Significance
Bacteriochlorophyll–protein complexes, such as LH2 and LH3 from Rhodoblastus acidophilus, represent the core of bacterial photosynthesis. While similar in many aspects, these two prototypical complexes show different absorption wavelengths, being the latter blue-shifted ca. 30 nm in its major peak respect to the former. These systems have been intensively studied so as to understand the underlying structural mechanisms controlling the difference in absorption and, ultimately, the photosynthetic process. Through highest level computational tools, we here demonstrate that, somewhat contrary to common belief, the major device steering the absorption of bacteriochlorophylls is the direction of their macrocycle ring curvature axis. This finding could lead to the design of novel, easily, and highly tunable chromophores, for future light-harvesting artificial systems.
Keywords: bacteriochlorophyll, LH3, MS-RASPT2, macrocycle ring deformation, chromophore mimics
Abstract
Natural light-harvesting is performed by pigment–protein complexes, which collect and funnel the solar energy at the start of photosynthesis. The identity and arrangement of pigments largely define the absorption spectrum of the antenna complex, which is further regulated by a palette of structural factors. Small alterations are induced by pigment–protein interactions. In light-harvesting systems 2 and 3 from Rhodoblastus acidophilus, the pigments are arranged identically, yet the former has an absorption peak at 850 nm that is blue-shifted to 820 nm in the latter. While the shift has previously been attributed to the removal of hydrogen bonds, which brings changes in the acetyl moiety of the bacteriochlorophyll, recent work has shown that other mechanisms are also present. Using computational and modeling tools on the corresponding crystal structures, we reach a different conclusion: The most critical factor for the shift is the curvature of the macrocycle ring. The bending of the planar part of the pigment is identified as the second-most important design principle for the function of pigment–protein complexes—a finding that can inspire the design of novel artificial systems.
Uncovering the inner workings of photosynthetic light-harvesting can inspire artificial mimics, offering a new avenue for clean solar energy. The current understanding of the initial processes, light absorption, energy transfer, and charge separation is built on the marriage of crystal structures and ultrafast spectroscopic experiments (1–3), yet disentangling the rich data material into structure–function relationships, using theoretical models and simulations, is highly nontrivial due to the complexity of the systems. The light-harvesting complexes are composed of chromophores held together by a protein framework, with the first design principle being the arrangement of the chromophores. The relative positions of the chromophores affect their couplings and determine to which degree excitation energy may be shared between several chromophores in a quantum delocalization, thus red-shifting the absorption band (4).
It has been suggested that environment fluctuations can enhance energy transport (5, 6).
In any case, a full understanding of the energy transfer process is only possible with a correct description of the interaction between the chromophores and their fluctuating protein environment. A lot of effort has been devoted to this subject recently (7–16). The environment affects both site energies and couplings and is a constant source of noise; this has a smearing effect on experimental spectra and poses great challenges to theoretical treatments. The interactions include electronic–vibrational couplings and electrostatic effects, which are quantities that may be difficult to extract from both computation and experiment.
Fortunately, some hints are provided by nature. Light-harvesting systems 2 and 3 (LH2 and LH3) from the purple bacterium Rhodoblastus acidophilus have nearly identical arrangements of bacteriochlorophyll a (BChl) chromophores, although their proteins differ in the primary structure (see Fig. 1). As a result, LH3, found in a low light-adapted strain of the bacterium (18), harvests light more efficiently than LH2, which is grown under normal conditions. The absorption spectrum reveals that the changes in the protein mainly affect the BChls in the 18-membered ring, which absorbs at about 850 and 820 nm in LH2 and LH3 (19, 20), respectively, whereas the 9-membered ring absorbs at around 800 nm in both complexes. By comparing the crystal structures, one notices that the protein residues adjacent to the BChls in LH2, tyrosine and tryptophan, are replaced by phenylalanine and leucine in LH3. While the former ones form hydrogen bonds with the acetyl moiety of the BChl, the latter ones do not. A similar blue-shift is obtained by site-directed mutagenesis of the corresponding protein residues in LH2 from Rhodobacter Sphaeroides, further supporting the importance of these particular sites on the protein. Unfortunately, no crystal structures are available for the wild-type nor any of its variants of LH2 from R. Sphaeroides. Interestingly, both blue-shifts are accompanied by a reduction of the peak height relative to the 800-nm peak, which, to our knowledge, has yet to be explained (17, 21).
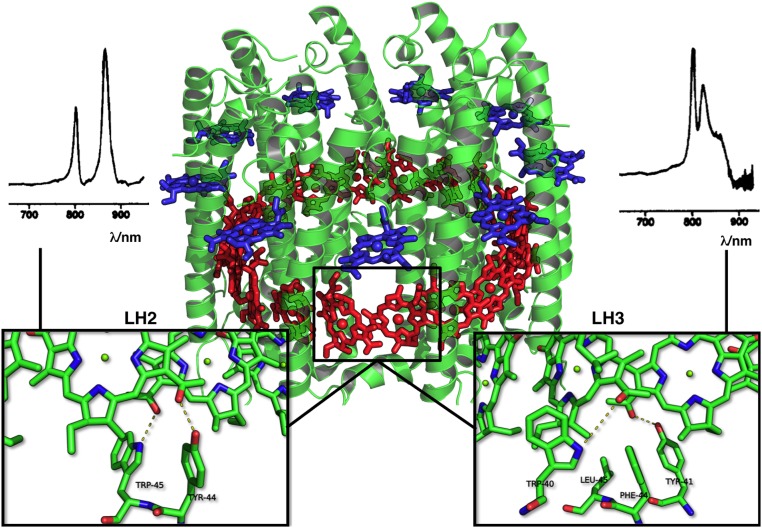
Fig. 1.
Structure of LH2 and LH3. The overall structure of LH2 and LH3 (as shown in the middle) is similar: A barrel of alpha helices (green) keeps in position the various bacteriochlorophyll units, shown in blue (the B800 ring) and in red (the B850 ring). The two rings are also bridged by carotenoid molecules, which were omitted for clarity. The frame highlights BChl units 304 and 305 from B850. The differences in hydrogen bonding are shown in the two lower Insets. In LH2 (Bottom Left Inset), tyrosine 44 (Y44) and tryptophan 45 (W45) provide hydrogen bonds (dashed lines) to the acetyl moieties, while these residues are substituted by phenylalanine (F44) and leucine (L45) in LH3 (Bottom Right Inset). Possibly, in LH3, W40 and Y41 of two different chains provide hydrogen bonds to the acetyl moieties. The corresponding experimental spectra are shown above the two Insets. Reprinted from ref. 17. Copyright (1986), with permission from Elsevier.
While it seems likely that the blue-shift is linked to the residues, the underlying mechanism is an open question. It has previously been proposed that the rotation of the acetyl moiety (torsion d in Fig. 2) is responsible for the energy modulation (22–24), but according to ref. 16, this effect is much too small to account for the 30-nm shift. On general grounds, substituting a bulky residue such as tyrosine with a smaller residue like leucine is expected to relax the structure. For this to bring a blue-shift, the energy of the ground state must be lowered more than the energy of the excited state. We have previously demonstrated that the BChl ground state is affected by the curvature of its macrocycle ring (MCR). (24) Importantly, the MCR also affects the transition dipole moment (TDM), while the same cannot be said of the acetyl rotation.
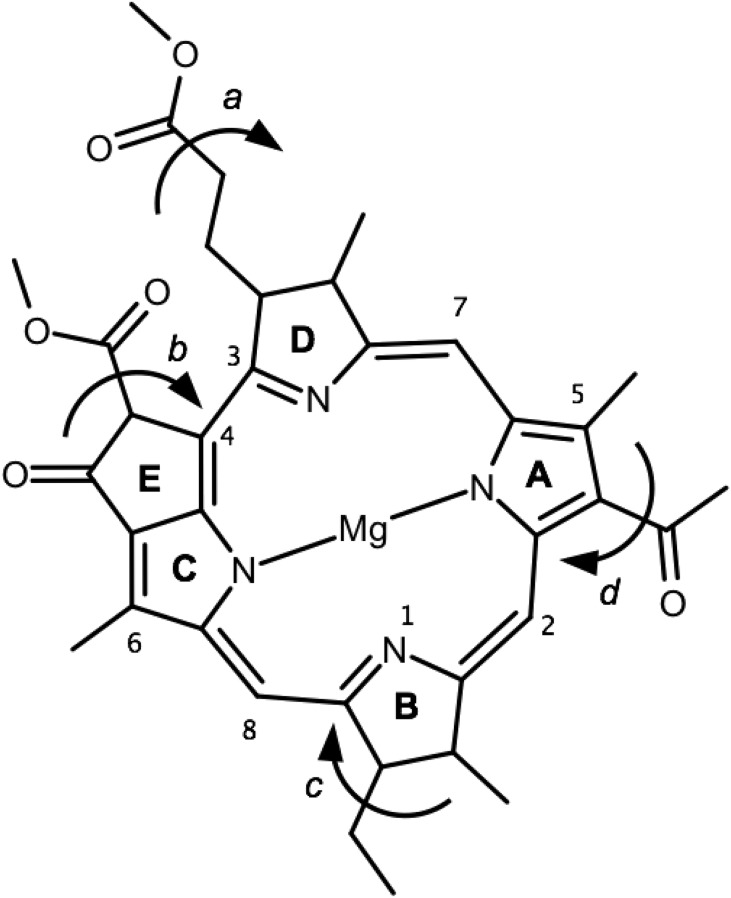
Fig. 2.
Model structure of bacteriochlorophyll a. Shown is the structure of the model of bacteriochlorophyll a used in this work, with standard ring naming (A, B, C, D, E) and the investigated dihedral torsions (a, b, c, d). Numbers indicate various atoms mentioned in the text.
Already, the work of Cogdell et al. recognized various possible structure-based mechanisms as possible sources of modulation of the first absorbance band wavelength of bacteriochlorophylls (22). Some of the more recent work has placed a stronger emphasis on the protein-induced deformations of the chromophores. Bednarczyk found that changing the position of a nearby tryptophan residue induced deformation of a tetrapyrrole MCR in water-soluble chlorophyll binding proteins, which accounted for two-thirds of the observed spectral shift between two natural variants (25). MacGowan and Senge used a factorial relaxation procedure to relate the effects of various protein-induced BChl deformations on the site energies and found reasonably strong correlations, which was surprising because the site-energy differences had previously been accounted for in their entirety by the electrostatic environments of the pigments (26). However, it should also be noted that porphyrins, and related molecules, show a reduced excited state lifetime when their geometry is strongly distorted. This reducing effect has been noted in both free and metal-containing porphyrins and on both singlet and triplet excited states (27–31).
Even more recently, Nottoli et al. (32) demonstrated that, once again, the dihedral torsion of the acetyl moiety alone cannot be held accountable for the B820 blue-shift. In their work, they find that changes in the tuning of charge-transfer states, rather than chromophore–chromophore couplings, are involved in the blue-shift. In this work, we demonstrate how MCR deformations have, possibly, an even larger influence on absorbance spectra and can act also on a single bacteriochlorophyll unit.
Similar conclusions were reached by Montemayor et al. (33). They demonstrated that differences in dihedral torsional angles were minimal between LH2 and LH3, in an ensemble of BChls obtained through molecular dynamics simulations. The authors conclude that the torsional angle cannot be considered as the sole responsibility for the blue-shift.
In this work, we continue in this vein and compute excitation energies of BChls interpolated from the LH2 to the LH3 geometry as well as selected dihedral torsions. The results are inserted into a model for the absorption spectrum to determine which geometrical variations are important for the blue-shift. Our target is not so much an accurate description of the LH2 and LH3 spectra per se but is rather an expert analysis of possible, BChl structure-related causes for the experimental blue-shift. Based on our findings, we identify a secondary design principle for artificial light-harvesters and suggest concrete strategies on how this can be exploited for BChls.
To shed light on the real causes of the observed blue-shift, we present highest-quality multistate multiconfiguration restricted active space with second-order perturbation theory correction (MS-RASPT2) computed excitation energies of selected BChls from LH3. Furthermore, we analyze, through time-dependent density functional theory (TD-DFT) calculations, the structural changes in one BChl when going from LH2 to LH3. The aim is to locate which geometrical change is relevant for an effective blue-shift, thus providing rationalization to the shifting effect itself. Finally, we use a theoretical model to simulate the spectra of both LH2 and LH3, based on both MS-RASPT2 and TD-DFT energetics data. Through the simulated spectra, we investigate if the computed LH3 BChl excitation energies are sufficient to generate the experimental blue-shift, if different geometrical modes were capable of obtaining the same effect, or if something else were necessary.
Results
The high-quality MS-RASPT2 computed spectroscopical properties for the alpha and beta units of LH3, as shown in Table 1, give a dual indication. On the one hand, comparing these data with those we previously obtained for LH2 (24), we can observe a clear blue-shift (i.e., higher excitation energies and longer wave lengths). Such blue-shift is very pronounced (ca. 0.23 eV, −90 nm) and unexpected, when considering that the LH2-to-LH3 experimental shift of the overall system is only 30 nm. On the other hand, it is possible to see also a diminishing of the TDM of the BChls from LH3 with respect to BChls from LH2, which could be an indication for the cause of the lower recorded 820 peaks in LH3.
Table 1.
MS-RASPT2 excitation energies (site energies), oscillator strengths, and TDMs of 2 LH3 BChl units (this work)* and corresponding LH2 average values (ref. 24)
| Excitation energy | ||||||
| System | Site | Type | eV | nm | Oscillator strength | TDM, eÅ |
| LH3 | 303 | 1.91 | 650 | 0.30 | 1.34 | |
| LH3 | 304 | 1.87 | 661 | 0.30 | 1.34 | |
| LH2 | Average | 1.67 | 741 | 0.30 | 1.43 | |
| LH2 | Average | 1.65 | 750 | 0.31 | 1.45 | |
Complete energetics are reported in SI Appendix, Table S6
We decided to perform an adiabatic scan between the geometries of two BChl units, both corresponding to the beta unit 304 but one extracted from the crystal structure relative to LH2 (herein indicated as BChl-1) and the other from LH3 (herein indicated as BChl-47). Further details are provided in Materials and Methods. The scan was divided so as to analyze separately the differences in dihedral angles of four peripheral moieties (among which is the acetyl moiety; see Fig. 2) and the difference in curvature of the MCR. The results are reported in Fig. 3.
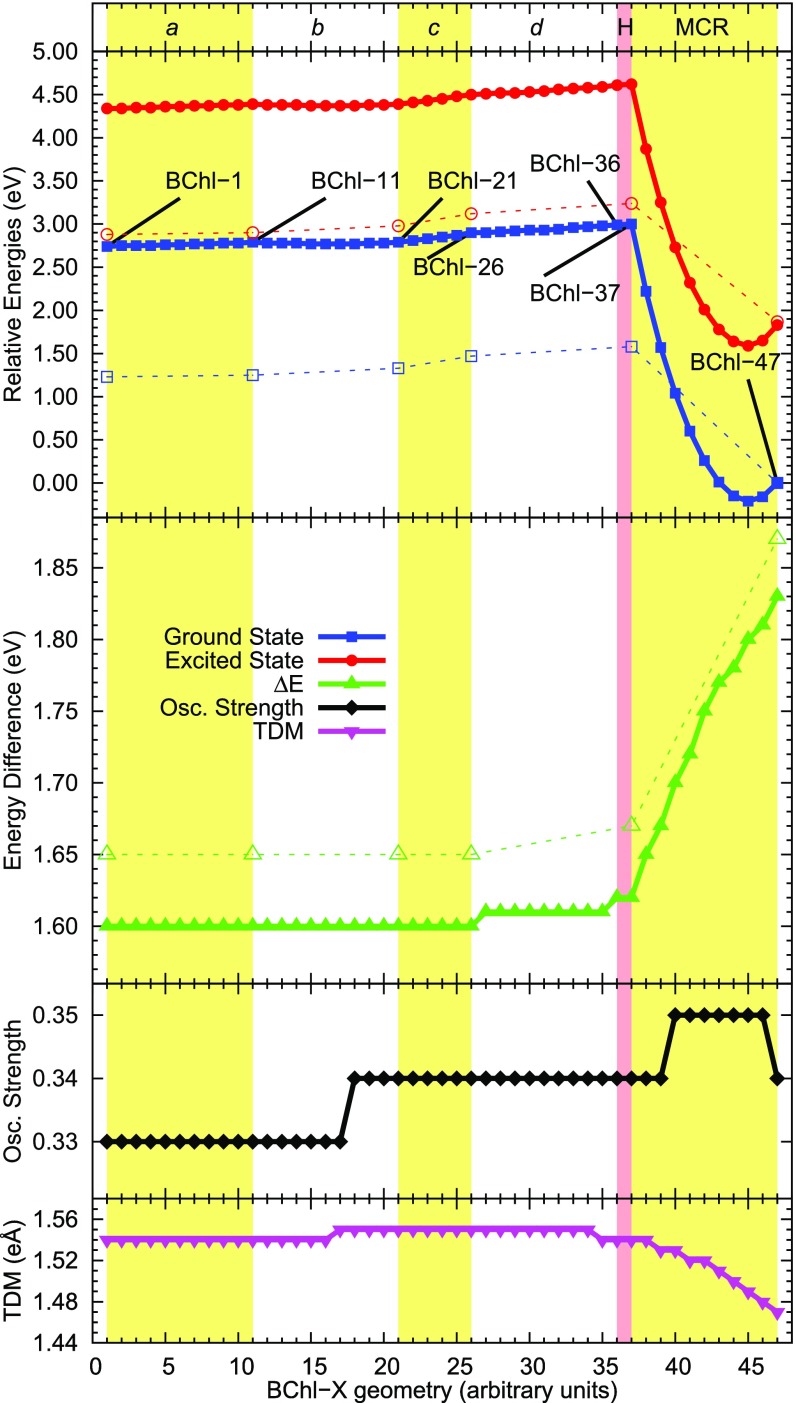
Fig. 3.
Computed energetics profiles of unit 304. From the top, TD-DFT (full symbols and lines) and MS-RASPT2 (open symbols and dashed lines) computed potential energy surfaces, excitation energy, oscillator strength, and TDM changes along the interpolation scan relative to unit 304, where BChl-1 and BChl-47 are the geometries obtained from LH2 and LH3 crystal structures, respectively. Relative energies were computed with respect to BChl-47. The vertical bands divide the geometries relative to torsion around dihedral angles a, b, c, and d; the adjustment of hydrogen atom position (H); and the change in the MCR curvature. A similar landscape was also found for unit 303, as reported in SI Appendix, Fig. S5. SI Appendix, Table S4 reports relative energetics along the scan for selected geometries.
TD-DFT data for the entire scan and MS-RASPT2 data for selected geometries qualitatively agree on computed excitation energies, oscillator strengths, and TDMs, and the following analysis is based on such values. The two sets differ on relative energies. We will comment on this in Discussion.
As expected, modifications to the dihedral angles a (BChl-1 to BChl-11), b (BChl-11 to BChl-121), and c (BChl-21 to BChl-26) do not change the excitation energy of BChl, since they do not participate to the extended conjugated system. The change in the dihedral angle d (BChl-26 to BChl-36) produces a change in excitation energy of ca. 0.02 eV. This is in line with what was expected, considering the change in the dihedral angle previously seen when comparing different BChl units in LH2 (24).
The change in the MCR curvature (BChl-37 to BChl-47) has dramatic effects on the energies of bacteriochlorophyll. While BChl-37 and BChl-47 differ by only 0.24 Å (only heavy atoms) (34, 35), the ground state energy is greatly stabilized (lower energy). Conversely, the excited state is less stabilized, leading to the increase in excitation energy of ca. 0.2 eV previously noted. The curvature change leads also to a small increase in oscillator strength and a constant decrease of the TDM. Similar results were obtained relative to the alpha unit 303.
The results of Fig. 3 show that the difference in absorption wavelengths of BChls from LH2 or LH3 has a dual origin, although the extent of such effects is overall different. (i) A small change in the dihedral angle d causes a small change in excitation energy (right-most white band in Fig. 3). (ii) A small change in the MCR curvature causes a huge change in excitation energy (right-most yellow band in Fig. 3). For this reason, we decided to further analyze what the magnitudes of such changes can be, considered separately, on the absorption spectrum of the overall protein–BChl complex.
To achieve this, we used a recently coded utility to simulate the spectrum of LH2 and LH3, based on computed, high-quality excitation energies and TDMs (36). The utility is capable of reproducing the spectrum of LH2 in its two peaks—namely, 800 and 850 nm. See black line in Fig. 4B (BChl-1).
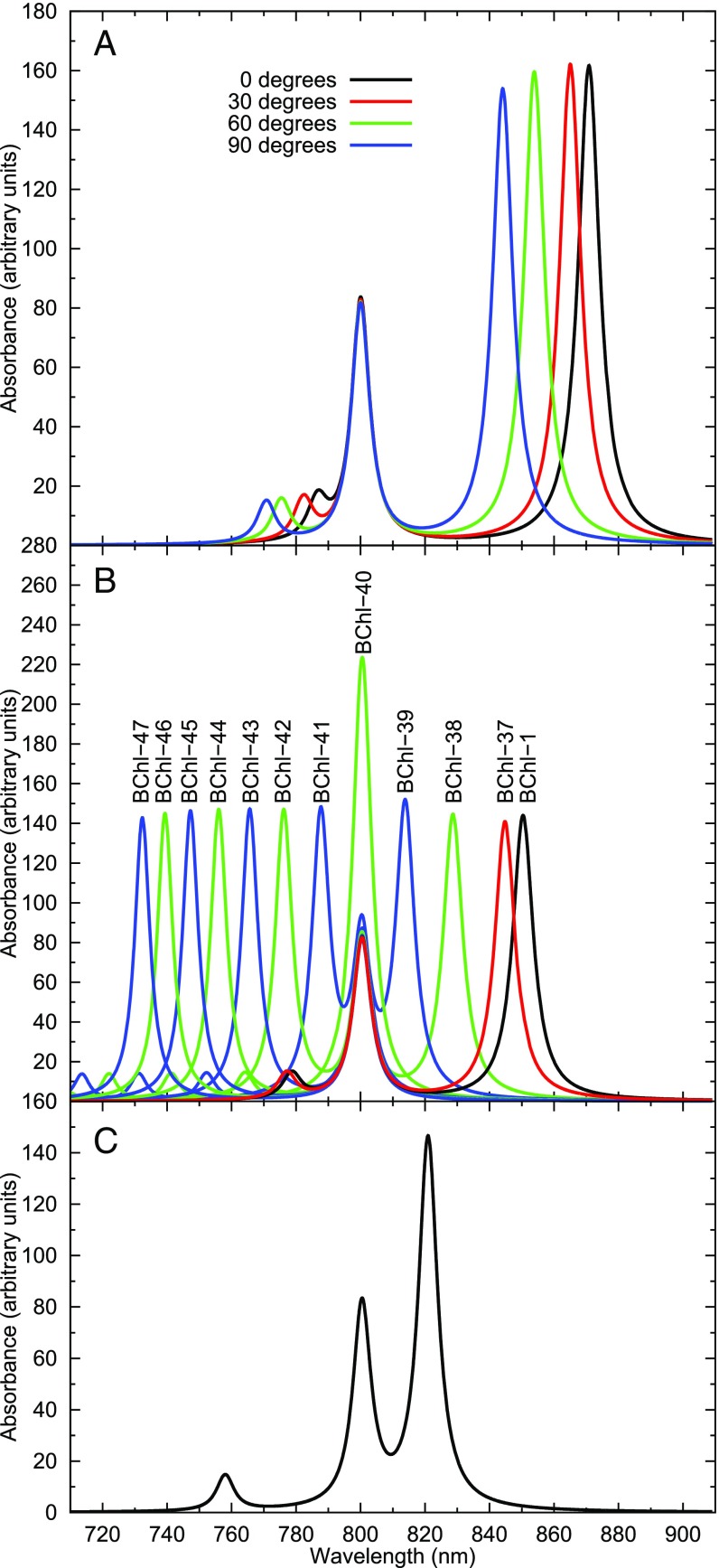
Fig. 4.
Simulated spectra. (A) Simulated spectra for a hypothetical torsion from 0 to 90 degrees of the acetyl moiety. The spectra are based on TD-DFT–obtained excitation energies and TDM values, and both alpha and beta units were assigned the same excitations. (B) Changes in simulated spectra along the LH2 to LH3 scan. Underlying data are obtained from interpolated MS-RASPT2 excitation energies and TDM, for both alpha and beta units. The numbering of the spectra corresponds to the various BChl structures, as reported in the text. (C) Simulated spectrum using the excitation energies and TDM of alpha and beta units denoted 38.5 as the best approximation of the experimental spectrum of LH3. All simulated spectra are the result of considering 18 BChl units and their various couplings, as described in ref. 36.
The simulated spectrum when considering only the torsion around d, but no MCR curvature change, obtained from the computed MS-RASPT2 data relative to BChl-37 for unit 304 and the corresponding structure for unit 303, is reported as a red line in Fig. 4B. The observed blue-shift is only 5 nm.
To further elucidate the role of the torsion of the acetyl moiety, we simulated the spectra relative to a 0–90° full rigid rotation (SI Appendix, Fig. S4), assuming the same energies for both alpha and beta units. The results are reported in Fig. 4A. While the values of the peak positions are only qualitative, nevertheless a maximum blue-shift of only ca. 30 nm can be expected from the acetyl torsion. However, only the most extreme change in acetyl torsion dihedral (from 0 to 90°) could be responsible for the experimental blue-shift of LH3. It is clearly asserted in the literature that the acetyl moiety of BChl units in LH2 is not planar (37). Therefore, the 30-nm blue-shift of LH3 cannot be associated only with a change in dihedral angle.
Fig. 4B shows the simulated spectra along the geometrical scan. The data are based on MS-RASPT2 interpolated energies and TDM for alpha and beta units. Complete details are given in SI Appendix. Clearly, the energies relative to the geometries obtained from the LH3 crystal structure produce a spectrum that is too much blue-shifted (BChl-47). Intermediate geometries between BChl-38 and BChl-39 (denoted as BChl-38.5) for both alpha and beta units are expected to reproduce a spectrum close to the experimental one. This is shown in Fig. 4C. Surprisingly, even if the overall TDM of BChls from LH3 is lower than that of LH2, the simulated spectrum does not show significant lowering of the corresponding absorption peak.
Discussion
The LH2 and LH3 structures used in this work were retrieved from the Protein Data Bank (ID codes 1NKZ and 1IJD) (23, 37). The LH2 data extend to 2 Å, while the LH3 system is only available at the lower resolution of 3 Å. Both structures were refined by imposing noncrystallographic symmetry (ncs) restraints on the three crystallographically independent B850/B820 units.
The restrained refinements of the bacteriochlorophyll units in LH2 and LH3 use a flat conformation as the optimal geometry. Deviations from this geometry are therefore introduced by the refinement against the experimental structure factor amplitudes. The limited resolution of the LH3 structure, in combination with the chosen ncs restrained refinement strategy, allows a discussion of trends and differences in conformation, whereas one should put less confidence in the actual magnitudes of the curvatures. However, the quality of the omit maps presented in figure 6 of ref. 23 is an indication of a well-determined electron density.
As shown in Fig. 4, a distortion of the MCR is fundamental to reproduce the experimental absorption peak at 820 nm of LH3. This can be rationalized as follows: As shown in table 2 of ref. 24, the main contribution to the electronic excitation in BChls comes from molecular orbitals residing on the MCR. In other words, these are the molecular orbitals mostly depleted or enriched of electronic population upon photo excitation. Therefore, it is clear that a change of the torsion of the acetyl moiety (not covered by the aforementioned molecular orbitals) is contributing only marginally to the overall absorption spectrum. Conversely, a change in curvature of the MCR directly affects these molecular orbitals and thus the excitation energy.
In the LH2 experimental spectra, the 850 peak is higher than the 800 nm, because it is generated by the absorption of twice the number of BChl units (Fig. 1, Top Left) (17). The same should have been true also for LH3, but the experimental peak is actually lower and relatively broader (Fig. 1, Top Right) (17). We found that BChl units from LH3 have computed lower TDM values than their LH2 counterparts (Table 1). However, this did not translate into a corresponding lower absorption peak in the simulated spectra (Fig. 4C).
The broadening of the 820-nm peak may be due to the acetyl torsion. We suggest that the majority of the BChl units in LH3 have their acetyl moieties kept in place by hydrogen bonds, as shown in Fig. 1, Bottom Right, thus producing the sharp feature of the 820 peak. However, few BChl acetyl moieties may be able to return to their planar, more electronically stable conformation. This may be due to the fact that at least one of the two residues possibly responsible for the hydrogen bonds in LH3 (W40 and Y41) is relatively more distant from the BChl acetyl oxygen atoms (2.74 and 4.48 Å, respectively) (23) than Y44 and W45 in LH2 (2.68 and 2.88 Å, respectively) (37). A planar acetyl moiety produces a red-shifted absorption (Fig. 4A). Therefore, the broad, red-shifted shoulder of the 820-nm absorption peak of LH3 may be due to BChl units where the acetyl moiety is planar, and thus reducing the overall height of the absorption peak, with respect to the 800-nm peak.
Fig. 4B clearly shows that using the computed excitation energies of the LH3 crystal structure (BChl-47) leads to a simulated spectrum that is too blue-shifted. Since the simulation of the LH2 spectrum (e.g., BChl-1 black line in Fig. 4B) is reliable (36), we believe this is due to the different refinement of the LH3 structure, as previously noted. As seen in Fig. 3, the blue-shift in LH3 is mainly due to relaxation of the chromophore’s ground state (BChl-37–BChl-47). Thus, we make the hypothesis that the LH3 crystal structure refinement put too much emphasis on relaxing the geometries of the chromophores toward a lower energy.
A possible way to verify this hypothesis would be through a quantum mechanic/molecular mechanic (QM/MM) reoptimization of the entire LH3 complex. For example, we chose our TD-DFT methodology (in terms of functional and basis set) because, as previously found, it gives reliable differences in excitation energies (38). While this is confirmed by our results (Fig. 3, Middle), we also saw a large discrepancy with the relative ground state energies (Fig. 3, Upper) computed at the MS-RASPT2 level of theory (a most reliable methodology) (24). Clearly, an extensive preliminary study is necessary to choose an appropriate methodology that is able to provide reliable structures and energies, compared with MS-RASPT2 data, coupled with the computational ease necessary for a massive QM/MM optimization of the entire LH3 complex. To the best of our knowledge, such a comparative study specific for bacteriochlorophylls is not yet available and will be part of our future endeavors.
Based on our results, we here try to make a list, in order of importance, of the ways that the protein matrix controls the absorption of light-harvesting complexes based on rings of BChl molecules:
-
i)
The primary way of controlling the absorption peaks is, clearly, by aggregation—that is, guiding the relative position of the BChl units. The major division into two separate peaks (e.g., at ca. 800 and 850 nm in LH2) is due to the relative positions of the BChl units in the two rings (Fig. 1). The relative positions of the TDM vectors (coplanar for the B800 ring, nearly antiparallel for the B850) (24) and the BChl–BChl distances are the major players in defining the absorption peaks (36).
-
ii)
The secondary role of the protein matrix is that of guiding the conformation of the MCR portion of the BChl units. As seen in table 2 of ref. 24 and as previously discussed, the MCR curvature controls the shape of the molecular orbitals mostly involved in the electronic excitation of the BChl units. The protein matrix can strongly influence the position of the absorption peaks by imposing a different or more/less pronounced curvature to the MCR. This is what happens in LH3: By allowing a partial relaxation of the BChl units (Fig. 3), the protein complex blue-shifts the lower energy absorption peak.
It is possible to see the action of the protein matrix also in terms of promoting different conformers of the MCR. As shown in SI Appendix, the BChl geometries of unit 304 extracted from the LH2 and LH3 crystal structures (BChl-1 and BChl-47) can be associated with two different conformers, through normal structure decomposition (39). Since, as shown in Fig. 3, Upper, BChl-1 and BChl-47 have clearly different relative ground state energies, it is up to the protein matrix to promote one conformer or the other.
The recognition of the importance of the MCR curvature in defining the position of the absorption peaks, and how the protein matrix can exploit it, represents the major outcome of the present work.
-
iii)
The third role of the protein matrix is that of fine-tuning the absorption peaks, by twisting the acetyl moiety. As shown in Fig. 4A, changing the acetyl moiety torsion from 0° to 90° allows a maximum possible shift of ca. 30 nm. According to our hypothesis, it is possible that such an effect is responsible for, for example, the lower height of the B820 peak of LH3.
-
iv)
A fourth, and less evident, effect of the protein matrix is the direct, dynamical tuning of the BChl electronic excitation by their surrounding residues. As we recently demonstrated (16), the residues in the immediate vicinity of each BChl unit can influence the excitation energies according to certain residue–BChl vibrational modes. Most obviously, these effects are not visible in a static, simple absorption spectrum but could be responsible for some of the features of time-dependent, 2D electronic spectra (40, 41).
-
v)
Finally, the protein complex provides an electrostatic overall environment, which is responsible for a systematic shift of the peaks, through a dielectric screening.
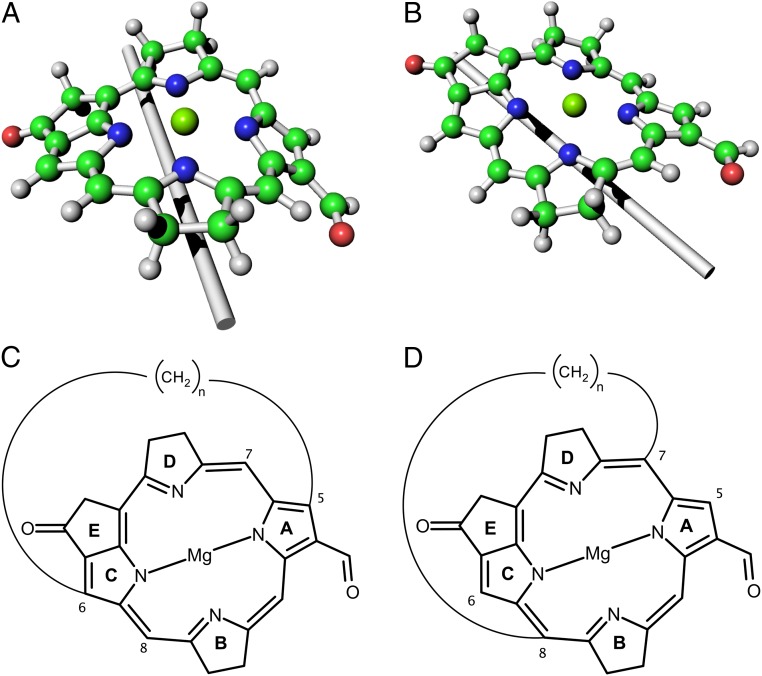
Analyzing the geometrical structures of BChl-1 and BChl-47, it is possible to notice that their MCRs are differently curved in the following manner. Following the atom numbering in Fig. 2, in LH2 (BChl-1) the MCR is curved along an axis that can be thought of as running between atoms 1 and 3, while atoms 5 and 6 are pulled in the opposite direction of the central Mg atom (Fig. 5A). In LH3 (BChl-47), the MCR is curved along an axis running between atoms 2 and 4, while atoms 7 and 8 are pulled against the direction of the Mg atom (Fig. 5B).
Fig. 5.
Bacteriochlorophyll curvatures. Simplified models of BChl, exemplifying the differences in MCR torsion between (A) LH2 and (B) LH3. A fictitious axis is added to both structures to guide the eye. Also shown are examples of bacteriochlorophyll-inspired chromophores, in which the ()n bridge should induce an MCR torsion similar to that of (C) LH2 or (D) LH3, depending on the connecting atoms on the ring.
While the models reported in Fig. 5 A and B are only an exemplification of the different MCR curvature in LH2 and LH3, they can be taken as templates for devising bacteriochlorophyll mimetic, photoactive compounds with tunable absorption maxima. Fig. 5 C and D show two possible classes of chromophores, where a ()n bridge forces the MCR ring to assume a curvature similar to that of naturally occurring BChls in their protein environment. Different curvatures can be obtained depending on the MCR atoms to which the bridge is connected, although one has to keep in mind that, as previously noted, changes in the overall BChl structures may lead to reduction of excited state lifetimes. If the bridge were connected to the atoms corresponding to 7 and 8 in BChl (Fig. 5D), the resulting chromophore should show a blue-shifted absorbance in respect to the chromophore with the same bridge length (number of units) but linked to atoms 5 and 6 (Fig. 5C). Preliminary computational results (SI Appendix, Table S10 and relative discussion) seem to indicate that that is the case.
Conclusions
In this work, we performed excitation energy calculations on BChl geometries obtained by interpolating the LH2 and LH3 crystal structures as well as selected dihedral torsions. By combining MS-RASPT2 and TD-DFT calculations, we get the best of two worlds: accuracy and speed. The trend is clear: The change in geometry from LH2 to LH3, essentially a change in the MCR curvature, brings a huge blue-shift of 90 nm, while the dihedral torsions only modestly affect the excitation energy. Using a simplified model, which is capable of reproducing the 850–820-nm blue-shift, we find that the geometry that best reproduces the 820-nm absorption peak in LH3 is an intermediate between the LH2 and LH3 geometries. We also find that the differences in acetyl moiety torsion alone, as shown in Fig. 4A, are not sufficient to explain the blue-shift for the mutants of LH2 from R. sphaeroides, which is contrary to previous hypotheses. Auxiliary mechanisms have to be in place and warrant further future investigation.
The different MCR curvatures are the results of different protein residues in the immediate environment, which push and pull the MCR, thus regulating its excitation energy. We envision that the same effect can be achieved in synthetic BChl-mimicking chromophores by attaching bridges at specific sites on the BChl analogue. Ongoing studies will test this hypothesis, with particular emphasis on possible modifications of the computed excited state lifetime.
Future work on geometry optimizations of light-harvesting complexes such as LH2 and LH3 will need to address the large discrepancies between MS-RASPT2 and DFT ground state energies. If DFT energies are used in a QM/MM scheme-based optimization, one runs the risk of artificially preferring a LH3-like BChl chromophore.
The next step in understanding the 850–820 blue-shift will be to perform accurate calculations of the BChl–BChl couplings in the LH2 and LH3 complexes, to see whether differences in the couplings, as due to, for example, different chromophore–chromophore distances, are also involved in the absorbance shift.
Materials and Methods
BChl geometries were extracted from LH2 and LH3 crystal structures (PDB ID codes 1NKZ and 1IJD, respectively) (23, 37). Energetics were computed at the MS-RASPT2/SA-RASSCF (42–45) with ANO-RCC double zeta basis set (46) and at the TD-DFT (47–49) PBE0 functional (50) 6-311+G (51–53) basis set with df-def2 auxiliary basis (54) levels of theory using the programs MOLCAS (version 7.8 and 8.1) (55, 56) and LS-DALTON (version 2015.0) (57), respectively. Complete details can be found in SI Appendix.
Supplementary Material
Acknowledgments
Tõnu Pullerits is acknowledged for fruitful discussion about LH2 and LH3. T.H. is grateful for financial support from the Lundbeck Foundation. L.D.V. acknowledges MIUR Grant “Dipartimento di Eccellenza 2018–2022.” A.A. was supported by the Australian Research Council under the Centre of Excellence funding scheme (Grant CE170100026). All calculations were conducted using the resources available at the Danish Center for Scientific Computing at the University of Copenhagen. The Center for Exploitation of Solar Energy is acknowledged for providing computational resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719355115/-/DCSupplemental.
References
- 1.Dostál J, Pšenčík J, Zigmantas D. In situ mapping of the energy flow through the entire photosynthetic apparatus. Nat Chem. 2016;8:705–710. doi: 10.1038/nchem.2525. [DOI] [PubMed] [Google Scholar]
- 2.Fuller FD, et al. Vibronic coherence in oxygenic photosynthesis. Nat Chem. 2014;6:706–711. doi: 10.1038/nchem.2005. [DOI] [PubMed] [Google Scholar]
- 3.Romero E, et al. Quantum coherence in photosynthesis for efficient solar-energy conversion. Nat Phys. 2014;10:676–682. doi: 10.1038/nphys3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timpmann K, et al. Short-range exciton couplings in LH2 photosynthetic antenna proteins studied by high hydrostatic pressure absorption spectroscopy. J Phys Chem B. 2001;105:8436–8444. [Google Scholar]
- 5.Plenio MB, Huelga SF. Dephasing-assisted transport: Quantum networks and biomolecules. New J Phys. 2008;10:113019. [Google Scholar]
- 6.Rebentrost P, et al. Environment-assisted quantum transport. New J Phys. 2009;11:033003. [Google Scholar]
- 7.Curutchet C, Mennucci B. Quantum chemical studies of light harvesting. Chem Rev. 2017;117:294–343. doi: 10.1021/acs.chemrev.5b00700. [DOI] [PubMed] [Google Scholar]
- 8.Jurinovich S, Viani L, Curutchet C, Mennucci B. Limits and potentials of quantum chemical methods in modelling photosynthetic antennae. Phys Chem Chem Phys. 2015;17:30783–30792. doi: 10.1039/c5cp00986c. [DOI] [PubMed] [Google Scholar]
- 9.Jurinovich S, Curutchet C, Mennucci B. The Fenna-Matthews-Olson protein revisited: A fully polarizable (TD)DFT/MM description. Chem Phys Chem. 2014;15:3194–3204. doi: 10.1002/cphc.201402244. [DOI] [PubMed] [Google Scholar]
- 10.Daday C, Curutchet C, Sinicropi A, Mennucci B, Filippi C. Chromophore-protein coupling beyond Nonpolarizable models: Understanding absorption in green fluorescent protein. J Chem Theor Comput. 2015;11:4825–4839. doi: 10.1021/acs.jctc.5b00650. [DOI] [PubMed] [Google Scholar]
- 11.Rosnik AM, Curutchet C. Theoretical characterization of the spectral density of the water-soluble chlorophyll-binding protein from combined quantum mechanics/molecular mechanics molecular dynamics simulations. J Chem Theor Comput. 2015;11:5826–5837. doi: 10.1021/acs.jctc.5b00891. [DOI] [PubMed] [Google Scholar]
- 12.van der Vegte CP, Prajapati JD, Kleinekathöfer U, Knoester J, Jansen TLC. Atomistic modeling of two-dimensional electronic spectra and excited-state dynamics for a light harvesting 2 complex. J Phys Chem B. 2015;119:1302–1313. doi: 10.1021/jp509247p. [DOI] [PubMed] [Google Scholar]
- 13.Kyung Lee Mi, Coker DF. Modeling electronic-nuclear interactions for excitation energy transfer processes in light-harvesting complexes. J Phys Chem Lett. 2016;7:3171–3178. doi: 10.1021/acs.jpclett.6b01440. [DOI] [PubMed] [Google Scholar]
- 14.König C, Neugebauer J. Protein effects on the optical spectrum of the Fenna-Matthews-Olson complex from fully quantum chemical calculations. J Chem Theor Comput. 2013;9:1808–1820. doi: 10.1021/ct301111q. [DOI] [PubMed] [Google Scholar]
- 15.Renger T, et al. Normal mode analysis of the spectral density of the Fenna-Matthews-Olson light-harvesting protein: How the protein dissipates the excess energy of excitons. J Phys Chem B. 2012;116:14565–14580. doi: 10.1021/jp3094935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anda A, De Vico L, Hansen T. Intermolecular modes between LH2 bacteriochlorophylls and protein residues: The effect on the excitation energies. J Phys Chem B. 2017;121:5499–5508. doi: 10.1021/acs.jpcb.7b02071. [DOI] [PubMed] [Google Scholar]
- 17.Angerhofer A, Cogdell RJ, Hipkins MF. A spectral characterisation of the light-harvesting pigment-protein complexes from Rhodopseudomonas acidophila. Biochim Biophys Acta Bioenerg. 1986;848:333–341. [Google Scholar]
- 18.Cogdell RJ, Durant I, Valentine J, Lindsay JG, Schmidt K. The isolation and partial characterisation of the light-harvesting pigment-protein complement of Rhodopseudomonas acidophila. Biochim Biophys Acta Bioenerg. 1983;722:427–435. [Google Scholar]
- 19.Georgakopoulou S, et al. Absorption and CD spectroscopy and modeling of various LH2 complexes from purple bacteria. Biophys J. 2002;82:2184–2197. doi: 10.1016/S0006-3495(02)75565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardiner AT, Cogdell RJ, Takaichi S. The effect of growth conditions on the light-harvesting apparatus in Rhodopseudomonas acidophila. Photosynth Res. 1993;38:159–167. doi: 10.1007/BF00146415. [DOI] [PubMed] [Google Scholar]
- 21.Fowler GJS, Visschers RW, Grief GG, van Grondelle R, Hunter CN. Genetically modified photosynthetic antenna complexes with blueshifted absorbance bands. Nature. 1992;355:848–850. doi: 10.1038/355848a0. [DOI] [PubMed] [Google Scholar]
- 22.Cogdell RJ, Howard TD, Isaacs NW, McLuskey K, Gardiner AT. Structural factors which control the position of the Qy absorption band of bacteriochlorophyll a in purple bacterial antenna complexes. Photosynth Res. 2002;74:135–141. doi: 10.1023/A:1020995224156. [DOI] [PubMed] [Google Scholar]
- 23.McLuskey K, Prince SM, Cogdell RJ, Isaacs NW. The crystallographic structure of the B800-820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050. Biochemistry. 2001;40:8783–8789. doi: 10.1021/bi010309a. [DOI] [PubMed] [Google Scholar]
- 24.Anda A, Hansen T, Vico LD. Multireference excitation energies for bacteriochlorophylls a within light harvesting system 2. J Chem Theor Comput. 2016;12:1305–1313. doi: 10.1021/acs.jctc.5b01104. [DOI] [PubMed] [Google Scholar]
- 25.Bednarczyk D. Fine tuning of chlorophyll spectra by protein-induced ring deformation. Angew Chem Int Ed. 2016;55:6901–6905. doi: 10.1002/anie.201512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacGowan SA, Senge MO. Contribution of bacteriochlorophyll conformation to the distribution of site-energies in the FMO protein. Biochim Biophys Acta. 2016;1857:427–442. doi: 10.1016/j.bbabio.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Gentemann S, et al. Photophysical properties of conformationally distorted metal-free porphyrins. Investigation into the deactivation mechanisms of the lowest excited singlet state. J Am Chem Soc. 1994;116:7363–7368. [Google Scholar]
- 28.Gentemann S, et al. Variations and temperature dependence of the excited state properties of conformationally and electronically perturbed zinc and free base porphyrins. J Phys Chem B. 1997;101:1247–1254. [Google Scholar]
- 29.Bai F-Q, Nakatani N, Nakayama A, Hasegawa J-y. Excited states of a significantly ruffled porphyrin: Computational study on structure-induced rapid decay mechanism via intersystem crossing. J Phys Chem A. 2014;118:4184–4194. doi: 10.1021/jp502349h. [DOI] [PubMed] [Google Scholar]
- 30.Karcz D, et al. Lessons from chlorophylls: Modifications of porphyrinoids towards optimized solar energy conversion. Molecules. 2014;19:15938–15954. doi: 10.3390/molecules191015938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivashin NV, Shchupak EE, Panarin AY, Sagun EI. Photophysical properties of porphyrins with sterically distorted and partially screened macrocycles. Opt Spectrosc. 2015;118:882–892. [Google Scholar]
- 32.Nottoli M, et al. The role of charge-transfer states in the spectral tuning of antenna complexes of purple bacteria. Photosynth Res. 2018;137:215–226. doi: 10.1007/s11120-018-0492-1. [DOI] [PubMed] [Google Scholar]
- 33.Montemayor D, Rivera E, Jang SJ. Computational modeling of exciton-bath Hamiltonians for light harvesting 2 and light harvesting 3 complexes of purple photosynthetic bacteria at room temperature. J Phys Chem B. 2018;122:3815–3825. doi: 10.1021/acs.jpcb.8b00358. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Cryst. 1976;32:922–923. [Google Scholar]
- 35.Kromann JC, Steinman C, Bratholm L, aandi, Lauritzen KP. 2016 doi: 10.5281/zenodo.46697. Data from “Calculate RMSD for two XYZ structures.” Zenodo. . Accessed November 4, 2017. [DOI]
- 36.Osipov VA, De Vico L, Anda A, Hansen T. 2017. The minimal model of light harvesting complex with dipole-quadrupole interaction derived from ab initio calculation. arXiv:1707.09053v1. Preprint, posted July 27, 2017.
- 37.Papiz MZ, Prince SM, Howard T, Cogdell RJ, Isaacs NW. The structure and thermal motion of the b800-850 LH2 complex from Rps. acidophila at 2.0 Å resolution and 100 k: New structural features and functionally relevant motions. J Mol Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 38.List NH, Curutchet C, Knecht S, Mennucci B, Kongsted J. Toward reliable prediction of the energy ladder in multichromophoric systems: A benchmark study on the FMO light-harvesting complex. J Chem Theor Comput. 2013;9:4928–4938. doi: 10.1021/ct400560m. [DOI] [PubMed] [Google Scholar]
- 39.Jentzen W, Song X-Z, Shelnutt JA. Structural characterization of synthetic and protein-bound porphyrins in terms of the lowest-frequency normal coordinates of the macrocycle. J Phys Chem B. 1997;101:1684–1699. [Google Scholar]
- 40.Engel GS, et al. Evidence for wavelike energy transfer through quantum coherence in photosynthetic systems. Nature. 2007;446:782–786. doi: 10.1038/nature05678. [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Cheng Y-C, Fleming GR. Coherence dynamics in photosynthesis: Protein protection of excitonic coherence. Science. 2007;316:1462–1465. doi: 10.1126/science.1142188. [DOI] [PubMed] [Google Scholar]
- 42.Roos BO, Taylor PR, Siegbahn PEM. A complete active space SCF method (CASSCF) using a density matrix formulated super-CI approach. Chem Phys. 1980;48:157–173. [Google Scholar]
- 43.Malmqvist P-Å, Pierloot K, Moughal Shahi AR, Cramer CJ, Gagliardi L. The restricted active space followed by second-order perturbation theory method: Theory and application to the study of CuO2 and Cu2O2 systems. J Chem Phys. 2008;128:204109. doi: 10.1063/1.2920188. [DOI] [PubMed] [Google Scholar]
- 44.Roos BO, et al. 1996. Multiconfigurational perturbation theory: Applications in electronic spectroscopy. Advances in Chemical Physics: New Methods in Computational Quantum Mechanics, eds Prigogine I, Rice SA (John Wiley & Sons, New York), pp 219–332.
- 45.Finley J, Malmqvist P-Å, Roos BO, Serrano-Andrés L. The multi-state CASPT2 method. Chem Phys Lett. 1998;288:299–306. [Google Scholar]
- 46.Roos BO, Lindh R, Malmqvist P-Å, Veryazov V, Widmark P-O. Main group atoms and dimers studied with a new relativistic ANO basis set. J Phys Chem A. 2004;108:2851–2858. [Google Scholar]
- 47.Runge E, Gross EKU. Density-functional theory for time-dependent systems. Phys Rev Lett. 1984;52:997–1000. [Google Scholar]
- 48.Dreuw A, Head-Gordon M. Single-reference ab initio methods for the calculation of excited states of large molecules. Chem Rev. 2005;105:4009–4037. doi: 10.1021/cr0505627. [DOI] [PubMed] [Google Scholar]
- 49.Casida ME, Huix-Rotllant M. Progress in time-dependent density-functional theory. Annu Rev Phys Chem. 2012;63:287–323. doi: 10.1146/annurev-physchem-032511-143803. [DOI] [PubMed] [Google Scholar]
- 50.Adamo C, Barone V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J Chem Phys. 1999;110:6158–6170. [Google Scholar]
- 51.Krishnan R, Binkley JS, Seeger R, Pople JA. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys. 1980;72:650–654. [Google Scholar]
- 52.McLean AD, Chandler GS. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z=11–18. J Chem Phys. 1980;72:5639–5648. [Google Scholar]
- 53.Clark T, Chandrasekhar J, Spitznagel GW, Schleyer PVR. Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21+g basis set for first-row elements, Li-F. J Comput Chem. 1983;4:294–301. [Google Scholar]
- 54.Tanaka M, Katouda M, Nagase S. Optimization of RI-MP2 auxiliary basis functions for 6-31g** and 6-311g** basis sets for first-, second-, and third-row elements. J Comput Chem. 2013;34:2568–2575. doi: 10.1002/jcc.23430. [DOI] [PubMed] [Google Scholar]
- 55.Aquilante F, et al. Molcas 7: The next generation. J Comput Chem. 2010;31:224–247. doi: 10.1002/jcc.21318. [DOI] [PubMed] [Google Scholar]
- 56.Aquilante F, et al. MOLCAS 8: New capabilities for multiconfigurational quantum chemical calculations across the periodic table. J Comput Chem. 2016;37:506–541. doi: 10.1002/jcc.24221. [DOI] [PubMed] [Google Scholar]
- 57.Aidas K, et al. The Dalton quantum chemistry program system. Wiley Interdiscip Rev: Comput Mol Sci. 2014;4:269–284. doi: 10.1002/wcms.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.