Abstract
Hsp72 is a member of the 70-kD heat shock family of molecular chaperones (Hsp70s) that comprise a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD) connected by a linker that couples the exchange of adenosine diphosphate (ADP) for adenosine triphosphate (ATP) with the release of the protein substrate. Mitotic phosphorylation of Hsp72 by the kinase NEK6 at Thr66 located in the NBD promotes the localization of Hsp72 to the mitotic spindle and is required for efficient spindle assembly and chromosome congression and segregation. Here, we determined the crystal structure of the Hsp72 NBD containing a genetically encoded phosphoserine at position 66. This revealed structural changes that stabilized interactions between subdomains within the NBD. ATP binding to the NBD of unmodified Hsp72 resulted in the release of substrate from the SBD, but phosphorylated Hsp72 retained substrate in the presence of ATP. Mutations that prevented phosphorylation-dependent subdomain interactions restored the connection between ATP binding and substrate release. Thus, phosphorylation of Thr66 is a reversible mechanism that decouples the allosteric connection between nucleotide binding and substrate release, providing further insight into the regulation of the Hsp70 family. We propose that phosphorylation of Hsp72 on Thr66 by NEK6 during mitosis promotes its localization to the spindle by stabilizing its interactions with components of the mitotic spindle.
Introduction
Proliferating human cells experience a peak of protein phosphorylation during mitosis, a phase of the cell cycle in which many protein kinases are active (1–3). Kinase activity regulates the spectacular reorganisation of cellular structures that are hallmarks of mitotic cells, such as the formation of the mitotic spindle through the coordinated action of microtubule-associated proteins and motor proteins (4). The specificity of signaling through mitotic kinases occurs in part through the recognition of different phosphorylation sites and in part through the discrete localization patterns of these kinases (5). Mitotic kinases are activated at specific locations through protein-protein interactions and through the action of kinase cascades, such as the Aurora-A – PLK1 – NEK9 – NEK6/NEK7 network that governs early mitotic events, including centrosome separation and mitotic spindle assembly (6, 7).
Heat shock protein 72 (Hsp72), the major stress-inducible isoform of the Hsp70 family of molecular chaperones, is a mitotic substrate of the NEK6 kinase (8). NEK6 phosphorylates Hsp72 at Thr66, which targets the chaperone to the mitotic spindle, promoting efficient spindle assembly, chromosome congression and segregation (8–10). NEK6 is activated specifically during mitosis; the phosphorylation of Hsp72 at Thr66 is not detected at other phases of the cell cycle (9, 11, 12). Mitotic phosphorylation of Hsp72 contributes to proper spindle assembly, and the expression of Hsp72-T66A or depletion of NEK6 causes defects in spindle assembly, while a Hsp72-T66E phosphomimetic mutant can rescue the defects in spindle assembly due to NEK6 depletion (9). Here, we investigated the effect of Thr66 phosphorylation on Hsp72 activity and the mechanism by which this confers spindle localization.
The 70kDa Heat shock proteins (Hsp70s) are ubiquitous molecular chaperones that play critical roles in protein folding, translocation and assembly of multi-protein complexes (13, 14). All Hsp70s contain two functional domains: a nucleotide binding domain (NBD) and a substrate-binding domain (SBD) that are connected by a highly conserved flexible linker (15). The NBD contains four subdomains (IA, IB, IIA and IIB) harboring a central nucleotide-binding cleft (16). The SBD is composed of two subdomains: SBDβ that contains the peptide-binding pocket and SBDα that acts as a helical lid covering the pocket (17). ATP-bound Hsp70 has low affinity for the substrate, whereas the ADP-bound form has high affinity (18). The allosteric mechanism by which the NBD and SBD are coupled has been determined, and involves structural rearrangements transmitted through a linker region that connects the two domains (19).
The importance of phosphorylation in the modulation of Hsp70 activity is highlighted by other studies (20–22). However, the inability to produce homogenously phosphorylated proteins has hampered structural and biochemical attempts to unveil the molecular mechanisms of regulation by phosphorylation (23). Therefore, to reconstitute the NEK6-mediated phosphorylation of Hsp72, we incorporated pSer at position 66 by means of genetic code expansion (24). This method had not yet been optimized for pThr, but we reasoned that pSer would be an acceptable mimic based on its chemical similarity and ability to substitute for Thr in the context of kinase regulation (25). We considered using a standard phosphomimic mutation (Glu), which does provide a partial rescue of function, but because the carboxylic acid group is not an isostere for the phosphate we did not think this would be appropriate for a high-resolution structural study of the mechanism (9). This approach enabled us to characterize the consequence of phosphorylation on the structural and biochemical mechanisms of Hsp72.
Results
Phosphorylation of Hsp72 NBD on residue 66 stabilizes an inter-subdomain interaction
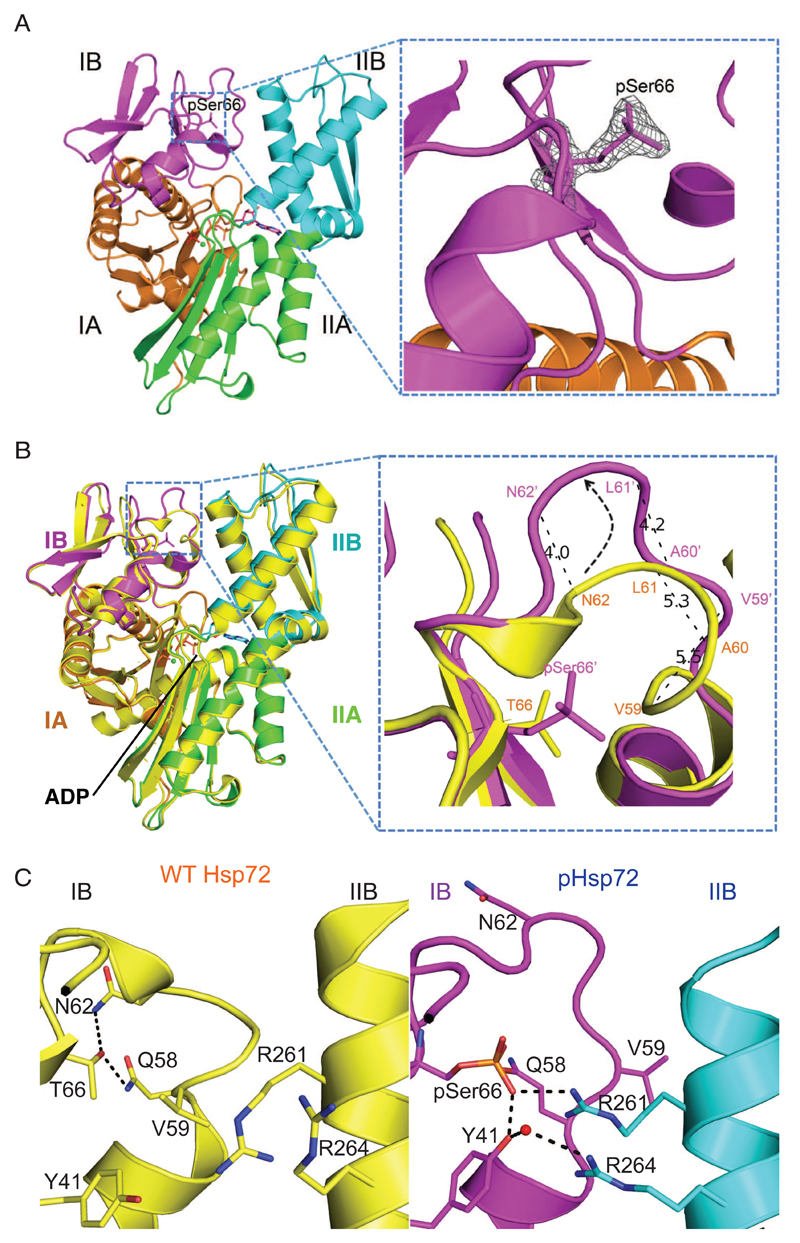
We produced multi-milligram quantities of pure, homogeneous pSer66-Hsp72 (pHsp72)-NBD (Fig. S1) and solved the crystal structure in complex with ADP at 1.66Å resolution (Table 1). The phosphorylated residue was clearly defined in the electron density map (Fig. 1A). To understand the structural changes arising as a result of phosphorylation, we superimposed the structure of pHsp72-NBD with that of unphosphorylated Hsp72-NBD obtained from the Protein Data Bank [PDB 3AY9 (26)] by aligning the corresponding Cα positions of the subdomain IB (Fig. 1B, Movie S1). The loop consisting of residues 58-65 was unwound in the phosphorylated version, thereby exposing pSer66. This resembles the opening of a lid covering the phosphosite by about 45°, resulting in the displacement of the equivalent Cα atoms by 4-5.5 Å in an anti-clockwise direction (Fig. 1B). Phosphorylation induces this conformational change both through disruption of interactions that are present in the unphosphorylated protein and through the formation of new interactions. In the unphosphorylated Hsp72 structure (PDB code 3AY9) (26), the side chain of Thr66 forms H-bonds with the main chain O atom and side chain NH group of Asn62 and the side chain NH group of Gln58 (Fig. 1C). These interactions may serve to pin together the loop region that spans residues 58-65. There is insufficient space to accommodate a phosphate group on Thr66 without a conformational change in the protein, and the position of the phosphate in the structure of pHsp72-NBD clashes with the positions of atoms in the unphosphorylated protein comprising the main chain of residues 62-63 and the side chain of Val59. Notably, the side chain of Val59 is flipped from a buried position in the unphosphorylated Hsp72-NBD structure to a solvent-exposed position in the structure of pHsp72-NBD (Fig. 1C). The space vacated by this side chain is filled by the guanidine group on the side chain of Arg261, which juts out from subdomain IIB, as well as the phosphate group on residue 66. The side chain of Tyr41 in subdomain IB also makes a striking conformational change, pivoting around to form H-bonds with the phosphate group and a water-mediated H-bond with the side chain of Arg264 (Fig. 1C). These interactions contribute to an enhanced interaction between subdomains IB and IIB in the pHsp72-NBD structure (Fig. 1C and Movie S2), which might lock together these two structural elements. The key residues of Hsp72 in this regulatory mechanism are conserved in a subset of other Hsp70 family members (Fig. S6). Notably, while Thr66 is conserved in E. coli Hsp70 (DnaK) the other three critical residues are replaced, whereas all but one are conserved in BiP and Mortalin (Fig. S6A). All four key residues are conserved in the yeast Hsp70 homologues, Ssa3 and Ssa4, suggesting that these homologues have the potential to be regulated by a Ser/Thr kinase (Fig. S6B).
Table 1. Data collection and refinement statistics.
The table provides parameters that define the crystal system and quality of the data used to determine the structure of the phosphorylated Hsp72 NBD, and an assessment of the quality of the structural model. *Values in parentheses are for highest-resolution shell.
| pHsp72-NBD (PDB: 6FHK) |
|
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 51.63, 110.88, 63.48 |
| α, β, γ (°) | 90.00, 90.41, 90.00 |
| Resolution (Å) | 55.09 - 1.657 (1.662- 1.657)* |
| Rmerge | 0.074 (0.646) |
| Rpim | 0.048 (0.473) |
| I/σI | 13.2 (2.0) |
| CC1/2 | 0.997 (0.592) |
| Completeness (%) | 98.30 (91.51) |
| Redundancy | 6.4 (5.0) |
| Refinement | |
| Resolution (Å) | 55.09 - 1.657 |
| No. reflections | 83152 |
| Rwork / Rfree | 0.1617/0.1942 |
| No. atoms | |
| Protein | 5874 |
| Ligand/ion | 68 |
| Water | 579 |
| B-factors | |
| Wilson | 20.56 |
| Average | 24.40 |
| Protein | 23.60 |
| Water | 34.10 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.39 |
| Ramachandran favored (%) | 99 |
| Ramachandran outliers (%) | 0 |
Figure 1. Phosphorylation of Hsp72 at residue 66 increases the interaction with domain IIB.
(A) Left: Structure of phosphorylated Hsp72 nucleotide binding domain (pHsp72-NBD) comprises two lobes embracing the central nucleotide-binding site and is divided into four sub-domains IA, IB, IIA and IIB, color-coded in orange (IA), magenta (IB), green (IIA) and cyan (IIB). Right: 2Fo-Fc map contoured at 2 σ showing electron density of pSer66. (B) Left: Structural superposition of pHsp72-NBD with unphosphorylated Hsp72-NBD (PDB 3AY9, shown in yellow) highlighting the local changes around pSer66. The structural superposition was done by alignment of the Cα atoms from subdomain IB of the two structures. Right: The loop surrounding the residue 66 in the subdomain IB undergoes a conformational change resulting in a movement of the equivalent Cα atoms by 4-5.5 Å. (C) Comparison of the inter-subdomain interactions within the IB-IIB interface in unphosphorylated Hsp72-NBD (left image) and pHsp72-NBD (right image). In pHsp72-NBD, pSer66 and Tyr41 located in subdomain IB are engaged in salt bridge and water mediated H-bonding interactions with Arg261 and Arg264 located in subdomain IIB respectively. The water molecule is shown as a red sphere.
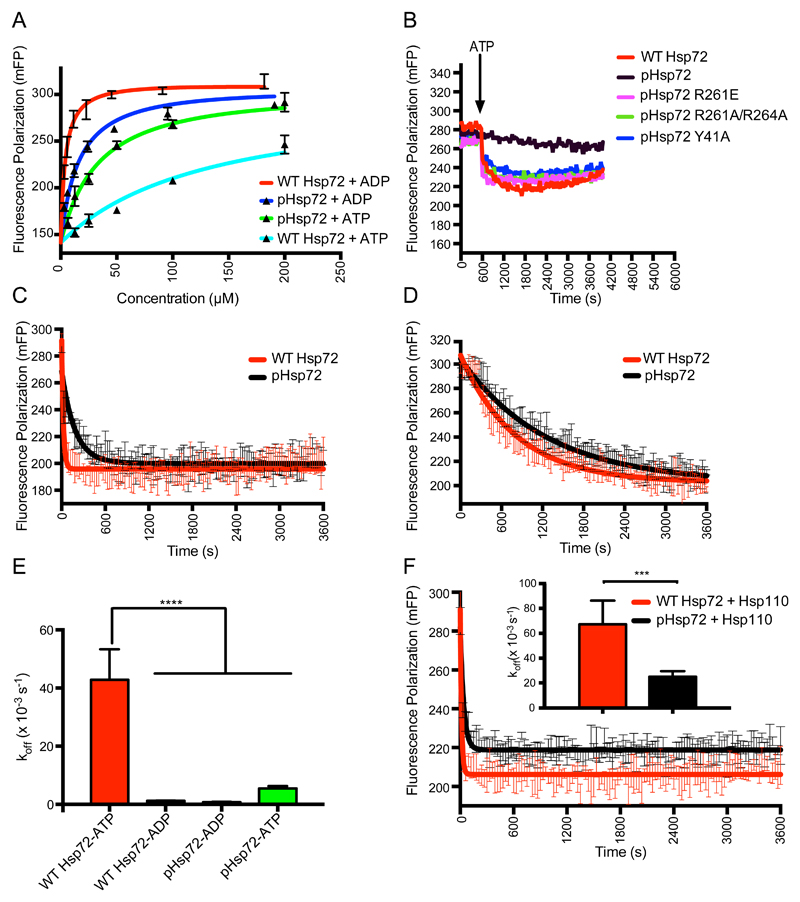
We investigated the biochemical consequences of phosphorylation of residue 66 using full-length Hsp72 proteins with or without incorporation of pSer66. First, we used isothermal titration calorimetry (ITC) to measure the affinity for nucleotide, using samples of Hsp72 purified under conditions in which nucleotide had been stripped out using ethylenediaminetetraacetic acid (EDTA) (Fig. 2) (26). We found that pHsp72 binds ADP ten times more tightly than does unphosphorylated Hsp72 [dissociation constant (Kd) 31 nM vs 303 nM], and ATP twice as tightly (Kd 0.49 μM vs 0.93 μM) (Fig. 2). Similarly, pHsp72 binds AMP-PNP (adenylyl-imidodiphosphate) 3.6-fold stronger than does unphosphorylated Hsp72 (Kd 2.47 μM vs 9 μM) (Fig. 2). Altogether, these phosphorylation-dependent effects are consistent with the known conformational changes that Hsp70 family proteins undergo when they bind nucleotide: the nucleotide-free enzyme has a conformation in which subdomains IB and IIB are separated, whereas these sub-domains come together in nucleotide-bound states (27).
Figure 2. Phosphorylation of Hsp72 enhances nucleotide binding.
(A) Isothermal titration calorimetry (ITC) analysis of the interaction of ADP, ATP and AMP-PNP with Hsp72, unphosphorylated (top images) and phosphorylated (bottom images). (B) ITC data for interactions between Hsp72 proteins with various nucleotides.
Hsp72 phosphorylation uncouples ATP binding by the NBD from substrate release by the SBD
In general, ADP-bound Hsp70 proteins have high affinity for substrate, whereas ATP-bound Hsp70 proteins have low affinity for substrate, as a consequence of the allosteric coupling of the NBD and SBD (18, 19). We investigated the consequences of phosphorylation at residue 66 of Hsp72 on the binding affinity of fluorescein-labeled NR (F-NR) peptide, using a fluorescence polarization assay. In the presence of ADP, unphosphorylated Hsp72 bound to the peptide substrate with a Kd of 3.95 μM, whereas the equivalent interaction of pHsp72 was 3.1-fold weaker (Fig. 3A). ATP weakened the interaction of unphosphorylated Hsp72 by a factor of 40 (Kd 157.1 μM) (Fig. 3A), consistent with the role of ATP in promoting substrate release. The binding of pHsp72 to substrate was much less sensitive to nucleotide state, with only a 2.2-fold difference in affinity (Kd values of 12.4 μM and 27.6 μM for ADP and ATP states, respectively) (Fig. 3A). This suggests that ATP would only have a limited effect on stimulating the release of peptide from pHsp72. We also carried out the binding experiments with an additional fluorescein-labeled substrate peptide (F-ALLQ) (28)(Fig. S2A). The results for F-ALLQ mirrors those obtained for the NR peptide substrate, suggesting a general feature of pHsp72 irrespective of the substrate (Fig. S2A).
Figure 3. Phosphorylated Hsp72 binds the substrate irrespective of its nucleotide state.
(A) Binding affinity of NR-peptide with unphosphorylated and phosphorylated Hsp72 proteins calculated using fluorescence polarization assays. Kd values were calculated to be: WT Hsp72 + ADP: 3.95 ± 0.42 μM;WT Hsp72 + ATP: 157.1 ± 16.48 μM; pHsp72 + ADP: 12.36 ± 2.66 μM; and pHsp72 + ATP: 27.55 ± 5.36 μM. (B) Hsp72 proteins were incubated with F-NR peptide in presence of ADP. WT Hsp72 is unphosphorylated, pHsp72 proteins incorporate pS66 and mutations are indicated in the key. After binding reached equilibrium, ATP was added (indicated by an arrow), and the release of fluorescently-labeled - NR was monitored over time. (C) Kinetics of fluorescently-labeled NR peptide release upon addition of ATP and an excess of unlabeled peptide, compared between unphosphorylated Hsp72 and pHsp72. Calculated values of koff (× 10-3 s-1): 42.82 ± 4.7 (unphosphorylated Hsp72); 5.47 ± 0.35 (pHsp72). Results are shown as mean ± SEM, based on data from five assays. (D) Kinetics of fluorescently-labeled NR peptide release upon addition of an excess of unlabeled peptide, compared between unphosphorylated Hsp72 and pHsp72. Calculated values of koff (× 10-3 s-1): 1.23 ± 0.04 (unphosphorylated Hsp72); 0.79 ± 0.03 (pHsp72). Results are shown as mean ± SEM, based on data from four experiments. (E) Comparison of peptide release rates as calculated in (C and D). Results are shown as mean ± SEM, ****P < 0.0001 by one-way ANOVA with Newman-Keuls post-hoc analysis. (F) Kinetics of fluorescently-labeled NR peptide release upon addition of ATP and an excess of unlabeled peptide, compared between unphosphorylated Hsp72 and pHsp72 in the presence of Hsp110. Calculated values of koff (× 10-3 s-1): 67.3 ± 7.7 (unphosphorylated Hsp72); 25.2 ± 1.7 (pHsp72). Results are shown as mean ± SEM, based on data from six experiments. Inset: Comparison of peptide release rates, shown as mean ± SEM based on data from six experiments, ***P < 0.001 by unpaired t-test.
We next probed the consequences of phosphorylation on the allosteric coupling between the nucleotide-bound state of NBD and substrate binding to the SBD. F-NR peptide was first bound to Hsp72 proteins (unphosphorylated WT, pHsp72 and mutated versions of pHsp72) in the presence of ADP and incubated for several hours to allow binding to reach equilibrium (29). ATP was then added, and fluorescence polarization recorded to follow the status of F-NR substrate binding (Fig. 3B). While the majority of the bound F-NR peptide was released spontaneously upon ATP addition to the sample of unphosphorylated Hsp72, the release of bound peptide from pHsp72 was significantly reduced (Fig. S7, A and B). We also tested F-ALLQ peptide in the same experimental setup and the results were similar to those observed with the F-NR peptide (Fig. S2B). This suggests that phosphorylation of pSer66 locks Hsp72 in a substrate-bound conformation regardless of the ATP or ADP status of the NBD. Allosteric coupling of substrate release to ATP binding is rescued by the mutation of residues Tyr41, Arg261 or Arg264, which are involved in the stapling of the sub-domains IB and IIB in pHsp72. Furthermore, we measured the kinetics of release of the fluorescent substrate in the presence of the unlabeled peptide for the ATP and ADP states (Fig. 3C-D). In the ATP state, the release rate was ~8 times slower for pHsp72 than the unphosphorylated chaperone, while it was only ~1.5 times slower in the ADP state (Fig. 3E). To test if the slow rate of substrate release by pHsp72 in the presence of ATP is due to loss of allosteric coupling between the NBD and SBD, we carried out the release assays with previously identified mutations that block the allostery between the NBD and the SBD (30). All these residues tested for the mutational analysis (Leu391, Leu392, Leu393 and Asp395) are located on the inter-domain linker, and are critical for inter-domain communication. We found that pHsp72 behaves similar to the mutants which lack allosteric coupling between the NBD and SBD (Fig. S3A-C). Thus, we conclude that the reduced kinetics of the substrate release by pHsp72 occurs due to the loss of allosteric coupling.
Nucleotide exchange factors (NEFs), such as Hsp110, stimulate the release of substrate from ADP-bound Hsp72, by facilitating the re-binding of ATP (31) To test the effect of NEF, we performed the ATP mediated substrate release experiment in the presence of Hsp110 and unlabeled substrate peptide (Fig. 3F). We found that the rate of peptide release in the presence of Hsp110 was ~2.7 fold slower in pHsp72 compared to Hsp72.
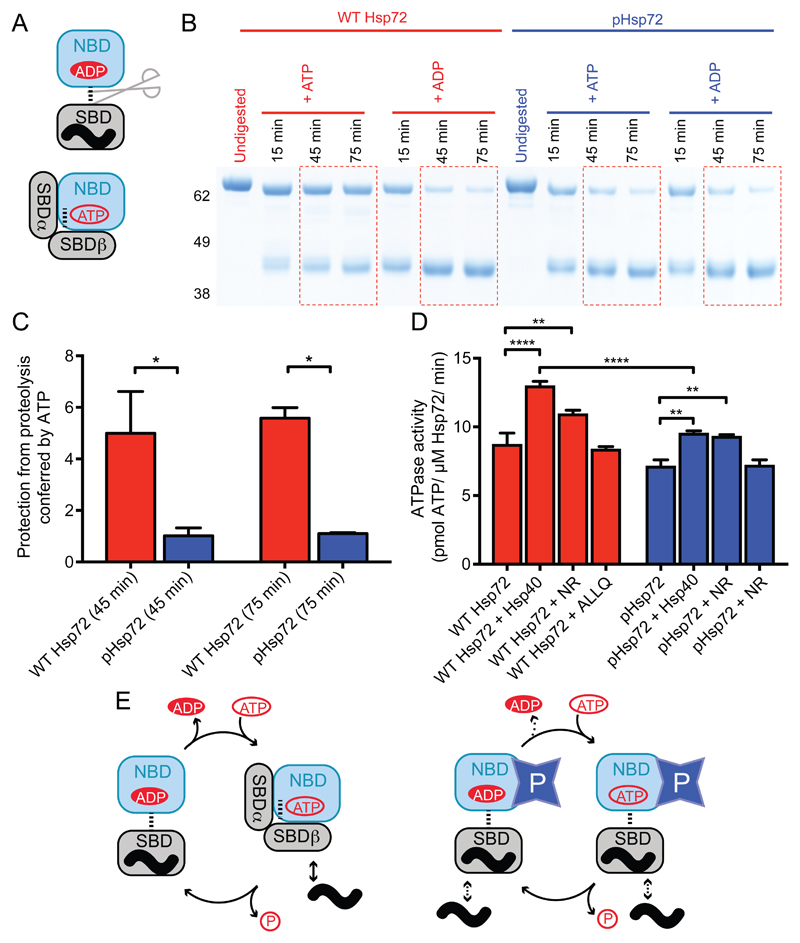
Substrate binding is coupled to nucleotide state through the linker region that connects the NBD and SBD. When ADP is bound, the linker is accessible and the SBD adopts a conformation that encloses the substrate (Fig. S4A) (15, 32). ATP binding to the NBD generates a binding site for the linker with the NBD, causing a conformational change in the SBD that releases the substrate (33, 34). The two states of the linker can be probed using proteinase K digestion, which cleaves the exposed linker of ADP-bound Hsp72 but not the buried linker of ATP-bound Hsp72 (Fig. 4A) (32). Here, we found that pHsp72 was sensitive to proteinase K in the presence of ADP or ATP, whereas the linker of unphosphorylated Hsp72 was protected from digestion by ATP as expected (Fig. 4B,C).
Figure 4. Phosphorylated Hsp72 has an undocked NBD-SBD linker irrespective of nucleotide state.
(A) Schematic illustration of the connection between proteinase K sensitivity and nucleotide state in Hsp72. When ADP is bound to the NBD, the SBD has high affinity for substrate and the linker is in an exposed, undocked conformation and sensitive to digestion by proteinase K. When ATP is bound to the NBD, the linker is docked and protected from digestion. (B) Representative SDS-PAGE gel showing the proteolysis of unphosphorylated Hsp72 and pHsp72 by proteinase K in the presence of ADP or ATP at various time points. (C) Degree of protection from proteolysis conferred by ATP is presented as the ratio of the fraction of 70 kDa species in presence of ATP to the fraction in the presence of ADP ([%70 kDa(+ATP)]/[%70 kDa(+ADP)]) at equivalent time points of 45 minutes and 75 minutes respectively. Results are shown as mean ± SEM based on 3 experiments, * p< 0.05 by one-way ANOVA with Newman-Keuls post-hoc analysis. (D) ATPase assay to quantify the effect of phosphorylation on Hsp72 activity. Results are shown as mean ± SEM from five experiments. ** p< 0.01, ****p <0.0001 by one-way ANOVA with Newman-Keuls post-hoc analysis. (E) Schematic diagrams showing the canonical ATPase cycle in Hsp72 (left), and how we propose this to be altered by phosphorylation of residue 66 (right). Phosphorylation stabilizes Hsp72 in a conformational state in which the linker is exposed and thus sensitive to digestion by proteinase K irrespective of whether ADP or ATP is bound.
To test the effect of phosphorylation on the catalytic activity of Hsp72, we measured the ATPase activity of unphosphorylated Hsp72 and pHsp72 in the absence and presence of a J-domain protein, Hsp40, as well as two substrate peptides, NR and ALLQ (Fig. 4D). The ATPase rate of unphosphorylated and pHsp72 were comparable, with the latter being ~1.2 fold higher than the former. We noted that Hsp40 stimulated the ATPase activity of unphosphorylated Hsp72 more significantly than pHsp72 (Fig. 4D). The J-domain of Hsp40 enhances the ATPase activity of Hsp70 through a highly dynamic interaction (35–37). The addition of NR peptide stimulated the ATPase activity of both unphosphorylated as well as phosphorylated Hsp72 (Fig. 4D). However, there was no significant enhancement in the presence of ALLQ peptide. This indicates that length and sequence of the substrate peptide plays a significant role in stimulation of ATPase activity, as shown in a previous study (38). Putting our biochemical and structural observations together, we sketched out a model to illustrate how phosphorylation of residue 66 of Hsp72 affects the ATPase/substrate cycle. Unphosphorylated Hsp72 cycles between an ADP-bound/high-affinity state and an ATP-bound/low-affinity state (Fig. 4E). Phosphorylation of Hsp72 reduces the ATPase activity and increases the affinity for ADP, both of which will reduce the efficiency of the ATPase cycle. Most striking of all, allosteric coupling between nucleotide binding in the NBD and substrate binding in the SBD is lost in pHsp72, which is in a linker-exposed, substrate-binding conformation regardless of whether ADP or ATP is present (Fig. 4E, fig. S4B).
The decoupling of ATP binding from substrate release enhances spindle pole localization of Hsp72
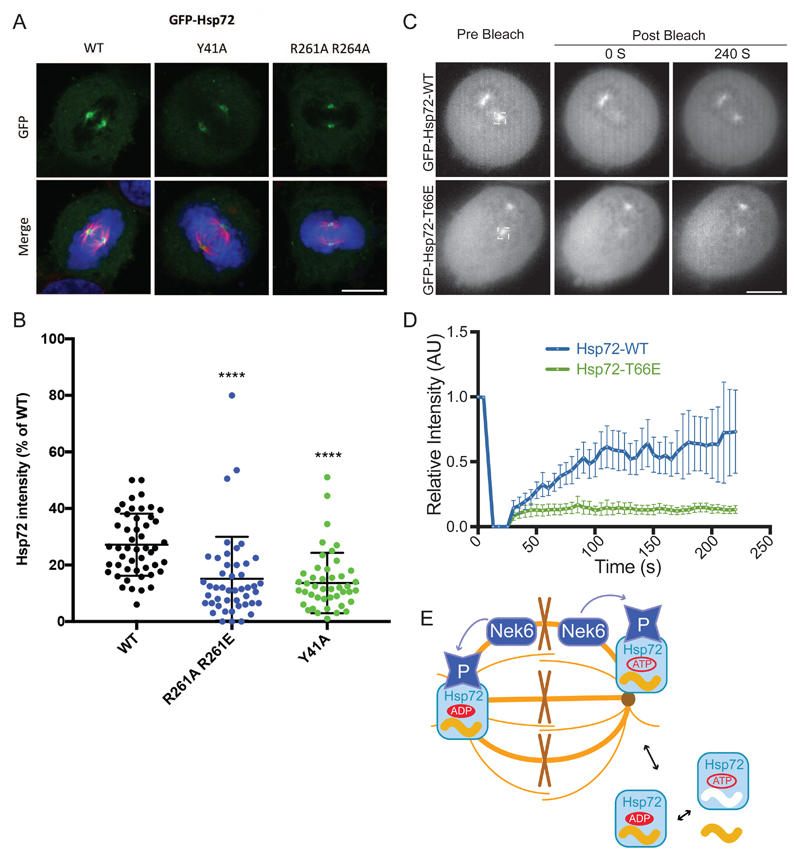
We previously showed that phosphorylation by Nek6 targets Hsp72 to the mitotic spindle, mainly to the spindle poles (9). Among other observations, we showed that siRNA knockdown of Nek6 depleted spindle-associated Hsp72 to 20% of control, and that a T66A mutant did not localize to the spindle (9). Here, we investigated the hypothesis that the spindle localization of pHsp72 is due to stabilized interactions of the SBD with spindle-associated substrates that result from uncoupling of ATP binding from substrate release. To test this hypothesis, we analyzed the localization of Hsp72 proteins with mutations in residues Tyr41, Arg261 and Arg264 that restore this coupling in the context of pHsp72. The localization of Green Fluorescent Protein (GFP)-tagged Hsp72 proteins was determined by confocal microscopy in mitotic cells following synchronization using a standard, double-thymidine block and release protocol (Fig. 5A, fig. S5A). We expressed GFP-tagged Hsp72 proteins in HeLa cells and verified that the mutants had the same abundance as wild-type protein and were phosphorylated to the same extent on Thr66 (Fig. S5B). Unfortunately, the expression of the R261A mutant was reduced, and so we focused on the Y41A single-point mutant and R261A/R264A double-point mutant. We further verified that the mutants were functionally active as they bind NR-peptide at similar affinities compare to WT (Fig. S5C). Both Hsp72 mutants showed localization to the mitotic spindle poles, but at only of 13-14% of total Hsp72, compared to 27% of wild-type protein (Fig. 5B, fig. S5D). Thus, we conclude that efficient localization of Hsp72 to the mitotic spindle is a consequence of uncoupling ATP binding from substrate release, which stabilizes the interactions with substrate proteins on the spindle. We investigated the dynamics of Hsp72 association with spindle poles using fluorescence recovery after photobleaching (FRAP; Fig. 5C,D). Wild-type GFP-Hsp72 exhibited a moderate rate of recovery, reaching ~50% of the initial intensity after 100 s. In contrast, the phosphomimic Hsp72 T66E mutant exhibited very little recovery. These results are consistent with spindle localisation of Hsp72 being dependent on phosphorylation of Thr66 and suggest that release from the spindle requires dephosphorylation of Hsp72 (Fig. 5E). Previous work showed that the Hsp72 inhibitor VER-155008 disrupted spindle assembly (9). Here we probed the biochemical mechanism, and found that the inhibitor did not affect the binding of peptide substrate but reduced the ATP substrate release rate by ~ 8.6 fold in unphosphorylated Hsp72, while ATP-mediated rate of release of substrate peptide was increased by ~ 2.6 (Fig. S7, C to E). This suggests that VER-155008 might decrease the spindle localization of pHsp72 by increasing the rate of release of pHsp72 from its client protein substrates on the spindle. Further studies are required to investigate this hypothesis and to identify the relevant substrates.
Figure 5. Localization of pThr66-Hsp72 to the mitotic spindle is dependent on the interaction between IB-IIB.
(A) HeLa cells were subjected to a double thymidine block and transfected with GFP-Hsp72 constructs as indicated during the first thymidine release. Cells were pre-extracted and fixed with ice-cold methanol 8 hours after release from the second thymidine block. Cells were immunostained with α-tubulin (red). Merge image includes DNA stained with Hoechst 33258 (blue). Scale bars, 10 μm. (B) Dot plot shows the intensity of GFP-tagged WT and mutant Hsp72 at individual spindle poles as a percentage of total cellular GFP intensity. Upper and lower lines show the interquartile range and the middle line represents the median. Data represent measurements taken from at least 60 cells. ****, P<0.0001 in comparison to GFP-Hsp72 WT spindle pole intensities by unpaired t-test. (C) FRAP was performed on HeLa:GFP-Hsp72-WT and –T66A as indicated. A region of interest corresponding to one spindle pole was bleached per cell and stills from representative videos of cells immediately prior to and after bleaching are shown. Scale bar, 10 μm. (D) The relative GFP intensity within the bleached area was recorded as a measure of time. Data are average intensity ± SD for 4-5 cells from 2 experiments. (E) Model showing how phosphorylation of Hsp72 by Nek6 is related to spindle localization. Cytoplasmic Hsp72 cycles between substrates bound and free forms, coupled to ATP binding and hydrolysis. Phosphorylation of Hsp72 on Thr66 by Nek6 in the vicinity of the spindle stabilizes its interactions with spindle-associated proteins. We postulate that release of Hsp72 from the spindle requires dephosphorylation.
Discussion
The importance of subdomain interactions and closure of the nucleotide binding site have been previously linked to the ADP-ATP exchange cycle and communication with the substrate binding domain in Hsp70 family proteins, as exemplified by studies on bacterial DnaK (33, 34, 39, 40). In the ADP-bound state, the NBD and SBD of Hsp70 act as two more-or-less independent domains, connected by a flexible linker. The SBD adopts a closed conformation with high affinity for substrate. Upon the exchange of ADP for ATP, the linker docks onto the NBD and brings the SBD into close association with the NBD. SBDα and SBDβ are separated, releasing substrate, and SBDα forms an extensive interaction with the NBD, notably including with subdomain IB. Solution-based studies have clarified that this is not a completely stable conformation, and that a significant proportion of ATP-bound Hsp70 resembles the ADP-bound state. Our working hypothesis is that phosphorylation of Thr66 increases this proportion by disfavoring the interactions between the linker and the NBD, which in turn decreases the interaction between the SDB with the NBD. These changes occur through allosteric mechanisms propagated from the altered IB conformation and stabilized IB-IIB interface. Furthermore, several lines of evidence reveal that ATP binding leads to rotations of lobe I relative to lobe II, which transduce the allosteric signal from NBD to SBD via the linker (34, 41). Phosphorylation of Thr66 stabilizes the interaction between the two lobes and antagonizes the transduction of the allosteric signal between NBD and SBD. On the other hand, it is less clear to us how phosphorylation might decrease the binding of substrate in the ADP-bound state of pHsp72 relative to the unphosphorylated protein. These questions, which relate to the dynamics of Hsp72, are perhaps best resolved using a combination of solution-based biophysical methods.
ATP is maintained at very high concentrations of ~5 mM in cells (42). At such high concentrations of ATP, unphosphorylated Hsp72 will undergo the canonical ATPase cycle resulting in rapid substrate binding and release phases coupled with the exchange of ADP for ATP. On the contrary, pHsp72 is likely to have more stable interactions with its substrates in the presence of high concentrations of ATP due to the uncoupling of ATP binding from substrate release. This possibly explains the increased localization of pHsp72 on the spindle. Hsp72 contributes to robust mitotic spindle assembly by enabling the recruitment of proteins such as TACC3 and ch-TOG that form intermicrotubule bridges and inhibition of Hsp72 leads to mitotic defects (9, 43). The mechanism by which pHsp72 contributes to this process is presently unclear, but we speculate that it stabilizes the unfolded state of hydrophobic peptides within spindle-associated proteins, and perhaps assists in the assembly of their protein complexes.
The function of protein phosphorylation in cell regulation is perhaps best understood in terms of regulating protein-protein interactions with modules such as SH2, FHA, 14-3-3 or BRCT domains that directly recognize the phosphorylated side chain (44, 45). Phosphorylation has a distinct role in the stimulation of catalytic activity of protein kinases, most commonly through phosphorylation of the activation loop to stabilize a catalytically-competent state (46, 47). Although at least 313 phosphosites have been identified within Hsp70 proteins across the 11 species, the purpose behind only a few of these sites have been experimentally investigated, and the mechanistic consequences of these events has not been probed through structural and biochemical approaches on proteins with defined phosphorylation states (20–22). The capability to genetically encode phosphorylation allows us to precisely address such questions, opening up new avenues to explore the phospho-regulation of Hsp70, or indeed any other protein that can be generated through recombinant methods. Here, we have discovered a role for phosphorylation in the regulation of a molecular chaperone, Hsp72, which decouples the binding of nucleotide and substrate, and enhances nucleotide binding, through stabilization of the interface between two subdomains.
Materials and methods
DNA manipulation
For site-specific incorporation of phosphoserine at the residue 66 on Hsp72, we applied the genetic code expansion method that utilizes an orthogonal aminoacyl-tRNA synthetase/tRNACUA pair to incorporate phosphoserine at the amber codon on the coding DNA sequence (24). An amber codon was introduced at residue-66 position by site-directed mutagenesis. Rhinovirus 3C protease-cleavable N-terminal His6 tagged versions of Hsp72 (full-length or NBD (aa 1-381)) containing an amber codon at position 66 were then amplified by PCR and subcloned in pNHD plasmid between BsrG1 and Xho1. The Hsp72 mutations, viz., Y41A, R261A, R261E, R261A/R264A were introduced by standard site-directed mutagenesis protocols. The expression constructs of unphosphorylated Hsp72 were made in pTwo-E plasmid that contains a Rhinovirus 3C protease-cleavable N-terminal His6 tag.
Expression and purification
For overexpression of Hsp72 and mutants with incorporation of phosphoserine at position 66, pNHD-His6-Hsp72 constructs were transformed in BL21 ΔserB(DE3) cells containing pKW2-EF-Sep (a chloramphenicol resistant plasmid containing SepRS 2, pSer tRNAB4CUA and EF-Sep) (24). Colonies were then grown over-night in Terrific broth (TB) containing 30 μg/ml Chloramphenicol and 15 μg/ml Tetracycline. The over-night cultures were used to inoculate 6 x 1 L of TB media containing appropriate antibiotics and 2 mM phosphoserine. The cultures were expressed at 37°C, 22 0 rpm and induced by adding 450 μM isopropyl β-D-1-thiogalactopyranoside and 2 mM phosphoserine. They were further incubated for 16 hours at 18°C, 220 rpm. The cells were harvested by centrifugation. Non-phosphorylated His-tagged Hsp72 proteins were expressed in E.coli using standard procedures. His-tagged Hsp72 proteins were initially purified using a HisTrap (GE Healthcare) affinity column equilibrated in 50 mM Hepes-KOH pH 7.5, 250 mM NaCl, 5% glycerol, 1 mM β-mercaptoethanol (BME) and an imidazole gradient (20-400 mM). The protein was extensively dialysed against 20 mM Hepes-KOH pH 7.5, 300 mM NaCl, 5 mM EDTA, 8% glycerol, 1 mM BME at 4°C and the N - terminal His6-tag removed by glutathione-S-transferase (GST)-tagged 3C Protease. The 3C protease was removed by pass-back over GST resin, and the protein further purified by anion exchange using 5ml Mono-Q (GE Healthcare) and size-exclusion chromatography using Superdex-200 16/60 (GE Healthcare). His-tagged Hsp40 and Hsp110 were expressed in E.coli and purified by Ni-affinity chromatography followed by size-exclusion chromatography using Superdex-200 16/60 (GE Healthcare). pET28a-Hsp110 plasmid was a gift from Dr. E. Lafer, Department of Biochemistry, The University of Texas Health Science Center, USA. The final storage buffer was 20 mM Hepes-KOH, 400 mM KCl, 5% glycerol, 10 mM MgCl2, 5 mM DTT.
Molecular mass measurement by LC-MS
The intact mass of pHsp72-NBD was confirmed by liquid chromatography-mass spectrometry. Briefly, 1 μL of 5 μM sample was loaded onto a MassPREP protein desalting column (Waters UK, Manchester, UK), washed with 10% acetonitrile in water for 5 min and then eluted into the mass spectrometer (Xevo G2-XS Q-TOF, Waters UK, Manchester, UK) in 90% acetonitrile in water. The MS was operated in positive TOF mode.
Gel processing, tryptic digestion, LC-MS/MS and database searching
The phosphorylation site was confirmed by trypsin digestion of the protein followed by LC-MS/MS analysis, database searching and manual confirmation. Briefly, gel bands of pHsp72-NBD were excised, destained in 30% ethanol, reduced with DTT and alkylated with iodoacetamide. Trypsin digestion was performed for 18 h at 37 °C. The tryptic peptides were separated by LC us ing a C18 stationary phase and a water/acetonitrile gradient (1%-60% acetonitrile in 40 min). Eluting peptides were directed to the mass spectrometer (Xevo G2-XS Q-TOF, Waters UK, Manchester, UK) for mass measurement and fragmentation. The MS and MS/MS data was searched against the Uniprot database (release 2016_09) with the sequences of pHsp72-NBD manually appended. Carbamiodomethylation was selected as a fixed modification, variable modifications were set for oxidation of methionine and deamidation of glutamine and asparagine and phosphorylation at S, T and Y residues. MS/MS spectra for candidate phosphorylated peptides identified by the database search were manually interpreted and annotated to confirm the phosphorylation site.
Protein crystallization
pHsp72-NBD (10mg/ml) was mixed with 5mM ADP (Calbiochem) prior to crystallization by sitting drop vapour diffusion method. Diffraction quality crystals were obtained in 0.1 M Succinic acid pH 7.0, 15% w/v Polyethylene glycol 3,350. Crystals were briefly soaked in reservoir solution supplemented with 20% ethylene glycol and flash frozen in liquid nitrogen.
Crystallography
Diffraction data were collected at Diamond Light Source, Didcot, UK at beamline I04 at a temperature of 100 K and a wavelength of 0.9795Å. Auto-processed data were used directly for molecular replacement using PHASER (48) with Hsp72 NBD (PDB 3AY9) (26) as the search model. Two molecules were located in the asymmetric unit. Subsequent iterative refinement cycles were performed using PHENIX (49) and COOT (50). Geometry was good (99% Ramachandran favoured, 0% outliers) as assessed using MolProbity (51).
Isothermal Titration Calorimetry
Unphosphorylated Hsp72 and pHsp72 proteins were extensively dialyzed in 20 mM Hepes-KOH pH 7.5, 300 mM KCl, 5 mM EDTA, 5% glycerol and 5 mM BME. For binding measurements with ADP, the Hsp72 proteins were buffer exchanged into the ITC buffer (30 mM Hepes-KOH pH 7.5, 350 mM KCl, 5% glycerol, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 5 mM MgCl2) using Superdex-200 16/60 column. For testing the binding with ATP, Hsp72 proteins were buffer exchanged into Mg2+ free ITC buffer to minimize the hydrolysis effect. The binding measurements with AMP-PNP were carried out similarly to replicate the ATP binding conditions. ITC binding studies were performed with an iTC200 instrument (MicroCal, GE Healthcare). 200-400 μM nucleotides (syringe) was titrated against 15-30 μM Hsp72 proteins (cell) over 20 injections and the data fitted to a one-site binding model using the Origin software package v7.0 supplied by Microcal.
Malachite Green ATPase assays
Experiments were performed according to the published protocol (52, 53). A final concentration of 6 μM Hsp72 proteins in the absence or presence of 400 μM NR or ALLQ –peptide or 30 μM Hsp40 were used for assaying activity. Reactions were carried out in clear 96-well plates, and 1 mM ATP used to initiate the reactions. The reactions proceeded for 1 h at 37 °C, before developing with malachite green reagent and quenc hing with sodium citrate: plate absorbance was measured at 620 nm. A phosphate standard curve was used to calculate pmol ATP hydrolysed/μM Hsp72/min.
Kinetics of substrate peptide release
50 μM of Hsp72 proteins were incubated with 40 nM F-NR at room temperature in the presence of 125 μM ADP in Buffer A (25 mM Hepes–KOH pH 7.5, 150 mMKCl, 10% glycerol, 10 mM MgCl2, 2 mM DTT) for more than 5 hours to allow binding to reach equilibrium. Then, unlabelled NR was added to a final concentration of 150 μM to compete off the bound F-NR. Fluorescence polarization was measured over time to record the release of F-NR using a Victor X5 plate reader (PerkinElmer) at 25°C with excitation at 490 nm and emission at 535 nm. Kinetic rates were calculated using Prism 7 (GraphPad).
ATP mediated substrate release assay
To assay peptide release upon ATP binding, we incubated 50 μM of Hsp72 proteins with 20 nM F-NR or F-ALLQ peptides at room temperature in the presence of 125 μM ADP in Buffer A. After several hours, binding reached equilibrium. Then, ATP was added to a final concentration of 2 mM. Further experiments included 150 μM substrate peptide, 125 μM Hsp110, 100 μM VER-155008. Fluorescence polarization was measured before ATP addition and then every 30s for 4000s after ATP addition. Fluorescence polarization measurements were made as above.
Proteinase K digestion assay
50 μl reaction mixtures containing 10 μg of unphosphorylated or pHsp72 and 2 μg of proteinase K were incubated in the presence of 200 μM ATP or ADP at room temperature at time points of 15 minutes, 45 minutes and 75 minutes. The reactions were stopped by adding 1 μl of 0.1 M PMSF followed by 1-hour incubation on ice before SDS-PAGE analysis.
Peptide binding assays
For measuring the binding in the presence of ADP, Hsp72 proteins were serially diluted in Buffer A containing 125 μM ADP. For measuring the binding in the presence of VER-155008 or DMSO, Hsp72 proteins were serially diluted in Buffer A containing a final concentration of 100 μM VER-155008. Control assays were carried out in with Hsp72 protein containing DMSO alone. F-NR or F-ALLQ peptide was added to a final concentration of 20 nM and incubated for several hours at room temperature to reach equilibrium. Fluorescence polarization measurements were made using the Victor X5 plate reader (Perkin Elmer) at 25°C with excitation at 490 nm and emission at 535 nm. The dissociation constant Kd was calculated by fitting the fluorescence anisotropy data to a one-site binding equation by nonlinear regression analysis using Prism 7 (GraphPad). Binding studies in the presence of ATP were performed according to published protocols (54). Hsp72 proteins were first serially diluted in Buffer A. ATP was added to a final concentration of 2 mM, and incubated for 2 min to allow ATP binding. F-NR or F-ALLQ peptide was added to a final concentration of 20 nM, and fluorescence polarization measurements were carried out after 10 minutes incubation since it usually takes 10 min for the binding to reach equilibrium but before any significant ATP hydrolysis occurs (54).
Cell culture, synchronisation and transfection
HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with GlutaMAX (Invitrogen) containing 10% heat-inactivated Fetal Bovine Serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 atmosphere. Cell synchronisation was carried out using a double-thymidine block. Briefly, cells at 30% confluency were treated with 2 mM thymidine in DMEM supplemented with 10% FBS for 18 hours. Cells were released in normal growth media for 9 hours before addition of 2 mM thymidine for a further 17 hours. Transfections were carried out with FuGENE HD (Promega) according to the manufacturer’s instructions. For localisation studies during mitosis, transfection mix was added to the cells after release from the first thymidine block.
Indirect immunofluorescence microscopy
Cells grown on acid-etched glass coverslips were pre-extracted for 15 seconds (1 mM MgCl2, 1 mM egtazic acid, 80 mM piperazine-N,N’-bis(2-ethanesulfonic acid), 0.1% Triton X-100), washed with PBS and fixed with ice-cold methanol and incubated at -20°C for at least 15 minutes. Cells were rehydrated with phosphate-buffered saline (PBS) and blocked with 1% bovine serum albumin (BSA) in PBS for 30 minutes before incubation at room temperature for 1 hour, with an antibody that recognises alpha tubulin (1.2 μg/ml, Sigma) diluted in 3% BSA in PBS. After three 5 minute washes with PBS, coverslips were incubated with secondary antibody Alexa Fluor 594 goat antibody that recognises mouse IgG (10 μg/ml; Invitrogen) and Hoechst 33258 (0.8 μg/ml) diluted in PBS with 3% BSA. Imaging was performed on the Zeiss LSM880 upright confocal microscope using a 40x oil objective (numerical aperture, 1.4). Z-stacks comprising 15 0.45 μm sections were acquired using Zen 2.1 software (Zeiss).
Intensity measurements and data analysis
Intensity of Hsp72 at the spindle pole was measured using maximum intensity projections obtained using constant exposure times and gain settings for each experimental repeat. A 50 by 50 pixel region of interest was positioned over the spindle pole and whole cell background was measured using a 256 pixels x 256 pixels circular box, and the values used to calculate the mean fluorescence intensity at the spindle pole as a percentage of the total. Measurements were performed using Image J software.
Turnover of Hsp72 at spindles determined by FRAP
GFP-Hsp72-WT and T66E constructs were generated by sub-cloning Hsp72-WT and Hsp72-T66E PCR fragments into pLEICS-138. HeLa:GFP-Hsp72-WT and T66A cell lines were generated by transfecting the relevant construct and selecting cells in media supplemented with 2 mg/ml G418 over several days. FRAP was performed with a VisiTech Infinity3 confocal laser scanning microscope. Regions of interest (ROIs) of 3 μm by 3 μm were bleached with 2 iterations and 100% laser power. Two images were captured prior to bleaching and 40 images were obtained after bleaching at intervals of 5 s.
Statistical Analysis
Comparison of peptide release rates (Fig. 3E, S3C, S7B, S7E), proteolysis protection (Fig. 4C) and ATPase rates (Fig. 4D) were done by one-way ANOVA with Newman-Keuls post-hoc analysis (* p< 0.05, ** p< 0.01, ***p < 0.001, ****p < 0.0001. Comparison of peptide release rates (Fig. 3F) and intensities of GFP fluorescence (Fig. 5B) were done by unpaired t-test.
Supplementary Material
One-sentence summary.
Phosphorylation uncouples the functions of the chaperone Hsp72 during mitosis.
Editor’s summary.
Modified Hsp72 structure-function in mitosis
Chaperone proteins support protein folding, mediate the formation of multiprotein complexes, and enable protein degradation. The functions of the chaperone Hsp72 are coupled; that is, nucleotide exchange triggers the release of bound substrate proteins. Upon phosphorylation, Hsp72 is localized to the mitotic spindle in dividing cells. Changes in Hsp72 abundance or activity are implicated in aging, inflammation and cancer. Mukherjee et al. used a modified Hsp72 construct to examine structure-function relationships affecting nucleotide exchange and substrate binding during mitosis. Crystal structure analysis showed that phosphorylation induced conformational changes in Hsp72 that enabled nucleotide exchange but maintained substrate binding, which were critical to its function at the mitotic spindle. Exploring ways to mimic or disrupt phosphorylation-dependent changes in Hsp72 may lead to new research and therapeutic tools.
Acknowledgments
We thank the beamline scientists of Diamond I04, Astbury support scientists Iain Manfield and Chi Trinh for technical assistance, Sharon Yeoh for assistance with manuscript and figure preparation and Dr. E. Lafer (UTHSCSA) for her kind gift of pET28a-Hsp110 plasmid.
Funding: This work was supported by grants from BBSRC and CRUK grants to RB (BB/L023113/1 and C24461/A12772), and Worldwide Cancer Research to AMF (16-0119). The Xevo G2-XS Q-TOF mass spectrometer and liquid chromatography equipment was purchased with an Advanced Life Sciences Research Technology Initiative (ALERT 2014) grant from the BBSRC (BB/M012573/1).
Footnotes
Author contributions: MM, AF, AZ and RB designed the project. MM, SS, LOR, JS, MWR and JRA performed experiments and analysed data. NH-D and JWC designed methods and reagents to generate phosphoproteins. MM, SS, LOR, JS, MWR, AZ, AMF and RB contributed to writing the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Structure coordinates and data are deposited in the Protein Data Bank with accession code 6FHK. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 2.Ma HT, Poon RY. How protein kinases co-ordinate mitosis in animal cells. Biochem J. 2011;435:17–31. doi: 10.1042/BJ20100284. [DOI] [PubMed] [Google Scholar]
- 3.Caron D, Byrne DP, Thebault P, Soulet D, Landry CR, Eyers PA, Elowe S. Mitotic phosphotyrosine network analysis reveals that tyrosine phosphorylation regulates Polo-like kinase 1 (PLK1) Sci Signal. 2016;9:rs14. doi: 10.1126/scisignal.aah3525. [DOI] [PubMed] [Google Scholar]
- 4.Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18:187–201. doi: 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- 5.Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, Ivins F, Sessa F, Hudecz O, Nigg EA, Fry AM, et al. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayliss R, Fry A, Haq T, Yeoh S. On the molecular mechanisms of mitotic kinase activation. Open Biol. 2012;2:120136. doi: 10.1098/rsob.120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry AM, Bayliss R, Roig J. Mitotic Regulation by NEK Kinase Networks. Front Cell Dev Biol. 2017;5:102. doi: 10.3389/fcell.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Regan L, Sampson J, Fry AM. Hsp70 proteins in mitosis and disease. Oncotarget. 2015;6:32293–32294. doi: 10.18632/oncotarget.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Regan L, Sampson J, Richards MW, Knebel A, Roth D, Hood FE, Straube A, Royle SJ, Bayliss R, Fry AM. Hsp72 is targeted to the mitotic spindle by Nek6 to promote K-fiber assembly and mitotic progression. J Cell Biol. 2015;209:349–358. doi: 10.1083/jcb.201409151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prosser SL, O'Regan L, Fry AM. Novel insights into the mechanisms of mitotic spindle assembly by NEK kinases. Mol Cell Oncol. 2016;3:e1062952. doi: 10.1080/23723556.2015.1062952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem. 2003;278:34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- 12.O'Regan L, Fry AM. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol Cell Biol. 2009;29:3975–3990. doi: 10.1128/MCB.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 14.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 19.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim A, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- 22.Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, Kron SJ. CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell. 2012;151:1308–1318. doi: 10.1016/j.cell.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strack R. Efficient Generation of Proteins with Site-Specific Phosphoserines. Nat Methods. 2015;12:702–703. doi: 10.1038/nmeth.3512. [DOI] [PubMed] [Google Scholar]
- 24.Rogerson DT, Sachdeva A, Wang K, Haq T, Kazlauskaite A, Hancock SM, Huguenin-Dezot N, Muqit MM, Fry AM, Bayliss R, Chin JW. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowan FC, Richards M, Bibby RA, Thompson A, Bayliss R, Blagg J. Insights into Aurora-A kinase activation using unnatural amino acids incorporated by chemical modification. ACS Chem Biol. 2013;8:2184–2191. doi: 10.1021/cb400425t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arakawa A, Handa N, Shirouzu M, Yokoyama S. Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 2011;20:1367–1379. doi: 10.1002/pro.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci U S A. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci L, Williams KP. Development of fluorescence polarization assays for the molecular chaperone Hsp70 family members: Hsp72 and DnaK. Curr Chem Genomics. 2008;2:90–95. doi: 10.2174/1875397300802010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar DP, Vorvis C, Sarbeng EB, Cabra Ledesma VC, Willis JE, Liu Q. The four hydrophobic residues on the Hsp70 inter-domain linker have two distinct roles. J Mol Biol. 2011;411:1099–1113. doi: 10.1016/j.jmb.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 31.Rampelt H, Mayer MP, Bukau B. Nucleotide Exchange Factors for Hsp70 Chaperones. Methods Mol Biol. 2018;1709:179–188. doi: 10.1007/978-1-4939-7477-1_13. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. Pathways of allosteric regulation in Hsp70 chaperones. Nat Commun. 2015;6:8308. doi: 10.1038/ncomms9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kityk R, Kopp J, Mayer MP. Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol Cell. 2018;69:227–237 e224. doi: 10.1016/j.molcel.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Malinverni D, Jost Lopez A, De Los Rios P, Hummer G, Barducci A. Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis. Elife. 2017;6 doi: 10.7554/eLife.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan R, McMacken R. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- 39.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai AL, Clerico EM, Blackburn ME, Patel NA, Robinson CV, Borbat PP, Freed JH, Gierasch LM. Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J Biol Chem. 2017;292:8773–8785. doi: 10.1074/jbc.M116.770404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharya A, Kurochkin AV, Yip GN, Zhang Y, Bertelsen EB, Zuiderweg ER. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 43.Hood FE, Williams SJ, Burgess SG, Richards MW, Roth D, Straube A, Pfuhl M, Bayliss R, Royle SJ. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J Cell Biol. 2013;202:463–478. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can't pSERious? Structure. 2001;9:R33–38. doi: 10.1016/s0969-2126(01)00580-9. [DOI] [PubMed] [Google Scholar]
- 45.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr Opin Cell Biol. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 47.Steichen JM, Kuchinskas M, Keshwani MM, Yang J, Adams JA, Taylor SS. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J Biol Chem. 2012;287:14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 53.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J Biol Chem. 2014;289:1402–1414. doi: 10.1074/jbc.M113.521997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Nune M, Zong Y, Zhou L, Liu Q. Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP. Structure. 2015;23:2191–2203. doi: 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.