Abstract
Type-A γ-aminobutyric acid receptors (GABAARs) are the principal mediators of inhibitory neurotransmission in the human brain. Endogenous neurosteroids interact with GABAARs to regulate acute and chronic anxiety and are potent sedative, analgesic, anticonvulsant and anaesthetic agents. Their mode of binding, and mechanism of receptor potentiation remain, however, unknown. Here we report crystal structures of a chimeric GABAAR construct, in apo and pregnanolone-bound states. The neurosteroid-binding site is mechanically coupled to the helices lining the ion channel pore, and modulates the desensitization gate conformation. We demonstrate that the equivalent site is responsible for physiological, heteromeric, GABAAR potentiation and explain the contrasting modulatory properties of 3α versus 3β neurosteroid epimers. These results illustrate how peripheral lipid ligands can regulate the desensitization gate of GABAARs, a process of broad relevance to pentameric ligand-gated ion channels.
Introduction
GABAARs are pentameric ligand-gated ion channels (pLGICs)1. Binding of the neurotransmitter GABA to the extracellular domain (ECD) opens an intrinsic pore, surrounded by a ring of five M2 transmembrane domain (TMD) helices, one from each subunit, to permit the passage of negatively charged chloride ions2. The pore-opening event is subject to modulation by endogenous ligands such as Zn2+ and neurosteroids including pregnanolone and its 5α epimer allopregnanolone3. Through modulation of GABAARs, neurosteroids are key regulators of the stress response and anxiety4–6. Over the ovarian cycle, fluctuations in neurosteroid levels impact on the GABAAR inhibitory drive affecting neuronal excitability, raising susceptibility to seizure and anxiety7. Changes to GABAAR subunit expression levels in the dentate gyrus due to fluctuating neurosteroid concentrations are linked to postpartum depression following pregnancy8. Most GABAARs in the human brain are heteromers, typically comprising two α (α1-6), two β (β1-3) and one γ (γ1-3) or δ subunits, out of 19 possible types9. The only GABAAR structure solved to date, the β3 homopentamer crystallized in an agonist-bound desensitised conformation, offers the best available guide to a physiological receptor10. However, the other principal components of GABAARs, the α subunits, are the ones conveying neurosteroid potentiation, through a key glutamine residue within the M1 helices3,11. In the absence of structural information it is impossible to understand how neurosteroids modulate GABAergic neurotransmission in mechanistic terms. To address this, we sought to express, characterize and solve structures of apo and neurosteroid-bound GABAAR constructs containing α subunit TMDs.
Results
Neurosteroid potentiation of a β3-α5 chimeric GABAAR construct
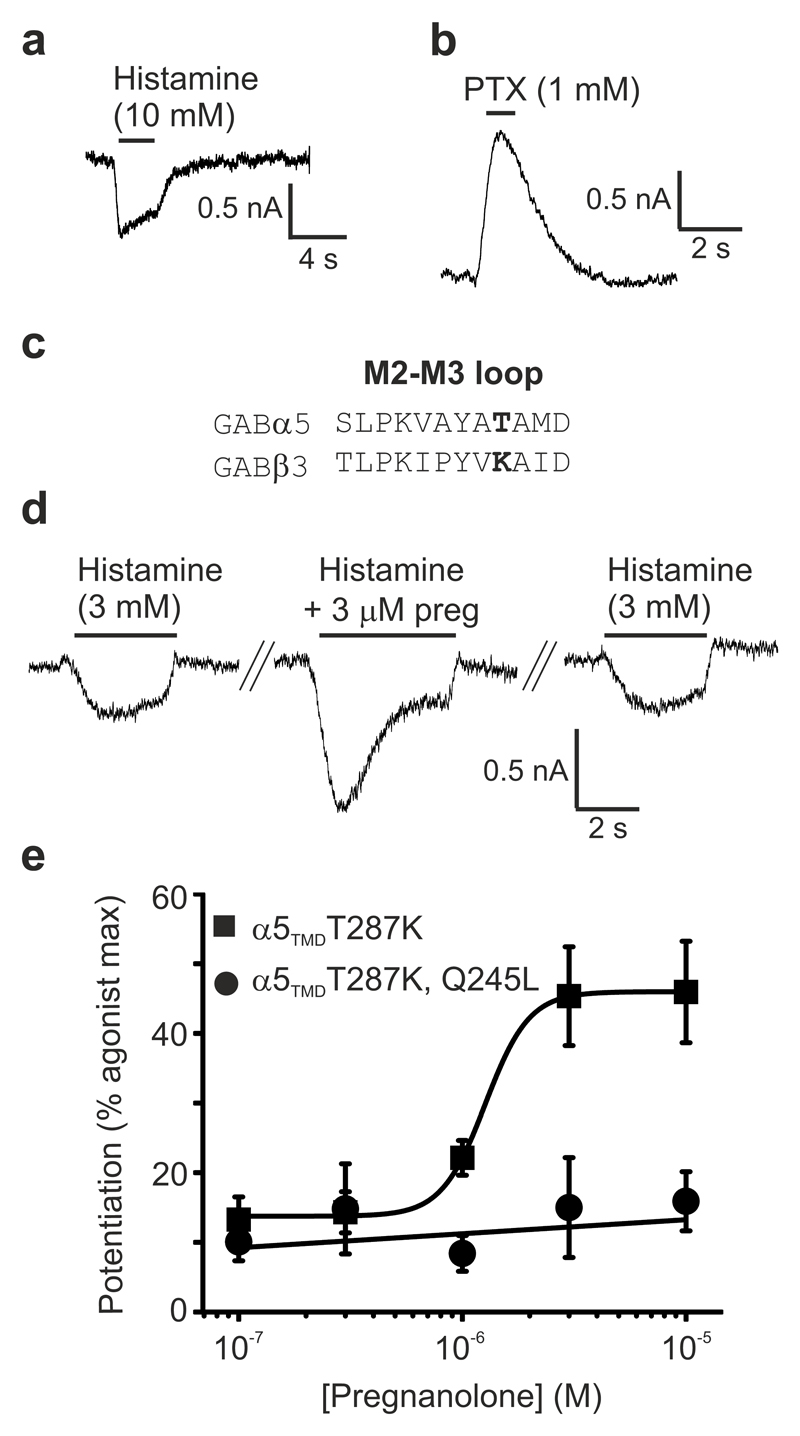
Heteromeric GABAARs have, thus far, proved intransigent to structural determination, and α subunits expressed alone do not form homopentameric assemblies. To overcome these limitations, we generated a chimeric human receptor by fusing the α5 subunit TMD region to the β3 subunit ECD. GABAAR-β3 homopentamers are gated by the natural agonist histamine10,12 (rather than GABA) and the chimera, referred to as α5TMD, was also activated and then desensitised by histamine during whole-cell patch-clamp recordings from transfected HEK-293T cells (Fig. 1a). As reported for GABAAR-β312, α5TMD channels also mediate a picrotoxin-sensitive leak current (IPTX) caused by spontaneous (agonist-independent) gating which accounts for 67 ± 3 % (n = 8) of the total current (IhistamineMAX + IPTX) (Fig. 1b). Thus, α5TMD assembles into a functional gating unit. To measure pregnanolone potentiation of submaximal histamine responses it was necessary to reduce the very high level of spontaneous activity in α5TMD because neurosteroids potentiate low efficacy activated states, as observed in heteromeric GABAARs13. To achieve this we introduced a point mutation, T287K, into the M2-M3 loop critical for receptor gating (identified from screening residue swaps between the α5 and β3 subunits; see alignment in Fig. 1c) which reduced spontaneous gating from 67 ± 3 % to 21 ± 3 % (n = 16). On this background, pregnanolone strongly potentiated submaximal (EC10-15) histamine doses, by 325 ± 104 %, n = 6 (EC50 = 1.3 ± 0.1 μM, n = 6) (Fig. 1d,e). Critically, this effect was ablated by leucine substitution of the key M1 Gln (α5 Gln245) previously identified to be essential for neurosteroid potentiation in heteromeric αβγ/δ GABAARs (Fig. 1e). Furthermore, purified α5TMD in detergent was thermostabilised by addition of pregnanolone (EC50 = 5.5 ± 0.3 μM, n = 3) and this thermostabilisation was strongly attenuated by Q245L or Q245W mutations (tryptophan is the equivalent residue in β1-3 subunits) (EC50 > 30 μM, n = 3, in both cases) (Supplementary Fig. 1). Thus, whether in cell membranes or in detergent, α5TMD houses a neurosteroid potentiation site that requires the same key M1 glutamine for function as reported for heteromeric GABAARs.
Figure 1.
α5TMD forms a functional gating unit potentiated by pregnanolone at the Gln245 site.
(a) HEK-293T whole-cell patch-clamp recording of the agonist histamine activating an inward current through α5TMD, that subsequently desensitizes. (b) The classical GABAAR channel blocker picrotoxin blocks a spontaneous inward leak current generated by α5TMD. (c) Sequence alignment of the M2-M3 loop from GABAAR α5 and β3 subunits, highlighting in bold the T287K substitution. (d) Raw whole-cell patch-clamp EC10 histamine responses from HEK cells transfected with α5TMDT287K, before and after co-application with 3 μM pregnanolone. (e) Pregnanolone dose-response curve for potentiation of histamine EC10-15 responses in α5TMD T287K, and the ablation of potentiation upon introduction of a Q245L mutation in the M1 helix. Each data point represents mean ± s.e.m. of n = 6 experiments, each measurement being from different cells.
The α5TMD crystal structure
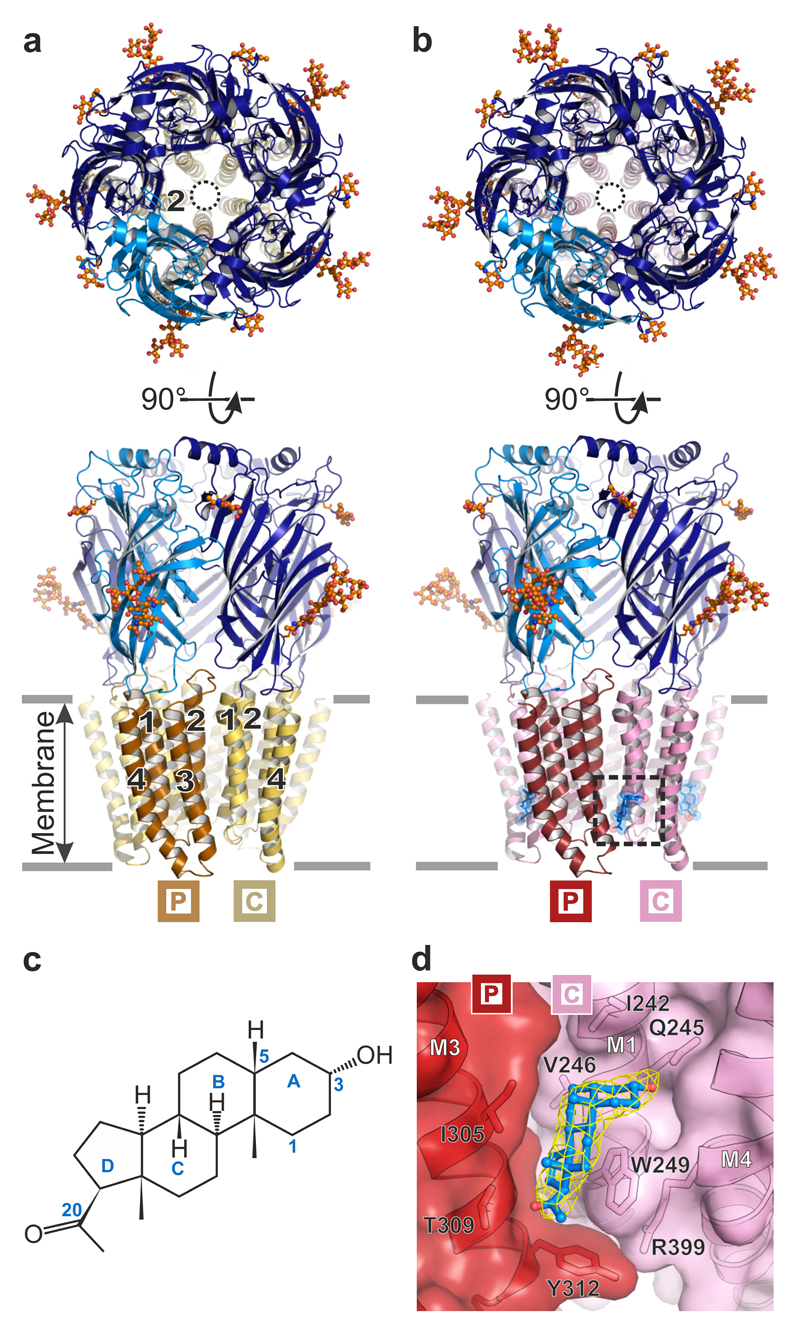
We next crystallized and determined the X-ray structures of α5TMD in the absence and presence of pregnanolone, to 3.3 and 3.2 Å resolution, respectively (Fig. 2a-d, Table 1 and Supplementary Video 1). In each case α5TMD was bound to a nanobody (Nb25) raised against the β3 ECD, which facilitated crystallisation (Supplementary Fig. 2a). α5TMD retains the hallmark architecture of all eukaryotic pentameric ligand-gated ion channels (pLGICs) characterized to date including nicotinic acetylcholine, serotonin type 3, glycine and GluCl receptors1,14–21. Each subunit comprises an N-terminal β3 ECD folded into a twisted β-sandwich followed by an α5 four-helical bundle TMD (Fig. 2a,b). The principal face (P) of each subunit intercalates with the complementary face (C) of its neighbour to form a ring of five subunits surrounding a central vestibule at the ECD level, which flows into a five-fold pseudo-symmetric pore in the TMD region. Each Nb25 straddles the neurotransmitter binding site located under the loop-C between adjacent subunits (Supplementary Fig. 2a-d). The ECDs of the apo and pregnanolone-bound α5TMD structures are structurally very similar (RMSD in the 0.16-0.21 Å range between different chains, over 204 equivalent Cα positions). Both adopt the same overall activated conformation as the agonist-bound GABAAR-β310, exhibiting a closed loop-C that contrasts with the antagonist-bound open loop-C in glycine receptors16,17 (Supplementary Fig. 2c,e,f). When applied to α5TMDT287K transfected HEK-293T cells in whole cell patch-clamp recordings, Nb25 alone did not induce responses. Moreover, Nb25 did not affect maximal potentiation in the presence of 10 μM pregnanolone, the concentration used in crystallisation experiments (Supplementary Fig. 2g). However, the activated state adopted by α5TMD ECDs in crystal structures, in the absence of agonist, is consistent with the strong propensity of this construct to be spontaneously active in a cellular context (Fig. 1b).
Figure 2.
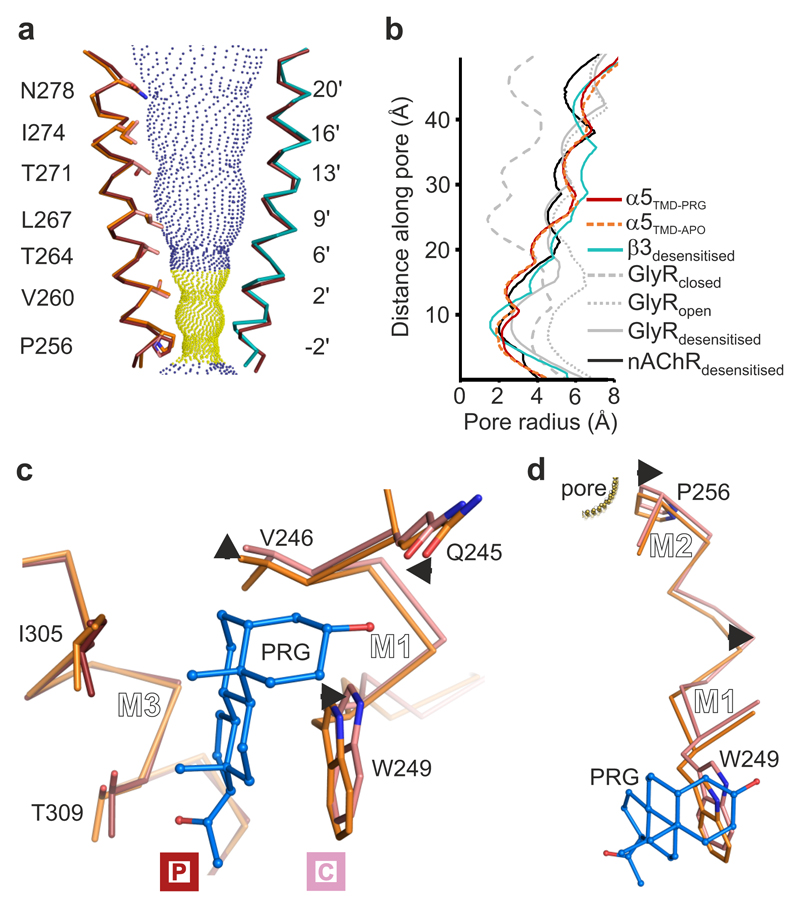
Architecture of GABAAR α5TMD. (a) and (b) Top-down and side-on views of the α5TMD pentamer in apo and pregnanolone-bound forms, respectively. Subunits each comprise the β3 extracellular domain (dark and light blue) and α5 transmembrane domain (α-helices M1-M4 shown in beige/yellow or red/pink). In top-down views, the pore is indicated by a black circle, lined by five M2 helices (one per subunit). Side-on views highlight the helical organisation (M1-M4, numbered 1 to 4 per subunit, where visible) in (a), and the presence of pregnanolone in (b) (blue stick and space-fill, inside dashed box). N-linked glycans are shown as spheres coloured by atom type (carbon atoms in orange, oxygen atoms in red). The principal (P) and complementary (C) subunits, for the visible inter-subunit interface, are indicated by boxed labels. (c) Structural formula of pregnanolone with rings labelled A to D. Carbon atoms mentioned in the text are numbered. (d) Close-up of pregnanolone ("ball-and-stick" representation) bound to α5TMD (cartoon and surface representation). The 2Fo-Fc electron density map surrounding the ligand is contoured at 1.3σ (yellow mesh).
Table 1. Crystallographic data collection and refinement statistics.
| Pregnanolone-bound α5TMD | Apo α5TMD |
||
|---|---|---|---|
| Uncorrected | Anisotropy corrected | ||
| (PDB 5O8F) | (PDB 5OJM) | ||
| Data collection | |||
| Space group | I23 | C2 | |
| Cell dimensions | |||
| a, b, c (Å) | 290.1, 290.1, 290.1 | 177.3, 139.9, 191.5 | |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 102.2, 90.0 | |
| Resolution (Å) | 205.13-3.15 (3.31-3.15)a | 187.14-3.28 (3.42-3.28) | |
| Rmerge | 0.728 (3.107) | 0.352 (2.807) | 0.352 (2.792) |
| Rmeas | 0.741 (3.169) | 0.371 (2.960) | 0.371 (2.944) |
| Rpim | 0.135 (0.621) | 0.117 (0.936) | 0.117 (0.930) |
| I/σ(I) | 7.55 (1.26) | 7.85 (1.08) | 7.86 (1.09) |
| CC1/2 | 0.988 (0.215) | 0.998 (0.395) | 0.998 (0.400) |
| Completeness (%) | 99.94 (100.00) | 99.78 (99.71) | 73.86 (6.89) |
| Redundancy | 30.4 (25.6) | 10.0 (9.9) | 10.0 (9.9) |
| Refinement | |||
| Resolution (Å) | 49.75-3.19 (3.31-3.19) |
48.88-3.30 (3.42-3.30) |
48.88-3.30 (3.42-3.30) |
| No. reflections | 66647 (6629) | 68726 (6866) | 50816 (473) |
| Rwork/Rfree | 0.232/0.244 (0.360/0.367) |
0.255/0.282 (0.389/0.395) |
0.236/0.256 (0.244/0.261) |
| No. atoms | |||
| Protein | 18217 | - | 18141 |
| N-linked glycans | 485 | - | 397 |
| Pregnanolone | 115 | - | - |
| B factors (Å2) | |||
| Protein | 72.1 | - | 71.3 |
| N-linked glycans | 90.7 | - | 99.1 |
| Pregnanolone | 59.6 | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | - | 0.009 |
| Bond angles (°) | 1.22 | - | 0.71 |
One crystal was used per structure.
Values in parentheses are for the highest-resolution shell.
Neurosteroid binding modes
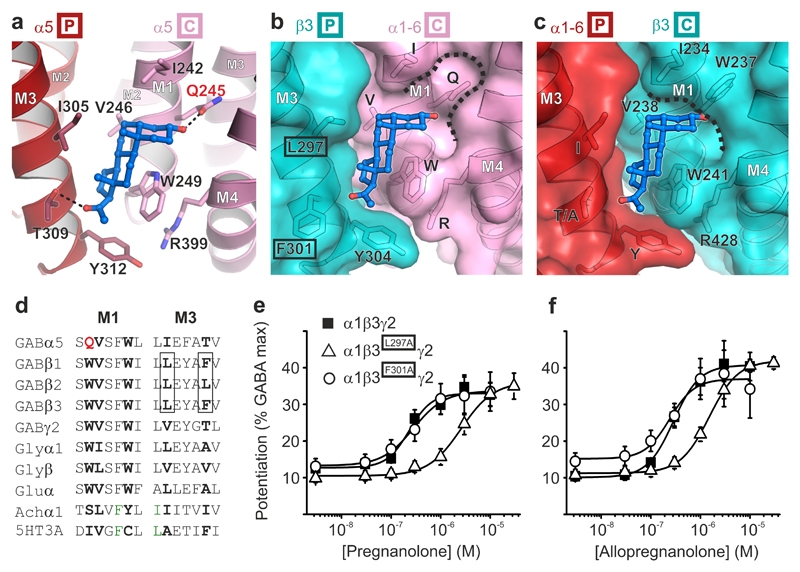
Surrounding the TMD pentamer of the pregnanolone-bound structure, but not the apo one, five large peaks were observed in the Fo–Fc omit electron density map. Each of these occupies a hydrophobic cavity at the interface between the principal M3 and complementary M1 helices of adjacent subunits (Supplementary Fig. 3). These were accounted for by pregnanolone molecules with half-chair geometry due to a cis junction between the A and B rings (Fig. 2c,d). The hydrophobic cavities contain the critical Gln245 residue, whose ε-oxygen contributes a putative hydrogen bond from the top of the site to anchor the pregnanolone A ring 3α-hydroxyl (Fig. 3a). The neurosteroid A ring is positioned flat beneath the M1 Ile242 side chain, perpendicular to and above the indole rings of M1 Trp249 (Fig. 3a). The B-D rings angle downwards, sandwiched between the side chains of M3 Ile305 and M1 Trp249. Tryptophan residues are highly prevalent in protein steroid binding sites22 and the stacking interaction observed is consistent with a previous study showing that an aliphatic Leu substitution of this M1 Trp ablated potentiation in GABAAR heteromers11. Pregnanolone B-D rings also make side-on van der Waals contacts with M1 Val246, which demarcates the deepest point of the pocket. Polar interactions anchor the neurosteroid at both ends. The D ring C20 ketone anchors the bottom end of the ligand by acting as a hydrogen bond acceptor for the M3 Thr309 hydroxyl. Mutation of each of these residues individually, Q245S, V246A, W249L, I305A and T309A, all reduced sensitivity to pregnanolone potentiation of histamine responses on the T287K background by between 3- and 10-fold, with W249L reducing sensitivity the most (from 0.8 ± 0.2 μM, n = 4 to > 10 μM, n = 8) and T309A reducing sensitivity the least (to 2.2 ± 0.3 μM, n = 3)(Supplementary Fig. 4a). Structural alignment of the β3 TMD pentamer10 over the α5 TMD pentamer and subsequent examination of α/β interfaces reveal that pregnanolone is well accommodated in the heteromer site at the βP-αC interface containing the highly conserved α-subunit M1 glutamine (Gln245 in α5TMD, Fig. 3b). However, at the αP-βC interface, the equivalent M1 position is occupied by β3 Trp237, removing the potential for 3α-hydroxyl coordination, clashing with pregnanolone and closing the top of the pocket (Fig. 3c,d). Although no γ subunit structure is currently available, sequence alignment also reveals an M1 tryptophan in place of M1 glutamine, predicting that the αP-γC pocket would be similarly occluded. Thus, α5TMD possesses a neurosteroid binding site that correctly predicts: (i) an involvement for the key M1 glutamine and tryptophan residues in heteromeric GABAARs3,11 and (ii) that receptor potentiation by neurosteroids occurs only via the βP-αC interface23 (Fig. 3b versus 3c).
Figure 3.
The neurosteroid potentiation site. (a) Pregnanolone (ball-and-stick representation, carbon atoms in blue, oxygen atoms in red) bound to an inter-subunit site between M3 residues on the principal face of one subunit (P in box) and M1 residues on the complementary face of the next (C in box). Putative hydrogen bonds between the pregnanolone 3α-hydroxyl and the Gln245 amide (2.4 Å) and C20 ketone and Thr309 hydroxyl (3.0 Å) are indicated by dashed lines. (b) and (c) Modelled heteromeric βP-αC and αP-βC interfaces respectively, in cartoon and surface representation, in which the relevant α5TMD face is substituted by the equivalent β3 subunit structure (PDB ID: 4COF). Note that α5 residues in these two panels are labelled by letters only, without numbers, to reflect their conservation across the α subunits (α1-6) within this site. Pregnanolone is well accommodated in the heteromeric βP-αC site (b) but not in the αC-βP site (c) due to the replacement of α M1 Gln with the β3 M1 Trp237, which closes the top of the pocket (dashed line). (d) Sequence alignment of the M1 and M3 motifs containing the neurosteroid binding site residues (bold) in the GABAAR α5 subunit and equivalent regions in other human pLGIC superfamily members. M1 Gln245, unique to GABAAR α-subunits, is highlighted in red. GABAAR β-subunit residues contributed from the complementary face in heteromeric receptors are boxed. (e) Pregnanolone and (f) Allopregnanolone dose-response curves for potentiation of GABA EC10-15 responses. Alanine substitution of β3 principal face residues Leu297, but not Phe301, reduces neurosteroid sensitivity. These residues are highlighted by black boxes in b. Each data point represents mean ± s.e.m. Pregnanolone EC50 n number: EC50: WT n = 13, L297A n = 11, F301A n = 8. Allopregnanolone EC50: WT n = 5, L297A n = 9, F301A n = 4. Each n EC50 measurement is from different cells.
A previous mutagenesis study, guided by homology modelling, placed the α-subunit M1 Gln close to M4 residues to create an intra-subunit pocket3, rather than the inter-subunit one described here. In α5TMD, the equivalent M4 residues Asn410 and Tyr413 are located outside the neurosteroid binding site and are incompatible with ligand coordination by Gln245 (Supplementary Fig. 4b,c). Therefore, the previously reported effects of mutations to these residues on neurosteroid binding were, most likely, indirect. Moreover, (3α,5β)-6-azi-pregnanolone photolabels β3 homopentameric GABAARs at Phe30124, the equivalent residue to α5TMD M3 Thr309 (Fig. 3a,b), consistent with the inter-subunit site we have identified. To functionally validate whether the neurosteroid-binding site observed crystallographically is equivalent to an inter-subunit potentiation site in the heteromeric α1β3γ2 GABAAR, we introduced mutations into the β-subunit face that opposes the key heteromer α-subunit M1 glutamine and tryptophan residues11. We compared the sensitivity to pregnanolone potentiation of wild-type GABAAR heteromers and heteromers containing β3 subunits with alanine substitutions at either M3 Leu297 (corresponding to α5TMD Ile305) or M3 Phe301 (corresponding to α5TMD Thr309) in whole-cell patch clamp recordings from transfected HEK-293T cells (Fig. 3e). Both α1β3L297Aγ2 and α1β3F301Aγ2 GABAARs retained wild-type sensitivity to the neurotransmitter GABA (wild type EC50 = 3.0 ± 1 μM, n = 8; L297A EC50 = 5.0 ± 2 μM, n = 3; F301A EC50 = 4.2 ± 2 μM, n = 3). However, α1β3L297Aγ2 GABAARs were less sensitive to potentiation by both pregnanolone and its 5α epimer allopregnanolone (a trans isomer with a "flat" geometry) by 10-fold and 5-fold, respectively (pregnanolone WT EC50 = 240 ± 20 nM, n = 13; L297A EC50 = 2500 ± 200 nM, n = 11; allopregnanolone WT EC50 = 280 ± 40 nM, n = 5; L297A EC50 = 1600 ± 200 nM, n = 9)(Fig. 3e,f). In contrast, α1β3F301Aγ2 GABAARs retained wild-type sensitivity to both pregnanolone and allopregnanolone potentiation (pregnanolone EC50 = 280 ± 40 nM, n = 8; allopregnanonlone EC50 = 260 ± 60 nM, n = 4). This is consistent with the more distal location of Phe301 on the border of the pocket, and also with its lack of conservation between human β-subunit subtypes at this position and the fact that the β-subunit subtype does not impact neurosteroid potentiation25. However, the strong impact of the β3 L297A mutation confirms that the inter-subunit location of the neurosteroid site observed in α5TMD is retained in α1β3γ2 heteromeric GABAARs.
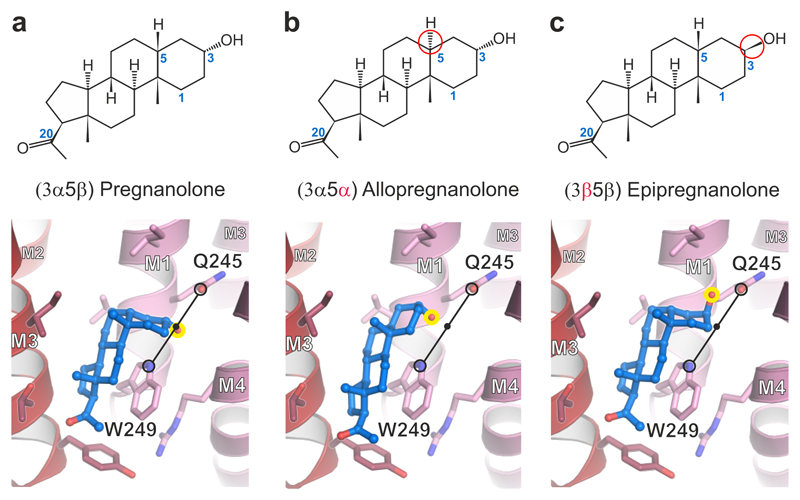
To explore the α5TMD neurosteroid binding mechanism further, we performed in silico analyses using the quantum mechanics polarized ligand docking (QMPLD)26 algorithm implemented in the Schrödinger suite (http://www.schrodinger.com). The binding mode observed upon pregnanolone docking into the α5TMD neurosteroid pocket was in agreement with the crystal structure (RMSD of 1.7 Å, calculated by VMD over all pregnanolone atoms) (Fig. 4a and Supplementary Fig. 5a). Van der Waals contacts were predicted to be the predominant driving force of interaction (binding energy of -36.6 Kcal mol-1), consistent with other steroid binding site analyses27. The electrostatic contribution was only -1.2 Kcal mol-1. Despite an elongated flat rather than half-chair geometry, allopregnanolone was also well accommodated into the site (Fig. 4b), with a similar binding energy (Eevdw = -35.4 Kcal mol-1; Eecoul = -1.4 Kcal mol-1). Moreover, pregnanolone and allopregnanolone potentiated the α5TMDT287K histamine responses similarly (Supplementary Fig. 5b). Allopregnanolone retained the same binding mode for its 3α-hydroxyl, positioned equidistantly between the M1 Gln245 εO and the Trp249 εN (3.5 Å for pregnanolone; 3.6 Å for allopregnanolone). Due to its elongated flat geometry, this imposed a 1.2 Å downwards translation on the allopregnanolone B-D rings. In contrast to pregnanolone, its 3β epimer, epipregnanolone, is a weak potentiator of GABAAR heteromers28. Epipregnanolone has been shown to non-competitively antagonise pregnanolone potentiation, raising the hypothesis that it acts through an alternative site28. However, our epipregnanolone docking in the α5TMD neurosteroid pocket led to similar binding energy estimates (Eevdw = -35.5 Kcal mol-1; Eecoul = -1.8 Kcal mol-1), suggesting that it can bind the same site as pregnanolone and allopregnanolone. These alternative scenarios can be reconciled if epipregnanolone binding to one GABAA heteromer site allosterically prevents potentiation by pregnanolone bound at the other. However, the possibility also remains that GABAAR heteromers contain additional, non-overlapping, sites that bind epipregnanolone and mediate non-competitive inhibition. Notably, our docking studies reveal that the 3-hydroxyl group of epipregnanolone points "upwards" rather than "downwards", losing the potential coordination with Trp249 εN, although potential coordination with Gln245 εO is retained (Fig. 4c). We propose that positioning of the 3α-hydroxyl between the Gln245 and Trp249 side chains is a contributing factor in determining whether transduction is potentiating or inhibitory in GABAAR heteromers. In contrast to the very weak potentiation on GABAAR heteromers, epipregnanolone triggers significant α5TMDT287K potentiation (Supplementary Fig. 5b). This might be due to its cumulative action through the five neurosteroid-binding sites present in α5TMDT287K, as opposed to only two equivalent sites in GABAAR heteromers, resulting in a stronger effect. Nevertheless, in these circumstances, the proposal that an "upwards" position of the 3-hydroxyl reduces neurosteroid potentiation cannot be validated in the α5TMDT287K chimera.
Figure 4.
Computational docking of pregnanolone and related neurosteroids. Structural formulae and α5TMD structures showing binding modes of computationally docked (a) pregnanolone, (b) allopregnanolone and (c) epipregnanolone. Stereoisomeric differences from pregnanolone are indicated in formulae by red circles. In the 3D models, the critical C3 hydroxyl group is highlighted by a yellow circle. Potentiators possess a 3α-hydroxyl which is computationally docked equidistantly between the Gln245 εO Gln and Trp249 εN (3.5 Å for pregnanolone; 3.6 Å for allopregnanolone; indicated by a black line) regardless of whether the rings assume a half-chair or flat geometry. The C3 hydroxyl of 3β pregnanolone can only share a putative hydrogen bond with the Gln245 amide (2.8 Å distance).
Impact of neurosteroid binding on TMD conformation
To examine the impact of pregnanolone binding on the pore conformation, we next compared the TMD regions of the apo and pregnanolone-bound α5TMD structures. Individual subunit TMDs superpose closely (RMSD in the 0.43-0.55 Å range between different chains over 123 equivalent Cα positions), adopting similar pore conformations to the desensitized GABAAR-β3 homopentamer10 (Fig. 5a), rather than the open, desensitized (or partly open) and closed conformations of the related glycine receptor (GlyR)16,17 (Fig. 5b). In both apo and pregnanolone-bound states, the five M2 helices lining the pore, one from each subunit, taper inwards to a constriction at the intracellular end. The side chains of Val260 and Pro256, occupying the 2' and -2' positions (pLGIC pore numbering convention), demarcate a hydrophobic desensitization gate (Fig. 5a). However, while the pore diameter at 2' valine appears to be state-independent (4.8 Å), pregnanolone binding leads to pore enlargement, from 3.7 Å to 4.3 Å diameter, at -2’ proline (Fig. 5b and Supplementary Video 2). This contrasts with the GABAAR-β3 homopentamer, in which the desensitization gate comprises only a single ring, at -2’ (Ala248) with a 3.1 Å diameter (Supplementary Fig. 6), but is reminiscent of the cation-conducting α4β2 nicotinic acetylcholine receptor (nAChR) in which two rings of residues, at 2' and -1' form a similar, albeit negatively charged, gate21 (Fig. 5b). Although the α5TMD pore in both states is theoretically wide enough to conduct dehydrated Cl- ions (Pauling radius 1.8 Å) the gate contains no polar groups to substitute for the hydration shell and is highly likely to impede conductance29.
Figure 5.
Pore conformation and neurosteroid induced motions in α5TMD. (a) View of two opposing pore-lining M2 helices, the other three being removed for clarity. The pregnanolone-bound α5TMD state (ruby) is shown on both sides. For comparison, overlays of α5TMD apo state (on the left side, orange), and of GABAAR-β3 (on the right side, teal) M2 helices are shown. Blue and yellow spheres define pregnanolone-bound α5TMD pore radii greater than or less than 3.2 Å respectively (radius of a hydrated Cl- ion). (b) A plot of pore radii for α5TMD structures compared to GABAAR-β3 in the desensitised state (PDB ID: 4COF), the glycine receptor in the resting closed, open and desensitised states (PDB IDs: 5JAD, 5JAE and 5JAF), and α4β2 nAChR (PDB ID: 5KXI). (c) Superposition of apo (orange) versus pregnanolone-bound (ruby) α5TMD principal faces reveals pregnanolone-induced motions on the complementary face (pink) of the neurosteroid pocket (indicated by short black arrows). (d) The same superposition as in c, zooming in on the pregnanolone-induced motions along the M1-M2 linker of a complementary face subunit, which impact on the Pro256 desensitization gate.
The increased -2’ Pro ring diameter is the result of neurosteroid pocket expansion, required to accommodate pregnanolone (Fig. 5c,d and Supplementary Video 2). Neurosteroid binding displaces the complementary face away from the principal one, in particular at M1 Val246 (Cα displaced 0.9 Å) and Trp249 (Cα displaced 0.7 Å). Contrasting with this expansion, the polar interaction from the 3α-hydroxyl draws the side chain of Gln245 into the pocket by 0.6 Å and down towards the Trp249 εN by 0.7 Å (measurements corresponding to Gln245 εO in both cases). The resultant torque from this rearrangement at the base of the M1 helix, which twists sideways and outwards, drives the translation of the M1-M2 intracellular linker (Fig. 5d and Supplementary Video 2; mean Cα displacement of Leu250, Asn251, Arg252, Glu253, Ser254, Val255 is 0.95 Å), not involved in crystal packing (Supplementary Fig. 7).
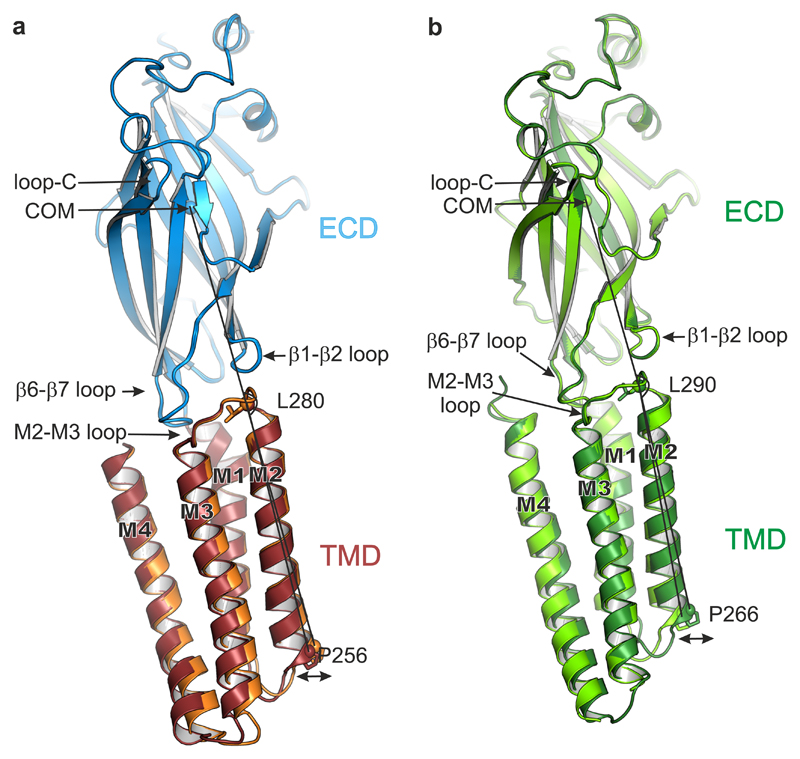
Besides pore dilation, which partly relieves its hydrophobic constriction, pregnanolone binding has longer-range effects, extending upwards through the TMD helical bundles and destabilizing their desensitized-state arrangement. Notably, straightening of the M2 helices drives inwards their extracellular ends and the contiguous M2-M3 loop, a key component of the TMD-ECD interface (Supplementary Video 3). This leads to an overall straightening of the subunits, with the ECD and TMD undergoing a rocking motion about their shared interface as α5TMD switches between apo and pregnanolone-bound states (Fig. 6a and Supplementary Video 3). The type and magnitude of motions observed here are consistent with those previously proposed to support transitions between gating states in the heteromeric Torpedo nAChR, studied within a membrane environment30, as well as in the detergent-solubilised homopentameric zebrafish α1 GlyR16 (Fig. 6b) and C. elegans GluClα receptor19, suggesting that all these constructs operate in the same allosteric framework, broadly applicable to pLGICs1.
Figure 6.
TMD motions relative to the ECD. (a) Side-on view of a single subunit (B chains), from apo α5TMD (orange TMD) and pregnanolone-bound α5TMD (ruby TMD), superposed by their ECDs (blue). The ECD conformations are very similar (RMSD of 0.19 Å over 204 equivalent Cα positions). However, the lower half of the pregnanolone-bound TMD flexes such that the desensitisation gate at Pro256 swings outwards from the pore (indicated by double-headed arrow). The angles measured between the ECD centre of mass (COM) and the Cα atoms of Leu280 at the top of M2 and Pro256 at the bottom of M2 are 162.5° (apo α5TMD) and 164.4° (pregnanolone-bound α5TMD), respectively, i.e. a 1.9° swing. (b) Equivalent view and superposition between open GlyR (light green, PDB ID: 3JAE) and desensitized (or partially open) GlyR (dark green, PDB ID: 3JAF). The narrowest pore diameter is at -2' (Pro266) position in both cases (8.8 Å and 5 Å, respectively). The GlyR ECD conformations are also very similar (RMSD of 0.31 Å over 210 equivalent Cα positions in chains B). As observed in α5TMD, relative to the superposed ECDs the lower half of the GlyR TMD flexes such that the desensitization gate at Pro266 is displaced outwards from the pore to expand and open the channel. The angles measured between the ECD COM and the Cα atoms of Leu290 at the top of M2 and Pro266 at the bottom of M2 are 161.4° (desensitized GlyR) and 164.1° (open GlyR), respectively, i.e. a 2.7° swing.
The observed outward pull on the desensitisation gate in response to binding pregnanolone, and the impact of structure-guided mutagenesis, offer a molecular mechanism by which neurosteroids support pore opening in heteromeric receptors. Consistent with this model, the M1-M2 linker controls desensitisation in heteromeric GABAARs31. Expansion of the TMD inter-subunit space as a mechanism of potentiation has previously been described for the anti-parasitic agent ivermectin binding to the related C. elegans receptor GluCl18,19. However, ivermectin binds GluCl to a pocket higher up the TMD, not observed in GABAARs, and penetrates deeper between the subunits (Supplementary Fig. 8a,b).
Discussion
Here, we present structures of a homomeric GABAAR construct, α5TMD, in apo form and bound to a potentiation site by the endogenous neurosteroid pregnanolone. The binding site for neurosteroid corresponds to the one in heteromeric αβγ/δ GABAARs, which constitute the vast majority of physiological GABAARs9. These structures reveal the mode of neurosteroid binding and the motions induced in response to binding. Pregnanolone binds at the inter-subunit interface between the M3 helix on the principal face and the M1 helix on the complementary face. Consistent with the high hydrophobicity of neurosteroids, binding is driven by interactions with hydrophobic residues (-36.6 Kcal mol-1 of the total -37.8 Kcal mol-1 binding energy), in particular by a stacking interaction between the pregnanolone B-D rings and the α5 M1 Trp249. The critical M1 Gln245 residue, forms a putative hydrogen bond via its ε-oxygen to the pregnanolone A ring 3α-hydroxyl. Expansion of Trp249 away from M3 to accommodate pregnanolone within the site, combined with attraction of Gln245 towards pregnanolone stacked on Trp249 imparts torque on the lower segment of M1, which triggers a sideways motion on the M1-M2 linker and withdraws M2 at the -2’ ring to expand the desensitisation gate. The structure of the α5TMD homopentamer potentiation site, compared with GABAAR-β310, also explains why in heteromeric GABAARs neurosteroids are well accommodated at the two βP-αC interfaces (Fig. 3a-c) but not at the two αP-βC interfaces23. In the latter, the key αC M1 Gln245 is replaced by βC Trp237, which occludes the site and will sterically clash with the neurosteroid.
In a physiological context, extrasynaptic GABAARs are key targets for endogenous neurosteroids32. In particular, neurosteroids increase tonic inhibition by promoting opening of GABAARs that are only partially occupied by the low ambient concentrations of GABA present in the extrasynaptic regions32. The structural framework provided by α5TMD in a desensitised state, bound by pregnanolone, reveals that the molecular basis of this physiological process can be accounted for by the neurosteroid drawing the equilibrium away from the closed resting state towards open and desensitized states. The degree by which neurosteroid binding supports the open state versus the desensitized state likely depends on the surrounding lipid environment. Neurosteroid potentiated the open state of α5TMD in whole-cell patch-clamp recordings in HEK cells, i.e. within a membrane environment, but was bound to desensitised α5TMD in detergent micelles in the crystal structure. The α5TMD structure also reveals the exposed location of the neurosteroid site on the outer face of the TMD, meaning that pregnanolone will bridge interactions between the GABAAR and the lipid bilayer. Thus, it is not surprising that the broader lipidic context of the protein will affect the extent to which neurosteroid binding expands the desensitization gate and so potentiates the open state.
Overall, this study provides key insight into the mechanical coupling between peripheral lipid binding sites and the channel desensitization gate, and illustrates a novel form of modulation within the pLGIC superfamily.
Online Methods
Construct design
The α5TMD construct was designed by fusing the extracellular domain of the human GABAAR β3 subunit (mature polypeptide numbering 1-217, QSVN…LKRN; Uniprot P28472) to the transmembrane domain of the human GABAAR α5 subunit (mature polypeptide numbering 226-431, IGYF…GAASPK; Uniprot P31644). The α5 intracellular M3-M4 loop amino acids 316-392 (RGWA…NSIS) were substituted by the SQPARAA sequence10,33 to enhance the recombinant protein yield and facilitate crystallisation. This construct was cloned into the pHLsec vector34, between the N-terminal secretion signal sequence and a C-terminal 1D4 purification tag derived from bovine rhodopsin (TETSQVAPA) that is recognised by the Rho-1D4 monoclonal antibody (University of British Columbia)35,36. Point mutations were introduced to constructs by overlap PCR of whole construct cDNAs and subsequent ligation into the pHLsec vector34.
Large-scale expression and purification of α5TMD
Twenty-litre batches of HEK293S-GnTI- cells (ATCC Cat# CRL-3022, RRID: CVCL_A785); which yield proteins with truncated N-linked glycans, Man5GlcNAc237,38) were grown in suspension to densities of 2 x 106 cells ml-1 in Protein Expression Media (PEM, Invitrogen) supplemented with L-glutamine, non-essential amino-acids (Gibco) and 1% foetal calf serum (Sigma-Aldrich) in 200 ml volumes each in empty 600 ml DMEM bottles (Gibco) with lids loose shaking at 130 rpm, 37°C, 8 % CO2. For transient transfection, cells from 1 litre of culture were collected by centrifugation (200 g for 5 mins) and resuspended in 150 ml Freestyle medium (Invitrogen) containing 3 mg PEI Max (Polysciences) and 1 mg plasmid DNA, followed by a 4 h shaker-incubation in a 2 litre conical flask at 160 rpm. Subsequently, culture media were topped up to 1 litre with PEM containing 1 mM valproic acid and returned to empty DMEM bottles. Typically, 40-70 % transfection efficiencies were achieved, as assessed by control transfections with a monoVenus-expressing plasmid39,40. 72 h post-transfection cell pellets were collected, snap-frozen in liquid N2 and stored at -80 °C.
Cell pellets (approx. 200g) were solubilised in 600 ml buffer containing 20 mM HEPES pH 7.2, 300 mM NaCl, 1.5 % (w/v) dodecyl 1-thio-β-maltoside (DDTM, Anatrace), 1 % (v/v) mammalian protease inhibitor cocktail (Sigma-Aldrich, cat. P8340) for 2 hours at 4 °C. Insoluble material was removed by centrifugation (10,000 g, 15 min). The supernatant was diluted 2-fold in a buffer containing 20 mM HEPES pH 7.2, 300 mM NaCl and incubated for 2 hr at 4 °C with 10 ml CNBr-activated sepharose beads (GE Healthcare) pre-coated with 50 mg Rho-1D4 antibody (3.3 g dry powdered beads expand during antibody coupling to approximately 10 ml). Affinity-bound samples were washed slowly by gravity flow over 2 hours at 4 °C with 200 ml buffer containing 20 mM HEPES pH 7.2, 300 mM NaCl, 0.1 % (w/v) DDTM (approximately 20 x CMC), then 200 ml buffer containing 20 mM HEPES pH 7.2, 300 mM NaCl, 0.01 % (w/v) DDTM. α5TMD was eluted overnight in 15 ml buffer containing 15 mM HEPES pH 7.2, 225 mM NaCl, 0.007 % (w/v) DDTM, 500 μM TETSQVAPA peptide (Genscript). The eluate was centrifuged (30,000g, 15 min) and the supernatant was concentrated by ultrafiltration to 1-2 ml at 1-5 mg/ml using 100-kDa cut-off membranes (Millipore). The concentrated sample was centrifuged (30,000 g, 15 min) and the supernatant was aliquoted in 0.5-1.5 mg protein per 0.7 ml aliquots and either snap-frozen for storage at -80 °C or gel filtrated as appropriate. A single aliquot was loaded onto a Superose 6 10/300 Increase gel filtration column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.2, 150 mM NaCl, 0.007 % (w/v) DDTM. The nanobody Nb25, expressed and purified as described below, was added to α5TMD at 10-fold molar excess and the complex was concentrated by ultrafiltration to 4 mg/ml, using 100 kDa cut-off membranes (Millipore), for crystallisation trials. Typical final yields were 0.2 mg α5TMD per litre of cells grown in suspension (10 g cell pellet). For the production of neurosteroid-bound α5TMD, the gel filtration buffer described above was supplemented with pregnanolone, 30 μM final concentration.
Large-scale expression and purification of heteromeric α1β3 GABAARs for generation of nanobody libraries
Human GABAAR α1 and β3 subunits wild-type mature protein sequences were used (α1 Uniprot P14867 entry Gln28 is Gln1 1-429 QPSL…PTPHQ; β3 Uniprot P28472 isoform 1 entry Gln26 is Gln1 1-448 QVSN…LYYVN), except that the intracellular loop between transmembrane helices 3 and 4 (M3-M4) was substituted, for α1 from Arg313 to Ser390, and for β3 from Gly308 to Asn421, with the linker sequence, SQPARAA, to increase expression10. These constructs were cloned into the pHLsec vector34, between the N-terminal secretion signal sequence and for β3 a double stop codon, or for α1 a C-terminal 1D4 purification tag. The expression and purification procedure was as described above for α5TMD but with the following differences. Plasmids encoding the α1 and β3 subunits were co-transfected in a 1:1 ratio. The detergent used for solubilization was decyl maltose neopentyl glycol (DMNG, Anatrace), supplemented with cholesterol hemisuccinate (CHS, Anatrace) at 10:1 molar ratio. The 15 ml eluted samples were loaded in 5 ml batches onto a Superdex 200 16/600 column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.2, 150 mM NaCl, 0.007 % (w/v) DMNG10:1CHS, 50 μM GABA. Peak fraction concentrations were typically 0.1 mg/ml and were further concentrated by ultrafiltration to 1 mg/ml using 100 kDa cut-off membranes (Millipore), aliquoted and snap-frozen at -80 °C. The final α1β3 GABAAR yield was 0.05 mg per litre of suspension culture (10 g cell pellet).
Nanobody generation and purification
Camelid nanobodies against α1β3 GABAARs were generated using established protocols41. One animal was immunized six times with the recombinant 1D4 tagged human α1β3 GABAAR pentamer (described above). Subsequent to library generation, nanobodies were selected by phage display in two different ways: either by passive absorption of the GABAA-α1β3 pentamer, or by trapping the receptor using the 1D4 antibody. After two rounds of selections, 96 clones were screened and positive clones were sequenced, revealing 13 nanobody families. The nanobody Nb25, binding to the GABAA β3 subunit, was produced and purified in milligram quantities from WK6su– E. coli bacteria. Bacteria were transformed with ~200 ng of nanobody expression plasmid pMESy4 containing the nanobody of interest and selected on Lysogeny broth (LB)-agar plates containing 2% glucose and 100 μg/ml ampicillin. 2-3 colonies were used for preparing a pre-culture, which was used to inoculate 0.5 L Terrific broth (TB) cultures supplemented with 0.1 % glucose, 2 mM MgCl2 and 100 μg/mL ampicillin. Cultures were grown at 37 °C until their OD600 reached 0.7, at which point Nb25 expression was induced with 1 mM IPTG. After induction cells were grown at 28 °C overnight and harvested by centrifugation (20 mins, 5000 g). Nanobodies were released from the bacterial periplasm by incubating cell pellets with an osmotic shock buffer containing 0.2 M Tris pH 8.0, 0.5 mM EDTA, and 0.5 M sucrose. The C-terminally His6-tagged Nb25 was purified using nickel affinity chromatography (binding buffer: 50 mM HEPES pH 7.2, 1 M NaCl, 10 mM imidazole; elution buffer: 50 mM HEPES pH 7.2, 0.2 M NaCl, 0.5 M imidazole), followed by size-exclusion chromatography on a Superdex 75 16/600 column (GE Healthcare) in 10 mM Hepes pH 7.2, 150 mM NaCl. Nb25 stocks were concentrated to 5-10 mg/mL, snap-frozen in liquid nitrogen and stored at -80 °C.
Crystallization and X-ray data collection
The α5TMD construct contains 15 N-linked glycosylation sites, bringing a considerable extra volume, flexibility and potential occupancy heterogeneity. Therefore, prior to crystallization, concentrated α5TMD samples, with and without pregnanolone, were incubated with 0.01 mg ml-1 endoglycosidase F142 for 2h at room temperature. Sitting drop vapour diffusion crystallization trials were performed in 96-well Swisssci 3 well crystallisation plates (Hampton Research), at three ratios: 200 nl protein plus 100 nl reservoir, 100 nl protein plus 100 nl reservoir, 100 nl protein plus 200 nl reservoir. Drops were dispensed by a Cartesian Technologies robot43, and plates were maintained at 6.5 °C in a Formulatrix storage and imaging system. Initial crystals grew within 1-7 days in a range of conditions44, and diffracted up to intermediate resolution (>5 Å). Following additive-based optimization (MemAdvantage, Molecular Dimensions), pregnanolone-bound α5TMD crystals diffracting to 3.2 Å resolution were grown in 14 % poly-ethylene (PEG) 6000, 0.1 M N-(2-Acetamido)iminodiacetic acid (ADA), pH 5.6, 7.6 mM 4-Cyclohexyl-1-Butyl-β-D-Maltoside (CYMAL-4) and apo α5TMD crystals diffracting to 3.3 Å resolution were grown in 14 % poly-ethylene (PEG) 5000, 0.08 M magnesium acetate, 0.1 M sodium citrate, pH 5.8, 1 mM ethylene glycol tetraacetic acid (EGTA). Crystals were cryoprotected by soaking in reservoir solution supplemented with 30 % ethylene glycol, and then were flash-frozen in liquid nitrogen. Diffraction images of 0.2° oscillation were collected at the Diamond Light Source, beamlines I03 (pregnanolone-bound α5TMD, λ=0.9763 Å) and I04 (apo α5TMD, λ=0.9795 Å), on Pilatus3 6M and Pilatus 6M-F detectors, respectively. X-ray data were indexed, integrated and scaled using the HKL2000 package45 and merged using Aimless46,47. Data collection statistics are shown in Table 1.
Structure determination, refinement and analysis
Pregnanolone-bound and apo α5TMD structures were solved by molecular replacement using the human GABAAR-β3cryst homopentamer10 (PDB ID: 4COF) and the gelsolin nanobody48 (PDB ID: 2X1Q) as a search model in Phaser49. Polypeptide chains were traced using iterative rounds of manual model building in Coot50 and refinement in Refmac51 and Phenix52,53. Automated X-ray and atomic displacement parameter (ADP) weight optimisation, and torsion angle non-crystallographic symmetry (NCS) restraints, were applied. The pregnanolone coordinates and geometry restraints were generated using the Grade Web Server (http://grade.globalphasing.org). Diffraction from the apo α5TMD crystal was strongly anisotropic. Ellipsoidal truncation, with resolution limits along the reciprocal cell directions a*, b* and c* being 3.9 Å, 3.8 Å and 3.3 Å respectively, anisotropic scaling and a negative isotropic B-factor correction (-49.29 Å2) were performed using the UCLA-DOE Lab Diffraction Anisotropy server (https://services.mbi.ucla.edu/anisoscale/)54. Anisotropy correction led to improvements in the electron density map quality and facilitated model building. Both datasets (uncorrected and anisotropy-corrected) were deposited in the Protein Data Bank. The model deposited was refined against the anisotropy-corrected dataset, however we also provide refinement statistics against the full (uncorrected) dataset in Table 1.
The final models contain one α5TMD homopentamer, bound to five Nb25 nanobodies, per asymmetric unit. The complete polypeptide chains could be built, with the exception of thirteen N-terminal (QSVNDPGNMSFVK) and thirteen C-terminal (NREPVIKGAASPK) GABAA residues, the final Ser residue in nanobody chains L, M and N and the C-terminal purification tags, presumably disordered. Furthermore, very strong additional electron density peaks were clearly visible in the outer parts of inter-subunit TMD regions in the α5TMD maps. These could be unambiguously assigned to five pregnanolone molecules, one per inter-subunit interface, based on shape, coordination and refinement statistics. The α5TMD extracellular region is heavily glycosylated, and we could observe clear electron density for 10 out of the 15 N-linked glycosylation sites, the remaining five being located in the N-terminal disordered regions. Glycans attached to Asn149 in each chain were protected from endoglycosidase F1 cleavage due to extensive interactions with both α5TMD and the Nb25 nanobody molecules, underlying their important structural role10. Stereochemical properties of the models were assessed in Coot50 and Molprobity55. Protein geometry analyses revealed no Ramachandran outliers, with 97.88% residues in favoured regions, 2.12% residues in allowed regions, for pregnanolone-bound α5TMD and 96.76% residues in favoured regions, 3.24% residues in allowed regions, for apo α5TMD. The overall Molprobity scores are 1.24 and 1.74, respectively (100th percentile).
Structural alignments were performed in SHP56. Protein interfaces were analysed using the PDBePISA web server at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html)57 and pore dimensions were analysed using the Coot implementation of Hole58, with a probe radius of 1.4 Å. Structural figures were prepared with the PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC. Molecular videos and analyses were performed with the UCSF Chimera package, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311)59.
Cell preparation and electrophysiology
One day prior to experiments, 8 ml of Dulbecco’s Modified Eagle Medium (DMEM) was pre-incubated for 10 mins at room temperature with 96 μl lipofectamine2000 (Thermofisher) and 48 μg plasmid DNA, then added to a single T175 cm2 flask containing HEK293T cells (30-50 % confluency; HEK 293T, ATCC Cat# CRL-3216, RRID: CVCL_0063) and 2 ml DMEM (supplemented with 10 % fetal calf serum, L-Gln and non-essential amino acids). After 3 hrs this media was removed and replaced by DMEM supplemented with 10 % fetal calf serum. For GABAAR heteromers, pHLsec plasmids containing human full-length cDNA constructs were mixed in 1:1:2 ratio (α:β:γ), supplemented with 3% plasmid encoding enhanced green fluorescent protein (EGFP) to assess transfection efficiency. Specifically, the cDNA inserts used for heteromeric receptor expression were as follows: human GABAAR α1 mature protein sequence (α1 Uniprot P14867 entry Gln28 is Gln1 1-429 QPSL…PTPHQ) and human β3 mature protein sequence (β3 Uniprot P28472 entry isoform 1 Gln26 is Gln1 1-448 QVSN…LYYVN) cloned into the pHLsec vector34 between the N-terminal secretion signal sequence and a double stop codon; Human GABAAR γ2 mature protein sequence (γ2 Uniprot P18507 isoform 1 entry Gln40 is Gln1 1-428 QKSDD…SYLYL) cloned into the pHLsec vector34 between the N-terminal secretion signal sequence followed by streptavidin binding protein (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREP) and a C-terminal 1D4 purification tag. Transfection efficiencies were typically 50-80 % (cells expressing EGFP, as estimated by fluorescence microscopy). Eighteen to twenty-four hours later cells were washed with phosphate buffered saline, incubated in 4 ml TrypLE (Gibco) for 7 mins at 37 °C, suspended in 21 ml DMEM supplemented with 10 % fetal calf serum and L-Gln, centrifuged at 100 g for 1.5 mins, then suspended in 50 ml Freestyle 293 Expression Medium (Gibco) and placed in a shaking incubator (130 rpm, 37°C, 8 % CO2) for 30 mins. 25 ml cell suspension was then centrifuged at 100 g for 1.5 mins, and suspended in 4 ml external recording solution. This solution contained (mM): 137 NaCl, 4 KCl, 1 MgCl2, 1.8 CaCl2, 10 HEPES, and 10 D-Glucose, pH 7.4 (≈ 305 mOsm). The internal recording solution contained (mM): 140 CsCl, 5 NaCl, 1 MgCl2, 10 HEPES, 5 EGTA, 0.2 ATP, pH 7.35 (≈ 295 mOsm). Electrophysiological recordings were performed at room temperature using an Ionflux16 (Molecular Devices) in ensemble mode, with series resistance compensation set at 80 % and cells held at -60 mV. Pregnanolone and allopregnanolone (Sigma-Aldrich) were dissolved in DMSO as 100 mM stocks prior to dilution in external recording solution. These neurosteroids were co-applied with EC10-15 GABA or histamine doses to generate dose response curves for heteromeric GABAARs and α5TMD. Expression of heteromeric receptors as assemblies of αβγ subunits was confirmed by response to GABA, which requires co-assembly of α1 and β3 subunits, and efficient inclusion of the γ2 subunit into αβγ heteromers was verified by testing for Zn2+ sensitivity, with heteromeric constructs exhibiting low sensitivity to Zn2+, defined as less than 50 % inhibition of an EC50 GABA response60.
Molecular docking and binding-energy calculation
Small-molecule coordinates were generated by eLBOW and energy minimized with Ligprep in the Schrödinger suite at pH 7.0 with the OPLS_2005 force field61. The standard conversion procedure with full hydrogen optimization was applied with the Protein Preparation workflow to the neurosteroid binding pocket between chains B and C in α5TMD. These processed coordinates were used for the subsequent grid generation and ligand-docking procedures. The Glide Grid62,63 (Schrödinger suite) was built using an inner box (the centroid of the pregnanolone molecule) of 10 × 10 × 10 Å3 and an outer box (within which all the ligand atoms must be contained) that extended 20 Å in each direction from the inner one. Default values were used for all other parameters. The hydrogen bonds between the 3α-hydroxyl group and the 20-carbonyl group of the pregnanolone molecule respectively with the carbonyl group of Gln245 and hydroxyl group of Thr309 were used as positional constraints. Furthermore, the atoms C03, C20 and C06 of pregnanolone molecule were used as additional positional constraints. For docking, the QMPLD26 protocol (Schrödinger suite, http://www.schrodinger.com/) was used. The most reliable binding pose for each small molecule was selected on the basis of calculated van der Waals and electrostatic interactions. RMSD values were calculated with VMD64.
Thermostability binding experiments
Information regarding the thermostability of a detergent-solubilised protein can be determined by heating protein samples over a range of temperatures for equal time periods and then measuring the reduction in the intensity of the monodisperse SEC profile for each protein sample65. With increasing temperature an increased proportion of protein is denatured, aggregates and is lost from the monodisperse peak when the protein is subsequently run on SEC. A measure of protein stability can then be obtained by plotting the decay in peak UV absorbance against increasing temperature, for example to obtain a 50 % melting temperature (Tm). Purified GABAAR α5TMD or α5TMD mutants at 0.02 mg ml-1 (100 nM) in 150 mM NaCl, 10 mM HEPES pH 7.2, 0.007 % DDTM (w/v) were separated into 50 μl aliquots in PCR tubes, and heated at a range of temperatures from 40-80 °C for 1 hour. Samples were run on a high-performance liquid chromatography system with automated micro-volume loader (Shimadzu) through a Superdex 200 Increase 3.2/300 column (GE Healthcare) maintained in 300 mM NaCl, 10 mM HEPES, 0.007 % DDTM (w/v). Monodisperse peak reduction with increasing temperature was measured relative to an unheated control sample maintained at 4 °C.
Importantly, because some drugs when bound thermostabilise detergent-solubilised protein65, the thermostability assay offers an efficient strategy to measure protein sensitivity to drugs in the detergent-solubilised environment. Purified GABAAR α5TMD or α5TMD mutants were separated into PCR tubes, supplemented with pregnanolone at a range of concentrations and heated at Tm30 % (the temperature at which the monodisperse peak was reduced by 70 %) for 1 hour. Afterwards samples were run on a high-performance liquid chromatography system with automated micro-volume loader (Shimadzu) through a Superdex 200 3.2/300 column (GE Healthcare) maintained in 300 mM NaCl, 10 mM HEPES, 0.007 % DDTM. Drug dose-response curves were generated by plotting UV absorbance against drug concentration.
Cell lines used
HEK293S-GnTI- cells (ATCC Cat# CRL-3022, RRID: CVCL_A785) and HEK 293T, (ATCC Cat# CRL-3216, RRID: CVCL_0063), both confirmed free from mycoplasma contamination.
Supplementary Material
Acknowledgements
We thank staff at Diamond Light Source beamlines I03 and I04 for assistance at the synchrotron; K. Harlos and T. Walter for technical support with crystallization; J. Kammonen and staff at Pfizer for very kind time and assistance with electrophysiology; E. Beke for technical assistence during nanobody discovery; Y. Zhao for technical support with tissue culture; members of STRUBI for helpful discussions; G. Sutton and T. Malinauskas for feedback regarding the manuscript. This work was supported by the UK Biotechnology and Biological Sciences Research Council grant BB/M024709/1 (A.R.A. and P.S.M.), the UK Medical Research Council grants MR/L009609/1 and MC_UP_1201/15 (A.R.A.), the Human Frontier Science Program grant RGP0065/2014 (A.R.A.) and the Wellcome Trust studentships 105247/Z/14/Z (S.S.) and (S.M.). We also thank INSTRUCT, part of the European Strategy Forum on Research Infrastructures and the Hercules Foundation Flanders for their nanobody discovery support. Further support from the Wellcome Trust Core Award 090532/Z/09/Z is acknowledged.
Footnotes
A Life Sciences Reporting Summary for this article is available online.
Data availability. The atomic coordinates and the structure factors are available from the Protein Data Bank under accession codes 5O8F (pregnanolone-bound α5TMD) and 5OJM (apo α5TMD). Source data for figures 1e, 3e and 3f are available with the paper online.
Author Contributions
Experimental work was performed by P.S.M. and S.S. (protein expression, purification, crystallization, electrophysiology), S.M. (protein expression, purification), L.D.C. (docking experiments), E.P. and J.S. (nanobody generation), A.R.A. (crystallography). The manuscript was written by P.S.M. and A.R.A., with input from all co-authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Nemecz A, Prevost MS, Menny A, Corringer PJ. Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron. 2016;90:452–70. doi: 10.1016/j.neuron.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Miller PS, Smart TG. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol Sci. 2010;31:161–74. doi: 10.1016/j.tips.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Hosie AM, Wilkins ME, da Silva HM, Smart TG. Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature. 2006;444:486–9. doi: 10.1038/nature05324. [DOI] [PubMed] [Google Scholar]
- 4.Majewska MD, Harrison NL, Schwartz RD, Barker JL, Paul SM. Steroid hormone metabolites are barbiturate-like modulators of the GABA receptor. Science. 1986;232:1004–7. doi: 10.1126/science.2422758. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar J, Wakefield S, MacKenzie G, Moss SJ, Maguire J. Neurosteroidogenesis is required for the physiological response to stress: role of neurosteroid-sensitive GABAA receptors. J Neurosci. 2011;31:18198–210. doi: 10.1523/JNEUROSCI.2560-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunn BG, Brown AR, Lambert JJ, Belelli D. Neurosteroids and GABA(A) Receptor Interactions: A Focus on Stress. Front Neurosci. 2011;5:131. doi: 10.3389/fnins.2011.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maguire JL, Stell BM, Rafizadeh M, Mody I. Ovarian cycle-linked changes in GABA(A) receptors mediating tonic inhibition alter seizure susceptibility and anxiety. Nat Neurosci. 2005;8:797–804. doi: 10.1038/nn1469. [DOI] [PubMed] [Google Scholar]
- 8.Maguire J, Mody I. GABA(A)R plasticity during pregnancy: relevance to postpartum depression. Neuron. 2008;59:207–13. doi: 10.1016/j.neuron.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sigel E, Steinmann ME. Structure, function, and modulation of GABA(A) receptors. J Biol Chem. 2012;287:40224–31. doi: 10.1074/jbc.R112.386664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–5. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akk G, et al. Mutations of the GABA-A receptor alpha1 subunit M1 domain reveal unexpected complexity for modulation by neuroactive steroids. Mol Pharmacol. 2008;74:614–27. doi: 10.1124/mol.108.048520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saras A, et al. Histamine action on vertebrate GABAA receptors: direct channel gating and potentiation of GABA responses. J Biol Chem. 2008;283:10470–5. doi: 10.1074/jbc.M709993200. [DOI] [PubMed] [Google Scholar]
- 13.Bianchi MT, Macdonald RL. Neurosteroids shift partial agonist activation of GABA(A) receptor channels from low- to high-efficacy gating patterns. J Neurosci. 2003;23:10934–43. doi: 10.1523/JNEUROSCI.23-34-10934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–81. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 15.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–55. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 16.Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–9. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature. 2015;526:277–80. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- 18.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–7. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–89. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Morales-Perez CL, Noviello CM, Hibbs RE. X-ray structure of the human alpha4beta2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori-Tanaka Y, Yura K, Takai-Igarashi T, Tanaka H. Structural classification of steroid-binding sites on proteins by coarse-grained atomic environment and its correlation with their biological function. Steroids. 2015;96:81–8. doi: 10.1016/j.steroids.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Bracamontes JR, Li P, Akk G, Steinbach JH. A neurosteroid potentiation site can be moved among GABAA receptor subunits. J Physiol. 2012;590:5739–47. doi: 10.1113/jphysiol.2012.237255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen ZW, et al. Neurosteroid analog photolabeling of a site in the third transmembrane domain of the beta3 subunit of the GABA(A) receptor. Mol Pharmacol. 2012;82:408–19. doi: 10.1124/mol.112.078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belelli D, Casula A, Ling A, Lambert JJ. The influence of subunit composition on the interaction of neurosteroids with GABA(A) receptors. Neuropharmacology. 2002;43:651–61. doi: 10.1016/s0028-3908(02)00172-7. [DOI] [PubMed] [Google Scholar]
- 26.Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–31. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eick GN, Thornton JW. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol. 2011;334:31–8. doi: 10.1016/j.mce.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Wang M, et al. 3beta -hydroxypregnane steroids are pregnenolone sulfate-like GABA(A) receptor antagonists. J Neurosci. 2002;22:3366–75. doi: 10.1523/JNEUROSCI.22-09-03366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aryal P, Sansom MS, Tucker SJ. Hydrophobic gating in ion channels. J Mol Biol. 2015;427:121–30. doi: 10.1016/j.jmb.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unwin N, Fujiyoshi Y. Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol. 2012;422:617–34. doi: 10.1016/j.jmb.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gielen M, Thomas P, Smart TG. The desensitization gate of inhibitory Cys-loop receptors. Nat Commun. 2015;6:6829. doi: 10.1038/ncomms7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brickley SG, Mody I. Extrasynaptic GABA(A) receptors: their function in the CNS and implications for disease. Neuron. 2012;73:23–34. doi: 10.1016/j.neuron.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. J Gen Physiol. 2008;131:137–46. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–50. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 35.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–60. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 36.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci USA. 1987;84:8874–8. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–24. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aricescu AR, Owens RJ. Expression of recombinant glycoproteins in mammalian cells: towards an integrative approach to structural biology. Curr Opin Struct Biol. 2013;23:345–56. doi: 10.1016/j.sbi.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–6. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 40.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 41.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–93. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang VT, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–73. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61:651–7. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker JL, Newstead S. Current trends in alpha-helical membrane protein crystallization: an update. Protein Sci. 2012;21:1358–65. doi: 10.1002/pro.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van den Abbeele A, et al. A llama-derived gelsolin single-domain antibody blocks gelsolin-G-actin interaction. Cell Mol Life Sci. 2010;67:1519–35. doi: 10.1007/s00018-010-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–5. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979;134:109–42. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 57.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–60. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Hosie AM, Dunne EL, Harvey RJ, Smart TG. Zinc-mediated inhibition of GABA(A) receptors: discrete binding sites underlie subtype specificity. Nat Neurosci. 2003;6:362–9. doi: 10.1038/nn1030. [DOI] [PubMed] [Google Scholar]
- 61.Banks JL, et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT) J Comput Chem. 2005;26:1752–80. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 63.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–9. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 64.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. 27-8. [DOI] [PubMed] [Google Scholar]
- 65.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–9. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–42. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.