Significance
Antibiotic tolerance causes antibiotic treatment failure and promotes the emergence of genotypic resistance in chronic infections, such as those caused by the pathogen Pseudomonas aeruginosa. Laboratory stationary-phase bacteria exhibit a slow growing and metabolically quiescent state associated with high levels of multidrug tolerance likely analogous to the in vivo environment during chronic infection. We demonstrate that superoxide dismutases confer multidrug tolerance in stationary-phase bacteria, and identify a link between (p)ppGpp-mediated stress responses, superoxide metabolism, and membrane permeability to antibiotics. Inhibition of superoxide dismutase activity may overcome multidrug tolerance and potentiate current bactericidal antibiotics in the treatment of P. aeruginosa chronic infections.
Keywords: (p)ppGpp stringent response, Pseudomonas aeruginosa, antibiotic tolerance, superoxide dismutase, stationary phase
Abstract
Metabolically quiescent bacteria represent a large proportion of those in natural and host environments, and they are often refractory to antibiotic treatment. Such drug tolerance is also observed in the laboratory during stationary phase, when bacteria face stress and starvation-induced growth arrest. Tolerance requires (p)ppGpp signaling, which mediates the stress and starvation stringent response (SR), but the downstream effectors that confer tolerance are unclear. We previously demonstrated that the SR is linked to increased antioxidant defenses in Pseudomonas aeruginosa. We now demonstrate that superoxide dismutase (SOD) activity is a key factor in SR-mediated multidrug tolerance in stationary-phase P. aeruginosa. Inactivation of the SR leads to loss of SOD activity and decreased multidrug tolerance during stationary phase. Genetic or chemical complementation of SOD activity of the ΔrelA spoT mutant (ΔSR) is sufficient to restore antibiotic tolerance to WT levels. Remarkably, we observe high membrane permeability and increased drug internalization upon ablation of SOD activity. Combined, our results highlight an unprecedented mode of SR-mediated multidrug tolerance in stationary-phase P. aeruginosa and suggest that inhibition of SOD activity may potentiate current antibiotics.
Treatment of chronic bacterial infections in the clinic often results in failure or relapses. A drug refractory state, commonly referred to as antibiotic tolerance, occurs even when the infecting organisms harbor no genotypic (heritable) resistance to the antibiotic, and renders many chronic infections difficult to eradicate (1–3). Medically important examples include chronic Pseudomonas aeruginosa lung infections in individuals with the genetic disease cystic fibrosis. Tolerance can also promote the emergence of genotypic drug resistance, thereby posing a major public health challenge (4). Bacteria develop drug tolerance during growth-limiting conditions when they adopt a slow or nonreplicating state, and a fraction of the population survives bactericidal drugs (5). In fact, a large proportion of microbes found in natural environments and in vivo during chronic human infections are likely metabolically quiescent (2, 6).
Laboratory stationary-phase bacteria provide a valuable window into the metabolically quiescent organisms widely observed in nature. The physiology of exponentially growing bacteria change remarkably as they enter stationary phase, yet little is known about the survival strategies of slow or nongrowing cells (7, 8). Stationary-phase bacteria must respond and adapt to a variety of growth-limiting stress and starvation cues (e.g., nutrient exhaustion, pH changes, oxidative or nitrosative stress) through processes regulated by the alternative σ-factor RpoS and (p)ppGpp signaling in Escherichia coli and P. aeruginosa (9, 10). The alarmone (p)ppGpp accumulates upon stress and starvation, leading to a global reorganization of cellular and metabolic functions that promote stress adaptation and cell survival, a process termed the stringent response (SR) (11, 12).
Antibiotic tolerance among metabolically quiescent bacteria is widely attributed to the notion that drug targets are unavailable or inactive when cellular replication and macromolecule synthesis are shut down. Although antibiotic killing typically correlates with bacterial growth rate (13, 14), the lack of replication alone in the absence of (p)ppGpp signaling and downstream adaptive responses is often insufficient to confer tolerance (15–17). The downstream cellular processes that protect against antibiotic toxicity remain poorly understood. We previously observed that SR inactivation in the (p)ppGpp-null ΔrelA spoT mutant of P. aeruginosa (ΔSR) impairs multidrug tolerance in nutrient-limited, biofilm and stationary-phase bacteria (16, 18). Notably, the ΔSR mutant exhibited impaired superoxide dismutase (SOD) and catalase activities, leading us to propose that SR-mediated multidrug tolerance is linked to enhanced antioxidant defenses (16, 18).
Superoxide radicals are by-products of aerobic metabolism and a primary source of intracellular oxidative stress (19). Superoxide causes toxicity through direct damage of iron-containing enzymes, and indirectly through highly reactive hydroxyl radicals generated by Fenton chemistry (20). SODs rapidly disproportionate superoxide to oxygen and hydrogen peroxide, and the latter is detoxified by catalases and peroxidases. P. aeruginosa encodes two different SODs, SodA and SodB. The Fe-cofactored SodB is the most abundant in iron-replete conditions, while the Mn-cofactored SodA is under iron-dependent repression and only expressed under iron limitation (21, 22). In this study, we demonstrate that SOD activity is a critical effector of SR-mediated multidrug tolerance in stationary-phase P. aeruginosa, and that SOD activity is correlated with membrane permeability and drug internalization. Our data demonstrate a link between antioxidant defenses, drug permeability, and SR-mediated drug tolerance when P. aeruginosa are metabolically quiescent.
Materials and Methods
Experimental details can be found in the SI Appendix.
Bacterial Strains and Plasmids.
All strains and plasmids used are listed in SI Appendix, Tables S1 and S2. The P. aeruginosa laboratory strain PAO1 is the parental WT strain. The (p)ppGpp-null isogenic ΔSR mutant carries unmarked deletions of both (p)ppGpp synthetases relA and spoT, and the +SR strain is the ΔSR mutant complemented for the relA and spoT genes (16).
Media and Growth Conditions.
Bacteria were grown in LB Miller medium as described in SI Appendix. Exponential phase cells were grown for ∼2 h to an OD600 = 0.2, and stationary-phase cells for 16 h to an OD600 = ∼3.5.
SOD Activity Assays.
Total SOD and Sod-specific activities of cell lysates were measured using an in-solution and an in-gel activity assay respectively as described previously (23).
Antibiotic Killing Assays.
Exponential or stationary-phase cultures were challenged with antibiotics without addition of fresh medium, and incubated in 96-well plates at 37 °C with shaking at 250 rpm. At specific time points, cells were mixed with 1:1 activated charcoal to bind free drug, and viable bacteria were measured by serial microdilution and overnight growth of CFU on LB agar plates.
Results
(p)ppGpp Signaling Is Required for the Multidrug Tolerance of Stationary-Phase P. aeruginosa.
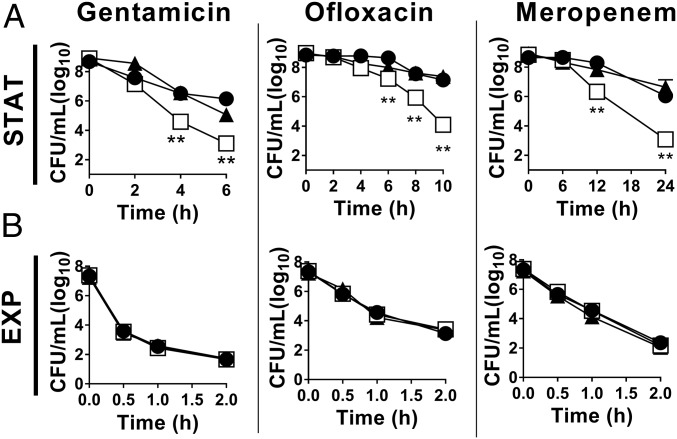
Stationary-phase bacteria are highly drug tolerant compared with their exponentially growing counterparts. To examine the contribution of (p)ppGpp signaling in P. aeruginosa stationary-phase drug tolerance, we challenged the (p)ppGpp-null ΔSR mutant to multiple distinct classes of bactericidal antibiotics and compared it to its WT isogenic parental strain. Stationary-phase ΔSR mutant cells are highly impaired for tolerance compared with WT (Fig. 1A), with 3–4 log10 greater antibiotic killing by the aminoglycoside gentamicin (5.9- vs. 2.9-log10 killing of ΔSR vs. WT, respectively, at t = 6 h), the fluoroquinolone ofloxacin (4.9- vs. 1.9-log10 killing at t = 10 h), and the β-lactam meropenem (6.0- vs. 2.9-log10 killing at t = 24 h). In contrast, exponential phase WT and ΔSR bacteria are equally susceptible to all three drugs and undergo rapid killing (Fig. 1B). Multidrug tolerance is fully restored to WT levels upon complementation of the ΔSR mutant with the relA and spoT genes (+SR), confirming that the loss of tolerance is attributable to relA and spoT mutations. Notably, the bacterial viability in stationary phase and growth rate in exponential phase are similar between the WT, ΔSR, and +SR strains (SI Appendix, Fig. S1 A and B). Deletion of relA and spoT in two additional P. aeruginosa clinical strains to abrogate (p)ppGpp synthesis also results in loss of stationary-phase multidrug tolerance, although the magnitude of this effect differs in different P. aeruginosa genetic backgrounds (SI Appendix, Fig. S2A).
Fig. 1.
SR inactivation impairs multidrug tolerance in stationary phase P. aeruginosa. (A) Stationary phase (STAT) or (B) exponential phase (EXP) cells of (●) WT, (□) ΔSR, and (▲) +SR challenged with 50 µg/mL gentamicin, 5 µg/mL ofloxacin, or 300 µg/mL meropenem in antibiotic killing assays. Note the different time scale in A and B. Results are mean ± SEM (n = 6). **P < 0.01 vs. WT.
SOD Activity Is Induced During Stationary Phase and Requires (p)ppGpp Signaling.
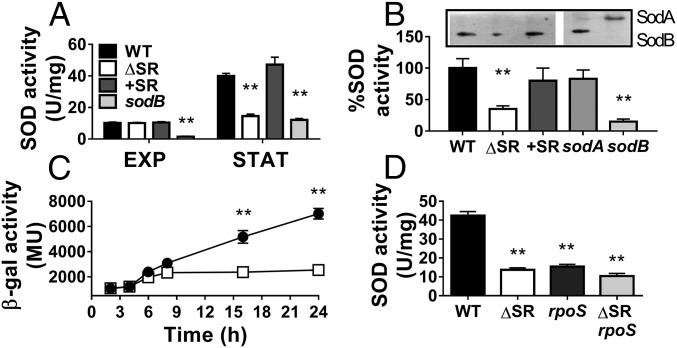
Our recent observation that ΔSR mutant biofilms exhibit low SOD activity led us to examine whether SOD activity requires (p)ppGpp signaling in planktonic stationary-phase bacteria. We first note a fourfold increase in SOD activity upon transition of the WT from exponential to stationary phase. However, this induction is largely abrogated in the ΔSR mutant and the SOD activity in stationary-phase ΔSR cells is reduced to 35% of WT levels (40 vs. 14.5 U/mg, P < 0.01) (Fig. 2A). We also observed similar effects upon inactivation of the relA and spoT genes in two P. aeruginosa clinical isolates (SI Appendix, Fig. S1B). Notably, differences in SOD activity are not due to disparities in bacterial growth rate, viability, or total cellular protein content (SI Appendix, Fig. S1 B and C). In addition, complementation of the ΔSR mutant with the relA and spoT genes restores SOD activity to WT levels, and confirms that the SOD defect is attributable to the relA and spoT mutations. These observations thus indicate that (p)ppGpp signaling is required for full SOD activity during stationary phase.
Fig. 2.
SOD activity and sodB expression are induced during stationary phase in the WT but not the ΔSR mutant. (A) Total SOD activity of exponential (EXP) or stationary phase (STAT) WT, ΔSR, +SR, and sodB strains. (B) SodA and SodB specific activities in stationary phase cells measured by in-gel SOD activity assays, with a representative gel image shown. SodB activity in the sodA mutant and SodA activity in the sodB mutant were confirmed as control. (C) sodB-lacZ reporter activities in the WT (●) or ΔSR (□) cells during growth in LB medium. (D) Total SOD activity in stationary-phase WT, ΔSR, rpoS, and ΔSR rpoS strains measured by in-solution assay. All results are shown as mean ± SEM (n ≥ 5). **P < 0.01 vs. WT in the corresponding growth phase.
SodB Is the Dominant SOD in Stationary Phase and Is Regulated by the SR and RpoS.
We showed by in-gel SOD activity assay that SodB confers all SOD activity in stationary-phase WT, ΔSR, and +SR cells, while SodA activity is undetectable under these conditions (Fig. 2B). Mirroring SodB activity, sodB expression, as measured by a sodB-lacZ transcriptional reporter, is induced in WT cells but not in the ΔSR mutant once cells enter stationary phase (t = 8 h). For example, sodB expression is 2.2- to 2.8-fold lower in stationary phase ΔSR compared with WT cells (Fig. 2C) in the absence of any differences in growth rate (SI Appendix, Fig. S1D).
The SR and the alternative σ-factor RpoS regulatory networks significantly overlap to control gene expression during stationary phase (24), and here we show that SodB activity is under both SR and RpoS control. The ΔSR and rpoS mutants show equally reduced SOD activity compared with WT (∼15 vs. 42 U/mg, P < 0.01) (Fig. 2D). Furthermore, the modest reduction in SOD activity in the ΔSR rpoS triple mutant compared with the rpoS mutant (10 vs. 14 U/mg, respectively, P < 0.05) further supports the overlapping SR- and RpoS-dependent regulation of SodB activity, and presumably sodB expression.
Stationary-Phase ΔSR Cells Have Higher Superoxide Levels and Lower Paraquat Tolerance than WT Cells.
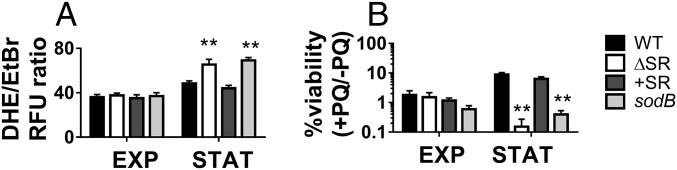
Because of reduced SOD activity, we predicted that ΔSR cells would be impaired in superoxide detoxification, leading to elevated superoxide levels and greater susceptibility to paraquat, a superoxide-generating compound. We therefore monitored relative intracellular superoxide levels using dihydroethidium (DHE), a cell-permeable probe that, when oxidized by superoxide, is converted to the fluorescent product 2-hydroxyethidium (25). DHE fluorescence intensity is ∼2.5-fold higher in the ΔSR and sodB mutants compared with WT and +SR strains in stationary, but not exponential phase (SI Appendix, Fig. S3 A and B). To account for potential differences in DHE probe loading, we used structurally similar ethidium bromide (EtBr) as a loading control and calculated DHE/EtBr fluorescence ratios as an estimate of relative superoxide levels. The DHE/EtBr ratio is 1.4-fold higher in the ΔSR and sodB mutants compared with the WT and +SR in stationary phase, but no differences are seen in exponential phase (Fig. 3A). Because aerobic respiration is a major source of superoxide (26), we also estimated the respiratory activity using resazurin reduction (27) and found no differences between the strains (SI Appendix, Fig. S1E). This leads us to infer that the excess accumulation of superoxide in the ΔSR mutant is likely attributable to its SOD defect, although other nonrespiratory sources of superoxide cannot be excluded. Additionally, we tested susceptibility to paraquat, a superoxide-generating compound, and found that killing by paraquat correlated with the SOD activity profiles across strains and growth phases (Fig. 3B). Combined, these results suggest that inactivation of the SR impairs protection against superoxide-mediated toxicity during stationary phase.
Fig. 3.
Inactivation of the SR is associated with increased superoxide levels and paraquat killing. (A) Relative intracellular superoxide levels using the DHE/EtBr fluorescence ratio. (B) Paraquat (PQ) killing of exponential (EXP) or stationary-phase (STAT) cells, calculated as percent bacterial survival after 6-h challenge with 10 mM PQ, compared with similar conditions without PQ. Results are shown as mean ± SEM (n ≥ 6). **P < 0.01 vs. WT.
Genetic or Biochemical Restoration of SOD Activity Rescues Stationary-Phase Multidrug Tolerance.
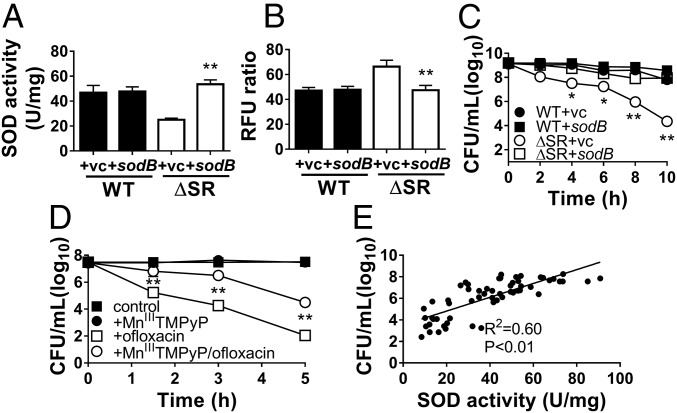
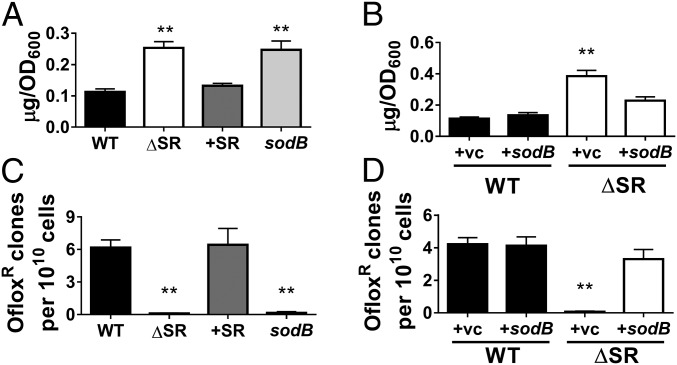
We first expressed pBAD-sodB in the ΔSR mutant (ΔSR +sodB), which restores its SOD activity (Fig. 4A) and DHE fluorescence levels (Fig. 4B) to WT levels. Notably, sodB expression in the ΔSR mutant also restores ofloxacin (Fig. 4C), gentamicin, and meropenem tolerance (SI Appendix, Fig. S4 A and B) to WT levels. Expression of sodA, the Mn-dependent SOD, similarly restores SOD activity and drug tolerance (SI Appendix, Fig. S5). Next, we chemically complemented the ΔSR mutant with 100 µM MnIIITMPyP, a cell-permeable SOD mimetic that catalytically dismutates superoxide at rates ∼10-fold lower than Mn-SodA and Fe-SodB (4 × 107 M−1s−1 vs. 7 × 108 M−1s−1) (28, 29). MnIIITMPyP confers protection against antibiotic killing, with 2- to 3-log10 higher viable CFU counts following challenges with ofloxacin (Fig. 4D), gentamicin, and meropenem (SI Appendix, Fig. S4 C and D). These results thus suggest that restoring SOD activity in the ΔSR mutant is sufficient to rescue its stationary-phase tolerance defect.
Fig. 4.
Complementation of SOD activity restores superoxide levels and rescues ofloxacin tolerance in the ΔSR mutant. Stationary-phase WT and ΔSR cells carrying the pBAD-sodB (+sodB) or control (+vc) vector were assayed for (A) total SOD activity, (B) relative superoxide levels (DHE/EtBr fluorescence ratio), or (C) killing by 5 µg/mL ofloxacin. (D) Stationary-phase ΔSR cells pretreated ± the SOD mimetic MnIIITMPyP before challenge with 5 µg/mL ofloxacin. All results are shown as mean ± SEM (n ≥ 6). *P < 0.05 and **P < 0.01 vs. WT +vc (in A to C) or vs. “+ofloxacin” (in D). (E) Correlation between bacterial viability of stationary-phase cells after ofloxacin challenge and SOD activity. The SOD activity was measured in stationary-phase cultures before ofloxacin challenge, and bacterial viability of the same culture was measured after 10-h challenge of 5 µg/mL ofloxacin. Data from different strains (WT, ΔSR, ΔSR +sodB, ΔSR +sodA, +SR) were combined and each data point represents an independent replicate (n ≥ 40). The correlation coefficient R2 was calculated using linear regression.
SOD Activity Correlates with Multidrug Tolerance in a Dose-Dependent Manner.
We exploited the biological variability of SOD activity in independent replicate batch cultures to assess the dose-dependent relationship between SOD activity and antibiotic tolerance. We measured SOD activity of a culture before antibiotic challenge and its bacterial viability following antibiotic challenge, and found a strong positive correlation with ofloxacin (R2 = 0.60, P < 0.01) (Fig. 4E), gentamicin (R2 = 0.62, P < 0.01), and meropenem tolerance (R2 = 0.64, P < 0.01) (SI Appendix, Fig. S4 E and F).
Stationary-Phase ΔSR Cells Have Increased Membrane Permeability, Which Correlates with Antibiotic Killing.
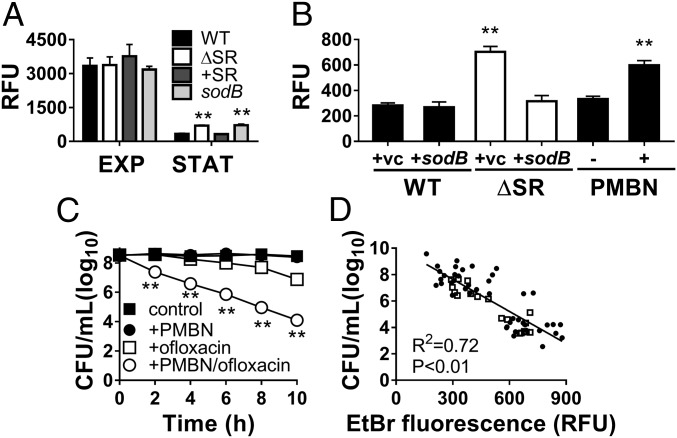
Internalization EtBr, which was used as a loading control for the DHE probe, also provides a well-established measure of global membrane permeability (30, 31). As expected, we first observed that EtBr internalization is 5- to 10-fold higher in exponential vs. stationary phase for all strains (Fig. 5A and SI Appendix, Fig. S3C). However, EtBr internalization is ∼twofold higher in the ΔSR and sodB mutants compared with WT and +SR strains during stationary phase, while no differences are seen during exponential phase. Furthermore, genetic restoration of SOD activity with pBAD-sodB expression in the ΔSR mutant restores its EtBr internalization to WT levels (Fig. 5B). Given that EtBr is a substrate for efflux pumps, which are highly expressed in P. aeruginosa, we wanted to confirm that differences in EtBr internalization were not due to differential efflux. To estimate of efflux activity, we measured the ratio of EtBr fluorescence with and without CCCP, a proton ionophore that inactivates efflux pumps, and found no differences between WT, ΔSR, and +SR cells (SI Appendix, Fig. S6 A and B). Finally, to further assess the outer membrane permeability, we carried out a periplasmic β-lactamase leakage assay (32) and found that ΔSR cells have leakier outer membranes than WT cells (SI Appendix, Fig. S6C), consistent with the EtBr results. Together, our results demonstrate that membrane permeability decreases in the WT upon transition from exponential to stationary phase, and this process is impaired in the ΔSR and sodB mutants.
Fig. 5.
The SR and SOD activity confers membrane impermeability, a key determinant of stationary-phase tolerance. EtBr internalization in the presence of CCCP in A of exponential (EXP) or stationary-phase (STAT) cells, (B) stationary-phase WT and ΔSR cells carrying the pBAD-sodB (+sodB) or control (+vc) vector, or in stationary-phase WT cells ±50 µg/mL PMBN. (C) Killing of stationary-phase WT with 5 µg/mL ofloxacin ± PMBN pretreatment. All results are shown as mean ± SEM (n ≥ 6). **P < 0.01 vs. “WT+vc” or vs. “+PMBN”. (D) Correlation between bacterial viability of stationary-phase cells after ofloxacin challenge and membrane permeability, as measured by the EtBr internalization assay of the same culture before ofloxacin challenge (5 µg/mL for 10 h). Data from different strains (WT, ΔSR, ΔSR +sodB, ΔSR +sodA, +SR) noted as ● or WT +50 µg/mL PMBN noted as □, were combined and each data point (n ≥ 40) represents an independent replicate. The correlation coefficient R2 was calculated using linear regression.
To further validate the contribution of membrane permeability to drug tolerance, we permeabilized stationary-phase cells using a chemical approach with polymyxin B nonapeptide (PMBN). PMBN is a cationic peptide that binds outer membrane lipopolysaccharides, leading to permeabilization of the outer membrane without intrinsic bactericidal activity (33). Stationary-phase WT cells challenged with 50 µg/mL PMBN alone show an ∼twofold increase in EtBr fluorescence (Fig. 5B) without loss of viability (Fig. 5C). Importantly, pretreatment of stationary-phase WT cells with PMBN significantly enhances ofloxacin killing (Fig. 5C) (4.4- vs. 1.4-log10 killing at t = 10 h), as well as gentamicin and meropenem killing (SI Appendix, Fig. S7 A and B). Finally, we observed a significant linear correlation between EtBr fluorescence and tolerance to all three drugs (Fig. 5D and SI Appendix, Fig. S7 C and D), suggesting that membrane permeability is a major determinant of SR- and SOD-dependent antibiotic tolerance in stationary-phase P. aeruginosa.
Drug Penetration Is Enhanced by the ΔSR and sodB Mutant Permeability Defect.
Membrane permeability is a major determinant of drug uptake and internalization. We thus directly assessed intracellular drug levels in stationary-phase cells using ofloxacin (which is intrinsically fluorescent) (SI Appendix, Fig. S6 A and B), Texas Red-labeled gentamicin (SI Appendix, Fig. S8 A and B), and FITC-labeled meropenem (SI Appendix, Fig. S8 C and D). Stationary-phase ΔSR and sodB cells exhibit 2- to 2.5-fold higher drug uptake than WT or +SR complemented cells for all three drugs: ofloxacin (Fig. 6A), Texas Red gentamicin (SI Appendix, Fig. S8A), and FITC-meropenem (SI Appendix, Fig. S8C). Furthermore, sodB expression in the ΔSR mutant restores drug accumulation to WT levels (Fig. 6B and SI Appendix, Fig. S8 B and D). As controls, we confirmed that uptake of unconjugated Texas Red and FITC fluorophores is minimal and that the bactericidal activity of labeled drugs is similar to that of unconjugated ones (SI Appendix, Fig. S9).
Fig. 6.
Loss of (p)ppGpp signaling and SOD activity increase ofloxacin internalization and abrogates emergence of genotypic resistance to ofloxacin. Intracellular ofloxacin levels (RFU, Ex/Em 292/496 nm) in (A) stationary-phase WT, ΔSR, +SR, and sodB strains or (B) WT and ΔSR expressing the pBAD-sodB (+sodB) or control (+vc) vector. The number of ofloxacin-resistant colonies that arose between 3 and 5 d after incubation of 1011 cells of each strain on 12 µg/mL ofloxacin LB agar plates in (C) WT, ΔSR, +SR, and sodB strains and (D) WT and ΔSR expressing the pBAD-sodB (+sodB). Results are mean ± SEM (n ≥ 6). **P < 0.01 vs. WT (for A and C) or WT +vc (for B and D).
Taking these data together, we demonstrate that ablation of (p)ppGpp signaling or deletion of sodB compromises membrane permeability specifically during stationary phase, leading to enhanced drug penetration and thus drug killing. Because restoration of SOD activity in the ΔSR mutant is sufficient to rescue both membrane impermeability and drug accumulation, this suggests that SOD activity is critical to SR-mediated membrane impermeability and multidrug tolerance in stationary-phase cells.
Loss of SOD Activity Abrogates the Emergence of Drug Resistance.
Genotypically resistant mutants likely arise from tolerant bacterial populations that survive sustained antibiotic exposure (1, 4). Hence, the loss of tolerance should abrogate the emergence of genotypic resistance. To test this, we measured the emergence of drug-resistant colonies from cell suspensions (∼1011 CFU per plate) of stationary-phase bacteria spread and incubated for 5 d on agar plates containing ofloxacin. We enumerated newly emerging ofloxacin-resistant colonies after 72-h drug exposure to exclude those stemming from preexisting resistant cells. Consistent with our previous observations (16), we find that ablation of (p)ppGpp signaling all but eliminates the emergence of ofloxacin resistance, and this defect is restored in the complemented +SR strain (Fig. 6C). The development of ofloxacin resistance is also abrogated in sodB cells, while sodB expression in the ΔSR mutant restores the rate of ofloxacin-resistant mutants to WT levels (Fig. 6D). Hence, perturbations in SOD activity are sufficient to supress the emergence of ofloxacin resistance.
Discussion
Our group and others have shown that the SR and (p)ppGpp signaling mediate antibiotic tolerance, likely through several different mechanisms (15, 16, 34, 35). Among these, the most extensively studied mechanism is the formation of specialized drug-tolerant cells termed persister cells (36). In E. coli, (p)ppGpp signaling is central to the regulation of toxin–antitoxin module-dependent pathways involved in persister formation (15, 36). In P. aeruginosa, Verstraeten et al. (34) reported that the Obg GTPase induces generation of aminoglycoside-tolerant cells, but through mechanisms yet unclear in P. aeruginosa. Although such recent studies provide important insights into the molecular basis of persister formation, it remains unclear to what degree persister mechanisms overlap or differ from those implicated in stationary-phase tolerance.
P. aeruginosa remains viable in a nonreplicating state for prolonged periods of time (8) and displays a high level of multidrug tolerance during stationary phase. The SR acts as a major global regulator of bacterial stationary-phase physiology and stress responses that exerts widespread direct and indirect effects on gene transcription (11, 12). We show here that the SR modulates sodB transcription and total SOD activity in stationary-phase P. aeruginosa primarily through RpoS, which underscores the significant overlap between SR- and RpoS-dependent gene regulation, as we and others have previously noted (18, 37, 38). The link between the SR and SOD regulation is not widely conserved across different bacterial species (39–41) but SodB expression in E. coli is SR-dependent (42), and carbon starvation induces both SodA and SodB synthesis (43).
Despite the pleiotropic effects of the SR, genetic and chemical complementation of SOD activity is sufficient to restore multidrug tolerance of the ΔSR mutant to WT levels. This suggests that SOD activity is protective against antibiotic toxicity and plays a key role in (p)ppGpp-dependent multidrug tolerance during stationary phase. We observed a strong correlation between SOD activity and antibiotic survival to all three classes of drugs, across different isogenic strains constructed in the PAO1 genetic background. The role of the SR and SOD appears conserved in other P. aeruginosa clinical strains, although the magnitude of their effect likely varies based on the genetic background.
We recognize that our measurements of SOD activity and stationary-phase survival following antibiotic challenge reflect population rather than single-cell phenotypes. Gene expression is highly heterogeneous during stationary phase (8), and phenotypic heterogeneity allows bacterial populations to persist in the face of fluctuating environments and lethal stress, including antibiotics (17). Future studies with single-cell analyses would be informative in demonstrating whether variations in SOD activity confers survival to antibiotic stress at the individual cell level.
We used DHE to probe superoxide levels and acknowledge its lack of specificity because it can also react with hydroxyl radicals (44). However, in our experiments DHE fluorescence intensity varied with SOD activity in the sodB mutant (which has ∼30% of the SOD activity of WT cells) (Fig. 2A) and in the sodB expression constructs (Fig. 4B), suggesting that the DHE signal reflects relative superoxide levels.
Although superoxide radicals are considered a primary source of intracellular oxidative stress and likely contribute to the lethal effects of antibiotics (45–47), the biological consequences of elevated superoxide are not fully understood. Whether SOD activity confers tolerance directly through its ability to detoxify superoxide radicals, or indirectly through downstream cellular processes responsive to SOD activity, remains to be determined. The relationship between SOD activity, SOD-dependent effects, and superoxide metabolism is complex. SODs appear to have moonlighting functions in eukaryotic cells. For example, SOD1 in the yeast Saccharomyces cerevisiae also functions in signal transduction as its SOD activity stabilizes kinases involved in oxidative and metabolic responses (48), and it can act as a nuclear transcription factor (49). Whether such unorthodox SOD functions exist in bacteria remains unknown.
To date, studies that examined the role of SODs in antibiotic lethality in various bacteria report divergent conclusions. Sod mutants show increased susceptibility to bactericidal antibiotics in Campylobacter jejuni (50), Staphylococcus aureus (51), and Enterococcus faecalis (52), but not Acinetobacter baumannii (53). A sodA sodB mutant of E. coli is reportedly no more susceptible than WT to ampicillin, gentamicin, and norfloxacin killing (54, 55). Furthermore, expression of sodA or sodB did not mitigate ampicillin and ofloxacin killing of E. coli (56). However, several important biological and experimental differences likely account for the different observations on the relationship between SOD and antibiotic susceptibility. First, many metabolic and antioxidant responses differ between E. coli and P. aeruginosa, including RpoS (57) and SoxR (58), particularly during growth arrest (8). Second, experimental conditions to assess antibiotic lethality differ significantly between the different studies. Most notably, antibiotic killing assays in the E. coli studies (54–56) were carried out on actively replicating (exponential phase) cells, or cells that exit stationary phase to resume growth upon dilution into fresh medium. In contrast, we challenged stationary-phase cells under nongrowing conditions where new nutrients are not supplied, and our results demonstrate that SOD-mediated tolerance is growth phase-specific. Others have also reported that stationary-phase sodA sodB mutant E. coli is more susceptible to gentamicin killing when maintained in a nongrowing state (59). Combined, these results suggest that SOD is particularly important to antibiotic survival during stationary phase.
Interestingly, Dukan et al. (43) previously observed that SOD-deficient E. coli mutants exhibit increased protein oxidation, but only in stationary-phase cultures. They proposed that superoxide stress was a hallmark of respiring but nonreplicating stationary-phase cells (60). More recently, studies in E. coli and S. aureus reported that the antibiotic-tolerant stationary-phase cells and persister cells have low cellular ATP levels, and that manipulations that increase ATP levels also enhance antibiotic killing (61–63). Although increased respiration increases both ATP levels and superoxide generation, it is not known if SOD activity in turn alters ATP levels. Our results show that the differences in SOD activity and tolerance cannot be attributable to differences in respiration.
Here, we show that the P. aeruginosa cell envelope becomes less permeable during stationary phase, thus limiting drug penetration, and this process is SR- and SOD-dependent. Loss of cell permeability likely represents a common adaptive strategy for bacteria to survive under growth-limiting conditions (2). Alterations in cell envelope composition and permeability have been described in bacteria that transition into growth-limiting conditions, and those in stationary phase (64, 65). For example, the cell wall structure of stationary-phase S. aureus displays reduced cross-linking and increased peptidoglycan mass (66). Nutrient-starved and nonreplicating Mycobacterium tuberculosis have altered cell walls that display decreased permeability to chemically distinct classes of drugs, including fluoroquinolones (67). How SOD activity directly or indirectly contributes to the alterations in the cell envelope remains to be determined.
We measured intracellular drug concentrations, which result from the net effect of drug uptake and efflux. Because our results showed comparable efflux activity in the ΔSR and sodB mutants, this implies that increased drug accumulation is likely due to increased internalization. In Gram-negative bacteria, the outer membrane is widely recognized as the primary barrier to drug internalization, although the molecular mechanisms of drug uptake are often still poorly understood (68). We tested gentamicin, ofloxacin, and meropenem, three distinct classes of bactericidal antipseudomonal antibiotics targeting the bacterial ribosomal translational machinery, DNA gyrase, and peptidoglycan cell wall synthesis, respectively. We recognize that these chemically distinct drugs are likely internalized through different pathways. Aminoglycosides, such as gentamicin, require active uptake through a proton-gradient dependent process (69), while meropenem and ofloxacin likely diffuse passively through the membrane bilayer via outer membrane porins, such as oprD (70) and ompF (71), respectively. The mechanisms by which SR- and SOD-dependent tolerance influence these uptake processes remain unknown.
Our study uncovers an unprecedented link between SOD activity, SR-mediated multidrug tolerance, membrane permeability, and antibiotic internalization in stationary-phase P. aeruginosa. We also demonstrate that deletion of SOD-dependent pathways enhances antibiotic lethality and abrogates the emergence of genotypic resistance in stationary-phase P. aeruginosa. Thus, targeting the (p)ppGpp and SOD-dependent pathways may improve bactericidal activity against slow-growing bacteria and prevent the emergence of drug resistance in chronic infections.
Supplementary Material
Acknowledgments
We thank Dr. Joe Harrison (University of Calgary) for the Gateway vectors; Prof. Kazuhiro Iiyama (Kyushu University) for the sodA and sodB mutants; Prof. David Leonard (Grand Valley State University) for providing FITC-meropenem; and Drs. Pradeep K. Singh, Dianne Newman, and Lucas Hoffman for helpful discussions and review of our manuscript. This research was funded by the Burroughs Wellcome Fund (Award 1006827.01 to D.N.) and Canadian Institutes of Health Research (Grant MOP102727 to D.N.). D.N. is a Cystic Fibrosis Canada and Fonds de Recherche Santé Quebec Scholar, and D.M. is supported by a Canadian Institutes of Health Research and Cystic Fibrosis Canada fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804525115/-/DCSupplemental.
References
- 1.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 2.Rittershaus ESC, Baek S-H, Sassetti CM. The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meylan S, Andrews IW, Collins JJ. Targeting antibiotic tolerance, pathogen bypathogen. Cell. 2018;172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 4.Levin-Reisman I, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 5.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 6.Kopf SH, et al. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2016;113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegele DA, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergkessel M, Basta DW, Newman DK. The physiology of growth arrest: Uniting molecular and environmental microbiology. Nat Rev Microbiol. 2016;14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: An expanded stringent response model. Mol Microbiol. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- 10.Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 11.Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 14.Evans DJ, Allison DG, Brown MR, Gilbert P. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: Effect of specific growth rate. J Antimicrob Chemother. 1991;27:177–184. doi: 10.1093/jac/27.2.177. [DOI] [PubMed] [Google Scholar]
- 15.Germain E, Roghanian M, Gerdes K, Maisonneuve E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci USA. 2015;112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol. 2013;195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seaver LC, Imlay JA. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J Biol Chem. 2004;279:48742–48750. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- 20.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassett DJ, Schweizer HP, Ohman DE. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett DJ, et al. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc. 2010;5:51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvint K, Farewell A, Nyström T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s) J Biol Chem. 2000;275:14795–14798. doi: 10.1074/jbc.C000128200. [DOI] [PubMed] [Google Scholar]
- 25.Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Radic Biol Med. 2007;43:995–1022. doi: 10.1016/j.freeradbiomed.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Korshunov S, Imlay JA. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol. 2006;188:6326–6334. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman LR, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faulkner KM, Liochev SI, Fridovich I. Stable Mn(III) porphyrins mimic superoxide dismutase in vitro and substitute for it in vivo. J Biol Chem. 1994;269:23471–23476. [PubMed] [Google Scholar]
- 29.Gray B, Carmichael AJ. Kinetics of superoxide scavenging by dismutase enzymes and manganese mimics determined by electron spin resonance. Biochem J. 1992;281:795–802. doi: 10.1042/bj2810795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 31.Lee K, Jung J, Kim K, Bae D, Lim D. Overexpression of outer membrane protein OprT and increase of membrane permeability in phoU mutant of toluene-tolerant bacterium Pseudomonas putida GM730. J Microbiol. 2009;47:557–562. doi: 10.1007/s12275-009-0105-y. [DOI] [PubMed] [Google Scholar]
- 32.Nicas TI, Hancock RE. Pseudomonas aeruginosa outer membrane permeability: Isolation of a porin protein F-deficient mutant. J Bacteriol. 1983;153:281–285. doi: 10.1128/jb.153.1.281-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsubery H, Ofek I, Cohen S, Eisenstein M, Fridkin M. Modulation of the hydrophobic domain of polymyxin B nonapeptide: Effect on outer-membrane permeabilization and lipopolysaccharide neutralization. Mol Pharmacol. 2002;62:1036–1042. doi: 10.1124/mol.62.5.1036. [DOI] [PubMed] [Google Scholar]
- 34.Verstraeten N, et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell. 2015;59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Abranches J, et al. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 37.van Delden C, Comte R, Bally AM, Bally M. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun. 2004;72:5638–5645. doi: 10.1128/IAI.72.10.5638-5645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eymann C, Homuth G, Scharf C, Hecker M. Bacillus subtilis functional genomics: Global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol. 2002;184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- 42.Carneiro S, Villas-Bôas S, Ferreira EC, Rocha I. A comparative proteome analysis of Escherichia coli ΔrelA mutant cells. Front Bioeng Biotechnol. 2016;4:78. doi: 10.3389/fbioe.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dukan S, Nyström T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- 44.Kalyanaraman B, et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 46.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol. 2013;31:160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci USA. 2012;109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddi AR, Culotta VC. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152:224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XFS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang S, Ryu S, Jeon B. Roles of the superoxide dismutase SodB and the catalase KatA in the antibiotic resistance of Campylobacter jejuni. J Antibiot (Tokyo) 2013;66:351–353. doi: 10.1038/ja.2013.20. [DOI] [PubMed] [Google Scholar]
- 51.Ladjouzi R, et al. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J Antimicrob Chemother. 2013;68:2083–2091. doi: 10.1093/jac/dkt157. [DOI] [PubMed] [Google Scholar]
- 52.Bizzini A, Zhao C, Auffray Y, Hartke A. The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J Antimicrob Chemother. 2009;64:1196–1202. doi: 10.1093/jac/dkp369. [DOI] [PubMed] [Google Scholar]
- 53.Heindorf M, Kadari M, Heider C, Skiebe E, Wilharm G. Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One. 2014;9:e101033. doi: 10.1371/journal.pone.0101033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Zhao X. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother. 2009;53:1395–1402. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ezraty B, et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science. 2013;340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 56.Orman MA, Brynildsen MP. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat Commun. 2015;6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suh SJ, et al. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palma M, et al. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect Immun. 2005;73:2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang JH, et al. Sigma S-dependent antioxidant defense protects stationary-phase Escherichia coli against the bactericidal antibiotic gentamicin. Antimicrob Agents Chemother. 2014;58:5964–5975. doi: 10.1128/AAC.03683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dukan S, Nyström T. Bacterial senescence: Stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conlon BP, et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 62.Lobritz MA, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adolfsen KJ, Brynildsen MP. Futile cycling increases sensitivity toward oxidative stress in Escherichia coli. Metab Eng. 2015;29:26–35. doi: 10.1016/j.ymben.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kjelleberg S, Hermansson M, Mårdén P, Jones GW. The transient phase between growth and nongrowth of heterotrophic bacteria, with emphasis on the marine environment. Annu Rev Microbiol. 1987;41:25–49. doi: 10.1146/annurev.mi.41.100187.000325. [DOI] [PubMed] [Google Scholar]
- 65.Cronan JE., Jr Phospholipid alterations during growth of Escherichia coli. J Bacteriol. 1968;95:2054–2061. doi: 10.1128/jb.95.6.2054-2061.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou X, Cegelski L. Nutrient-dependent structural changes in S. aureus peptidoglycan revealed by solid-state NMR spectroscopy. Biochemistry. 2012;51:8143–8153. doi: 10.1021/bi3012115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarathy J, Dartois V, Dick T, Gengenbacher M. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eren E, et al. Toward understanding the outer membrane uptake of small molecules by Pseudomonas aeruginosa. J Biol Chem. 2013;288:12042–12053. doi: 10.1074/jbc.M113.463570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Isabella VM, et al. Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa. Chem Biol. 2015;22:535–547. doi: 10.1016/j.chembiol.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 71.Xia PY, Feng P, Zhong L, Lv XJ, Lei BJ. Accumulation of ofloxacin and tosufloxacin in fluoroquinolone-resistant E coli. Acta Pharmacol Sin. 2001;22:210–214. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.