Significance
Cohesin is an ATPase that organizes chromosome structure for chromosome segregation, gene expression, and DNA repair. Its function has been implicated in the prevention of cancer, birth defects, and several human disorders. Cohesin uses its ATPase, through elusive mechanisms, to tether DNA molecules and possibly translocate along them. Our analysis of cohesin with an active site mutation and an ATP analog suggest that a function of the ATPase is to generate an intermediate nucleotide state of cohesin ATPase, likely cohesinADP-Pi, that is competent to stably tether two DNA molecules. This state is potentially a trapped intermediate derived from a DNA translocation activity. These results suggest how the regulation of cohesin ATPase may interconvert its tethering and putative motor activity.
Keywords: cohesin, ATP binding cassette ATPase, cohesion, SMC, DNA binding
Abstract
Cohesin is a four-subunit ATPase in the family of structural maintenance of chromosomes (SMC). Cohesin promotes sister chromatid cohesion, chromosome condensation, DNA repair, and transcription regulation. Cohesin performs these functions as a DNA tether and potentially a DNA-based motor. At least one of its DNA binding activities involves entrapment of DNA within a lumen formed by its subunits. This activity can be reconstituted in vitro by incubating cohesin with DNA, ATP, and cohesin loader. Previously we showed that a mutant form of cohesin (DE-cohesin) possesses the ability to bind and tether DNA in vivo. Using in vitro reconstitution assays, we show that DE-cohesin can form stable complexes with DNA without ATP hydrolysis. We show that wild-type cohesin with ADP aluminum fluoride (cohesinADP/AlFx) can also form stable cohesin–DNA complexes. These results suggest that an intermediate nucleotide state of cohesin, likely cohesinADP-Pi, is capable of initially dissociating one interface between cohesin subunits to allow DNA entry into a cohesin lumen and subsequently interacting with the bound DNA to stabilize DNA entrapment. We also show that cohesinADP/AlFx binding to DNA is enhanced by cohesin loader, suggesting a function for loader other than stimulating the ATPase. Finally, we show that loader remains stably bound to cohesinADP/AlFx after DNA entrapment, potentially revealing a function for loader in tethering the second DNA substrate. These results provide important clues on how SMC complexes like cohesin can function as both DNA tethers and motors.
Higher order chromatin structure in prokaryotes and eukaryotes is orchestrated by the SMC (structural maintenance of chromosomes) family of protein complexes. Cohesin, one evolutionarily conserved member of this family, mediates sister chromatid cohesion and is a key contributor to condensation, DNA-damage repair, and regulation of gene expression (1–4). Cohesin performs these biological functions, at least in part, by tethering together two regions of DNA, either within a single DNA molecule or between two DNA molecules (3).
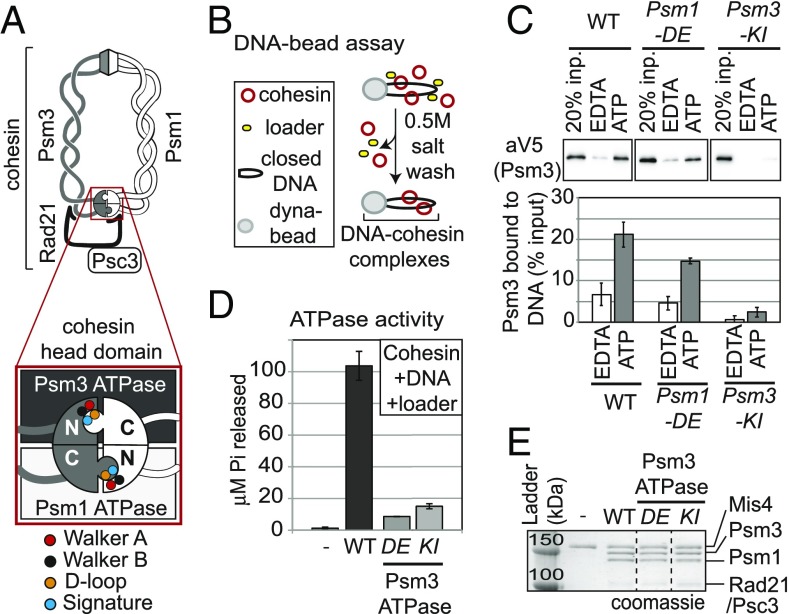
The complexity of cohesin’s DNA binding and tethering activities is inferred from its unusual architecture (5). The core of the cohesin complex from Schizosaccharomyces pombe is composed of Psm1 (Smc1) and Psm3 (Smc3). Each contains a globular hinge and head domain that lie at opposite ends of a long 40-nm coiled coil. The two Psm subunits dimerize via both head–head and hinge–hinge interactions. The interaction between the head domains is additionally bridged by Rad21 (Mcd1/Scc1) and Psc3 (Scc3) subunits (6). Dimerization of the head domains generates two composite ATPase active sites resembling those of ABC ATPases (7) (Fig. 1A). The composite Psm3 ATPase active site contains Walker A (ATP binding) and Walker B (ATP hydrolysis) motifs encoded by the Psm3 subunit, while the Psm1 subunit contributes the D loop and signature motifs (Fig. 1A). Conversely, the composite Psm1 ATPase active site contains Walker A and Walker B motifs from Psm1 while Psm3 encodes the D loop and signature motifs (8–10). Here, we investigate the molecular function of these ATPases in DNA binding.
Fig. 1.
DE-cohesin bound with ATP forms a salt-stable complex with DNA in absence of ATP hydrolysis. (A) Illustration of cohesin and its head domain (Inset) showing the two ATPase active sites. Psm1 ATPase Walker A mutant [Psm1-K38I abbreviated (abbr.) Psm1-KI] in red, Psm3 ATPase Walker A mutant (Psm3-K38I abbr. Psm3-KI) in red and Psm1 D-loop motif (Psm1-D1167E abbr. Psm1-DE) in orange are highlighted. As reference, Psm1/Psm3 Walker B (black) and signature (blue) motifs are shown. (B) Cartoon of the DNA-bead assay. Biotinylated DNA bearing CARC1 sequences was coupled to streptavidin-coated magnetic beads. Cohesin, loader, and depicted nucleotides were then added. Assembled cohesin–DNA complexes were washed in high salt (0.5 M KCl) to remove unstable cohesin–DNA complexes. (C) Effect of ATP on DNA binding of WT, Psm1 DE-, and Psm3 KI-cohesin. (Upper) Cohesin bound to DNA beads detected by Western Blot using anti-V5 antibodies (Psm3-3V5). (Lower) Quantitation of cohesin bound to DNA from Western blots in Upper. (D) Quantitation of ATPase activity of WT cohesin and Psm3 ATPase mutants (Psm1-DE and Psm3-KI) in the presence of loader and CARC1-containing plasmid DNA. (E) Coomassie-stained gel showing proteins present in ATPase experiments. Dotted lines represent where irrelevant lanes were removed.
A few important clues to cohesin’s ATPase molecular function have come from in vivo and in vitro experiments. Cohesin-harboring mutations in the Walker A or B sites of either cohesin ATPase active site are extremely compromised in their ability to bind chromosomes in vivo (9, 11). Consistent with these observations, neither ADP nor the nonhydrolyzable analog, ATPγS, can substitute for ATP in stimulating cohesin binding to DNA in vitro (12, 13). These results suggested that ATP binding and hydrolysis by both active sites were required for DNA binding in vivo and in vitro. An apparent exception to this conclusion was the observation that cohesin with Walker B mutations were found to bind DNA in vitro and to be bound to chromosomes immediately proximal to the centromeres in vivo (14, 15). However, recent in vitro experiments suggest that the Walker B mutations have residual ATPase activity (15). Thus, a requirement for cohesin ATPase activity in its DNA binding remains valid.
An explanation for the requirement of both ATP binding and hydrolysis came from the discovery that at least one of cohesin’s DNA binding activities involves topological entrapment of DNA within a lumen of a ring formed by cohesin subunits (16). Topological entrapment requires entry into the lumen by opening of at least one of the cohesin subunit interfaces, followed by closing the interface. A simple idea was that a nucleotide state (e.g., cohesinADP or the apo form) is open while another is closed (e.g., cohesinATP). ATP hydrolysis would serve to convert cohesin from the closed to open conformation (9, 11). Thus, interconversion of different nucleotide states of cohesin is thought to be required for loading onto DNA and stable binding. An auxiliary loader complex is required for cohesin’s DNA binding in vivo and enhances binding/ATPase activity in vitro (12, 17–19). Loader may facilitate DNA binding by modulating cohesin’s ATPase function (15).
This simple view of ATPase function was challenged first by the properties of a novel allele of the budding yeast Smc1 (13, 20, 21). This allele substitutes a glutamate (E) for the invariant aspartate (D) in the D loop of the Smc3 ATPase active site in all species. Cohesin with this mutation is referred henceforth as DE-cohesin. In vitro, purified DE-cohesin from S. pombe exhibits almost no ATPase activity, similar to that for cohesin with Walker A mutations (13). However, cells expressing only DE-cohesin are viable and have near normal cohesion (13). These results suggested that, in vivo, DE-cohesin entrapped DNA and tethered sister chromatids without continuous cycles of ATP hydrolysis, contradicting the apparent requirement for ATP hydrolysis deduced from studies of Walker A and B mutants (9, 22). These apparently contradictory results opened up the possibility that the role of cohesin’s ATPase in its DNA binding must be more complicated than simply interconverting cohesin between open (ADP bound or apo) and closed (ATP bound) states.
A more complex mechanism of DNA binding by cohesin has received further support by recent discoveries that other SMC complexes translocate along DNA in vivo and in vitro (23, 24). Translocation requires multiple independent DNA binding activities (23, 24). Furthermore, translocation requires a tight interaction between SMC complexes and DNA, very different from DNA simply floating within a lumen. Finally, to translocate, one DNA binding activity must persist while the other is transient, presumably driven by the ATPase. While motor activity for cohesin has yet to be demonstrated, several properties of cohesin required for translocation-associated activities have been reported. Once bound to DNA, wild-type (WT) cohesin can go through additional rounds of ATP hydrolysis without losing DNA binding (13). Also, tethering two DNA molecules by cohesin requires two independent DNA binding activities (25, 26). If these cohesin activities reflect a translocation function, then it raises the question: How could cohesin’s ATPase activity drive both the formation of stable tethers and translocation along DNA?
Here, we examine in vitro WT and DE-cohesin bound with an ATP transition analog, ADP aluminum fluoride (cohesinADP/AlFx). Our results suggest that an intermediate nucleotide state of cohesin, likely cohesinADP-Pi, allows DNA to enter the open cohesin lumen, then interact with the entrapped DNA to induce stable closure of the cohesin ring. These results support a model to reconcile cohesin’s ability to be both a tether and motor. Our results also suggest that loader (Mis4/Ssl3) promotes cohesin’s DNA binding beyond loader’s putative stimulation of cohesin’s ATPase to facilitate a cohesin function distinct from initial DNA binding.
Results
DE-Cohesin Bound with ATP Forms a Salt-Stable Complex with DNA in the Absence of ATP Hydrolysis.
Our previous study showed that DE-cohesin lacked ATPase activity in the absence of DNA (13). The DE mutation might have reduced DNA-independent ATPase activity of cohesin while retaining DNA-dependent ATPase activity. To test the potential impact of DNA on DE-cohesin ATPase activity, we first needed to reconstitute DE-cohesin binding to DNA in vitro. Two in vitro cohesin-DNA binding assays have been reported (12, 27). The first we referred to as the DNA-bead assay in which we monitored the formation of salt-resistant cohesin–DNA complexes on DNA coupled to beads by Western blotting (27) (Fig. 1B and SI Appendix, Fig. S1A). Salt-resistant DNA binding is a hallmark of topological entrapment (12, 27, 28).
In the presence of ATP, about 25% of WT cohesin was bound to DNA, which was about twofold greater than seen without the addition of nucleotide and three- to fivefold greater than with the addition of EDTA (SI Appendix, Fig. S1B). The difference between the binding of cohesin without exogenous nucleotide and EDTA could reflect ATP-independent DNA binding or cohesin that retained its ATP during purification. To distinguish between these two possibilities, we examined the level of DNA binding of cohesin with Psm3-KI, a Walker A mutation in the Psm3 ATPase that blocks ATP binding and DNA binding in vivo and in vitro (9, 11, 22). In the presence of ATP, no exogenous nucleotide, or EDTA the level of DNA binding of Psm3-KI cohesin, was similar to the very low level of DNA binding of WT cohesin in the presence of EDTA, suggesting that only this very low level of DNA binding was nucleotide independent. DE-cohesin–bound DNA on the beads at levels close to the WT protein (Fig. 1C). Like WT cohesin, the binding of the DE-cohesin to DNA in the presence of ATP was threefold higher than DE-cohesin in the presence of EDTA or cohesin with Psm3-KI in the presence of ATP (Fig. 1C). These results suggest that ATP promotes efficient binding of DE-cohesin to DNA.
Having established DE-cohesin binding to DNA in vitro, we assessed the impact of DNA binding on the ATPase activity. We followed ATP hydrolysis in assays containing DNA, loader, and equal amounts of either WT cohesin or DE-cohesin (Fig. 1D). As a negative control, we tested Psm3-KI cohesin. As expected from previous studies, the Psm3-KI cohesin shows dramatically reduced ATPase activity (20-fold) (Fig. 1D) (12, 13). We observed a similar reduction in ATPase activity for the DE-cohesin (Fig. 1D). Thus, neither the presence of loader nor the DNA binding can stimulate ATPase activity in DE-cohesin. Furthermore, the DE mutation in the Psm3 active site must, like the KI mutation, compromise the ATPase activity of the Psm1 active site. Together, these results suggest that DE-cohesin must bind ATP but does not need to hydrolyze it to form salt-stable cohesin–DNA complexes in vitro. We suggest that the combination of ATP binding and the DE mutation is sufficient to mimic a hydrolysis-induced transition state, bypassing the need for hydrolysis.
ADP/AlFx Promotes Stable Cohesin Binding to DNA.
Our results with the DE-cohesin suggested that binding of an appropriate ATP analog to WT cohesin might allow loading and stable binding of DNA without ATP hydrolysis. Previously, a number of nonhydrolysable ATP analogs had been tested in cohesin-DNA binding assays and failed to promote the formation of cohesin–DNA complexes. However, one hydrolysis transition state analog that was not tried was ADP/AlFx.
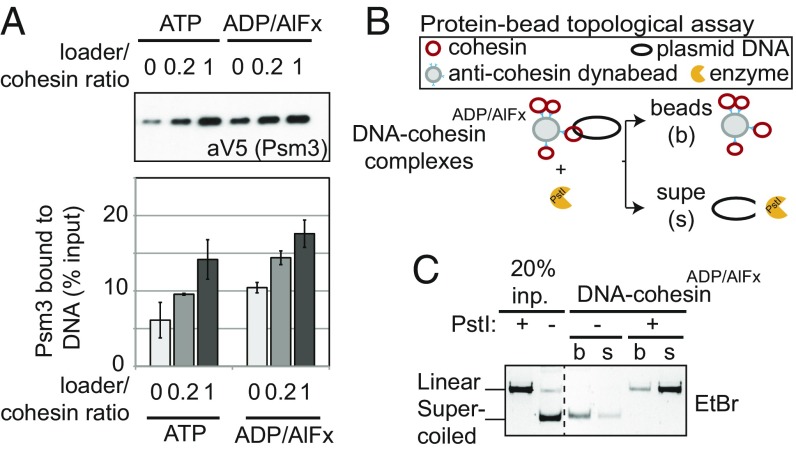
Due to the similarity in the structure of ADP-Pi and ADP/AlFx, complexes with ADP/AlFx have been used successfully to study transition states in phosphoryl transfer reactions (29, 30). To test the ability of ADP/AlFx to promote the formation of cohesin–DNA complexes, we substituted it for ATP in our DNA-bead assay (Fig. 2). The binding of cohesinADP/AlFx to the beads was dependent upon the presence of DNA (Fig. 2A and SI Appendix, Fig. S2). The level of binding of cohesinADP/AlFx to DNA was not only six- to sevenfold better than cohesin with EDTA but also better than cohesinATP (Fig. 2A). Therefore, ADP/AlFx induced the formation of salt-resistant, stable cohesin–DNA complexes. This conclusion was supported further by the fact that cohesinADP/AlFx–DNA complexes were stable for an hour in the presence of excess competitor DNA similar to that seen for WT cohesin in the presence of ATP (Fig. 2B).
Fig. 2.
ADP/AlFx promotes stable cohesin binding to DNA. (A) Effect of nucleotides on DNA binding of cohesin using DNA-bead assay. WT cohesin complexes incubated with EDTA, ADP, ATP, or ADP/AlFx and loader in the absence or presence of DNA on beads as described in Fig. 1B. (Upper) Cohesin bound to DNA beads detected by Western blot using anti-V5 antibodies (Psm3-3V5). (Lower) Quantitation of Western blots. (B) Effect of competitor DNA on the stability of cohesin–DNA complexes. WT cohesin was assembled on DNA coupled to beads with ADP/AlFx. Competitor DNA was added and the amount of cohesin bound [pellet (p)] and eluted [supernatant (s)] were visualized by silver staining. (C) Schematic illustration of the protein-bead assay. Plasmid DNA bearing CARC1 sequences was incubated with cohesin and loader in the presence of EDTA or nucleotides depicted. Cohesin was immunoprecipitated at a high-salt concentration (0.5 M NaCl) and further washed in a high-salt buffer (0.75 M NaCl) to eliminate unstably bound DNA. Coimmunoprecipitated plasmid DNA was eluted and analyzed by agarose gel electrophoresis. (D) Effect of nucleotides on DNA binding of cohesin using the protein-bead assay. WT cohesin complexes incubated with EDTA, ATP, ADP, ADP/AlFx, or ATPγS nucleotides as described in Fig. 2C. (Upper) DNA bound to immunoprecipitated cohesin complexes detected using ethidium bromide-stained agarose gels. (Lower) Quantitation of DNA. (E) Time-course showing the kinetics of DNA binding by WT cohesin assembled with ATP or ADP/AlFx using DNA-bead assay. (Upper) Cohesin bound to DNA beads detected by Western blot using anti-V5 antibodies (Psm3-3V5). (Lower) Quantitation of Western blots. Dotted line represents where irrelevant lanes were removed.
We noted that the binding of cohesin to DNA in the presence of ADP (Fig. 2A) was better than that reported previously using a second cohesin–DNA binding assay, henceforth referred to as the protein-bead assay (12) (Fig. 2C). In this assay, plasmid DNA, purified cohesin, and loader were incubated in solution with different nucleotides. Beads coupled with anti-cohesin antibodies were added to these reactions and incubated for 15 h in the presence of high salt to purify salt-stable cohesin–DNA complexes. The amount of plasmid DNA that coimmunoprecipitated with cohesin was quantified to assess the assembly of cohesin–DNA complexes. A possible explanation for the difference in the stringency for the added nucleotide in the two assays was that some aspect of the conditions of the DNA-bead assay make the subset of cohesin copurifying with ATP less effective in binding before ATP hydrolysis and/or exchange with the added nucleotide. Consistent with this hypothesis in the protein-bead assay, the cohesin binding to DNA without a nucleotide was similar to binding in the presence of EDTA (SI Appendix, Fig. S1C).
With this caveat in mind, we investigated the impact of ADP/AlFx on cohesin binding to DNA in the more stringent protein-bead assay. We observed that ATP or ADP/AlFx increased the assembly of salt-stable cohesin–DNA complexes 15- to 20-fold more than in the presence of EDTA (Fig. 2D). The amount of DNA bound by cohesin with ADP or nonhydrolyzable ATPγS was near background levels, similar to that reported previously (12) (Fig. 2D). Importantly, even with this more-stringent protein-bead assay, ADP/AlFx triggered the assembly of cohesin–DNA complexes as efficiently as ATP, if not better.
Cohesin ATPase activity is very slow (31). If ADP/AlFx allows cohesin to bypass the cohesin ATPase, the formation of salt-resistant cohesin–DNA complexes should occur more rapidly with cohesinADP/AlFx than with cohesinATP. To test this prediction, we performed a time-course experiment to estimate the kinetics of DNA binding by cohesin incubated with ATP and ADP/AlFx. CohesinADP/AlFx was able to form stable complexes approximately threefold faster than cohesinATP–DNA (Fig. 2E). The faster kinetics of cohesinADP/AlFx in DNA binding is consistent with it bypassing the need for a slow ATPase activity.
Both DE-cohesin and WT cohesin with ADP/AlFx allow DNA binding without ATP hydrolysis. To test whether they bypass hydrolysis through similar mechanisms, we asked whether the binding of DE-cohesin to DNA was further enhanced by substituting ADP/AlFx for ATP in the DNA-bead assay. The binding of DE-cohesin to DNA was similar with either ATP or ADP/AlFx (SI Appendix, Fig. S1D). This result is consistent with the DE mutation and ADP/AlFx trapping a similar intermediate of cohesin in its ATP hydrolysis cycle.
Efficient Assembly of CohesinADP/AlFx–DNA Complexes Is Enhanced by the Cohesin Loader and Shows Properties of Topologically Entrapped DNA.
The salt stability of the cohesinATP–DNA complexes is one of several unique features that indicates the topological entrapment of DNA by cohesin. Efficient formation of topological cohesin–DNA complexes requires the cohesin loader complex (12, 13). We previously showed that loader increases cohesinATP–DNA binding using the DNA-bead assay (13). Assembly of cohesin–DNA complexes under both ATP and ADP/AlFx conditions was stimulated twofold by the presence of loader (Fig. 3A). These results suggest that at least one mechanism by which loader promotes cohesin binding to DNA is independent of stimulating the cohesin ATPase. Another indicator of topological entrapment is that cohesin remains stably bound to DNA unless the DNA is linearized (12, 13). CohesinADP/AlFx binding to DNA was greatly reduced by linearizing the plasmid DNA in the protein-bead assay or treatment with DNase in the DNA-bead assay (Fig. 3 B and C). Taken together, these results support a model where the opening and closing of cohesin to topologically entrap DNA can be promoted by an intermediate nucleotide state of cohesin.
Fig. 3.
Efficient assembly of cohesinADP/AlFx–DNA complexes is enhanced by the cohesin loader and shows properties of topologically entrapped DNA. (A) Effect of cohesin loader on DNA binding of cohesin using DNA-bead assay. WT cohesin was incubated with either ATP or ADP/AlFx nucleotides with increasing amounts of loader in the DNA-bead assay described in Fig. 1B. (Upper) Cohesin bound to DNA beads detected by Western blot using anti-V5 antibodies (Psm3-3V5). Loader/cohesin ratio shown refers to the molar ratio of loader to cohesin in experiments. (Lower) Quantitation of Western blots. (B) Schematic of modified protein-bead assay to assess topologically entrapped DNA by cohesinADP/AlFx. Cohesin–DNA complexes were assembled using the protein-bead assay described in Fig. 2C. After purification of cohesinADP/AlFx–DNA complexes by immunoprecipitation, the DNA was linearized with PstI. (C) Effect of plasmid linearization on the ability of cohesinADP/AlFx to bind DNA. CohesinADP/AlFx–DNA complexes were digested in the presence (+) or absence (−) of PstI. Cohesin was immunoprecipitated and DNA bound to cohesinADP/AlFx (beads) or released into the supernatant (supe) was analyzed by ethidium-stained agarose gel electrophoresis. Plasmid not incubated with cohesin, either supercoiled or linearized with PstI, was used as an electrophoresis control (left two lanes). Dotted line represents where irrelevant lanes were removed.
Efficient Assembly of CohesinADP/AlFx–DNA Complexes Requires Binding of ADP-AlFx in both Active Sites.
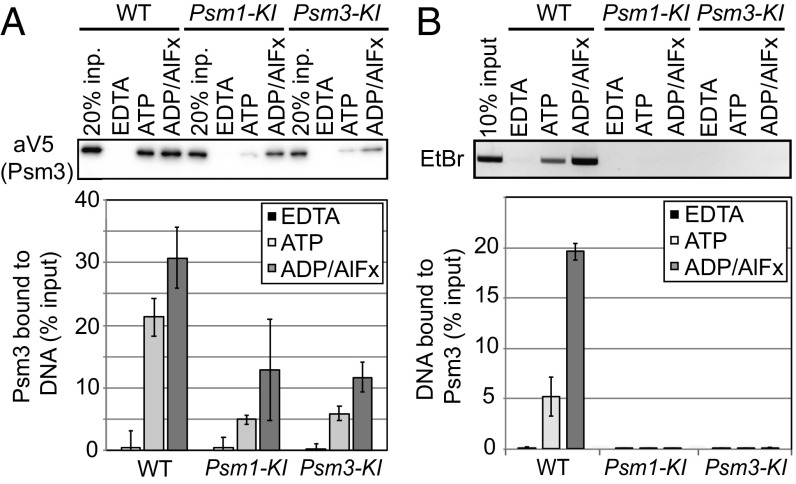
Cohesin’s binding to chromosomes in vivo and its ATPase activity in vitro can be inhibited by a KI substitution in either the Psm1 or Psm3 Walker A motif. This observation suggests potential cross-talk between the two ATPase active sites or that one functional site is not enough for proper ATPase function. With this in mind, we asked whether ADP/AlFx promotion of DNA binding requires binding to both ATPase active sites in vitro. To test this, we purified cohesin containing either the Psm1-KI or Psm3-KI subunit and assayed their ability to bind DNA. These mutant cohesins could presumably bind ADP/AlFx in the Psm3 ATPase or Psm1 ATPase active sites, respectively. We then compared their binding to DNA with the WT complex, using either DNA-bead or protein-bead assay (Fig. 4).
Fig. 4.
Efficient assembly of cohesinADP/AlFx–DNA complexes requires binding of ADP-AlFx in both active sites. (A) DNA-bead assay to assess Walker A cohesin mutant binding. (Upper) Cohesin bound to DNA beads incubated with EDTA, ATP, or ADP/AlFx and detected by Western blot using anti-V5 antibodies (Psm3-3V5). (Lower) Quantitation of Western blots. (B) Protein-bead assay to assess Walker A cohesin mutant binding. (Upper) DNA bound to cohesin incubated with EDTA, ATP, or ADP/AlFx, after cohesin immunoprecipitation, and detected using ethidium bromide-stained agarose gel. (Lower) Quantitation of DNA.
Overall, both assays showed that ADP/AlFx bound to only one of the two ATPase active sites was not sufficient to ensure efficient formation of stable cohesin–DNA complexes. In the DNA-bead assay, both KI-cohesin mutants exhibit greatly reduced DNA binding compared with WT cohesin when incubated in the presence of either ATP or ADP/AlFx (Fig. 4A). However, ADP/AlFx increased mutant cohesin binding to DNA about twofold compared with ATP, but this DNA binding level was still below WT cohesin with ADP/AlFx (Fig. 4A). These results suggest that in the DNA-bead assay, efficient binding of cohesin to DNA requires ADP/AlFx bound in both ATPase active sites, but some binding is supported by ADP/AlFx bound only to one site. In the protein-bead assay, the amount of KI-cohesin bound to DNA was reduced to background levels with either ADP/AlFx or ATP (Fig. 4B). Thus, as we observed before, binding using the protein-bead assay has more stringent nucleotide requirement than the DNA-bead assay. Our results suggest that both active sites must be capable of nucleotide binding and that at least one must also be trapped in an intermediate state of ATP hydrolysis to promote efficient cohesin binding to DNA.
Loader Remains Bound Stably and Stoichiometrically to CohesinADP/AlFx–DNA Complexes.
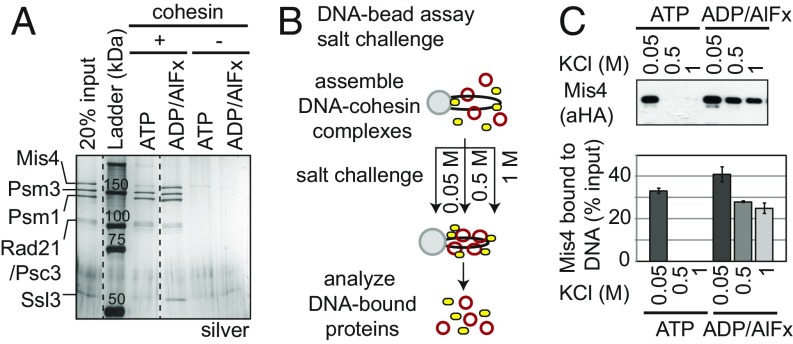
Protein staining and immunoblotting using the DNA-bead assay allowed us to assess the loader (Mis4/Ssl3) and cohesin binding to DNA. When we subjected cohesinATP–DNA complexes on the beads to a wash of 500 mM salt, very little loader was associated with cohesin–DNA complexes (Fig. 5A and SI Appendix, Fig. S3A). In contrast, the loader remained bound to cohesinADP/AlFx–DNA complexes in a one-to-one stoichiometry in 500 mM salt. No loader was detected on the DNA beads in the absence of cohesin or DNA, indicating that loader binding to beads was specific to the presence of cohesin–DNA complexes. Thus, cohesinADP/AlFx–DNA but not cohesinATP–DNA complexes form salt-stable complexes with loader.
Fig. 5.
Loader remains bound stably and stoichiometrically to cohesinADP/AlFx–DNA complexes. (A) Effect of ADP/AlFx on loader binding to cohesin using DNA-bead assay. WT cohesin and an equimolar amount of loader was assayed for binding to DNA beads in the presence or ATP or ADP/AlFx. Silver-stained gel to assess cohesin (Psm3, Psm1, Rad21/Psc3) and loader (Mis4, Ssl3) bound to DNA beads. Dotted line represents where irrelevant lanes were removed. (B) Schematic for assessing the stability of cohesin–DNA complexes in high-salt washes. (C) Effect of increasing salt concentration on loader binding to DNA beads in the presence of ATP or ADP/AlFx. (Upper) Loader bound to DNA at increased salt washes was analyzed by SDS/PAGE and Western blot using anti-HA antibodies (Mis4-HA). (Lower) Quantitation of Western blots.
To further characterize this loader–cohesin interaction, we repeated the DNA-bead assay but subjected the beads to increasing salt concentrations at the wash stage (after complex assembly was allowed to take place for 1 h at low salt) (Fig. 5B). At 50 mM salt wash, loader remained bound to both cohesinATP– and cohesinADP-AlFx–DNA complexes on the beads. Upon increasing the salt concentration in the wash to 500 mM or 1 M, cohesinATP and cohesinADP-AlFx remained bound to DNA (SI Appendix, Fig. S3B). However, loader behaved differently depending on the nucleotide bound to cohesin–DNA complex. For cohesinATP–DNA complexes, loader was no longer bound in the 500 mM wash, whereas loader remained bound to cohesinADP-AlFx–DNA (Fig. 5C). These results indicate that loader binds equally well to cohesinATP– and cohesinADP-AlFx–DNA complexes but dissociates more readily from the cohesinATP–DNA than cohesinADP-AlFx–DNA complexes.
Discussion
In this study, we interrogate how ATP binding and the ATPase cycle of cohesin influence its topological binding to DNA. We show that cohesin bearing a D-loop mutation in the Psm3 ATPase active site (DE-cohesin) requires ATP but not ATP hydrolysis to form stable complexes with DNA. Similarly, we show that WT cohesin with ADP aluminum fluoride (cohesinADP/AlFx), a transition analog for ATP hydrolysis, can also stably entrap DNA. These results suggest that an intermediate of the cohesin ATPase cycle can stably entrap DNA in vitro. DE-cohesin binds in vivo and the binding of cohesinADP/AlFx to DNA shares all of the characteristics of WT cohesin–DNA complexes assembled in vitro and in vivo (salt stability, loader dependence, and topological binding). Therefore, an intermediate in the cohesin ATPase cycle of cohesin can also likely stably entrap DNA in vivo. DNA tethering by cohesin (the capture of the second DNA strand) is also likely driven by this intermediate state since DE-cohesin is also competent to generate the tethering necessary for cohesion in vivo (13).
In the absence of bound DNA, one of the interfaces within cohesin, most likely the hinge dimerization interface (32), must be dissociated (at least transiently) to allow passage of DNA into the lumen of a ring. The binding of cohesinADP/AlFx or DE-cohesinATP to DNA could occur because the intermediate state is an equilibrium with hinge opening and closing. Alternatively, the nucleotide-bound forms of cohesin may be in equilibrium with apo cohesin that promotes hinge opening. Once DNA is bound, it must induce cohesin to undergo a conformation change that prevents subsequent release of the DNA, thereby generating its remarkable stable DNA binding. This conformation change either prevents the hinge from subsequent dissociation or traps the DNA in a subdomain of the lumen such that it cannot be released even if hinge dissociation occurs.
In order for DNA to induce a change in cohesin conformation and function, it must interact intimately with cohesin rather than simply float inside a lumen of a large ring as suggested by the original embrace model (5). Indeed, under physiological salt conditions, cohesin exhibits limited diffusion on DNA in vitro, consistent with intimate cohesin–DNA interactions beyond topological entrapment (28). Moreover, even under high-salt conditions when DNA-bound cohesin is more mobile, the DNA appears sequestered in a lumen far smaller than in the 40-nm one of an open ring conformation (28). Furthermore, a recent study reported nontopological interactions of cohesin with DNA in vivo, consistent with a number of studies of other SMC complexes that revealed direct interactions between SMC subunits and DNA (33–38).
This DNA-responsive form of cohesin likely is unique to an intermediate in the ATPase cycle (possibly ADP-Pi) since it can be mimicked only by ADP/AlFx and the DE mutation, but not by other ATP analogs nor Walker A mutations (this study). Thus, one function of cohesin ATPase is to generate a nucleotide state of cohesin that is competent to bind DNA, responsive to DNA to induce its stable binding, and capable of binding of a second DNA molecule. The subsequent completion of the ATPase cycle (ADP) may allow release of DNA from cohesin, first undoing tethering (release of the one DNA molecule) and then undoing DNA binding (release of the second DNA molecule). A two-step ATPase-dependent mechanism for the dissolution of tethering and DNA binding is supported from in vivo studies and its in vitro reconstitution, which is dependent on ATPase activity (12, 25, 26).
In this light, other biological motors like kinesin and myosin provide insight into the potential relationship of cohesin DNA tethering activity and the established DNA translocation activities of other SMC complexes. Kinesin has multiple microtubule binding activities. It translocates along microtubules by using ATP to change conformations and sequentially release microtubule binding (39). Furthermore, rigor binding of myosin to actin can be achieved by blocking myosin’s ATPase cycle (40). Thus, the inherent activities of a motor (ATP-dependent control of multiple interactions with its substrate) can be converted to generate a tether by controlling the ATPase cycle. Cohesin may be a specialized SMC complex in which the ATPase-dependent activities that are needed for translocation along DNA have been modified and regulated to make it a particularly good tether. A potential candidate for the regulation of cohesin ATPase is cohesin acetylation (13).
Our study also provides several interesting insights into the cohesin loader. We show that loader enhances binding of cohesinADP/AlFx to DNA. This result suggests that loader has a function in assembling cohesin–DNA complexes independent of ATPase activity. One possible model for loader is that it binds to the hinge domain promoting its dissociation, thereby enhancing DNA entry into the lumen. Indeed, while in vitro studies showed that loader stimulates cohesin ATPase activity in the presence of DNA, this stimulation could be a consequence of DNA binding rather than a prerequisite for destabilization of an interface to allow DNA binding. Interestingly, the association of loader with cohesinADP/AlFx–DNA complex is very salt stable while its binding to cohesinATP–DNA complex is not. An intriguing interpretation is that the stable complex of loader, DNA, and cohesinADP/AlFx is an intermediate trapped in the tethering conformation, with loader poised to help capture of the second DNA molecule. Indeed, the colocalization of the loader with cohesin on DNA was first reported in yeast by ChIP and more recently has been observed in vivo with single-molecule imaging (41, 42). In addition, Murayama et al. (43) recently reported that in an in vitro reconstitution assay, the capture of a second DNA molecule by the cohesin–DNA complex is loader dependent. In summary, the DE mutation or ADP/AlFx provide a means to trap cohesin in an active state that is competent for the capture of both DNA molecules. This tool likely will prove useful in elucidating the molecular basis for cohesin–DNA interactions that underlie its tethering activity and its remarkable biological functions.
Materials and Methods
Protein Expression and Purification.
S. pombe cohesin complex was overexpressed and purified from Saccharomyces cerevisiae as described (12). Minor modifications to the purifcation process were applied as described (13). Mutants of cohesin were generated by site-directed mutagenesis, and overexpression strains were generated as described (12). The loader complex from S. pombe was expressed and purified as described (12).
ATPase Assays.
For loader- and DNA-stimulated ATPase activity of cohesin, 80 nM cohesin, 80 nM loader, and 2.5 μg of plasmid DNA containing the sequence for CARC1 (pIO2) were incubated to measure loader-stimulated activity in solution as described (13).
DNA-Bead Assay.
CARC1 DNA substrates were prepared as described (27). For each binding reaction, 100 ng of biotin-labeled DNA was assembled on 20 μL of streptavidin-conjugated dynabeads (Invitrogen). Eighty nanomolar cohesin complex and 80 nM loader, with 1 mM ATP (Sigma) or other nucleotide analogs were added to beads. For ADP/AlFx complexes, 72 μM AlCl3 (Boston Bioproducts) and 12 mM NaF (Sigma) were mixed with cohesin-loader-DNA mixture, and 1 mM ADP (Sigma) was added to start the DNA binding reaction. Samples were analyzed by SDS/PAGE followed by Coomassie staining, silver staining, or Western blotting against Psm3-3V5-tag (Life Technologies) and Mis4-HA-tag (Roche).
Protein-Bead Assay.
Briefly, for each binding reaction, 700 ng of plasmid DNA bearing CARC1 sequences (pIO2) was mixed with 100 nM loader, 1 mM ATP (or depicted nucleotide analog), and 150 nM cohesin in 15 μL of CL1 buffer. For ADP/AlFx complexes, 1 mM ADP, 72 μM AlCl3, and 12 mM NaF was added. Mix was prepared and incubated on ice for 5 min and then shifted to 30 °C for 60 min. Binding to beads, washing steps, and elution were performed as described (12).
Elution of Cohesin from DNA Beads with PstI Digestion.
For restriction enzyme treatment, DNA–cohesin complexes were prepared as described above (protein-bead assay) in buffers lacking EDTA. Beads were resuspended in RE buffer (12) in the presence of 5U PstI (NEB) and incubated at 30 °C for 30 min. Supernatant was collected and remaining beads were boiled; both fractions were visualized by SDS/PAGE and silver staining.
Elution of Cohesin from DNA Beads with Competitor DNA.
DNA–cohesin complexes assembled in presence of ADP/AlFx as described above (DNA-bead assay) were resuspended in 20 μL of CL1 buffer, in the presence or absence of ADP/AlFx and plasmid DNA (5× excess in mass compared with DNA on beads). Supernatant and pellets were separated at the end of a 30-min incubation at 30 °C. Cohesin in supernatant and pellet fractions were subjected to SDS/PAGE and visualized by silver stain.
Quantitations.
Western blots and ethidium bromide-stained gels were quantified using Image Lab version 5.2 build 14 (BioRad). Provided numbers are the quantitated averages, and error bars represent SD of data from at least two independent experiments.
Kinetics Experiments.
DNA–cohesin complexes were assembled with ATP or ADP/AlFx as described above. Samples were incubated at 30 °C and then washed once in CL1 buffer, twice in CL1 with 500 mM KCl, and then once more in CL1 at indicated timepoints. Cohesin was then eluted and analyzed as described in the DNA-bead assay methods.
Supplementary Material
Acknowledgments
We thank Lorenzo Costantino, Vincent Guacci, and Rebecca Lamothe for critically reading the manuscript; other members of the D.K. laboratory for fruitful discussions; Dr. Frank Uhlmann for sharing cohesin and loader expression strains and plasmids; and the laboratory of David Drubin and Georjana Barnes for use of equipment. This work was funded by a National Institute of Health Grant 1R35 GM-118189-01 (to D.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807213115/-/DCSupplemental.
References
- 1.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 3.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 4.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 5.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 7.Löwe J, Cordell SC, van den Ent F. Crystal structure of the SMC head domain: An ABC ATPase with 900 residues antiparallel coiled-coil inserted. J Mol Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- 8.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 9.Arumugam P, et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Haering CH, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 12.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Çamdere G, Guacci V, Stricklin J, Koshland D. The ATPases of cohesin interface with regulators to modulate cohesin-mediated DNA tethering. eLife. 2015;4:13115. doi: 10.7554/eLife.11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murayama Y, Uhlmann F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015;163:1628–1640. doi: 10.1016/j.cell.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haering CH, Farcas A-M, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 17.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 19.Bernard P, et al. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Elbatsh AMO, et al. Cohesin releases DNA through asymmetric ATPase-driven ring opening. Mol Cell. 2016;61:575–588. doi: 10.1016/j.molcel.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber RG, et al. Impairing cohesin Smc1/3 head engagement compensates for the lack of Eco1 function. Structure. 2016;24:1991–1999. doi: 10.1016/j.str.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terakawa T, et al. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 2017;358:672–676. doi: 10.1126/science.aan6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Brandão HB, Le TBK, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eng T, Guacci V, Koshland D. ROCC, a conserved region in cohesin’s Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Mol Biol Cell. 2014;25:2351–2364. doi: 10.1091/mbc.E14-04-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birot A, et al. A second Wpl1 anti-cohesion pathway requires dephosphorylation of fission yeast kleisin Rad21 by PP4. EMBO J. 2017;36:1364–1378. doi: 10.15252/embj.201696050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onn I, Koshland D. In vitro assembly of physiological cohesin/DNA complexes. Proc Natl Acad Sci USA. 2011;108:12198–12205. doi: 10.1073/pnas.1107504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stigler J, Çamdere GÖ, Koshland DE, Greene EC. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 2016;15:988–998. doi: 10.1016/j.celrep.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 30.Ahmadian MR, Mittal R, Hall A, Wittinghofer A. Aluminum fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett. 1997;408:315–318. doi: 10.1016/s0014-5793(97)00422-5. [DOI] [PubMed] [Google Scholar]
- 31.Arumugam P, Nishino T, Haering CH, Gruber S, Nasmyth K. Cohesin’s ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr Biol. 2006;16:1998–2008. doi: 10.1016/j.cub.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Gruber S, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 33.Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner K-P. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kschonsak M, et al. Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell. 2017;171:588–600.e24. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016;35:743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano M, Hirano T. Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Soh Y-M, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivasan M, et al. The cohesin ring uses its hinge to organize DNA using non-topological as well as topological mechanisms. Cell. 2018;173:1508–1519.e18. doi: 10.1016/j.cell.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhodes J, Mazza D, Nasmyth K, Uphoff S. Scc2/Nipbl hops between chromosomal cohesin rings after loading. eLife. 2017;6:11202. doi: 10.7554/eLife.30000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. Establishment of DNA-DNA interactions by the cohesin ring. Cell. 2018;172:465–477.e15. doi: 10.1016/j.cell.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.