Abstract
Soil microorganisms control carbon losses from soils to the atmosphere1–3, yet their responses to climate warming are often short-lived and unpredictable4–7. Two mechanisms, microbial acclimation and substrate depletion, have been proposed to explain temporary warming effects on soil microbial activity8–10. However, empirical support for either mechanism is unconvincing. Here we used geothermal temperature gradients (> 50 years of field warming)11 and a short-term experiment to show that microbial activity (gross rates of growth, turnover, respiration and carbon uptake) is intrinsically temperature sensitive and does not acclimate to warming (+ 6 ºC) over weeks or decades. Permanently accelerated microbial activity caused carbon loss from soil. However, soil carbon loss was temporary because substrate depletion reduced microbial biomass and constrained the influence of microbes over the ecosystem. A microbial biogeochemical model12–14 showed that these observations are reproducible through a modest, but permanent, acceleration in microbial physiology. These findings reveal a mechanism by which intrinsic microbial temperature sensitivity and substrate depletion together dictate warming effects on soil carbon loss via their control over microbial biomass. We thus provide a framework for interpreting the links between temperature, microbial activity and soil carbon loss on timescales relevant to Earth’s climate system.
Soil-dwelling bacteria and fungi control the breakdown of organic matter in soil and its release as carbon dioxide (CO2) to the atmosphere1. Climate warming is expected to accelerate the activity of soil microbes, stimulating further CO2 release and a positive feedback to climate change2,3. A better understanding of microbial processes will likely improve climate change predictions15,16. Research in recent decades has thus sought to quantify the consequences of warming for soil microbes and the carbon cycle processes they govern, and to describe this using metrics such as microbial carbon use efficiency (CUE)17,18. However, the relationships between temperature and soil microbes remain apparently inconsistent in both space and time3–9, preventing consensus on the severity of feedbacks from microbial activity to future climate change.
Soil microbes degrade organic matter in soil, take up the carbon therein, allocate a portion to growth and release the remainder chiefly as CO2 through respiration19. The enzymatic reactions controlling these processes are intrinsically temperature sensitive20. However, warming effects on soil CO2 release are often short-lived5,9. First, warming begins a phase of accelerated respiration that causes excess CO2 release from soil. Then, within years of initiating warming, there is a deceleration of respiration and, in most cases (but see refs 4,7), a return to pre-warmed rates of soil CO2 release. The temporal dynamics of soil CO2 release will dictate the magnitude of soil carbon lost with climate warming. Nevertheless, the mechanisms behind it are the subject of intense scientific debate. Microbial communities may acclimate to sustained warming through physiological adjustments (e.g. CUE) or shifts in community composition9,21. Here we define microbial acclimation as a return of microbial activity towards pre-warmed rates over time. At the same time, accelerated microbial activity can cause substrate depletion, limiting resource availability and negatively impacting microbial processes8,10. Microbial acclimation and substrate depletion are not mutually exclusive and both may cause a deceleration of soil carbon loss under sustained warming. Despite this, microbial activity does not always attenuate to warming4,8, and neither mechanism can explain the variable patterns of warming-induced soil carbon loss observed at biome and global scales6,7. Indeed, while links between microbial activity and the carbon cycle have been repeatedly demonstrated, researchers have yet to quantify the interplay between temperature, microbial physiology and soil carbon loss over periods greater than hours to months. This is especially true for microbial growth, turnover and CUE, which until now have been estimated indirectly or using carbon substrates that bias the experimental system17,18,22. A mechanistic understanding is urgently needed to identify the role of microorganisms in warming-induced soil carbon loss and its importance over timescales relevant to the climate system.
We used the longest known in situ natural warming study (at least 50 years; ref 11) to determine the microbial mechanism responsible for warming-induced soil carbon loss. The study exploits natural geothermal activity in a sub-arctic grassland that has created gradients of warming from ambient temperature to + 6 ºC (n = 5). It provides a unique platform for assessing the long-term responses of microbes to warming in a region that holds large carbon stocks and is vulnerable to rapid temperature change1. We used direct, substrate independent, metrics of microbial physiology (gross rates of growth, respiration, turnover, organic carbon uptake and CUE; ref 23) to characterise microbial activity in ambient and warmed field plots (i.e. after at least 50 years of warming). We then used the same approach on a six-week laboratory warming experiment with soils from the same site to characterise microbial activity immediately after the onset of warming. By coupling measures on both timescales, we could determine whether microbial responses to warming were driven by microbial acclimation, community composition or substrate depletion. Finally, we used a biogeochemical model operating at the individual microbe scale12–14 to explore whether observations could be reproduced via changes to microbial physiology.
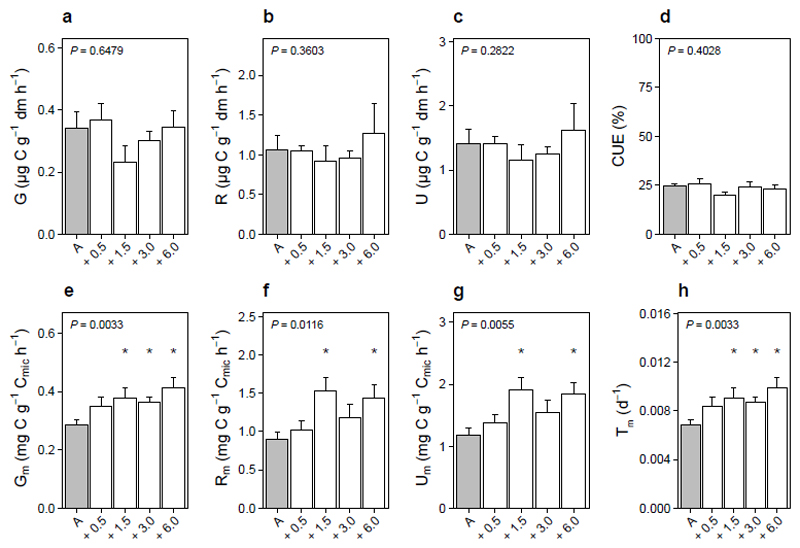
Soil microbial activity, expressed per unit of soil mass as microbial growth (Fig. 1a, P = 0.6479), respiration (Fig. 1b, P = 0.3603) and organic carbon uptake (Fig. 1c, P = 0.2822), did not differ between ambient and warmed field plots following at least 50 years of in situ warming. Microbial CUE, a metric linking microbial growth to soil carbon loss17,22, also remained unchanged between ambient and warmed field plots (Fig. 1d, P = 0.4028). This held true for warming of up to 6 ºC, thus encompassing the most severe IPCC climate projections (Scenario RCP8.5: 3.7 ± 0.7 by 2100; ref 1). Despite this, warmed soils contained up to 11.1 ± 3.5 % less carbon per ºC of temperature change (Supplementary Fig. S1a; P = 0.0001), corresponding to 1.2 ton ha-1 per ºC of the total soil organic carbon stock (data not shown). Given that microbial activity is the main vehicle of soil carbon loss1,3, we hypothesised that warming temporarily accelerated microbial activity, inducing a phase of CO2 release per unit of soil and the carbon loss observed at the field scale. This was confirmed by an incubation experiment, in which warming ambient temperature soils for six weeks accelerated microbial growth by 62 ± 22 % (Supplementary Fig. S2a, P = 0.0046), respiration by 40 ± 14 % (Supplementary Fig. S2b, P = 0.0200) and uptake by 38 ± 10 % (Supplementary Fig. S2c, P = 0.0097) per unit of soil. Taken together, our field and incubation data demonstrate two things. First, soil microbial physiology, when considered per unit of soil mass, is accelerated during a dynamic phase caused by the onset of warming, and this leads to greater soil carbon release5,9. Second, this dynamic phase ends within decades following warming, and the outcome for the ecosystem is a cessation of warming-induced CO2 release to the atmosphere.
Fig. 1. Soil microbial responses to long-term warming.
Mean (± SE, n = 5) microbial (a) growth (G; µg C g-1 soil dry mass h-1); (b) respiration (R; µg C g-1 soil dry mass h-1); (c) organic C uptake (U; µg C g-1 soil dry mass h-1); (d) carbon use efficiency (CUE; %); (e) mass-specific growth (Gm; mg C g-1 Cmic h-1); (f) mass-specific respiration (Rm; mg C g-1 Cmic h-1); (g) mass-specific organic C uptake (Um; mg C g-1 Cmic h-1); and (h) turnover (Tm; d-1) at ambient temperature (A; grey; 11 ºC) or after at least 50 years of warming (+ 0.5 to 6 ºC; white). Asterisks indicate significant differences (P < 0.05) between ambient and warmed temperatures.
The soil carbon cycle interacts with the climate system per unit of soil, with carbon cycle models expressing carbon fluxes on an area (m-2 surface) or mass (g-1 soil) scale16. At this scale, warming effects on microbial activity were temporary. However, physiological processes operate per unit of microbial biomass, not per unit of soil, and at this scale microbial activity never attenuated to warming. Mass-specific rates of microbial growth, respiration, uptake and turnover remained accelerated following six weeks (P = 0.0153, P = 0.0163, P = 0.0100, P = 0.0163) and at least 50 years (Fig. 1e-h, P = 0.0033, P = 0.0116, P = 0.0055, P = 0.0033) of warming. This occurred despite warming having no detectable influence over microbial community composition at the genus to operational taxonomic unit (OTU) level (i.e. OTU relative abundances; Supplementary Figs S3 & S4; ref 24). While it has been suggested that microbes acclimate to new thermal regimes9,22, we found no evidence to support this mechanism. Soil microbes did not adjust their growth rates, respiration rates or resource use strategies (e.g. C uptake, CUE) in response to warming. Indeed, our data alternatively show that microbial physiology does not acclimate to warming of up to 6 °C on timescales spanning weeks to at least 50 years, revealing an intrinsic temperature sensitivity of soil microbes and the processes dictating their influence over the soil system.
Substrate depletion has received extensive conceptual support as a mechanism to explain temporary warming effects on soil microbial activity7,8,10. In the absence of microbial acclimation, accelerated microbial activity under warming may deplete available substrate, creating a negative feedback on microbial processes that limits carbon loss from soil. Nevertheless, support for the substrate depletion mechanism is limited. This is because no study has convincingly shown that associations between microbial activity, substrate availability and microbial biomass persist regardless of warming intensity or duration, or that warming effects on microbial biomass lag behind (not just associate with) warming effects on microbial activity. Using incubation and field data, we discovered that microbial growth (r45 = 0.77, P < 0.0001), respiration (r44 = 0.53, P = 0.0001) and carbon uptake (r44 = 0.62, P < 0.0001) per unit of soil were positively correlated to microbial biomass irrespective of warming intensity or duration. Microbial biomass was similarly positively correlated with multiple soil substrate pools (Supplementary Fig. S5). Moreover, microbial biomass decreased by 22 ± 13 % under long-term warming (Supplementary Fig. S1b; P = 0.0038), and by only 6 ± 2 % after six weeks of warming (P = 0.0248). This illustrates that microbial biomass declined after an acceleration of microbial activity, leading to a temporary imbalance between turnover and growth. These results not only support the substrate depletion hypothesis, but also provide the first evidence that it acts via changes to microbial biomass.
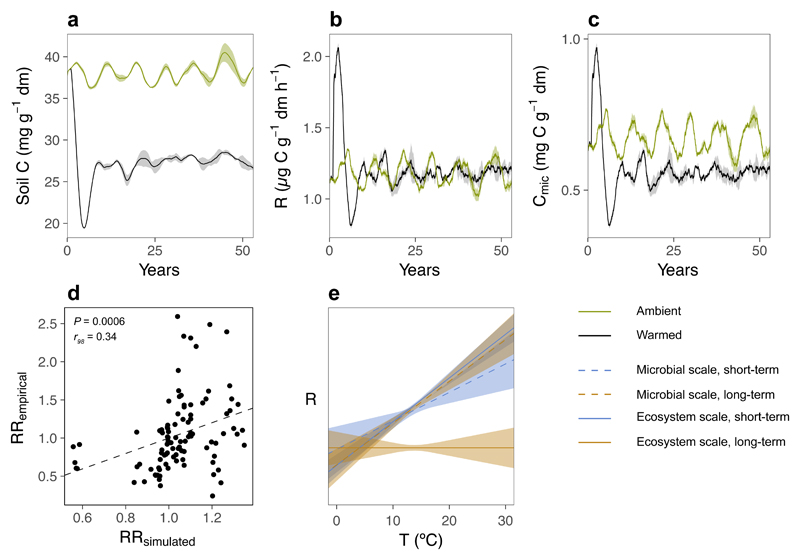
We used a microbial biogeochemical model12–14 to explore whether accelerated microbial physiology alone could explain empirical observations on both timescales. The model simulated warming through its direct effects on the physiology of individual microbes, with responses at higher organisational scales emerging as a consequence of these effects. Given that temperature controls multiple components of the microbial metabolism17–19,25,26, we mimicked warming using step changes in extracellular enzyme efficiency and substrate affinity, maintenance respiration, mortality and maximum uptake (Supplementary Table S3). In all scenarios, “warming” initiated a dynamic phase that shifted the system to a new steady state within 10 to 40 years (e.g. Supplementary Fig. S6, Fig. 2a-c). Increasing enzyme efficiency or substrate affinity reduced soil carbon, but did not accelerate mass-specific respiration or growth (Scenarios 1-7, Supplementary Fig. S6a-h). Increasing maintenance respiration decreased microbial biomass, but caused an accumulation, not a loss, of soil carbon (Scenarios 8-10, Supplementary Fig. S6i-l). Increasing maximum uptake and mortality reduced soil carbon and accelerated mass-specific growth, but not mass-specific respiration (Scenarios 11-13, Supplementary Fig. S6m-p). Only scenarios involving increases in enzyme efficiency, maintenance respiration, mortality and maximum uptake could reproduce empirical observations (Scenarios 18-27, Supplementary Fig. S7). These scenarios caused a permanent acceleration in mass-specific respiration, growth and turnover, no change to CUE and an ephemeral release of soil carbon that attenuated over time due to declining microbial biomass (e.g. Scenario 23, Table 1, Fig. 2a-c). Approximations from this example matched empirical responses to warming per unit of soil and biomass and on both timescales (Fig. 2d, r46 = 0.477, P = 0.0006). Our modelling exercise shows that intrinsic microbial temperature sensitivity has the capacity to cause ephemeral warming effects on the carbon cycle without microbial acclimation and to drive the ecosystem to a new steady state possessing pre-warmed rates of soil CO2 release (Fig. 2e).
Fig. 2. Simulated responses to warming.
Mean (± SE, n = 3) modelled responses of (a) soil carbon (mg g-1 soil dry mass), (b) microbial respiration (R; µg C g-1 soil dry mass h-1) and (c) microbial biomass C (Cmic; mg C g-1 soil dry mass) to 50 years of simulated warming (black) or a control scenario (green). (c) Relationship between empirical (RRe) and simulated (RRs) observations, displayed as response ratios irrespective of warming duration. (e) Relationships (± 95 % CIs) between microbial respiration (R) and temperature (T; ºC) from empirical data under short-term (blue) and long-term (orange) warming at microbial (dashed lines) and ecosystem (solid lines) scales.
Table 1. Modelled and empirical changes to soil carbon pools and fluxes under warming.
Mean responses of a model involving increases in microbial extracellular enzyme efficiency (15 %), maintenance respiration (10 %), mortality (10 %) and maximum uptake (5 %) and for empirical observations. Values show changes relative to the pre-warmed initiated model or ambient temperature field soil, respectively. The dynamic phase represents the model 40-50 days after perturbation or soil from ambient field plots after six weeks of warming. The warmed state represents the model 50 years after perturbation versus soil from field plots after at least 50 years of warming.
| Response | Dynamic phase |
Warmed state |
||
|---|---|---|---|---|
| Model | Empirical | Model | Empirical | |
| Soil C content | - 0.21 % | 2.94 % | - 30.86 % | - 27.10 % |
| Microbial biomass C | 19.42 % | - 5.53 % | -16.75 % | - 30.50 % |
| Total microbial growth | 43.34 % | 44.51 % | - 9.13 % | - 7.60 % |
| Total microbial respiration | 36.59 % | 33.45 % | 0.54 % | - 1.42 % |
| Mass-specific microbial growth | 20.04 % | 70.64 % | 9.15 % | 31.19 % |
| Mass-specific microbial respiration | 36.87 % | 32.78 % | 20.77 % | 41.97 % |
| Microbial CUE | 3.02 % | 5.96 % | - 6.17 % | - 4.16 % |
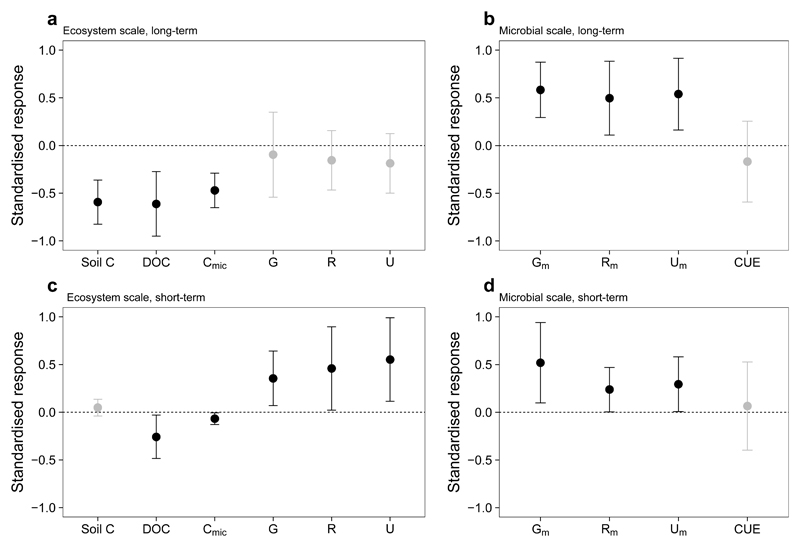
We show that intrinsic microbial temperature sensitivity can explain temporal variability in warming-induced soil carbon loss over periods of weeks to at least half a century. This is evidenced using direct physiological measurements from in situ gradients encompassing at least 50 years of warming, and is reinforced by a microbial biogeochemical model. From this, we propose a framework to explain warming effects on soil carbon loss over any timescale, which draws from the substrate depletion hypothesis8 but provides it with both a microbial mechanism and long-term empirical support. Warming permanently accelerates the growth, respiration and uptake of microbial communities (Fig. 3b,d), driving a dynamic phase of CO2 release at baseline amounts of microbial biomass (Fig. 3c). As soil carbon is lost from the ecosystem, substrates are depleted (Supplementary Fig. S5), causing a decline in microbial biomass (Fig. 3a). Thus, while microbial activity remains accelerated per unit of biomass (Fig. 3b), it declines per unit of soil. This causes an attenuation of warming-induced soil CO2 release (Fig. 3a) and a shift of the ecosystem to a new steady state. We suggest that such a “dynamic to steady state” response can explain not only attenuating warming effects on the soil carbon cycle5,22, but also examples where no attenuation occurs (i.e. where substrate does not become limiting within the observed timeframe)4,6–8,27,28. We thus offer a mechanism whereby an absence of microbial acclimation to warming can drive variable extents of ecosystem attenuation in soil carbon loss (Fig. 2e). This framework describes the interplay between temperature and soil microbial physiology on timescales relevant to Earth’s climate system, and provides a focus for future research to better constrain feedbacks from soils to climate change.
Fig. 3. Soil carbon cycle responses to climate warming.
Standardised empirical responses (± 95 % CIs) of the soil carbon cycle to (a,b) at least 50 years or (c,d) six weeks of warming (a,c) per unit of soil (g-1 soil) and (b,d) per unit of microbial biomass (g-1 microbial biomass C; Xm). G: microbial growth; R: microbial respiration; U: microbial organic C uptake; DOC: dissolved organic carbon; Cmic: microbial biomass C; CUE: microbial carbon use efficiency. Responses are from field plots (long-term) or laboratory incubations (short-term), and are presented as standardised effect sizes from linear mixed effects models including all levels of warming (i.e. a value of 0.5 represents a 50 % smaller response than a value of 1.0). Significant responses (P < 0.05) are shaded in black.
Materials & Methods
Site description & sampling
We took soil samples from the geothermal warming sites of the ForHot experiment11 near Hveragerdi in Iceland (64°00′01″ N, 21°11′09″ W), in August 2015. The experiment is in a fenced grassland, dominated by Agrostis capillaris, Ranunculus acris and Equiestum pratense, over a Brown Andosol. It consists of five replicated soil temperature gradients ranging from ambient (mean summer temperature from 2013 to 2015: 11.3 ± 0.4 ºC at 5 cm depth) to + 20 ºC above ambient, owing to geothermal activity that has been present for at least 50 years but probably since before 170811. While geothermal activity may have varied during this period, warming has been stable in the area since at least 1963 and warming intensity (i.e. ºC above ambient) has not varied since detailed measurements began in 2013 (2013-2015: ref 13; 2016-2018: data not shown). As such, we consider warmed field plots to represent a minimum of 50 years of sustained warming, with each gradient acting as a replicate block with its own ambient temperature control (inter-plot distance within blocks < 20 m). Between blocks, ambient temperature plots were similar in plant community composition, plant aboveground and belowground biomass, litter biomass, soil pH, soil moisture, pools of dissolved carbon and nitrogen (dissolved organic carbon, total dissolved nitrogen, amino acids, ammonium, nitrate) and soil carbon and nitrogen stocks (P = n.s. in all cases). Plant community composition, plant aboveground and belowground biomass, litter biomass, soil pH and soil moisture did not vary between temperatures (P = n.s. in all cases). We took soil samples (0 – 10 cm depth) at one time point from ambient, + 0.5 ºC, + 1.0 ºC, + 1.5 ºC, + 3 ºC and + 6 ºC plots of all replicate blocks (n = 5). Soils were sieved (2 mm mesh size), adjusted to 60 % of water holding capacity (WHC) and pre-incubated for four days at their respective temperatures prior to measurements.
Incubation experiment
We incubated ambient temperature field soils for six weeks at 11 ºC (i.e. at their field temperature; negative control), 14 ºC (+ 3 ºC) and 17 ºC (+ 6 ºC) to represent short-term warming. Given that soils were incubated in an artificial system, we additionally incubated warmed soils from + 3 ºC and + 6 ºC plots at their own field temperatures (i.e. 14 ºC and 17 ºC, respectively) as positive controls. In doing so, we were able to directly compare short-term and long-term warming effects on soil microbial processes using soils that had undergone the same treatment. Soils (100 g) were maintained in 500 ml glass vials under constant airflow (LI-COR-8150 multiplexer system; LI-COR Biosciences, Lincoln, USA) and at 60 % WHC. Measurements were taken at incubation temperature directly following the six-week incubation period.
Microbial physiology & carbon and nitrogen pools
We measured gross microbial growth rates using incorporation of 18O into microbial DNA. The method is direct (i.e. growth is measured as DNA replication, not the incorporation of carbon into biomass) and substrate independent, thus avoiding the addition of energy/nutrients that could alter the relationship between temperature and microbial growth17,18,22. Briefly, we incubated 500 mg soil for 24 h at field/incubation temperature with 18O-H2O to 20 at% enrichment and 80 % of WHC, alongside a duplicate containing the same volume of molecular grade non-labelled H2O as a natural abundance control. Microbial activity remained uninhibited by soil water 18O enrichments of up to 40 at%. DNA was then extracted (FastDNA™ SPIN Kit for Soil, MP Biomedicals, Santa Ana, USA), quantified (Quant-iT™ PicoGreen® dsDNA Assay Kit; Thermo Fisher, Waltham, USA) and analysed for 18O abundance and total O content using a Thermochemical Elemental Analyser (EA) coupled to a Delta V Advantage Isotope Ratio Mass Spectrometer (IRMS) via a Conflo III (Thermo Fisher, Waltham, USA). Microbial respiration (µg C g-1 soil dry mass h-1) was determined from gas samples taken at the start and end of the 24 h incubation period (analysed for CO2 concentration with a Trace GC Ultra; Thermo Fisher, Waltham, USA). Soil organic carbon and nitrogen concentrations (mg g-1 soil dry mass) were measured on dry soil (60 ºC for 48 h) with a Carlo Erba 1110 EA (CE Instruments, Wigan, UK) coupled to a Delta Plus IRMS via a Conflo III (Thermo Fisher, Waltham, USA). Microbial biomass carbon and nitrogen concentrations (mg g-1 soil dry mass) were measured via chloroform fumigation extraction followed by analysis on a TOC-VCPH/CPNTNM-1 analyser (Shimadzu, Kyoto, Japan; 48 h incubation period; 1 M KCl extraction for fumigated and non-fumigated samples; conversion factor 0.45). DNA production (µg DNA g-1 soil dry mass h-1) was then calculated and used to derive microbial community growth (µg C g-1 soil dry mass h-1), uptake (µg C g-1 soil dry mass h-1), turnover (d-1) and carbon use efficiency (%). We calculated DNA production (DNAp; µg DNA g-1 soil dry mass h-1) as:
where Oe is the 18O at% excess of the labelled sample (surplus 18O abundance (at%) relative to that of the corresponding natural abundance sample), Ol is the 18O enrichment (at%) of the labelled sample and the constant 31.21 is the proportional mass of O (%) in an average DNA molecule (data not shown). We then converted DNA production to equivalent microbial biomass carbon production, i.e. microbial growth (G; µg C g-1 soil dry mass h-1), for each sample separately using:
where Cmic and DNAmic are a sample’s microbial biomass carbon content (µg C g-1 soil dry mass) and DNA content (µg DNA g-1 soil dry mass). We calculated microbial carbon uptake (U; µg C g-1 soil dry mass h-1) as the sum of microbial growth (G) and microbial respiration (R):
We calculated microbial carbon use efficiency (CUE) as:
and microbial community turnover rate (T) as:
Microbial turnover rate is thus mathematically equivalent to mass-specific microbial growth (given as mg C g-1 Cmic h-1) at a system in steady state.
Microbial community composition
We investigated long-term (i.e. field plots) and short-term (i.e. incubated soils) warming effects (ambient, + 3 ºC, + 6 ºC) on soil microbial community composition (n = 5) by sequencing the 16S rRNA gene of bacteria and archaea and the fungal ITS1 region with an established multiplexed amplicon sequencing approach29. DNA was extracted from 500 mg soil samples (FastDNA™ SPIN Kit for Soil, MP Biomedicals, Santa Ana, USA) and purified with the OneStep™ PCR Inhibitor Removal kit (Zymo Research). Bacterial and archaeal 16S rRNA genes were amplified in triplicate PCR reactions using 25 cycles and the primer pair 515F_mod and 806R_mod. The fungal ITS1 region was amplified with 30 cycles and primers ITS1F and ITS2. Replicate PCR products were pooled, cleaned (ZR-96 DNA Clean-Up Kit™, Zymo Research, Irvine, USA), eluted in 30 µl nuclease-free water and used as a template in a second PCR reaction with primers containing sample-specific barcodes using 8 cycles29. PCR products were cleaned up, as above, quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Thermo Fisher, Waltham, USA) and pooled equimolarly (20 x 109 molecules per individual sample library) prior to sequencing on a MiSeq sequencing platform (Illumina, San Diego, USA) at Microsynth AG (Balgach, Switzerland). Bacterial/archaeal 16S rRNA gene sequence data were processed following ref 29. Fungal ITS1 sequence data were extracted from raw amplicon data using ITSx30, followed by detection of unique sequences and OTU identification. Fungal OTUs were assigned using a sequence similarity of 99% (based on OTU abundances of a mock community at different OTU clustering thresholds; data not shown). Taxonomic assignment was performed using a Bayesian classifier and the Warcup training set Version 231. Samples were rarefied to 1240 reads for bacterial samples and 1568 reads for fungal samples using the R package phyloseq. OTUs that were not present in 20% of the samples with a minimum of 10 reads were discarded from the further analysis.
Model construction
We used an soil biogeochemical model operating at the single-cell scale12–14 to investigate warming effects on soil carbon cycle processes through its influence solely over extracellular enzyme kinetics, microbial growth dynamics, substrate and consumer stoichiometry and microbial interactions on the microscale. The model is spatially explicit at the microscale: substrate turnover and microbial processes are calculated for individual soil microsites (5 x 5 x 5 µm) on a two-dimensional grid of 200 x 200 microsites. Microbes produce extracellular enzymes to degrade complex organic substrate (primary substrate: plant-derived organic matter, microbial-remains: organic matter formed by dead microbial biomass or products) in the microsite they inhabit. Enzymatic products (dissolved, bioavailable organic matter) can then be taken up by the microbe and used for maintenance, growth and further enzyme production. Elements that are in stoichiometric excess are either respired (for carbon: overflow respiration) or mineralized as inorganic nitrogen. If microbes reach a certain cell size they divide and one daughter cell populates a neighbouring microsite. The labile products of microbial and enzymatic activity (enzymatic products and inorganic nitrogen) are allowed to diffuse across the grid, which enables competitive and synergistic interactions between spatially proximate microbes. As a consequence, spatiotemporal microbial community dynamics emerge on the grid and feedback on carbon and nitrogen turnover rates12,13. The model can be set up with functionally different microbial groups, exhibiting different metabolic capabilities and cell stoichiometry12. Interactions between functionally different microbes were not the focus of this study, so we set up all model runs with one generalist microbial group able to synthesise every type of extracellular enzyme.
Model parameterisation
Model scenarios were parameterised to the same pre-warmed steady state using field data from ambient temperature plots (Supplementary Table S1). The model was parameterised and initiated through a spin-up phase of 1,600,000 time steps (approximately 90 years; one time step = 30 mins). Inputs of plant-derived primary substrate were held constant to run the model into a dynamic equilibrium where pools oscillated around a steady-state pool size14. Pre-warmed steady-state conditions established during the spin-up phase of the model were parameterised by varying parameters within a priori ranges derived from the literature and previous calibrations12 such that key pools (total soil and microbial biomass carbon, nitrogen), process rates (microbial respiration, growth, turnover) and ratios of output parameters (e.g. biomass-specific respiration, microbial biomass carbon per mass of soil carbon) were matching (as far as possible) those observed for the field experiment (spin-up parameter settings: Supplementary Table S2; steady-state conditions: Supplementary Table S1). Unlike data used for previous model applications, empirical data available in this study also included measurements of microbial growth and turnover, allowing us for the first time to parameterise the model also taking these output values into account. While the model approximated most rates, pools and ratios correctly, it underestimated soil C stocks slightly and overestimated microbial activity at the warmed steady state (Supplementary Table S1). This is likely because the model does not account for physico-chemical interactions between organic matter and soil minerals, which usually protect soil C from microbial decomposition. We used the spin-up model to test scenarios (Supplementary Table S3) in which we systematically manipulated key physiological parameters known to respond to warming, such as extracellular enzyme kinetics (efficiency, substrate affinity) and/or microbial activity (maintenance respiration, maximum uptake, mortality). Each scenario was run as a continuation from the spin-up model steady state for a total of 929,800 time steps (i.e. 53.1 years; n = 3). “Warmed” scenarios introduced a sudden change in one or more parameters after one further year of steady-state conditions, whereas an unchanged control scenario kept the spin-up parameter settings throughout the model run (deviations from spin-up parameters: Supplementary Table S3). For each scenario, three replicate runs were necessary and sufficient to account for the stochastic variability in the model (e.g. Fig. 2a-c; mean ± SE). We examined modelled carbon pools and fluxes at three time periods following the introduced physiological temperature response: (i) short-term (40 to 50 days, i.e. duration of six-week incubation experiment); (ii) peak short-term (1.5 to 3 years, i.e. peak of short-term responses observed during model runs); and (iii) long-term (49.5 to 50.5 years, i.e. minimum duration of field experiment). Following validation (see below), we selected the most representative scenario (#23) for a more detailed analysis of temporal dynamics (Fig. 2).
Model accuracy & development
Scenarios were examined against their ability to reproduce empirical observations following six-weeks (i.e. short-term incubation; scenario “short-term response”) and several decades (i.e. field experiment; scenario “long-term response”) of warming. Specifically, and further to correlations in the Main Text, we compared the accuracy of all model scenarios in reproducing soil carbon content (soil C), microbial biomass carbon (Cmic), carbon use efficiency (CUE), total respiration (R), total growth (G), biomass-specific respiration (Rm) and biomass-specific growth (Gm) as variables of key importance to interpretation of empirical data. For each scenario, a single accuracy value was calculated as the percentage of output values responding “similarly” to corresponding empirical values. For significant positive/negative empirical responses (e.g. biomass-specific respiration), corresponding scenario outputs were considered “similar” if they had a response of greater than 5 % (i.e. 0.05 in Supplementary Table S3) in absolute terms (i.e. same numerical sign). For unresponsive empirical observations (e.g. carbon use efficiency), corresponding scenario values were considered “similar” with a response of between -5 % and 5 % (i.e. -0.05 and 0.05 in Supplementary Table S3). We calculated one accuracy value per scenario (N = 27), and these data were used as a univariate response variable in linear models to determine which parameters had the greatest influence over model performance (i.e. with each scenario acting as a replicate). Scenarios involving multiple parameters (scenarios 14-27) were significantly more accurate (mean ± SE accuracy: 69 ± 3.7 %) than enzyme only (52 ± 2.4 %) and physiology only (51 ± 1.2 %) scenarios (P = 0.0011). For single-parameter scenarios (i.e. scenarios 1-13), enzyme efficiency was the most important parameter for improving accuracy (57.14 ± 0.00 %; P = 0.0101) and was thus included in all multiple parameter scenarios. By comparison, enzyme substrate affinity (51.79 ± 4.49 %; P = 0.0498), maintenance respiration (50.00 ± 0.00 %; P = 0.4926) and maximal uptake and mortality (52.38 ± 2.38 %; P = 0.7829) were less important determinants of model accuracy. Multiple-parameter scenarios containing extracellular enzyme efficiency, maintenance respiration, maximum uptake and mortality (76.53 ± 4.33 %), but not enzyme substrate affinity (71.43 ± 7.14 %), were most accurate (Supplementary Fig. S7; P = 0.0327). We thus selected scenario #23 (enzyme efficiency + 15 %, maintenance respiration + 10 %, maximum uptake + 5 %, mortality + 10 %; Supplementary Table S3) as an example scenario for analysis of temporal dynamics. Initial oscillations in this scenario immediately following perturbation were caused by the abrupt crash in microbial biomass following substrate depletion, which allowed substrate pools to partially recover prior to stabilising at a new equilibrium (Fig. 2a-c). Such oscillations are likely dampened in real soil, where a range of density-dependent processes, including competition for space, disease and predation, additionally regulate microbial abundance33.
Statistical analysis
Microbial physiology and carbon/nitrogen pools were analysed using standardised linear mixed effects models that included transect (i.e. block) as a random intercept term. Significance (P < 0.05) was determined using likelihood ratio (LR) tests between models including or excluding explanatory variables. Using field-collected soils, we tested for effects of warming treatment (ambient, + 0.5 ºC, + 1.5 ºC, + 3.0 ºC, + 6.0 ºC) on microbial physiology and carbon/nitrogen pools (Supplementary Table S4). Using incubated soils, we tested for effects of short-term warming treatment (ambient, + 3.0 ºC, + 6.0 ºC) on microbial physiology (Supplementary Table S4). Long-term and short-term warming effects on bacterial/archaeal and fungal community composition (relative OTU abundances, calculated as the abundance of a specific OTU relative to the total abundance of all OTUs in a sample) were assessed separately using PERMANOVAs and visualised using PCA plots and heatmaps of the 100 most abundant OTUs. The long timespan of the field warming experiment eliminated the potential for soil-borne relic DNA to influence the observed composition of living microbial communities32. We tested for associations between microbial biomass C (Cmic) and microbial growth, respiration and uptake, and between Cmic and concentrations of soil C, soil N, dissolved organic C, total dissolved N, nitrate N and ammonium N using Pearson Product Moment correlations. Simulated and empirical responses to warming were also compared using a Pearson Product Moment correlation between response ratios (warmed relative to control values calculated from means of all replicates; n = 5) of measured variables (soil C, microbial biomass C (Cmic), microbial respiration (R), growth (G), uptake, mass-specific respiration (Rm), mass-specific growth (Gm) and turnover), considered for short-term and long-term warming irrespective of warming duration. Finally, we plotted significant coefficients (P < 0.05) of linear regressions between microbial respiration and temperature using empirical data at microbial and ecosystem scales and at both measured timescales.
Supplementary Material
Acknowledgements
We thank K. Gavazov, M. Wagner and members of the Division of Terrestrial Ecosystem Research, University of Vienna, for discussions and comments on the manuscript. TWNW was funded by a JPI Climate Project (COUP-Austria; BMWFW-6.020/0008) awarded to AR. The study was additionally supported by a European Research Council Synergy Grant (IMBALANCE-P; ERC-2013-SyG 610028) awarded to IJ. FS was funded by a European Research Council Starting Grant (DormantMicrobes; 636928) awarded to DW. CWH was supported by a European Research Council Advanced Grant (NITRICARE; 294343) awarded to M. Wagner. BDS was supported by the Icelandic Research Council (ForHot-Forest; 163272-051) and the ClimMani COST Action (ES1308).
Footnotes
Data availability. DNA sequence data supporting the findings of this study have been deposited in the NCBI Short-Read Archive with the accession code SRP107216. Other data supporting the findings of this study are available from the corresponding authors upon request.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Author contributions
AR and TW conceived the study. NL, BS and IJ established the field sites. TW and AR did the fieldwork. TW performed the experiments, measurements and DNA extractions, and CK performed the modelling. FS and CH undertook the metagenomic investigations, with supervision from DW. TW analysed the data and wrote the manuscript in close collaboration with AR, CK and input from all co-authors.
References
- 1.IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2013. [Google Scholar]
- 2.Bardgett RD, Freeman C, Ostle NJ. Microbial contributions to climate change through carbon cycle feedbacks. Isme Journal. 2008;2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 3.Melillo JM, et al. Soil warming and carbon-cycle feedbacks to the climate system. Science. 2002;298:2173–2176. doi: 10.1126/science.1074153. [DOI] [PubMed] [Google Scholar]
- 4.Carey JC, et al. Temperature response of soil respiration largely unaltered with experimental warming. Proc Natl Acad Sci U S A. 2016;113:2–7. doi: 10.1073/pnas.1605365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo YQ, Wan SQ, Hui DF, Wallace LL. Acclimatization of soil respiration to warming in a tall grass prairie. Ecology Letters. 2001;413:622–625. doi: 10.1038/35098065. [DOI] [PubMed] [Google Scholar]
- 6.Crowther TW, et al. Quantifying global soil carbon losses in response to warming. Ecology Letters. 2016;104:104–108. doi: 10.1038/nature20150. [DOI] [PubMed] [Google Scholar]
- 7.Melillo JM, et al. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science. 2017;358:101–105. doi: 10.1126/science.aan2874. [DOI] [PubMed] [Google Scholar]
- 8.Hartley IP, Hopkins DW, Garnett MH, Sommerkorn M, Wookey PA. Soil microbial respiration in arctic soil does not acclimate to temperature. Ecology Letters. 2008;11:1092–1100. doi: 10.1111/j.1461-0248.2008.01223.x. [DOI] [PubMed] [Google Scholar]
- 9.Crowther TW, Bradford MA. Thermal acclimation in widespread heterotrophic soil microbes. Ecology Letters. 2013;16:469–477. doi: 10.1111/ele.12069. [DOI] [PubMed] [Google Scholar]
- 10.Kirschbaum MUF. Soil respiration under prolonged soil warming: Are rate reductions caused by acclimation or substrate loss? Global Change Biology. 2004;10:1870–1877. [Google Scholar]
- 11.Sigurdsson BD, et al. Geothermal ecosystems as natural climate change experiments : the FORHOT research site in Iceland as a case study. Iceland Agricultural Sciences. 2016;29:53–71. [Google Scholar]
- 12.Kaiser C, Franklin O, Dieckmann U, Richter A. Microbial community dynamics alleviate stoichiometric constraints during litter decay. Ecology Letters. 2014;17:680–690. doi: 10.1111/ele.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser C, Franklin O, Richter A, Dieckmann U. Social dynamics within decomposer communities lead to nitrogen retention and organic matter build-up in soils. Nature communications. 2015;6 doi: 10.1038/ncomms9960. 8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans S, Dieckmann U, Franklin O, Kaiser C. Synergistic effects of diffusion and microbial physiology reproduce the Birch effect in a micro-scale model. Soil Biology and Biochemistry. 2016;93:28–37. [Google Scholar]
- 15.Wieder WR, Bonan GB, Allison SD. Global soil carbon projections are improved by modelling microbial processes. Nature Climate Change. 2013;3:909–912. [Google Scholar]
- 16.Bradford MA, et al. Managing uncertainty in soil carbon feedbacks to climate change. Nature Climate Change. 2016;6:751–758. [Google Scholar]
- 17.Allison SD, Wallenstein MD, Bradford MA. Soil-carbon response to warming dependent on microbial physiology. Nature Geoscience. 2010;3:336–340. [Google Scholar]
- 18.Frey SD, Lee J, Melillo JM, Six J. The temperature response of soil microbial efficiency and its feedback to climate. Nature Climate Change. 2013;3:395–398. [Google Scholar]
- 19.Plante AF, Stone MM, McGill WB. Soil Microbiology, Ecology and Biochemistry. Elsevier; 2015. pp. 245–272. [DOI] [Google Scholar]
- 20.Bradford MA. Thermal adaptation of decomposer communities in warming soils. Front Microbiol. 2013;4:333. doi: 10.3389/fmicb.2013.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yergeau E, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. The ISME Journal. 2011;6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker CL, Bell J, Pendall E, Ogle K. Does declining carbon-use efficiency explain thermal acclimation of soil respiration with warming? Global Change Biology. 2013;19:252–263. doi: 10.1111/gcb.12036. [DOI] [PubMed] [Google Scholar]
- 23.Spohn M, Klaus K, Wanek W, Richter A. Microbial carbon use efficiency and biomass turnover times depending on soil depth - Implications for carbon cycling. Soil Biology and Biochemistry. 2016;96:74–81. [Google Scholar]
- 24.Radujkovic D, et al. Prolonged exposure does not increase soil microbial community response to warming along geothermal gradients. FEMS Microbiology Ecology. doi: 10.1101/102459. [DOI] [PubMed] [Google Scholar]
- 25.Blagodatskaya E, Blagodatsky S, Khomyakov N, Myachina O, Kuzyakov Y. Temperature sensitivity and enzymatic mechanisms of soil organic matter decomposition along an altitudinal gradient on Mount Kilimanjaro. Nature Scientific Reports. 2016;6 doi: 10.1038/srep22240. 22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manzoni S, Taylor P, Richter A, Porporato A, Ågren GI. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytologist. 2012;196:79–91. doi: 10.1111/j.1469-8137.2012.04225.x. [DOI] [PubMed] [Google Scholar]
- 27.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecology Letters. 2009;12:1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 28.Serna-Chavez HM, Fierer N, van Bodegom PM. Global drivers and patterns of microbial abundance in soil. Global Ecology and Biogeography. 2013;22:1162–1172. [Google Scholar]
- 29.Herbold CW, et al. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Ecology Letters. 2015;6:1–8. doi: 10.3389/fmicb.2015.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bengtsson-Palme J, et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods in Ecology and Evolution. 2013;4:914–919. [Google Scholar]
- 31.Deshpande V, et al. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia. 2016;108:1–5. doi: 10.3852/14-293. [DOI] [PubMed] [Google Scholar]
- 32.Carini P, et al. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Ecology Letters. 2016;53 doi: 10.1038/nmicrobiol.2016.242. 680840. [DOI] [PubMed] [Google Scholar]
- 33.Georgiou K, et al. Microbial community-level regulation explains soil carbon responses to long-term litter manipulations. Nature Communications. 8 doi: 10.1038/s41467-017-01116-z. 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.