using the comparative Ct method. Relative transcripts were determined by the formula 2−ΔCt. The ΔCt value was determined by subtracting the average GAPDH (as a reference) Ct value from the average target Ct value. These results were normalized as relative values (ΔCt) using GAPDH as a reference to compare mRNA expression, and the ratio of 2−ΔCt value in cancer cells/2−ΔCt value in nonmalignant epithelial cells was calculated.
Release of N-glycans and O-glycans from GlcNAc6ST-transfected cell lines
The cultured colonic cancer cell line SW480 was transfected with expression vector pIRESneo containing the gene for human GlcNAc6ST-1, -2, or -3 using Lipofectamine 2000 (Invitrogen) and screened in culture medium containing 400 μg/ml G418 as described previously (54).
20 million cultured cells were lysed in 0.1 m NH4HCO3 buffer, boiled for 10 min, and lyophilized. The lyophilized samples were first subjected to methanol/chloroform (1:2) delipidation, pelleted down by centrifugation at 2000 × g, and then extracted by 6 m guanidine chloride in 50 mm Tris-HCl with 1 mm CaCl2, pH 8.4, at 4 °C. Extracted proteins were reduced by 20 mm DTT (Sigma) in 6 m guanidine chloride at 37 °C for 4 h, followed by alkylation with 50 mm iodoacetamide (Sigma) at room temperature in the dark for another 4 h, and then dialyzed against double-distilled H2O. The dialyzed samples were digested with trypsin and chymotrypsin (Sigma), each for 4 h, in 50 mm ammonium bicarbonate buffer, pH 8, at 37 °C, followed by overnight treatment with peptide:N-glycosidase F (Roche Applied Science). Released N-glycans were isolated from the peptides by passing through a C18 Sep-Pak cartridge (Waters) in 5% acetic acid. Retained peptides were then eluted with 20–60% propanol, 5% acetic acid and used for subsequent release of O-glycans by reductive elimination (1 m NaBH4, 0.05 n NaOH, 37 °C, 3 days). After terminating the chemical reaction by pure acetic acid on ice, the solution was taken through a Dowex 50 × 8 column (H+ form; Bio-Rad) in 5% acetic acid, dried, and co-evaporated with 10% acetic acid in methanol to remove borates.
Permethylation of sulfated glycans and fractionation by amine tip
Glycan sample dried down in the glass tube was redissolved in a slurry of finely ground NaOH pellets in DMSO (∼0.2 ml), followed by the addition of 0.1 ml of methyl iodide. The reaction mixture was gently vortexed for 3 h at 4 °C and then quenched on ice with 0.2 ml of cold water, followed by careful neutralization with 30% aqueous acetic acid and then applied directly to a prewashed and equilibrated C18 Sep-Pak cartridge (Waters). Hydrophilic salts and contaminants were stepwise washed off with 5 ml of water and 2.5 and 10% acetonitrile. Subsequently, permethylated N-glycans or O-glycans were eluted with 3 ml of 25% acetonitrile, followed next by 3 ml of 50% acetonitrile, collected in a tube. Both sulfated and nonsulfated N-glycans or O-glycans were collected in a fraction and further fractionated by charge using amine beads. As reported previously (46), the amine beads (Nucleosil; 5-μm particle size, 100-Å pore size) were packed into a pipette tip with the tapered end plugged by filter paper. Depending on sample quantity, the volume of packed beads can range from as little as 0.5 μl to about 5 μl, using microtips of different sizes and different wash/elution volumes (50–200 μl). The packed amine microcolumn was first conditioned and washed sequentially with 5% acetonitrile plus 0.1% formic acid, 50% acetonitrile plus 0.1% formic acid, and 95% acetonitrile plus 0.1% formic acid. Permethylated sample was dissolved in 100% acetonitrile for loading. Nonsulfated permethylated glycans were collected in an unbound fraction and an additional 95% acetonitrile wash fraction. Mono- and disulfated permethylated glycans were eluted with 2.5 mm and 10 mm ammonium acetate in 50% acetonitrile, respectively.
For further sample clean up or concentration, when necessary, the permethylated glycans were dissolved in 20 μl of 10% acetonitrile and taken up by prewashed ZipTipC18 (Millipore). After washing several times with 0.1% TFA, the sulfated glycans were eluted by 10 μl of 50% acetonitrile, 0.1% TFA, or less volume, for collection into microtubes or direct spotting onto the MALDI target plate.
Synthetic sulfated Gal-GlcNAc standards
The synthetic sources of various sulfated Gal-GlcNAc (55) and sialylated sulfated LacNAc (41) standards, along with their detailed characterization by 1H and 13C NMR, optical rotation, and MS analyses have been described in previous publications.
MALDI-MS and MS/MS analysis
The permethylated sulfated glycan samples in acetonitrile were mixed 1:1 with 3,4-diaminobenzophenone matrix (10 mg/ml in 75% acetonitrile, 0.1% TFA) (Acros Organics) and spotted onto the target plate for negative ion mode MALDI-MS and MS/MS analyses on a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA) operated in the reflectron mode. The potential difference between the source acceleration voltage and the collision cell was set at 1 kV in the negative ion mode on TOF/TOF to obtain the desirable fragmentation pattern, as described before (46, 56).
Negative ion mode nanoLC-MS/MS analysis
Negative ion mode nanoLC-MS/MS analyses of the permethylated sulfated glycans were performed on a nanoACQUITY UPLC system (Waters) coupled to an LTQ-Orbitrap Elite hybrid mass spectrometer (Thermo Scientific). Sample was dissolved in 5% acetonitrile and separated at a constant flow rate of 300 nl/min, with a linear gradient of 30–60% acetonitrile (with 0.1% formic acid) over the course of 30 min and then increased to 80% acetonitrile over the course of 5 min and held isocratically for another 10 min. For each data-dependent acquisition cycle, the full-scan MS spectrum (m/z 700–2000) was acquired in the Orbitrap at 120,000 resolution (at m/z 400) with automatic gain control target value of 1 × 106. A target precursor inclusion list was applied to precede further selection of the five most intense ions with an intensity threshold of 5000 counts for both CID and HCD. The automatic gain control target value and normalized collision energy applied for CID and HCD experiments were set as 1 × 104, 40% and 5 × 104, 100%, respectively. MSn (n > 2) analyses were performed by selectively isolating and fragmenting target precursors written into an inclusion list. The mass isolation window was set at 2, and a normalized collision in the range of 35% was used. The MSn spectra were averaged over a period of retention time according to the elution profile of its precursor, and the spectra were interpreted manually. Additional mapping of the sulfated glycotopes based on MS2 diagnostic ions was performed using the in-house developed software, GlyPick (45). The mass tolerance for the target CID/HCD MS2 ions to be extracted from the acquired data sets was set at ±100 ppm, with their absolute and relative intensity threshold set at above 100 and 1%, respectively. The summed ion intensity for each of the extracted target ions was expressed as a percentage of the total of all selected MS2 ions, as described in detail previously (45).
Flow cytometry analysis
SW480 and transfected cells were maintained in Dulbecco's modified Eagle's medium (high-glucose). Cells were stained with MECA-79 mAb at 0.5 μg/ml (Biolegend) at 4 °C for 30 min. The cells were then washed three times with PBS containing 0.5% BSA, followed by stained with a 1:200 dilution of FITC-conjugated goat anti-rat IgM (Cappel Laboratories, Cochranville, PA) at 4 °C for 30 min. The stained cells were analyzed on a FACSCalibur (BD Biosciences).
Immunohistochemical staining
Frozen sections of 10-μm thickness for immunohistochemical examination were prepared from surgical specimens obtained from 31 patients with colorectal cancer operated at the Aichi Cancer Center Hospital. The avidin–biotin complex technique for the immunohistochemical study was performed as described in the instructions for the kits (Vectastain) provided by Vector Inc. (Burlingame, CA). The AG107 antibody (murine IgM), directed to nonsialylated forms of conventional 6-sulfated carbohydrate determinants, was prepared as described previously.
Author contributions
S.-Y. Y. performed all of the glycomic sample preparation and analysis, interpreted all MS data, and prepared the manuscript. C.-T. H. assisted with the manual processing of LC-MS/MS data sets and GlyPick analysis. M. I. did histochemical staining on tissue samples and performed statistical analyses. A. Y. performed RT-PCR analyses. H. I. and S. N. selected colonic tissue samples, and H. Y. cultured the gene-transfected cells. R. K. and K.-H. K. conceived the experimental design, reviewed the data, and wrote and edited the manuscript.
Supplementary Material
Acknowledgments
The MALDI-MS and MS/MS data were acquired at the previous Core Facilities for Protein Structural Analysis at Academia Sinica, supported under the Taiwan National Core Facility Program for Biotechnology, NSC 100-2325-B-001-029. We further acknowledge the use of Academia Sinica Common Mass Spectrometry Facilities at Institute of Biological Chemistry and technical assistance of Ming-Chieh Tsai and Dr. Shu-Yu Lin for nanoLC-MS/MS analysis. The synthetic sulfated glycan standards were generously provided by Prof. Chun-Hung Lin (Institute of Biological Chemistry, Academia Sinica) and Prof. James C. Paulson (Scripps Research Institute).
This work was supported by Academia Sinica and Taiwan National Science Council (NSC) Grants 99-2311-B-001-021-MY3 and 102-2311-B-001-026-MY3 (to K. K. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
- HEV
- high endothelial venules
- CID
- collision-induced dissociation
- HCD
- higher-energy collisional dissociation
- 6-sulfo LacNAc
- Galβ1–4GlcNAc(6-SO3−)β1-
- 6-sulfo Lewis X
- NeuAcα2-3Galβ1–4(Fucα1–3)GlcNAc(6-SO3−)β1-
- LacNAc
- N-acetyllactosamine
- Gal3S
- Gal-3-O-sulfate
- Gal3ST
- galactose-3-O-sulfotransferase
- GlcNAc6S
- GlcNAc-6-O-sulfate
- GlcNAc6ST
- N-acetylglucosamine-6-O-sulfotransferase
- HEC
- high endothelial cells
- nanoLC
- nano-liquid chromatography
- nanoESI
- nano-electrospray ionization
- pNP
- p-nitrophenyl
- TCGA
- the Cancer Genome Atlas
- Hex
- hexose
- HexNAc
- N-acetylhexosamine.
References
- 1. Fukuda M., Hiraoka N., Akama T. O., and Fukuda M. N. (2001) Carbohydrate-modifying sulfotransferases: structure, function, and pathophysiology. J. Biol. Chem. 276, 47747–47750 10.1074/jbc.R100049200 [DOI] [PubMed] [Google Scholar]
- 2. Rosen S. D. (2004) Ligands for L-selectin: homing, inflammation, and beyond. Annu. Rev. Immunol. 22, 129–156 10.1146/annurev.immunol.21.090501.080131 [DOI] [PubMed] [Google Scholar]
- 3. Kawashima H. (2006) Roles of sulfated glycans in lymphocyte homing. Biol. Pharm. Bull. 29, 2343–2349 10.1248/bpb.29.2343 [DOI] [PubMed] [Google Scholar]
- 4. Kobayashi M., Fukuda M., and Nakayama J. (2009) Role of sulfated O-glycans expressed by high endothelial venule-like vessels in pathogenesis of chronic inflammatory gastrointestinal diseases. Biol. Pharm. Bull. 32, 774–779 10.1248/bpb.32.774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitsuoka C., Sawada-Kasugai M., Ando-Furui K., Izawa M., Nakanishi H., Nakamura S., Ishida H., Kiso M., and Kannagi R. (1998) Identification of a major carbohydrate capping group of the L-selectin ligand on high endothelial venules in human lymph nodes as 6-sulfo sialyl Lewis X. J. Biol. Chem. 273, 11225–11233 10.1074/jbc.273.18.11225 [DOI] [PubMed] [Google Scholar]
- 6. Yeh J. C., Hiraoka N., Petryniak B., Nakayama J., Ellies L. G., Rabuka D., Hindsgaul O., Marth J. D., Lowe J. B., and Fukuda M. (2001) Novel sulfated lymphocyte homing receptors and their control by a Core1 extension β 1,3-N-acetylglucosaminyltransferase. Cell 105, 957–969 10.1016/S0092-8674(01)00394-4 [DOI] [PubMed] [Google Scholar]
- 7. Bochner B. S., Alvarez R. A., Mehta P., Bovin N. V., Blixt O., White J. R., and Schnaar R. L. (2005) Glycan array screening reveals a candidate ligand for Siglec-8. J. Biol. Chem. 280, 4307–4312 10.1074/jbc.M412378200 [DOI] [PubMed] [Google Scholar]
- 8. Campanero-Rhodes M. A., Childs R. A., Kiso M., Komba S., Le Narvor C., Warren J., Otto D., Crocker P. R., and Feizi T. (2006) Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem. Biophys. Res. Commun. 344, 1141–1146 10.1016/j.bbrc.2006.03.223 [DOI] [PubMed] [Google Scholar]
- 9. Carlsson S., Oberg C. T., Carlsson M. C., Sundin A., Nilsson U. J., Smith D., Cummings R. D., Almkvist J., Karlsson A., and Leffler H. (2007) Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology 17, 663–676 10.1093/glycob/cwm026 [DOI] [PubMed] [Google Scholar]
- 10. Tateno H., Ohnishi K., Yabe R., Hayatsu N., Sato T., Takeya M., Narimatsu H., and Hirabayashi J. (2010) Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J. Biol. Chem. 285, 6390–6400 10.1074/jbc.M109.041863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ideo H., Matsuzaka T., Nonaka T., Seko A., and Yamashita K. (2011) Galectin-8-N-domain recognition mechanism for sialylated and sulfated glycans. J. Biol. Chem. 286, 11346–11355 10.1074/jbc.M110.195925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kletter D., Singh S., Bern M., and Haab B. B. (2013) Global comparisons of lectin-glycan interactions using a database of analyzed glycan array data. Mol. Cell Proteomics 12, 1026–1035 10.1074/mcp.M112.026641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brockhausen I. (2003) Sulphotransferases acting on mucin-type oligosaccharides. Biochem. Soc. Trans. 31, 318–325 10.1042/bst0310318 [DOI] [PubMed] [Google Scholar]
- 14. Grunwell J. R., and Bertozzi C. R. (2002) Carbohydrate sulfotransferases of the GalNAc/Gal/GlcNAc6ST family. Biochemistry 41, 13117–13126 10.1021/bi020507h [DOI] [PubMed] [Google Scholar]
- 15. Kawashima H., Petryniak B., Hiraoka N., Mitoma J., Huckaby V., Nakayama J., Uchimura K., Kadomatsu K., Muramatsu T., Lowe J. B., and Fukuda M. (2005) N-Acetylglucosamine-6-O-sulfotransferases 1 and 2 cooperatively control lymphocyte homing through L-selectin ligand biosynthesis in high endothelial venules. Nat. Immunol. 6, 1096–1104 10.1038/ni1259 [DOI] [PubMed] [Google Scholar]
- 16. Uchimura K., Gauguet J. M., Singer M. S., Tsay D., Kannagi R., Muramatsu T., von Andrian U. H., and Rosen S. D. (2005) A major class of L-selectin ligands is eliminated in mice deficient in two sulfotransferases expressed in high endothelial venules. Nat. Immunol. 6, 1105–1113 10.1038/ni1258 [DOI] [PubMed] [Google Scholar]
- 17. Lee J. K., Bistrup A., van Zante A., and Rosen S. D. (2003) Activities and expression pattern of the carbohydrate sulfotransferase GlcNAc6ST-3 (I-GlcNAc6ST): functional implications. Glycobiology 13, 245–254 10.1093/glycob/cwg018 [DOI] [PubMed] [Google Scholar]
- 18. Uchimura K., El-Fasakhany F. M., Hori M., Hemmerich S., Blink S. E., Kansas G. S., Kanamori A., Kumamoto K., Kannagi R., and Muramatsu T. (2002) Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to L-selectin ligand synthesis and tumor-associated enzyme expression. J. Biol. Chem. 277, 3979–3984 10.1074/jbc.M106587200 [DOI] [PubMed] [Google Scholar]
- 19. Uchimura K., Fasakhany F., Kadomatsu K., Matsukawa T., Yamakawa T., Kurosawa N., and Muramatsu T. (2000) Diversity of N-acetylglucosamine-6-O-sulfotransferases: molecular cloning of a novel enzyme with different distribution and specificities. Biochem. Biophys. Res. Commun. 274, 291–296 10.1006/bbrc.2000.3141 [DOI] [PubMed] [Google Scholar]
- 20. Kitagawa H., Fujita M., Ito N., and Sugahara K. (2000) Molecular cloning and expression of a novel chondroitin 6-O-sulfotransferase. J. Biol. Chem. 275, 21075–21080 10.1074/jbc.M002101200 [DOI] [PubMed] [Google Scholar]
- 21. Hemmerich S., Lee J. K., Bhakta S., Bistrup A., Ruddle N. R., and Rosen S. D. (2001) Chromosomal localization and genomic organization for the galactose/N-acetylgalactosamine/N-acetylglucosamine 6-O-sulfotransferase gene family. Glycobiology 11, 75–87 10.1093/glycob/11.1.75 [DOI] [PubMed] [Google Scholar]
- 22. Akama T. O., Nishida K., Nakayama J., Watanabe H., Ozaki K., Nakamura T., Dota A., Kawasaki S., Inoue Y., Maeda N., Yamamoto S., Fujiwara T., Thonar E. J., Shimomura Y., Kinoshita S., Tanigami A., and Fukuda M. N. (2000) Macular corneal dystrophy type I and type II are caused by distinct mutations in a new sulphotransferase gene. Nat. Genet. 26, 237–241 10.1038/79987 [DOI] [PubMed] [Google Scholar]
- 23. Hiraoka N., Petryniak B., Nakayama J., Tsuboi S., Suzuki M., Yeh J. C., Izawa D., Tanaka T., Miyasaka M., Lowe J. B., and Fukuda M. (1999) A novel, high endothelial venule-specific sulfotransferase expresses 6-sulfo sialyl Lewis(x), an L-selectin ligand displayed by CD34. Immunity 11, 79–89 10.1016/S1074-7613(00)80083-7 [DOI] [PubMed] [Google Scholar]
- 24. Akama T. O., Misra A. K., Hindsgaul O., and Fukuda M. N. (2002) Enzymatic synthesis in vitro of the disulfated disaccharide unit of corneal keratan sulfate. J. Biol. Chem. 277, 42505–42513 10.1074/jbc.M207412200 [DOI] [PubMed] [Google Scholar]
- 25. Byrd J. C., and Bresalier R. S. (2004) Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 23, 77–99 10.1023/A:1025815113599 [DOI] [PubMed] [Google Scholar]
- 26. Yamori T., Kimura H., Stewart K., Ota D. M., Cleary K. R., and Irimura T. (1987) Differential production of high molecular weight sulfated glycoproteins in normal colonic mucosa, primary colon carcinoma, and metastases. Cancer Res. 47, 2741–2747 [PubMed] [Google Scholar]
- 27. Vavasseur F., Dole K., Yang J., Matta K. L., Myerscough N., Corfield A., Paraskeva C., and Brockhausen I. (1994) O-Glycan biosynthesis in human colorectal adenoma cells during progression to cancer. Eur. J. Biochem. 222, 415–424 10.1111/j.1432-1033.1994.tb18880.x [DOI] [PubMed] [Google Scholar]
- 28. Chandrasekaran E. V., Jain R. K., Vig R., and Matta K. L. (1997) The enzymatic sulfation of glycoprotein carbohydrate units: blood group T-hapten specific and two other distinct Gal:3-O-sulfotransferases as evident from specificities and kinetics and the influence of sulfate and fucose residues occurring in the carbohydrate chain on C-3 sulfation of terminal Gal. Glycobiology 7, 753–768 10.1093/glycob/7.6.753 [DOI] [PubMed] [Google Scholar]
- 29. Chandrasekaran E. V., Jain R. K., Rhodes J. M., Chawda R., Piskorz C., and Matta K. L. (1999) Characterization of distinct Gal:3-O-sulfotransferase activities in human tumor epithelial cell lines and of calf lymph node GlcNAc: 6-O-sulfotransferase activity. Glycoconj. J. 16, 523–536 10.1023/A:1007074005371 [DOI] [PubMed] [Google Scholar]
- 30. Seko A., Nagata K., Yonezawa S., and Yamashita K. (2002) Down-regulation of Gal 3-O-sulfotransferase-2 (Gal3ST-2) expression in human colonic non-mucinous adenocarcinoma. Jpn. J. Cancer Res. 93, 507–515 10.1111/j.1349-7006.2002.tb01285.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seko A., Sumiya J., Yonezawa S., Nagata K., and Yamashita K. (2000) Biochemical differences between two types of N-acetylglucosamine:→6sulfotransferases in human colonic adenocarcinomas and the adjacent normal mucosa: specific expression of a GlcNAc:→6sulfotransferase in mucinous adenocarcinoma. Glycobiology 10, 919–929 10.1093/glycob/10.9.919 [DOI] [PubMed] [Google Scholar]
- 32. Seko A., Nagata K., Yonezawa S., and Yamashita K. (2002) Ectopic expression of a GlcNAc 6-O-sulfotransferase, GlcNAc6ST-2, in colonic mucinous adenocarcinoma. Glycobiology 12, 379–388 10.1093/glycob/12.6.379 [DOI] [PubMed] [Google Scholar]
- 33. Izawa M., Kumamoto K., Mitsuoka C., Kanamori C., Kanamori A., Ohmori K., Ishida H., Nakamura S., Kurata-Miura K., Sasaki K., Nishi T., and Kannagi R. (2000) Expression of sialyl 6-sulfo Lewis X is inversely correlated with conventional sialyl Lewis X expression in human colorectal cancer. Cancer Res. 60, 1410–1416 [PubMed] [Google Scholar]
- 34. Bowman K. G., Cook B. N., de Graffenried C. L., and Bertozzi C. R. (2001) Biosynthesis of L-selectin ligands: sulfation of sialyl Lewis x-related oligosaccharides by a family of GlcNAc-6-sulfotransferases. Biochemistry 40, 5382–5391 10.1021/bi001750o [DOI] [PubMed] [Google Scholar]
- 35. Bistrup A., Bhakta S., Lee J. K., Belov Y. Y., Gunn M. D., Zuo F. R., Huang C. C., Kannagi R., Rosen S. D., and Hemmerich S. (1999) Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J. Cell Biol. 145, 899–910 10.1083/jcb.145.4.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khoo K. H., and Yu S. Y. (2010) Mass spectrometric analysis of sulfated N- and O-glycans. Methods Enzymol. 478, 3–26 10.1016/S0076-6879(10)78001-0 [DOI] [PubMed] [Google Scholar]
- 37. Kobayashi M., Mitoma J., Hoshino H., Yu S. Y., Shimojo Y., Suzawa K., Khoo K. H., Fukuda M., and Nakayama J. (2011) Prominent expression of sialyl Lewis X-capped core 2-branched O-glycans on high endothelial venule-like vessels in gastric MALT lymphoma. J. Pathol. 224, 67–77 10.1002/path.2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patnode M. L., Yu S. Y., Cheng C. W., Ho M. Y., Tegesjö L., Sakuma K., Uchimura K., Khoo K. H., Kannagi R., and Rosen S. D. (2013) KSGal6ST generates galactose-6-O-sulfate in high endothelial venules but does not contribute to L-selectin-dependent lymphocyte homing. Glycobiology 23, 381–394 10.1093/glycob/cws166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patnode M. L., Cheng C. W., Chou C. C., Singer M. S., Elin M. S., Uchimura K., Crocker P. R., Khoo K. H., and Rosen S. D. (2013) Galactose 6-O-sulfotransferases are not required for the generation of Siglec-F ligands in leukocytes or lung tissue. J. Biol. Chem. 288, 26533–26545 10.1074/jbc.M113.485409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng P. F., Snovida S., Ho M. Y., Cheng C. W., Wu A. M., and Khoo K. H. (2013) Increasing the depth of mass spectrometry-based glycomic coverage by additional dimensions of sulfoglycomics and target analysis of permethylated glycans. Anal. Bioanal. Chem. 405, 6683–6695 10.1007/s00216-013-7128-2 [DOI] [PubMed] [Google Scholar]
- 41. Cheng C. W., Chou C. C., Hsieh H. W., Tu Z., Lin C. H., Nycholat C., Fukuda M., and Khoo K. H. (2015) Efficient mapping of sulfated glycotopes by negative ion mode nanoLC-MS/MS-based sulfoglycomic analysis of permethylated glycans. Anal. Chem. 87, 6380–6388 10.1021/acs.analchem.5b01409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yagi H., Takahashi N., Yamaguchi Y., Kimura N., Uchimura K., Kannagi R., and Kato K. (2005) Development of structural analysis of sulfated N-glycans by multidimensional high performance liquid chromatography mapping methods. Glycobiology 15, 1051–1060 10.1093/glycob/cwi092 [DOI] [PubMed] [Google Scholar]
- 43. Broad Institute TCGA Genome Data Analysis Center (2016) Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. Dataset, 10.7908/C11G0KM9 (17 February 2016) [DOI] [Google Scholar]
- 44. Chandrasekaran E. V., Lakhaman S. S., Chawda R., Piskorz C. F., Neelamegham S., and Matta K. L. (2004) Identification of physiologically relevant substrates for cloned Gal:3-O-sulfotransferases (Gal3STs): distinct high affinity of Gal3ST-2 and LS180 sulfotransferase for the globo H backbone, Gal3ST-3 for N-glycan multiterminal Galβ1, 4GlcNAcβ units and 6-sulfoGalβ1, 4GlcNAcβ, and Gal3ST-4 for the mucin core-2 trisaccharide. J. Biol. Chem. 279, 10032–10041 10.1074/jbc.M311989200 [DOI] [PubMed] [Google Scholar]
- 45. Hsiao C. T., Wang P. W., Chang H. C., Chen Y. Y., Wang S. H., Chern Y., and Khoo K. H. (2017) Advancing a high throughput glycotope-centric glycomics workflow based on nanoLC-MS2-product dependent-MS3 analysis of permethylated glycans. Mol. Cell Proteomics 16, 2268–2280 10.1074/mcp.TIR117.000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu S. Y., Wu S. W., Hsiao H. H., and Khoo K. H. (2009) Enabling techniques and strategic workflow for sulfoglycomics based on mass spectrometry mapping and sequencing of permethylated sulfated glycans. Glycobiology 19, 1136–1149 10.1093/glycob/cwp113 [DOI] [PubMed] [Google Scholar]
- 47. Hiraoka N., Kawashima H., Petryniak B., Nakayama J., Mitoma J., Marth J. D., Lowe J. B., and Fukuda M. (2004) Core 2 branching β1,6-N-acetylglucosaminyltransferase and high endothelial venule-restricted sulfotransferase collaboratively control lymphocyte homing. J. Biol. Chem. 279, 3058–3067 10.1074/jbc.M311150200 [DOI] [PubMed] [Google Scholar]
- 48. Seko A., Sumiya J., and Yamashita K. (2005) Porcine, mouse and human galactose 3-O-sulphotransferase-2 enzymes have different substrate specificities: the porcine enzyme requires basic compounds for its catalytic activity. Biochem. J. 391, 77–85 10.1042/BJ20050362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Filipe M. I. (1969) Value of histochemical reactions for mucosubstances in the diagnosis of certain pathological conditions of the colon and rectum. Gut 10, 577–586 10.1136/gut.10.7.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuhns W., Jain R. K., Matta K. L., Paulsen H., Baker M. A., Geyer R., and Brockhausen I. (1995) Characterization of a novel mucin sulphotransferase activity synthesizing sulphated O-glycan core 1,3-sulphate-Galβ1–3GalNAcα-R. Glycobiology 5, 689–697 10.1093/glycob/5.7.689 [DOI] [PubMed] [Google Scholar]
- 51. Fukushima K., Ohkura T., Kanai M., Kuroki M., Matsuoka Y., Kobata A., and Yamashita K. (1995) Carbohydrate structures of a normal counterpart of the carcinoembryonic antigen produced by colon epithelial cells of normal adults. Glycobiology 5, 105–115 10.1093/glycob/5.1.105 [DOI] [PubMed] [Google Scholar]
- 52. Okayama H., Kumamoto K., Saitou K., Hayase S., Kofunato Y., Sato Y., Miyamoto K., Nakamura I., Ohki S., Koyama Y., Ishii Y., and Takenoshita S. (2011) Ectopic expression of MECA-79 as a novel prognostic indicator in gastric cancer. Cancer Sci. 102, 1088–1094 10.1111/j.1349-7006.2011.01895.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miyazaki K., Ohmori K., Izawa M., Koike T., Kumamoto K., Furukawa K., Ando T., Kiso M., Yamaji T., Hashimoto Y., Suzuki A., Yoshida A., Takeuchi M., and Kannagi R. (2004) Loss of disialyl Lewis(a), the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (Siglec-7) associated with increased sialyl Lewis(a) expression on human colon cancers. Cancer Res. 64, 4498–4505 10.1158/0008-5472.CAN-03-3614 [DOI] [PubMed] [Google Scholar]
- 54. Kimura N., Ohmori K., Miyazaki K., Izawa M., Matsuzaki Y., Yasuda Y., Takematsu H., Kozutsumi Y., Moriyama A., and Kannagi R. (2007) Human B-lymphocytes express α2–6-sialylated 6-sulfo-N-acetyllactosamine serving as a preferred ligand for CD22/Siglec-2. J. Biol. Chem. 282, 32200–32207 10.1074/jbc.M702341200 [DOI] [PubMed] [Google Scholar]
- 55. Tu Z., Hsieh H. W., Tsai C. M., Hsu C. W., Wang S. G., Wu K. J., Lin K. I., and Lin C. H. (2013) Synthesis and characterization of sulfated Gal-β-1,3/4-GlcNAc disaccharides through consecutive protection/glycosylation steps. Chem. Asian J. 8, 1536–1550 10.1002/asia.201201204 [DOI] [PubMed] [Google Scholar]
- 56. Yu S. Y., Wu S. W., and Khoo K. H. (2006) Distinctive characteristics of MALDI-Q/TOF and TOF/TOF tandem mass spectrometry for sequencing of permethylated complex type N-glycans. Glycoconj. J. 23, 355–369 10.1007/s10719-006-8492-3 [DOI] [PubMed] [Google Scholar]
- 57. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.