Significance
The mechanisms of norovirus capsid interactions with host receptors and the mechanisms by which soluble cofactors augment norovirus infection are not understood. We recently identified CD300lf as a cell surface receptor for murine norovirus (MNoV) and observed that a small molecule cofactor was critical for efficient binding of virus to CD300lf. Herein we identify the bile acid GCDCA as a cofactor enhancing MNoV infection and provide a biophysical characterization of the capsid–receptor and capsid–cofactor interactions, thereby providing a structure-based understanding of how noroviruses initiate cellular infection. This work has important implications for the design of norovirus therapeutics.
Keywords: norovirus, virus receptor, bile acid, crystallography
Abstract
Murine norovirus (MNoV) is closely related to human norovirus (HNoV), an infectious agent responsible for acute gastroenteritis worldwide. Here we report the X-ray crystal structure of the dimeric MNoV VP1 protruding (P) domain in complex with its cellular receptor CD300lf. CD300lf binds the P domain with a 2:2 stoichiometry, engaging a cleft between the AB and DE loops of the P2 subdomain at a site that overlaps the epitopes of neutralizing antibodies. We also identify that bile acids are cofactors enhancing MNoV cell-binding and infectivity. Structures of CD300lf–P domain in complex with glycochenodeoxycholic acid (GCDCA) and lithocholic acid (LCA) reveal two bile acid binding sites at the P domain dimer interface distant from receptor binding sites. The structural determinants for receptor and bile acid binding are supported by numerous biophysical assays utilizing interface residue mutations. We find that the monomeric affinity of CD300lf for the P domain is low and is divalent cation dependent. We have also determined the crystal structure of CD300lf in complex with phosphocholine, revealing that MNoV engages its receptor in a manner mimicking host ligands including similar metal coordination. Docking of the cocomplex structures onto a cryo-EM–derived model of MNoV suggests that each virion can make multiple CD300lf engagements, and thus, infection may be driven by the avidity of cell surface clustered CD300lf. These studies identify multiple potential modulators of norovirus infection that may act to regulate the interaction between the viral capsid P domain and its cognate cellular receptor.
Murine norovirus (MNoV) and human noroviruses (HNoVs) share biological similarities including genome structure, environmental stability, fecal–oral transmission, replication in the gastrointestinal tract, and prolonged viral shedding (1). MNoV is an important model system for noroviruses in part because it is the only member of the norovirus genus that can be efficiently cultured, has a reverse genetics system, and can be studied in a robust small animal model (1–4). Recently, we identified CD300lf as a proteinaceous cellular receptor for MNoV essential for MNoV binding and entry in cell lines, primary cells and mice (5). CD300lf is a critical determinant of MNoV species and cell tropism (5–7). The CD300 family contains type I transmembrane proteins with single Ig-V–like domains involved in immunoregulatory processes (8, 9). A general feature of this family appears to be the ability to bind lipids in a cleft on the extracellular Ig domain (10). Potential ligands for murine CD300lf include ceramide, phosphotidylserine, and phosphotidylcholine, with these ligands binding in a calcium-dependent manner (11–13).
Norovirus is a genus in the Caliciviridae family composed of nonenveloped positive-sense single-stranded RNA viruses (14). Caliciviruses are unique among animal viruses in possessing a capsid composed of a single outward-facing structural protein called VP1 (15, 16). The capsid consists of 180 VP1 molecules paired into 90 dimers that self-assemble to form a T = 3 icosahedral shell with a diameter of 35–39 nm. The first 225 N-terminal residues of VP1 encode the shell (S) domain, the interior surface of the capsid surrounding the viral RNA. The S domain is highly conserved and consists of an eight-stranded antiparallel β-sandwich with two α-helical decorations. The remainder of the VP1 protein, called the protruding (P) domain, can be divided into two subdomains, P1 and P2. The P1 subdomain consists of a single α helix and eight β strands and is connected to the S domain by a flexible stalk-like hinge (17). The P2 subdomain appears as an insertion in P1 (137 amino acids, 278–415) that forms a six-stranded antiparallel β-barrel–like structure. The P2 subdomain makes up the distal surface of the virion and is a target for neutralizing antibodies (18–29).
We previously reported that efficient binding of MNoV to cells required a <5-kD serum cofactor resistant to boiling, delipidation, and proteinase K treatment (5). It was recently found that bile can enhance HNoV replication in intestinal enteroids by an unknown mechanism (30). Bile acids are a diverse set of cholesterol-derived molecules synthesized by host enzymes in the liver (31). They consist of a hydrophobic core of carbon rings conjugated to a hydrophilic side chain, typically glycine or taurine. Bile acids are secreted into the proximal small intestine as primary bile acids but can be modified by intestinal bacteria into secondary bile acids. Bile acids are resorbed in the distal small intestine and colon and returned to the liver through the portal circulation. Herein we have undertaken a series of structural and biophysical experiments with the goal of better understanding the structural basis of NoV–receptor interactions and bile acid-mediated enhancement of viral attachment and infection.
Results
Crystal Structure of MNoV P Domain in Complex with Murine CD300lf.
To understand the structural basis of MNoV cell recognition, we obtained a crystal structure of murine CD300lf bound to the MNoV strain CW3 P domain at 2.0 Å resolution. Crystals of the P domain/CD300lf cocomplex were obtained in space group P212121 (a = 48.71 Å, b = 135.24 Å, c = 139.51 Å), and the structure was determined by molecular replacement. The final refined model has an Rwork of 18.5% with an Rfree of 22.5% (SI Appendix, Table S1). The atomic model spans P domain residues 229–531, with residues 532–541 missing due to poor electron density, and the entire CD300lf ectodomain (residues 19–131) with an N-terminal methionine added for protein synthesis. In the asymmetric unit, two CD300lf ectodomains bind to one P domain dimer with each CD300lf monomer contacting distantly positioned P2 subdomains (Fig. 1). The location of this binding site is in agreement with results reported for CD300lf with MNoV CR10 strain capsid (32). The CDR3 and CC′ loops of CD300lf (SI Appendix, Fig. S1) protrude deep into a cleft between the P2 subdomain AB and DE loops (SI Appendix, Fig. S2). The two P monomers in the CD300lf-bound structure adopt nearly identical conformations yielding a RMSD difference of only 0.22 Å in Cα over 303 aligned residues. Twenty-one P2 subdomain residues fall within the footprint of CD300lf, and most are entirely conserved in MNoV but not HNoV (SI Appendix, Fig. S2).
Fig. 1.
CD300lf binding site is located in the P2 domain of VP1. Ribbon model of the CD300lf–P domain complex structure with a schematic diagram of key interactions given in Bottom for orientation. The two chains of the P domain dimer are colored cyan and green. The secondary structure of the P domain is indicated on the green monomer. The receptor protein CD300lf is shown in yellow with the secondary structure labeled. The metal ions are shown as silver spheres, and the metal ions associated with one P domain are marked with arrows.
The Bile Acid GCDCA Enhances MNoV Binding and Infection and Binds the P Domain.
As reported (5), MNoV binding to BV2 cells in PBS was enhanced by adding 10% FBS (Fig. 2A). We found that addition of crude bovine bile acids or the individual bile acid GCDCA, but not taurocholic acid (TCA) or casein, rescued MNoV binding to cells (Fig. 2A). This enhanced binding of MNoV to cells translated to increased infectivity (Fig. 2B). No significant enhancement was observed with TCA, glycocholic acid, beta-muricholic acid, cholic acid, or tauroursodeoxycholic acid. We were unable to assess cofactor activity of chenodeoxycholic acid or deoxycholic acid due to cytotoxic effects or of lithocholic acid (LCA) due to insolubility. Of note, divalent cations, calcium plus magnesium, augmented infection when present during viral adsorption (Fig. 2B). Histo-blood group antigen (HBGA), which can bind HNoV, failed to increase infectivity (Fig. 2B). We next performed isothermal calorimetry (ITC) to assess bile acid binding to the MNoV P domain. Injection of GCDCA into soluble P domain yielded an average KD of ∼5.76 ± 1.26 μM for three independent experiments, with ∼1:1 binding stoichiometry (Fig. 2C and SI Appendix, Fig. S4). Consistent with the virus attachment and infectivity assays, TCA, which fails to enhance infection, did not bind the MNoV P domain (Fig. 2D), supporting the relevance of the observed binding to viral attachment and infection.
Fig. 2.
The bile acid GCDCA enhances MNoV–receptor interactions and binds the P domain. (A) MNoV was incubated with BV2 cells in PBS without divalent cations (PBS−/−) on ice. Unbound virus was washed off, and MNoV genomes were enumerated relative to host actin transcripts. Bovine bile and glycochenodeoxycholic acid (GCDCA) but not taurocholic acid (TCA) rescued MNoV binding. (B) MNoV was added to a confluent monolayer of BV2 cells in various buffer conditions at room temperature. Unbound MNoV was removed, and methylcellulose was overlaid. Infection was determined by plaque-forming units 2 d postbinding. FBS, divalent cations (Ca2+ and Mg2+), bovine bile, and GCDCA rescued infectivity, whereas other bile acids and HBGA did not (one-way ANOVA; means ± SEM; NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Data in A are pooled from at least two independent experiments each performed in at least triplicate. Data in B are from at least three independent experiments performed in at least duplicate. (C) Isothermal titration calorimetry measurements were performed in which buffer containing GCDCA was injected into the same buffer containing P domain protein. Fitting of the binding isotherms yielded an average KD of ∼5.76 ± 1.26 μM for GCDCA and close to 1:1 molar stoichiometry (SI Appendix, Fig. S4). The heat of dilution was determined to be negligible in separate titrations of GCDCA into buffer alone. (D) Despite being chemically similar, TCA did not enhance infection in tissue culture and did not bind P domain in the ITC assay.
Crystal Structure of P Domain in Complex with Bile Acids.
To explore bile acid enhancement of MNoV infection, we obtained crystals of GCDCA or LCA bound to the CD300lf–P domain complex (Fig. 3 and SI Appendix, Fig. S3). We chose LCA in addition to GCDCA as a secondary bile acid present in the intestine of mice and humans. The structures were solved by molecular replacement. GCDCA-bound crystals belong to space group P212121 (a = 49.11 Å, b = 134.82 Å, c = 139.59 Å), and the final refined model has an Rwork of 20.9% with an Rfree of 25.7% to 1.95 Å resolution (SI Appendix, Table S2). Similarly, the LCA-bound crystals belong to space group P21212 (a = 101.09 Å, b = 152.89 Å, c = 70.04 Å), and the final refined model has an Rwork of 17.4% with an Rfree of 20.4% to 1.80 Å resolution (SI Appendix, Table S2). These structures reveal binding pockets located at the dimer interface between the P1 and P2 subdomains such that each P dimer binds two bile acid molecules, a stoichiometry consistent with our ITC analysis (Fig. 2C).
Fig. 3.
Bile acids bind in a deep pocket on the MNoV P domain. (A) Ribbon diagram of the CD300lf–P domain complex loaded with GCDCA displayed as a ball and stick model with carbon atoms colored in magenta. The two chains of the P domain dimer are colored and oriented as in Fig. 1. (B) Solvent accessible surface of the P dimer with GCDCA bound. (C) The two panels show the dimer, opened like a book, to reveal contact residues along the pocket walls. P domain residues within 4.5 Å of GCDCA have carbon atoms colored white, and their surface contributions are mapped in white on the P domain surface. The ligand makes a salt bridge from the carboxyl terminus of the Gly side chain to Arg392, Arg437, and Trp245. The carbon ring core of the bile acid makes several hydrophobic contacts with the pocket. (D) Plot of GCDCA contacts within the pocket. (E) P domain variants Y250D and R437D display a greatly diminished ability to GCDCA as measured by ITC, whereas the R390Y variant displayed a roughly 50-fold enhancement in binding.
The GCDCA glycine is buried deep in a hydrophobic binding pocket where oxygens from the carboxyl terminus form a salt bridge to amino acids in both chains of the P dimer (Fig. 3C). This interaction involves Arg392, Arg437, and Trp245. The GCDCA carbon ring core contacts many hydrophobic residues lining sides of the pocket (Fig. 3D). Similarly, LCA binds into the same pocket, albeit not as deeply, with a carboxyl oxygen making a salt bridge to Arg392 but with the carbon ring core sitting in a different orientation relative to GCDCA (SI Appendix, Fig. S3 B and D). Thus, the hydrophobic nature of the pocket facilitates binding of diverse bile acids.
To assess whether residues lining the structurally identified ligand binding pocket control GCDCA binding, we created mutant proteins for ITC binding analysis. We chose to produce three variants (Y250D, R390Y, and R437D), avoiding substitutions predicted to affect the P dimer interface (Fig. 3). Bacterially produced P domain variants eluted from size exclusion chromatography similar to the wild-type protein. The Y250D and R437D variants completely lost the ability to bind GCDCA (Fig. 3E). Arg437 lies deep in the pocket close to the glycine of GCDCA, whereas Tyr250 forms one side of the pocket that supports the hydrophobic backbone of bile acid carbon rings (Fig. 3C). Mutation R390Y enhanced GCDCA binding by ∼50-fold (Fig. 3E). The Arg390 side chain makes numerous GCDCA contacts and adopts distinct rotamers in the GCDCA-bound, LCA-bound, and empty structures, with its position the most variable element of the ligand binding pocket (Fig. 3C and SI Appendix, Fig. S3). Taken together, the loss and gain of function mutations strongly support the structural definition of the bile acid binding pocket located at the P domain dimer interface.
Role of Metal Ions in CD300lf Binding.
The addition of magnesium and calcium increased MNoV infection (Fig. 2B), leading us to examine the role of divalent cations in virus–receptor interactions. Each CD300lf–P domain cocomplex structure contains three metal ions per P domain. Two are at the CD300lf–P domain interface, whereas a third binds midway between the interface metal location and bile acid pocket, held by residues Gln438 and Asp440. Crystals of the cocomplex between CD300lf and the P domain alone and of the cocomplex containing GCDCA were both grown in 100 mM MgCl2 (Figs. 1, 3A, and 4B). These structures refine well with magnesium, having geometry, bond angels, and fit of B factors to the environment of the metal atoms within acceptable limits as assessed via the CheckMyMetal server (33). The crystal of the cocomplex between CD300lf and the P domain containing LCA was grown in 100 mM CaCl2 and contains six Ca2+ ions located analogously to the Mg2+ in the other two complexes. In all three complexes a divalent cation (Ca2+ or Mg2+) situated between the CD300lf protein and the P domain coordinates three atoms in the CD300lf CDR3 loop (carbonyl oxygen atoms of Lys94 and Gly96 and a side chain oxygen of Asp98) with a carbonyl oxygen from Asn364 in the DE loop of the P domain. For the Mg2+-containing structures, a second divalent cation orients the side chain of P domain Asp366 allowing it to hydrogen bond across the interface to a backbone nitrogen of Gly96 in the CDR3 loop. However, in the CD300lf–P domain–LCA structure, the second Ca2+ at the interface enables the carboxylate oxygen of P domain Asp366 to coordinate to the CDR3 engaged Ca2+ by virtue of its longer bond distance capacity compared with Mg2+ (compare Asp366 in Fig. 4 B and C). The significant structural role of divalent cations at the receptor binding interface is consistent with the capacity of high concentrations of these ions to enhance infection.
Fig. 4.
CD300lf employs a similar strategy to bind PC and the P domain. (A) Model showing the orientation of the P2 subdomain relative to CD300lf in the empty (no bile salt) complex colored as in Fig. 1. (B) Close-up of the empty CD300lf–P domain complex showing the side chain of Asn364 from the P domain DE loop binding to CD300lf in a pocket made by the CC′ loop. The carbonyl oxygen of Asn364 coordinates with a magnesium ion at the binding interface. A second magnesium ion supports the P domain DE loop, holding it against the binding pocket, by bridging Asp410 and Asp366. (C) Close-up of the LCA loaded CD300lf–P domain complex showing a calcium ion at the interface. Here Asp366 completes the octahedral coordination of the interface calcium ion. (D) Close-up of the CD300lf with PC bound. The PC choline moiety (blue and green sticks) rests in a hydrophobic pocket, whereas the phosphate group (red and tan sticks) coordinates with a calcium ion held in place by atoms from the CD300lf CDR3 loop. CD300lf residues around the pocket are labeled. The expected position for the fatty acid lipid tail attachment is shown. The tails are drawn extending from one of the PC phosphate oxygen atoms.
MNoV Binds CD300lf as a Molecular Mimic of Phospholipids.
Phosphatidylcholine (PC) is one of several lipids reported to bind CD300lf (12). To identify the lipid-binding site on CD300lf, we obtained cocomplex crystals with the head group of PC belonging to space group P212121 (a = 32.64 Å, b = 40.06 Å, c = 68.29 Å). The structure was solved by molecular replacement to 1.35 Å resolution with an Rwork of 16.8% and an Rfree of 19.2% (SI Appendix, Table S1). The PC head group binds in a hydrophobic cleft between the CDR3 and CC′ loops (Fig. 4D) in a prominent protrusion extending out from the main Ig fold. The bottom of the cleft is composed of a disulfide bond (Cys36–Cys44) and the side chains of Met100, Tyr34, Trp89, Gln41, and Arg42, which hold the choline end of the PC. The backbone amide of Asp98 and side chain nitrogen of Arg42 orient the phosphate end of the head group that coordinates with a Ca2+ ion held by the CDR3 loop. This orientation would allow the fatty acid tail of phosphatidylcholine to protrude out of the binding site away from the CD300lf bearing cell, an orientation consistent with the role of CD300lf as a lipid receptor (11–13).
To understand the relationship between CD300lf interactions with phospholipids and the P domain, we superimposed the PC and P domain-bound costructures and compared the contact footprints of each ligand. Both PC and the P domain engage the same CD300lf surface cavity located between the CDR3 and CC′ loops (Fig. 4), and both coordinate with a similarly positioned metal ion. Although PC uses an oxygen from the phosphate group to engage the CDR3 coordinated metal, the P domain uses the carbonyl oxygen from Asn364. The choline group of PC makes extensive contact with the CC′ loop. In the P domain-bound structures, the side chain of Asn364 interacts with many of the same CC′ loop residues as PC. It is worth noting that Asn364 is nearly invariant among reported MNoV sequences (SI Appendix, Fig. S2). These data clearly provide evidence for viral mimicry, whereby MNoV chemically mimics phospholipid in engaging the CD300lf receptor.
The MNoV P Domain Binds CD300lf with Low Affinity.
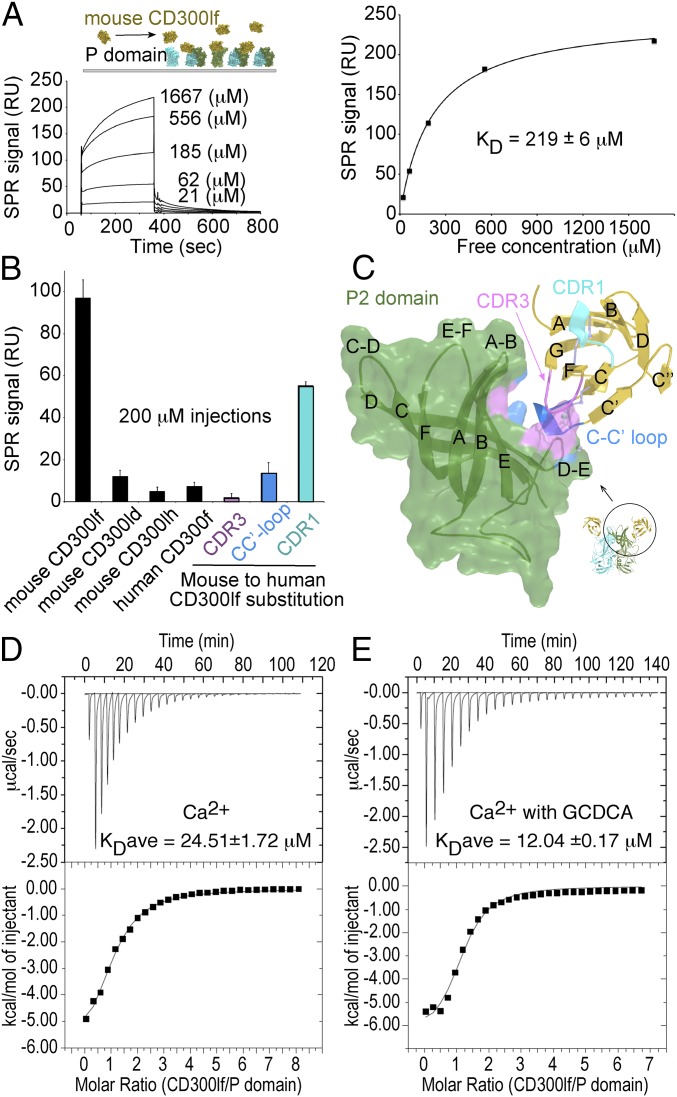
We next measured the binding affinity of CD300lf for the MNoV P domain using soluble recombinant proteins. Analysis of CD300lf binding to the P domain by surface plasmon resonance (SPR) over a 21–1,667 μM concentration range yielded a monomeric binding affinity of KD ∼ 219 μM (Fig. 5A). To ensure that this weak SPR binding signal was specific, we assessed binding of CD300 family members previously evaluated for a role in MNoV infection. Of the nine mouse CD300 family members, only CD300lf and, to a lesser extent, CD300ld conferred MNoV susceptibility to human HeLa cells, whereas mouse CD300lh and human CD300f did not (5). Only the soluble ectodomain of mouse CD300lf, and not mouse CD300ld or CD300lh, or human CD300f neutralizes MNoV infectivity (5). We therefore measured binding of these proteins to the MNoV P domain using SPR (Fig. 5B). Mouse CD300lf gave a strong signal for binding to the P domain after background subtraction, whereas very little binding was observed for mouse CD300ld or CD300lh or human CD300f. We then assessed if this interaction involved the receptor and PC-binding determinants of CD300lf identified in our crystal structures. CD300lf containing human sequence substitutions of the CC′ or CDR3 loop fail to support MNoV infection, and the soluble ectodomains of these proteins do not neutralize MNoV infection (5). Similarly, mouse CD300lf containing these human sequences or CDR3 loop exhibited greatly diminished binding to the P domain (Fig. 5 B and C). A control substitution, more distal from the binding cleft in the CDR1 loop, diminished the binding signal by about half (Fig. 5 B and C). These results verify our direct affinity measurements for the interaction between the P domain and CD300lf, and the concordance between viral infection and the binding assays strengthen the conclusion that our structural studies are directly related to viral receptor function.
Fig. 5.
CD300lf binds the P domain with low affinity. Recombinant soluble MNoV P domain protein was immobilized onto the surface of a CM5 chip for SPR analysis. (A) The response to various concentrations of soluble mouse CD300lf is shown. The binding was specific and saturable yielding a KD = 219 ± 5.66 μM, representing the mean ± SD from three independent experiments. The data were analyzed using Biaevaluation 3.1 (GE Healthcare). The equilibrium concentration curve values were fitted at steady-state assuming a 1:1 binding model. (B) Comparison of the P domain binding response against various recombinant ectodomains at 200 μM concentration. Mouse CD300ld, CD300lh, and human CD300f show very low binding. In magenta, blue, and cyan are the CRD3, CC′ loop, and CDR1 variants where the mouse residues have been replaced by the human equivalent residues in those loops. Bars represent the mean response ± SD from three injections. (C) Model showing where the CDR3, CC′ loop, and CDR1 variants map onto the P domain surface. (D) Binding affinity measured in the presence of 1 mM CaCl2 by isothermal titration calorimetry. (Top) The raw data from titration of CD300lf injected into the P domain. (Bottom) The integrated heats for each injection versus molar ratio of CD300lf to P domain fit using a one-site binding model. The KD = 24.51 ±1.72 μM and represents the average from two independent experiments (SI Appendix, Fig. S5). (E) Addition of 250 μM GCDCA gave about a twofold increase in binding affinity for an average KD = 12.04 ± 0.17 μM from two experiments (SI Appendix, Fig. S5).
Role of Divalent Cations in P Domain Interactions.
The cocomplex crystal structures contain metal ions: two at each P domain/CD300lf interface and one on the side of each P domain. The SPR binding was performed in buffer containing 40 μM EDTA, which sequesters metal ions. Attempts to add Mg2+ or Ca2+ to the SPR buffer resulted in elevated background preventing interpretation. This led us to develop an ITC assay for binding of CD300lf to the P domain in solution. We observed binding of P domain and CD300lf by ITC in buffer consisting of 1 mM CaCl2, 30 mM Hepes, pH 7.4, and 150 mM NaCl, where CD300lf was injected into P domain containing solution (Fig. 5D). The signal was specific for CD300lf binding because injection of CD300lf into the buffer alone (no P domain) gave no signal (SI Appendix, Fig. S5). The ITC experiments with Ca2+ yielded an average binding affinity from two experiments of KDave ∼ 24.51± 1.72 μM, a value ∼10-fold higher than we saw by SPR without Ca2+, further supporting the conclusion that the P domain–CD300lf interaction is of low affinity and potentially regulated by divalent cations. Consistent with this, removal of free calcium by addition of 0.1 M EGTA to the ITC buffer inhibited CD300lf binding. Addition of 0.1 M EGTA and 1 mM MgCl2 (to chelate Ca2+ under conditions of excess Mg2+) gave binding similar to that obtained in 1 mM CaCl2, yielding a KDave of ∼24.29 ± 3.55 μM from two experiments (SI Appendix, Fig. S5). This suggested that both divalent cations could play a role in physiologic receptor binding. However, it remains unclear whether they act directly at the protein–protein interface or through stabilization of the P domain. The availability of this assay allowed us to demonstrate that addition of GCDCA at a concentration of 250 μM had a small but reproducible effect on CD300lf binding, yielding an increase in affinity of approximately twofold, for a KDave of ∼12.1 ± 0.17 μM from two experiments (Fig. 5E and SI Appendix, Fig. S5). Although the importance of this change is unclear, these data support the concept that bile acids and divalent cations may regulate P domain:CD300lf interactions in a combinatorial fashion.
Mapping of P Domain Residues Involved in CD300lf Interactions.
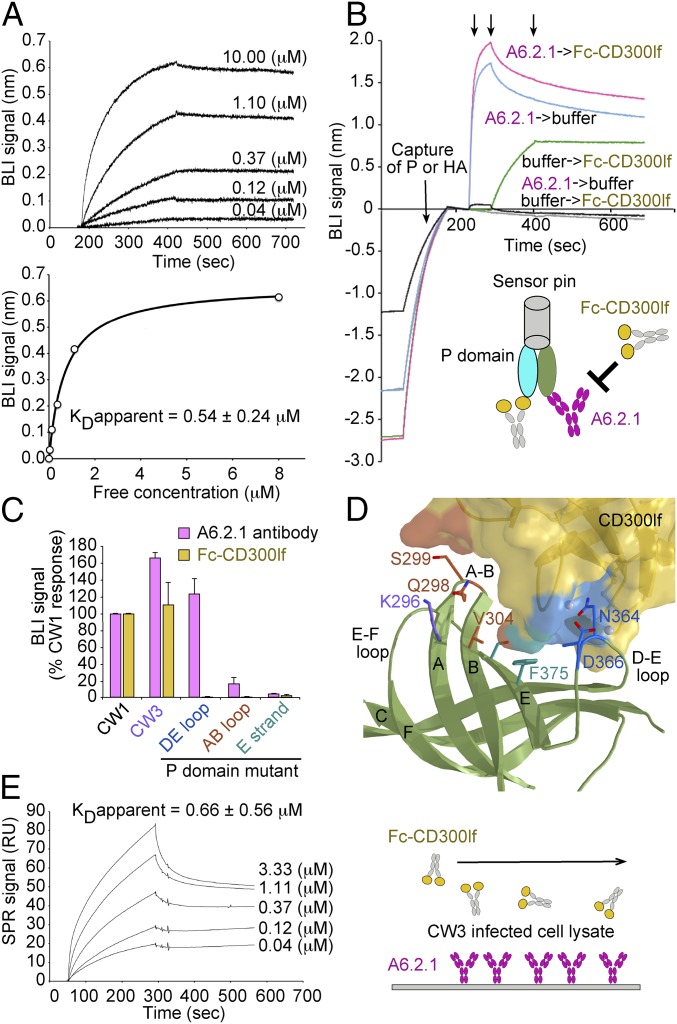
To enhance the avidity of binding and to allow mutational studies of the P domain:CD300lf interface, the CD300lf ectodomain was joined to a human Fc fragment (Fc–CD300lf), and the protein was transiently expressed in HEK293 cells. We confirmed the bivalent Fc–CD300lf fusion protein could recognize the P domain using biolayer interferometry (BLI). For this assay, streptavidin-coated biosensor pins loaded with biotin-labeled recombinant P domain were tested against wells containing various concentrations of Fc–CD300lf. Analysis of the response curves yielded an apparent avidity of ∼0.54 ± 0.24 μM (Fig. 6A). This binding was specific as it was blocked by the MNoV neutralizing antibody A6.2.1 (Fig. 6B) (2).
Fig. 6.
CD300lf binding is inhibited by neutralizing monoclonal antibody A6.2.1 and involves residues in the P2 subdomain DE and AB loops. Randomly biotin-labeled recombinant MNoV P domain was loaded onto streptavidin biosensors for BLI analysis. The pins were submerged in various concentrations of Fc-CD300lf to produce the binding curves shown (A). Analysis yielded a KD apparent = 0.54 ± 0.24 μM representing the mean ± SD from three independent experiments. The equilibrium concentration curves were fitted at steady-state assuming a 1:1 binding model. (B) Biosensor pins loaded with P domain or influenza HA control protein (Materials and Methods) were placed in A6.2.1 antibody or buffer. After equilibration against buffer alone, the pins were placed in wells containing Fc-CD300lf. A6.2.1 (magenta) inhibited binding of Fc-CD300lf compared with pins that had not been blocked but instead held in buffer (green). No binding was seen to the control HA-coated pins (black and gray). (C) P domain variants were assayed by BLI for Fc-CD300lf binding as in B. Streptavidin-coated pins loaded with biotin-labeled P domain, or P domain variants, were placed in wells containing 5 μM Fc-CD300lf or A6.2.1 antibody. The responses are reported as percentage of binding obtained with CW1 strain P domain. CD300lf binding was undetectable after deletion of the DE loop (Asn364, Ala365, and Asp368). (D) Position of variant residue alterations, color coded as in C, and with contacts mapped onto the CD300lf surface. The ribbon diagram shows the P domain in green with key side chains colored corresponding to the different variants. (E) CD300lf recognizes P domain protein produced in MNoVCW3-infected cells. The A6.2.1 antibody was immobilized onto a CM5 chip and used to capture P domain from infected cell lysates. Various concentrations of Fc-CD300lf were passed over the surface, and the SPR binding data were analyzed to obtain a KD apparent = 0.66 ± 0.56 μM, as the mean ± SD from three independent experiments.
To assess the role of P domain contacts with CD300lf seen in the cocrystal structures (Figs. 1 and 6D), variant P domains were tested for their ability to bind Fc–CD300lf using BLI pins coated with biotin-labeled CW1 P domain or variants dipped into wells containing 5 μM Fc–CD300lf. The MNoV strain CW3 P domain was also tested. It differs by a single amino acid from CW1, having a lysine instead of a glutamic acid at position 296 (4). CW3 binds A6.2.1 similarly to CW1 (34) and also binds well to Fc–CD300lf (Fig. 6C). In contrast, CD300lf binding was undetectable after deletion of three amino acids from the P domain DE loop (Asn364, Ala365, and Asp366). The DE loop variant also failed to bind monomeric CD300lf in the ITC binding assay (SI Appendix, Fig. S5), although it appears folded because it is recognized by A6.2.1, which is reported to not recognize denatured VP1 by immunoblot (2). Similarly, substitution of four residues in the AB loop region (Q298R, S299E, G300P, and V304K) diminished Fc–CD300lf binding to near background. Likewise, the substitution of two residues in the P domain E strand (F375D and S377K) also diminished CD300lf binding. Although the last two variants do not bind A6.2.1 as well as the wild-type P domain, they migrate similarly on size exclusion chromatography. The impaired ability to bind A6.2.1 is likely an effect of the AB loop and E strand mutations on the A6.2.1 epitope (SI Appendix, Fig. S6). This P domain mutational analysis clearly provides support for the structurally defined CD300lf binding determinants.
CD300lf Recognizes Lysates from MNoVCW3-Infected Cells.
To ensure the Fc–CD300lf fusion protein could recognize nonrecombinant P domain, we tested it against proteins produced in CW3-infected BV2 cells using an SPR binding assay (Fig. 6E). P domain proteins were captured from infected cell lysate on a chip coated with A6.2.1 antibody, then various concentrations of Fc–CD300lf were passed over the surface to observe binding. The Fc–cD300lf bound with an apparent avidity of 0.66 ± 0.56 μM, similar to that observed for recombinant protein (Fig. 6A). Fc–CD300lf binding to infected cell lysate was also inhibited by pretreatment with A6.2.1 antibody, and no binding of A6.2.1 or Fc–CD300lf was observed for mock-infected cell lysate (SI Appendix, Fig. S7). These data further support the validity of our quantitative assays for evaluating CD300lf interactions with viral P domains.
Discussion
The norovirus capsid protein VP1 contains determinants of genome encapsulation and particle assembly as well as membrane penetration and entry, thereby representing a portion of the molecular machinery required for infection. Here we have structurally defined the interaction between the MNoV P domain and the newly discovered entry receptor CD300lf, an interaction that appears to require interfacial coordination of metal ions. Further, we herein identify bile acids as cofactors for MNoV infection and establish the P domain structural determinants used in the engagement of the bile acids GCDCA and LCA. These results are supported by a series of biophysical assays that have incorporated analysis of structure-based mutations. These structural and functional studies have significant implications for our understanding of norovirus biology.
Interactions Between the P Domain and Its Cellular Receptor CD300lf.
Because of its position on the outer face of the viral capsid and its targeting by neutralizing antibodies (18–23, 29, 34, 35), the P2 subdomain was a prime candidate for CD300lf receptor engagement. Consistent with this, we show that CD300lf contacts a cleft in P2 formed by the AB and DE loops, with two receptors splayed away from the P domain dimer interface (Fig. 1). Feline calicivirus (FCV) and Hom-1 calicivirus employ junctional adhesion molecule A (JAM-A) and related proteins as receptors (36, 37). The cryo-EM structure of the FCV capsid in complex with the two Ig-like extracellular domains of fJAM-A indicates this receptor also binds to the P2 subdomain, although the receptors appear to engage the top of the P domain near the dimer interface (38). This same dimer interface proximal region of P2 is used by some human noroviruses to bind HBGA glycans (25, 28, 39, 40). Our structural data are robustly supported by quantitative binding analysis (SPR, BLI, and ITC), where mutation of critical determinants in the CD300lf CRD3 and CC′ loops along with mutations in the P domain AB loop, DE loop, and E strand all result in disruption of the interactions.
CD300lf binding to the P domain of MNoV was potently blocked using the neutralizing antibody A6.2.1. Cryo-EM models suggest that A6.2.1 binding site spans both the AB and EF loops of P2, a conclusion supported by both escape mutants and site-directed mutagenesis studies (21, 26, 34, 41). This predicts that the A6.2.1 contact site overlaps with the CD300lf contact site on the P domain seen in our cocomplex structure (SI Appendix, Fig. S6), strongly supporting the concept that A6.2.1 inhibits MNoV infection by blocking cell attachment. The monoclonal antibody 2D3.7 broadly neutralizes MNoV strains and by cryo-EM binds very close to the A6.2.1 site although at an angle that allows it to make more contact with the AB and EF loops (21). Based on our structural studies it seems likely that 2D3.7 also neutralizes MNoV by blocking the CD300lf binding site.
The Role of Multivalency in Cellular Attachment.
The MNoV P domain buries ∼630 Å2 of solvent accessible surface at the CD300lf contact interface, an area notably smaller than that buried in many other receptor–virus interfaces. For instance, the contact area buried by HIV gp120 and its receptor CD4 is ∼742 Å2 (42), between rhinovirus HRV14 and its receptor ICAM‐1 is ∼990 Å2 (43), between coxsackievirus A21 and its receptor ICAM-1 is ∼970 Å2 (44), and between poliovirus and the poliovirus receptor CD155 is ∼1,300 Å2 (45). Consistent with this small contact area, the affinity between the P domain and CD300lf is relatively weak with a KD of ∼25 μM for monotypic binding as measured in solution by ITC. The low-affinity binding observed between the P domain and CD300lf suggests that cell attachment might be avidity-driven, requiring that P domains on a virion simultaneously engage multiple CD300lf proteins on the cell surface. We therefore docked the X-ray structure of the complex onto a cryo-EM–derived model of the MNoV particle. We found that CD300lf domains cluster in threes on the virion surface (Fig. 7A). The distribution of CD300lf contacts can be seen by removing the CD300lf molecules from the model (Fig. 7B). This distribution makes clear that each CD300lf cluster is composed of receptors from three different P domain dimers (Fig. 7C). This docking model results in some minor overlap of CD300 domains (Fig. 7C), which may be resolved by mobility of P domains, which are flexibly connected to the shell domain of VP1. These results support a model in which multiple CD300lf engagements can be made by each virion to drive MNoV binding and entry. We undertook several binding studies using an Fc–CD300lf fusion protein, revealing that the bivalent reagent can bind recombinant P domain and MNoV from infected cell lysates with an avidity at least 50-fold higher than the monotypic binding affinities we observed. Because our data are most consistent with a need for multivalent interactions between the P domain and several cell surface CD300lf receptors, the strength of these bivalent interactions may be physiologically relevant.
Fig. 7.
Docking of CD300lf–P domain complex onto a cryo-EM–derived model of the MNoV virion. (A) The solvent accessible surfaces of CD300lf are shown in yellow, with the P domain subunits in blue and green. The CD300lf domains group in threes across the surface of the virion. Each CD300lf trimer is made up of receptors from three different P domain dimers. (B) The CD300lf contacts mapped in yellow on the model surface after removing CD300lf. Fivefold, threefold, and twofold symmetry is marked with a white pentagram, triangle, or ellipse, respectively. (C) (Right) A top-down view (same orientation as in A) for three P dimers in a trimer cluster with the shell region below in blue. (Left) A side view of the three P dimers. Note how the CD300lf C termini (marked by red cylinders) cluster at the center of each trimer and overlap slightly. The magenta ovals with a white letter B mark visible bile acid binding pockets.
Molecular Mimicry by MNoV in Receptor Engagement.
Our structure of CD300lf in complex with phosphocholine has defined the host ligand binding site used by the receptor. The ligand head group is orientated so that the fatty acid tail would extend out of the pocket away from the CD300lf-bearing cell surface, consistent with the function of CD300lf as a lipid receptor. The ligand orientation and metal coordination are very similar to that used by TIM-4, a related immune regulatory molecule, to bind phosphatidylserine (SI Appendix, Fig. S8) (46). By comparing the structures CD300lf bound to either phosphocholine or the MNoV P domain, we have revealed in precise atomic detail how the viral capsid mimics binding of a CD300lf ligand and the role of divalent cations in both of these processes. This form of molecular mimicry predicts that the interaction between the P domain and CD300lf during infection would be incompatible with simultaneous binding of ligands to the lipid-binding pocket of CD300lf, suggesting that infection might be regulated by high-affinity receptor ligands. Bacterial ligands may also bind to CD300 family members (10), raising the possibility that interactions between bacteria and the virus may involve binding of ligands to CD300lf that in turn compete for interactions with the P domain.
The Role of Bile in MNoV Infection.
We previously showed that efficient MNoV binding to cells and infection requires a small molecular weight heat stable serum cofactor (5). Here we show that the bile acid GCDCA can serve as a cofactor by increasing cell binding and infection. We do not know if GCDCA is responsible for the activity originally observed in serum. The role for this bile acid is likely to involve direct binding to the viral capsid protein because our crystal structures of the CD300lf–P domain complex bound with both GCDCA and LCA revealed two binding pockets per P domain dimer that can accommodate these bile acids. The location of the bile acid pocket was surprising for several reasons. First, the bile acid pocket is remote from the CD300lf–P domain interface, suggesting that if bile acids function by altering the CD300lf–P domain interaction, this must be via a long-distance allosteric effect. Second, the bile acid pocket is distinct from the glycan-binding sites observed on human and murine norovirus P domains (25, 40), suggesting that norovirus P domains are promiscuous in their ability to bind diverse biologic small molecules and further arguing that the interaction between noroviruses and target cells is regulated at multiple levels. It seems likely that the efficiency of norovirus infection of cells in culture requires that each of these different levels of control be perfectly matched to avoid abortive infection. Third, binding of GCDCA and LCA to MNoV P domain did not induce significant structural rearrangements in the P domain or in the CD300lf interface. We did find that addition of GCDCA had a small (approximately twofold) but reproducible effect on binding of soluble CD300lf to the P domain in the presence of divalent cations. It is possible that this small effect, summed over the many copies of the P domain on the virion, has biological significance. However, this is speculative, and the molecular mechanism of cofactor function remains an open question. It is intriguing to consider that bile acids, rather than directly modulating receptor:P domain interactions, instead alter the structural conversions that occur as the virus enters the cell to release its RNA into the cytoplasm or the conformation of the virion itself in ways not detected by biding studies of the P domain and CD300lf in solution.
We also found that addition of calcium and magnesium also augments infection. Our structural data indicate that there are at least three divalent cation binding sites associated with CD300lf binding to each P domain monomer. Two of these are in the binding interface between the P domain and CD300lf and likely facilitate the interaction. A third metal ion is located midway between the receptor and bile acid binding sites of the P domain, directly participating in neither interaction. Crystal structures and ITC binding of the MNoV P domain with CD300lf showed that either magnesium or calcium can support complex formation. Of particular interest, Ca2+ is present in high concentrations in bile (47), making it reasonable to propose that calcium might be present at sites of MNoV infection (48). It is intriguing to speculate that the availability of divalent cations in the body, which may not be available in the environment, might foster infectivity of the virus once it reaches its target.
Implications of These Studies.
These studies support the concept that noroviruses have evolved to be dependent on small molecules either by molecular mimicry of host lipids or by depending on cofactors such as bile acids and divalent cations for critical steps in infection. Physiologically, it is attractive to consider that the presence of bile acids and Ca2+ in the intestine downstream of the common bile duct, in combination with tropism of the virus for tuft cells as the sole epithelial cell expressing CD300lf (7), explains the tropism of the virus. This adds to the prior finding that tuft cell tropism is conferred by the combinatorial effects of the host cytokine IFNλ and the MNoV nonstructural protein NS1 (49). Further, although we could not reproducibly observe an effect of addition of LCA on binding or infectivity due to solubility issues, this secondary bile acid can also bind to the MNoV P domain in the pocket used by GCDCA. This indicates that secondary bile acids might serve a physiologically important role in vivo. However, enhancement appears to be rather specific because multiple other bile acids did not augment infection. Because we have not exhaustively sampled primary and secondary bile acids in the intestine, it is quite possible that additional cofactors or even inhibitors of norovirus infection will be discovered in the future.
In addition to identifying that the norovirus P domain interacts with a soluble cofactor, the structural studies presented here raise the possibility of designing effective orally bioavailable bile acid analogs for the treatment of human norovirus infection, which has also been shown to be enhanced by bile (30). Further, we previously showed that intestinal bacteria play a critical role in the establishment of persistent enteric MNoV infection (50). Because intestinal bacteria are involved in metabolizing primary bile acids into secondary bile acids, this raises the possibility that transkingdom interactions between norovirus and enteric bacteria are via alterations in bile acids in the intestine. Indeed, broad-spectrum antibiotics might alter norovirus infection via alterations in bile acid metabolism (50, 51).
Materials and Methods
Protein Expression and Purification.
A cDNA encoding the VP1 P domain of MNV1 strain CW3 (residues 229 through 540 of GenBank Sequence ABJ98944.1) was inserted into pET21 between the NdeI and XhoI sites to create a C-terminal 6His-tag. The P domain of MNV1 clone CW3, which differs from CW1 by a single amino acid substitution at E296K (which is not present at the interface between CD300lf and the P domain), proved more stable and was used for binding and structural studies except where indicated. The P domain was expressed as a soluble protein in BL21(DE3) cells (0.2 mM IPTG for 12 h at 25 °C). Cells were lysed in 50 mM Tris⋅HCl, pH 7.5, 500 mM sodium chloride, 10% glycerol. The supernatant was incubated on Ni-NTA agarose, washed, and protein eluted in 2× PBS at pH 8.0 containing 300 mM Imidazole. The P domain was purified over a Superdex column and was found to be monomeric (SI Appendix, Fig. S9). The CD300lf expression construct encodes residues 19–131 of NP_663609.2 inserted between the NdeI and XhoI sites of pET21. In brief, CD300lf protein was recovered from BL21(DE3) inclusion bodies, denatured in guanidine hydrochloride, and then oxidatively refolded by rapid dilution following standard methods (52). Refolded protein was purified over Superdex S200 and confirmed to be monomeric. The Fc–CD300lf construct hinge region was designed to be too short to allow both CD300lf domains to bind simultaneously to both sites on a P domain dimer. The construct joins residues 20–128 of NP_663609.2 with the Fc portion of human IgG2 in a pCDNA3.1 expression vector using an IL-2 signal peptide. The amino acid sequence of the Fc hinge region starts ASVECPPCP. The fusion protein was produced in HEK293 cells and purified on protein G followed by Superdex 200 gel filtration.
Crystallization and Structure Determination.
Cocomplexes were made by mixing equal molar amounts of MNV1.CW3 P domain with murine CD300lf in 25 mM Tris⋅HCl, pH 7.4, 150 mM NaCl. The mix was concentrated to 15 mg/mL and buffered exchanged to 20 mM Tris⋅HCl, pH 8.7, containing 20 mM NaCl. Crystals grew in hanging drops at 20 °C in 0.1 M Tris⋅HCl, pH 8.7, 0.1 M MgCl2, and 10% (wt/vol) PEG 8000. They were soaked in well solution containing 20% (vol/vol) ethylene glycol then flash-cooled in liquid nitrogen for data collection. The LCA containing CD300lf–P domain complex crystals were grown in 100 mM Tris⋅HCl, pH 8.7, 40 mM CaCl2, and 5.5% PEG 10,000. Because LCA is only slightly soluble in water, a drop of ethanol containing 0.1 μg of LCA was placed on the coverslip and allowed to air dry before the protein and mother liquor were added. Crystals were prepared for freezing by adjusting the concentration of PEG 10,000 in the drop to 25%. Similarly, the GCDCA crystals were grown in 100 mM Tris⋅HCl, pH 8.7, 110 mM MgCl2, and 11.0% PEG 10,000 with 0.1 mM GCDCA. These crystals were soaked in well buffer containing 15% (vol/vol) ethylene glycol and then flash-cooled in liquid nitrogen for data collection. CD300lf–phosphocholine crystals were grown in 33% (wt/vol) PEG 3350 using 100 mM phosphorylcholine chloride calcium salt adjusted to pH 7.4. Data were collected at 100°K on the Advanced Light Source (ALS) 4.2.2 beamline and processed with XDS (53). Initial phases were determined by molecular replacement using Phaser (54) with models from mouse CD300lf (PDB ID code 5FFL) and P domain (PDB ID code 3LQE) (26). Refinement was carried out in Phenix (55), with manual rebuilding in Coot (56). Data collection and refinement statistics are summarized in SI Appendix, Tables S1 and S2. The final coordinates were assessed using MolProbity (57). Solvent accessible area calculations used Naccess (58). Interface contacts were identified using the PISA server (59). Structure figures were rendered with PyMOL (60).
Virus Purification from Infected Cells.
BV2 cells grown to a density of 45 × 106 cells per 100 mL were infected with MNV1.CW3 (MOI of 0.05) and virus recovered 36 h later. Virus-infected cells were lysed by freezing at −80 °C and the lysate concentrated 10-fold over a 100-kDa membrane. After filtration (0.45 μm), 23 mL of concentrated lysate was pelleted through a 5-mL layer of 30% sucrose in 1× PBS for 3 h at 100,000 × g. The virus pellet was resuspended in 250 uL of PBS. A mock lysate was prepared from uninfected BV2 cells.
Surface Plasmon Resonance Binding Analysis.
Recombinant neutravidin was immobilized on a CM5 chip in 10 mM sodium acetate, pH 3.5, by standard amine coupling. Proteins were captured by random biotin labeling. Briefly, 100 μg of protein was mixed with biotin (EZ-Link-NHS-PEG4-Biotin; Thermo Fisher) at a molar ratio of 20:1 biotin:protein and incubated. Unreacted biotin was removed with a desalting column (Zeba Spin 7K; Thermo Fisher). The control lane was coated with biotin-labeled hemagglutinin (HA) extracellular domain from influenza A virus subtype H5N1 (A/Indonesia/5/2005). The HA control protein has an isoelectric point similar to the P domain of MNV1 and was made using baculovirus-infected High Five cells. The test lane contained biotin-labeled P domain. Both control and test lanes contained ∼3,000 RU of biotin-labeled protein. The binding buffer was 100 mM Sodium Phosphate (pH 7.4), 400 mM NaCl, 40 μM EDTA, and 0.005% (vol/vol) Tween-20 to minimize background. Monomeric CD300 family ectodomains were tested at 200 μM at a flow rate of 30 μL/min. The high protein concentrations required to show binding gave rise to higher than normal responses on the control lane, often approaching 60% of the total signal. We subtracted the HA control lane response from the P domain test lane response to obtain the specific binding. In the infected cell lysate assay, the antibody A6.2.1 was immobilized onto CM5 in 10 mM sodium acetate, pH 3.5, by standard amine coupling. CW3-infected BV2 cell lysate was passed over the chip in binding buffer consisting of PBS (pH 7.4) and 0.005% (vol/vol) Tween-20. P domain was detected by passing concentrations of Fc–CD300lf over the captured material. For the reference surface, chikungunya virus-specific antibody CHK-265 was immobilized onto the chip and used to capture recombinant chikungunya virus-like particles produced as described (61). We subtracted the CHK-265 control lane response from the infected cell lysate test lane response to obtain the specific binding. The saturation value was arbitrarily taken as 300 s after start of the injection (not equilibrium) and used to calculate the KD apparent by fitting to a 1:1 binding model. SPR experiments were performed using a Biacore T200 (GE Healthcare) at 25 °C.
Biolayer Interferometry Binding Analysis.
The apparent binding affinity of Fc–CD300lf was measured by BLI using an Octet-Red96 device (Pall ForteBio). Biotin-labeled P domain was loaded onto streptavidin biosensors until saturation, typically 10 μg/mL for 2 min in PBS (pH 7.4) and 0.005% (vol/vol) Tween-20. Association and dissociation were measured at 25 °C. The real-time data were analyzed using Biaevaluation 3.1. Steady-state equilibrium concentration curves were fitted using a 1:1 binding model to obtain an apparent KD (avidity). Negative control, streptavidin biosensors were loaded with biotin-labeled H5N1 influenza protein. For comparison, the binding of each P domain variant was normalized to the amount of binding by the CW1 wild-type P domain.
Isothermal Titration Calorimetry.
Titration calorimetry was performed with a VP-ITC calorimeter (Microcal). Proteins were dialyzed overnight against buffer containing 30 mM Hepes, pH 7.4, 150 mM NaCl with 1 mM CaCl2. For some experiments the CaCl2 was replaced with 0.1 mM EGTA, or 0.1 mM EGTA and MgCl2 as indicated. Aliquots of the CD300lf solution were added by means of a 270-μL rotating stirrer-syringe (at 134 rpm) to a cell, containing 1.4 mL of the P domain solution. The concentration of P domain or CD300lf used is given under the titration plots (SI Appendix, Figs. S4 and S5). P domain concentrations ranged from 50 to 294 μM, whereas CD300lf concentrations varied from 900 to 3,694 μM. The heat of dilution was found negligible in separate titrations of CD300lf into the buffer solution. The heat of reaction was obtained by integration of each peak obtained after injection of titrant. All titrations were performed at 25 °C. Calorimetric data was analyzed with ORIGIN 7.0 software. Binding parameters such as the number of binding sites (n), the binding constant (K, M−1), and the binding enthalpy (ΔH, kcal/mol of bound ligand) were determined by fitting the experimental binding isotherms to a one-site binding model. All parameters were allowed to float during the least-squares minimization process. The GCDCA bindings to P domain titrations were done with bile acid in the syringe dissolved in the ITC buffer used to dialyze the P domain protein. The buffer consisted of 30 mM Hepes, pH 7.4, 150 mM NaCl with 1 mM CaCl2. The GCDCA concentration in the syringe ranged from 1,000 to 2,500 μM, and P domain concentrations in the well were between 63 and 100 μM. The GCDCA data were fitted to a one-site binding model. The heat of dilution was determined to be negligible in separate titrations of GCDCA into the buffer solution. The calculated binding stoichiometries (n) for both CD300lf and GCDCA on the P domain were close to unity, consistent with the structural data showing two CD300lf and two GCDCA binding sites per P domain dimer. The data curve fits indicate that binding of GCDCA to the recombinant P domain could be more complex than a simple one-site model, for example, monomer/dimer equlibria.
MNoV Binding Assay.
MNoV binding to the surface of BV2 cells was performed as described (5). Briefly, MNoV was mixed with BV2 cells on ice in the presence of PBS in the absence of divalent cations (PBS−/−) and either 10% FBS, 0.025% W/V bovine bile, 500 μM sodium glycochenodeoxycholic acid (GCDCA), 500 μM taurocholic acid (TCA), or 0.07% W/V casein. Unbound virus was washed off after 30 min, and then the ratio of viral genomes to host actin was determined by qPCR.
MNoV Infection Assay.
Two million BV2 cells were plated per well in a six-well plate for culture overnight in DMEM containing 10% FBS to produce a confluent monolayer as described (5). The cells were washed with PBS and the medium replaced with PBS−/− containing either 10% FBS, 0.9 mM calcium and 0.5 mM magnesium (PBS+/+), 500 μM of various bile acids, or 0.5 μg/mL histo-blood group antigen HBGA. One thousand plaque-forming units (PFUs) of MNVCW3 were adsorbed at room temperature for 30 min in the presence of cations, bile acids, or HBGA as indicated. Unbound virus was then washed off, and a methycellulose overlay containing 10% FBS was applied to allow for quantification of PFUs 48 h postinfection. PFUs were determined by staining with crystal violet (5). Each experiment was performed at least in duplicate in three independent experiments. In each experiment, PFUs for a given condition were normalized to that of the 10% FBS condition.
Docking the Cocomplex Structure on the MNoV Virion.
The model for MNV-1/CD300lf complex was derived from the pseudoatomic model of MNV-1 as previously described (17, 62). Briefly, the atomic structures of the MNV-1 P domain (26) and the rNV shell domain (16) were fitted into the 8 Å MNV-1 image reconstruction density (17) and refined using Situs (63). To facilitate generation of the entire capsid, the cryo-EM map was first rotated from the EMAN2 (64) to the Virus Particle Explorer (VIPER) (65) convention for icosahedral orientation and an icosahedral protomer unit of A, B, and C subunits were built into the density. The atomic structure of the P domain/CD300lf complex was then fitted onto the A/B and C/C P domain dimers using Coot (56). The new hybrid protomeric unit was then expanded using VIPER (65).
Supplementary Material
Acknowledgments
We thank Jay Nix at ALS beamline 4.2.2 for help in data collection and processing. We thank Ian Harvey, Alexander Kozlov, and Tim Lohman for help with ITC. We thank Dr. Mary Estes for providing timely advice and unpublished data on bile acids and human norovirus replication that was critical to the work done herein, and also Nicholas Davidson and Elizabeth Newberry for reagents. This work was supported by NIH Grants U19 AI109725 (to H.W.V.), AI127552 (to H.W.V. and D.H.F.), K08AI128043 (to C.B.W.), and GM030498 (to Tim Lohman) and Contract HHSN272201700060C (to D.H.F.).
Footnotes
Conflict of interest statement: Washington University School of Medicine and C.A.N., C.B.W., R.C.O., H.W.V., and D.H.F. have patents pending on CD300 family members as a receptor for norovirus and on the use of bile acids or analogs for therapy or prevention of norovirus infection. H.W.V. is an employee of Vir Biotechnology, which does not have access to any patents held by Washington University unless negotiated by the school. An IP filing has been made.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Research Collaboratory for Structural Bioinformatics (PDB ID codes 6C6Q for the CD300lf–P domain complex, 6C74 for the CD300lf–PC complex, 6E47 for complex with GCDCA, and 6E48 for complex with LCA). The virion model coordinates were deposited in the Protein Data Bank (PDB ID code 6CRJ). The cryo-EM density map data were deposited in the Electron Microscopy Data Bank (accession no. EMD-7564).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805797115/-/DCSupplemental.
References
- 1.Wobus CE, Thackray LB, Virgin HW., 4th Murine norovirus: A model system to study norovirus biology and pathogenesis. J Virol. 2006;80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wobus CE, et al. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW., 4th STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 4.Thackray LB, et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orchard RC, et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science. 2016;353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haga K, et al. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc Natl Acad Sci USA. 2016;113:E6248–E6255. doi: 10.1073/pnas.1605575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilen CB, et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 2018;360:204–208. doi: 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrego F. The CD300 molecules: An emerging family of regulators of the immune system. Blood. 2013;121:1951–1960. doi: 10.1182/blood-2012-09-435057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niizuma K, Tahara-Hanaoka S, Noguchi E, Shibuya A. Identification and characterization of CD300H, a new member of the human CD300 immunoreceptor family. J Biol Chem. 2015;290:22298–22308. doi: 10.1074/jbc.M115.643361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon JP, O’Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012;64:39–47. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
- 11.Choi SC, et al. Cutting edge: Mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J Immunol. 2011;187:3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izawa K, et al. The receptor LMIR3 negatively regulates mast cell activation and allergic responses by binding to extracellular ceramide. Immunity. 2012;37:827–839. doi: 10.1016/j.immuni.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Matsukawa T, et al. Ceramide-CD300f binding suppresses experimental colitis by inhibiting ATP-mediated mast cell activation. Gut. 2016;65:777–787. doi: 10.1136/gutjnl-2014-308900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green KY. Caliciviridae: The noroviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Vol 1. Lippincott Williams & Wilkins, Wolters Kluwer Health; Philadelphia: 2013. pp. 582–608. [Google Scholar]
- 15.Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 16.Prasad BV, Rothnagel R, Jiang X, Estes MK. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katpally U, et al. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J Virol. 2010;84:5836–5841. doi: 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: Norovirus evasion of the human immune system. Nat Rev Microbiol. 2010;8:231–241. doi: 10.1038/nrmicro2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale AD, et al. Identification of an epitope common to genogroup 1 “Norwalk-like viruses”. J Clin Microbiol. 2000;38:1656–1660. doi: 10.1128/jcm.38.4.1656-1660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy ME, et al. Antigenic mapping of the recombinant Norwalk virus capsid protein using monoclonal antibodies. Virology. 1996;217:252–261. doi: 10.1006/viro.1996.0112. [DOI] [PubMed] [Google Scholar]
- 21.Kolawole AO, et al. Norovirus escape from broadly neutralizing antibodies is limited to allostery-like mechanisms. MSphere. 2017;2:e00334-17. doi: 10.1128/mSphere.00334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochridge VP, Jutila KL, Graff JW, Hardy ME. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J Gen Virol. 2005;86:2799–2806. doi: 10.1099/vir.0.81134-0. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson M, et al. Evolution of human calicivirus RNA in vivo: Accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J Virol. 2003;77:13117–13124. doi: 10.1128/JVI.77.24.13117-13124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan M, Hegde RS, Jiang X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol. 2004;78:6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan M, et al. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: Evidence for a binding pocket. J Virol. 2003;77:12562–12571. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taube S, et al. High-resolution x-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol. 2010;84:5695–5705. doi: 10.1128/JVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, et al. Development of Norwalk virus-specific monoclonal antibodies with therapeutic potential for the treatment of Norwalk virus gastroenteritis. J Virol. 2013;87:9547–9557. doi: 10.1128/JVI.01376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindesmith LC, et al. Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J Virol. 2012;86:873–883. doi: 10.1128/JVI.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tohya Y, Yokoyama N, Maeda K, Kawaguchi Y, Mikami T. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J Gen Virol. 1997;78:303–305. doi: 10.1099/0022-1317-78-2-303. [DOI] [PubMed] [Google Scholar]
- 30.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson LR, editor. Essential Medical Pysiology. Vol 2. Lippincott-Raven; New York: 1998. pp. 445–472. [Google Scholar]
- 32.Kilic T, Koromyslova A, Malak V, Hansman GS. Atomic structure of the murine norovirus protruding domain and soluble CD300lf receptor complex. J Virol. 2018;92:e00413-18. doi: 10.1128/JVI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H, et al. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9:156–170. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolawole AO, et al. Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization. J Virol. 2014;88:4543–4557. doi: 10.1128/JVI.03685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardy ME. Norovirus protein structure and function. FEMS Microbiol Lett. 2005;253:1–8. doi: 10.1016/j.femsle.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Makino A, et al. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J Virol. 2006;80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sosnovtsev SV, et al. Identification of human junctional adhesion molecule 1 as a functional receptor for the Hom-1 calicivirus on human cells. MBio. 2017;8:e00031-17. doi: 10.1128/mBio.00031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhella D, Goodfellow IG. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J Virol. 2011;85:11381–11390. doi: 10.1128/JVI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanker S, et al. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: Implications for epochal evolution. J Virol. 2011;85:8635–8645. doi: 10.1128/JVI.00848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh BK, Leuthold MM, Hansman GS. Structural constraints on human norovirus binding to histo-blood group antigens. MSphere. 2016;1:e00049-16. doi: 10.1128/mSphere.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lochridge VP, Hardy ME. A single-amino-acid substitution in the P2 domain of VP1 of murine norovirus is sufficient for escape from antibody neutralization. J Virol. 2007;81:12316–12322. doi: 10.1128/JVI.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolatkar PR, et al. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao C, et al. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure. 2005;13:1019–1033. doi: 10.1016/j.str.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 45.He Y, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santiago C, et al. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity. 2007;27:941–951. doi: 10.1016/j.immuni.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan M, et al. Variation of calcium, copper and iron levels in serum, bile and stone samples of patients having different types of gallstone: A comparative study. Clin Chim Acta. 2017;471:254–262. doi: 10.1016/j.cca.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Tordoff MG, Bachmanov AA, Reed DR. Forty mouse strain survey of voluntary calcium intake, blood calcium, and bone mineral content. Physiol Behav. 2007;91:632–643. doi: 10.1016/j.physbeh.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S, et al. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe. 2017;22:449–459.e4. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baldridge MT, et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277:138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson CA, Lee CA, Fremont DH. Oxidative refolding from inclusion bodies. Methods Mol Biol. 2014;1140:145–157. doi: 10.1007/978-1-4939-0354-2_11. [DOI] [PubMed] [Google Scholar]
- 53.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hubbard SJ, Thornton JM. 1993. Naccess Computer Program (Department of Biochemistry and Molecular Biology, University College London, London), Version 2.1.1. [Google Scholar]
- 59.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 60.Schrodinger, LLC 2015. The PyMOL Molecular Graphics System (Schrodinger, LLC, New York), Version 1.8.
- 61.Akahata W, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katpally U, Wobus CE, Dryden K, Virgin HW, 4th, Smith TJ. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J Virol. 2008;82:2079–2088. doi: 10.1128/JVI.02200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Carrillo-Tripp M, et al. VIPERdb2: An enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.