Significance
Subtle changes in RNA transcript isoform expression can have dramatic effects on cellular behavior in both health and disease. As such, comprehensive and quantitative analysis of isoform-level transcriptomes would open an entirely new window into cellular diversity in fields ranging from developmental to cancer biology. The Rolling Circle Amplification to Concatemeric Consensus (R2C2) method we are presenting here has sufficient throughput and accuracy to make the comprehensive and quantitative analysis of RNA transcript isoforms in bulk and single-cell samples economically feasible.
Keywords: full-length cDNA sequencing, isoforms, single-cell transcriptomics, B cells, nanopore sequencing
Abstract
High-throughput short-read sequencing has revolutionized how transcriptomes are quantified and annotated. However, while Illumina short-read sequencers can be used to analyze entire transcriptomes down to the level of individual splicing events with great accuracy, they fall short of analyzing how these individual events are combined into complete RNA transcript isoforms. Because of this shortfall, long-distance information is required to complement short-read sequencing to analyze transcriptomes on the level of full-length RNA transcript isoforms. While long-read sequencing technology can provide this long-distance information, there are issues with both Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) long-read sequencing technologies that prevent their widespread adoption. Briefly, PacBio sequencers produce low numbers of reads with high accuracy, while ONT sequencers produce higher numbers of reads with lower accuracy. Here, we introduce and validate a long-read ONT-based sequencing method. At the same cost, our Rolling Circle Amplification to Concatemeric Consensus (R2C2) method generates more accurate reads of full-length RNA transcript isoforms than any other available long-read sequencing method. These reads can then be used to generate isoform-level transcriptomes for both genome annotation and differential expression analysis in bulk or single-cell samples.
Short-read RNAseq has been used for the analysis of transcriptomes for over a decade (1). The massive read output of Illumina sequencers makes it possible to quantify gene expression accurately using this approach. However, to accommodate Illumina sequencers’ short read-length, RNA or cDNA has to be fragmented during sample preparation, thereby losing long-distance RNA transcript isoform information. Specialized protocols like synthetic long read (SLR) (2) or sparse isoform sequencing (spISO-seq) (3) have been used successfully to recover long-distance information, but they require either specialized instrumentation or complex workflows. The SLR method assembles mostly incomplete cDNA molecules and has limited throughput, while spISO-seq requires a 10x Genomics instrument and generates read clouds, which capture long-distance information and yet cannot assemble full-length cDNA molecules.
In contrast, long-read sequencing technology has the capability to sequence entire cDNA molecules end-to-end. Currently, the Pacific Biosciences (PacBio) Iso-Seq pipeline represents a powerful gold standard for cDNA sequencing (4) and has been used to investigate a wide range of transcriptomes (5, 6). The PacBio Sequel sequencer produces ∼200,000 accurate circular consensus reads of full-length cDNA molecules per run.
Oxford Nanopore Technologies (ONT) technology could present a valuable alternative for cDNA sequencing, because the ONT MinION can currently generate more than one million reads per run. We and others have shown that the ONT MinION can sequence cDNA at high throughput, but that data analysis is challenging (7, 8) due to its high error rate. Base-level identification of splice junction sequence is the main challenge.
One strategy to increase the base accuracy of cDNA sequences produced by the higher-output ONT MinION sequencer is to apply the circular consensus principle applied by PacBio sequencers. By sequencing 16S amplicon molecules, the INC-seq (9) method has shown that this is possible, in principle. However, the reported throughput of a few thousand reads per run would be insufficient for transcriptome analysis. Further, like PacBio technology, the INC-seq method uses blunt-end ligation to circularize double-stranded DNA molecules, which does not differentiate between full-length or fragmented DNA molecules. In summary, current technology produces reads that are either too inaccurate (ONT), potentially incomplete (Illumina, PacBio, ONT, INC-seq), or too low-throughput/expensive (PacBio, SLR, INC-seq) to enable high-throughput complete cDNA sequencing.
Here we introduce the Rolling Circle to Concatemeric Consensus (R2C2) method, which overcomes these limitations by leveraging the long-read length of the ONT technology to generate consensus sequences with increased base accuracy. First, we benchmark R2C2 against the PacBio Iso-Seq gold standard for the analysis of the same synthetic transcript mixture. Second, we apply R2C2 to analyze the transcriptomes of 96 single B cells derived from a healthy adult. We show that a single run of R2C2 can generate over 400,000 reads covering full-length cDNA molecules with a median base accuracy of 94%. Using an updated version of our Mandalorion pipeline, these reads can be used to identify high-confidence RNA transcript isoforms present in bulk or single-cell transcriptomes. Illustrating the power of this approach, we find that some of the B cells in our study express RNA transcript isoforms of the CD19 gene that lack the epitope targeted by chimeric antigen receptor (CAR) T cell therapy (10–12).
Results
R2C2 Improves the Base Accuracy of the ONT MinION.
To benchmark the R2C2 method, we analyzed Spike-in RNA Variant (SIRV) E2 synthetic spike in RNA. First, we reverse-transcribed and amplified the synthetic spike in RNA using the Tn5Prime (13) protocol, which is a modification of the Smart-seq2 protocol, which uses a distinct template switch oligo containing 7-nt sample indexes during reverse transcription. Amplification introduces an additional 8-nt index into the cDNA molecules. The amplified cDNA is then circularized using a DNA splint and the NEBuilder HiFi DNA Assembly Master Mix, a proprietary variant of Gibson Assembly. The DNA splint is designed to circularize only full-length cDNA terminating on both ends in sequences complementary to the primers used to amplify cDNA (Fig. 1). Circularized cDNA is then amplified using Phi29 and random hexamers to perform Rolling Circle Amplification (RCA). The resulting High Molecular Weight (HMW) DNA was then debranched using T7 Endonuclease and sequenced on the ONT MinION sequencer using the 1D sequencing kit (LSK108) kit and R9.5 flow cell (FLO-MIN107).
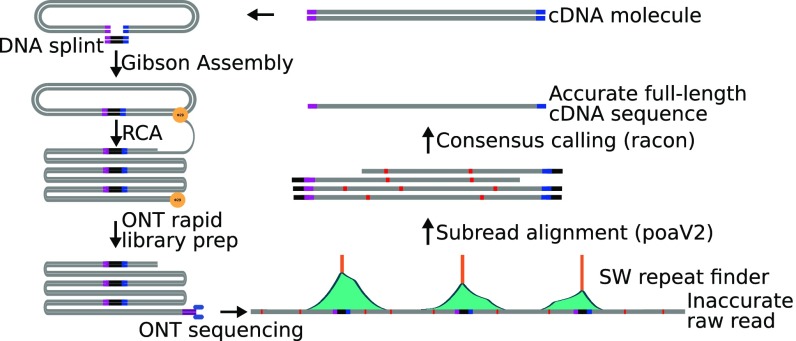
Fig. 1.
R2C2 method overview. cDNA is circularized using Gibson Assembly, amplified using RCA, and sequenced using the ONT MinION. The resulting raw reads are split into subreads containing full-length or partial cDNA sequences, which are combined into an accurate consensus sequence using our C3POa workflow, which relies on a custom algorithm to detect DNA splints as well as poaV2 and racon.
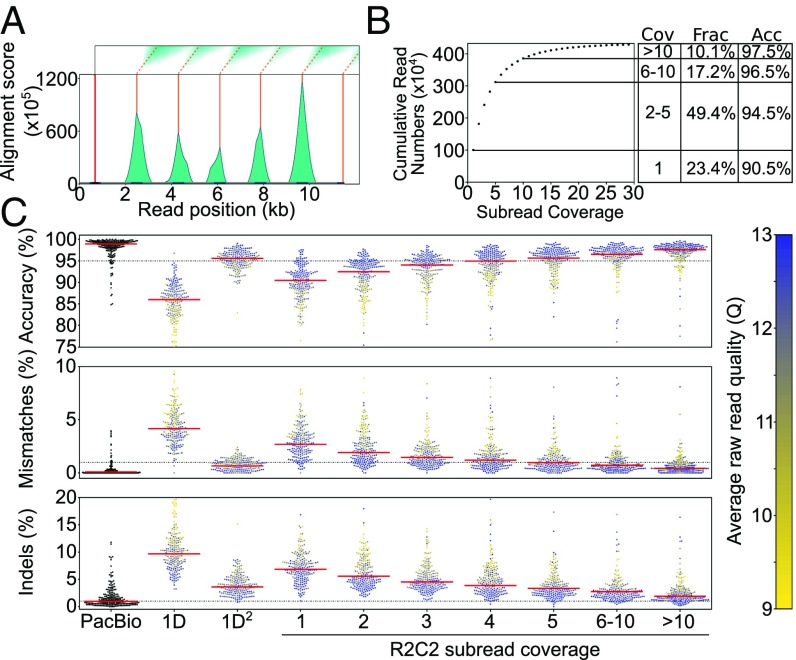
The sequencing run produced 828,684 reads with an average length of 5.0 kb, resulting in a total base output of 4.1 Gb. For downstream analysis we selected 621,970 of these reads that were longer than 1 kb and had a raw quality score (Q) ≥ 9. We next used our C3POa [Concatemeric Consensus Caller using partial order alignments (POA)] computational workflow to generate full-length cDNA consensus reads from the raw reads. C3POa detects DNA splint sequence raw reads using BLAST-Like Alignment Tool (BLAT) (14). Because BLAT is likely to miss DNA splint sequences in the noisy raw reads, we analyze each raw read for which BLAT found at least one DNA splint sequence with a custom repeat finder, which parses the score matrix of a modified Smith–Waterman self-to-self alignment (Figs. 1 and 2A). Repeats, or subreads, are then combined into a consensus and error-corrected using poaV2 (15) and racon (16), respectively. Finally, only reads containing known priming sites at both cDNA ends are retained as full-length consensus reads. In this way, C3POa generated 435,074 full-length cDNA consensus reads (and an additional 46,994 consensus reads from another multiplexed experiment) with varying subread coverage (Fig. 2 A and B and SI Appendix, Table S1).
Fig. 2.
Raw reads are processed into consensus reads of varying subread coverage. (A) Example of an 11.5-kb raw ONT read that was analyzed by our custom Smith–Waterman repeat finder. One initial splint (red line) is identified using the BLAT aligner, and then modified Smith–Waterman self-to-self alignments are performed starting from the location of the initial splint. The score matrices (Top) are then processed to generate alignment score histograms (teal). We then call peaks (orange) on these histograms. Complete subreads are then defined as the sequences between two peaks. (B) Cumulative number of SIRV E2 R2C2 consensus reads is plotted against their subread coverage. To the Right, coverage (Cov), fraction of all consensus reads (Frac), and accuracy (Acc) are given for four read bins. (C) PacBio Isoseq, standard ONT 1D, and 1D2 are compared with R2C2 at different subread coverages. Read accuracy is determined by minimap2 alignments to SIRV transcripts (see Methods). Median accuracy is shown as a red line. Accuracy distribution is shown as a swarm plot of 250 randomly subsampled reads. Average raw read quality of ONT reads is indicated by the color of the individual points.
We also analyzed the same cDNA pool using a standard, heavily multiplexed ONT 1D2 run generating 5,904 full-length 1D and 1,142 1D2 cDNA reads and the PacBio IsoSeq protocol generating 233,852 full-length cDNA Circular Consensus (CCS) reads. We aligned the resulting reads generated by each protocol to the SIRV transcript sequences using minimap2 and calculated percent identity (accuracy) using those alignments. The 1D2 run produced reads with a median accuracy of 87% (1D reads) or 95.6% (1D2 reads), while PacBio CCS reads had a median accuracy of 98.9%. R2C2 reads had a median accuracy of 94% (Fig. 2C) with the accuracy of individual R2C2 reads being highly correlated with average quality score of its underlying raw read as well as the numbers of subreads this raw read contained (Fig. 2C). While mismatch errors declined rapidly with increasing number of subreads, insertion and deletion errors declined more slowly. This might be explained by insertion and deletion errors not being entirely random but systematically appearing in stretches of the same base, i.e., homopolymers (8). Indeed, 4-mers (“AAAA,” “CCCC,” “TTTT,” and “GGGG”) were enriched around insertion and deletion errors in R2C2 consensus reads (SI Appendix, Fig. S1). Overall, more aggressive filtering of R2C2 reads based on raw read quality score and subread coverage could increase the median accuracy of the R2C2 method but would also reduce overall read output.
R2C2 Correlates Well with PacBio for the Quantification of SIRV Transcripts.
Next, we wanted to test whether R2C2 reads could be used to identify and quantify transcript isoforms. The SIRV E2 (Lexogen) transcript isoform mix presents a challenging test case for transcript isoform identification and quantification. The SIRV E2 mix contains a total of 69 polyadenylated transcript isoforms which are between 0.3 and 2.5 kb in length and belong to seven artificial gene loci. These gene loci and their associated transcript isoforms were designed to mirror highly complex gene loci in mammalian genomes and include alternative splicing events, transcription start sites (TSSs), and polyA sites as well as antisense transcripts. Each transcript isoform is present at one of four different concentrations (“1/32,” “1/4,” “1,” “4”) spanning two orders of magnitude.
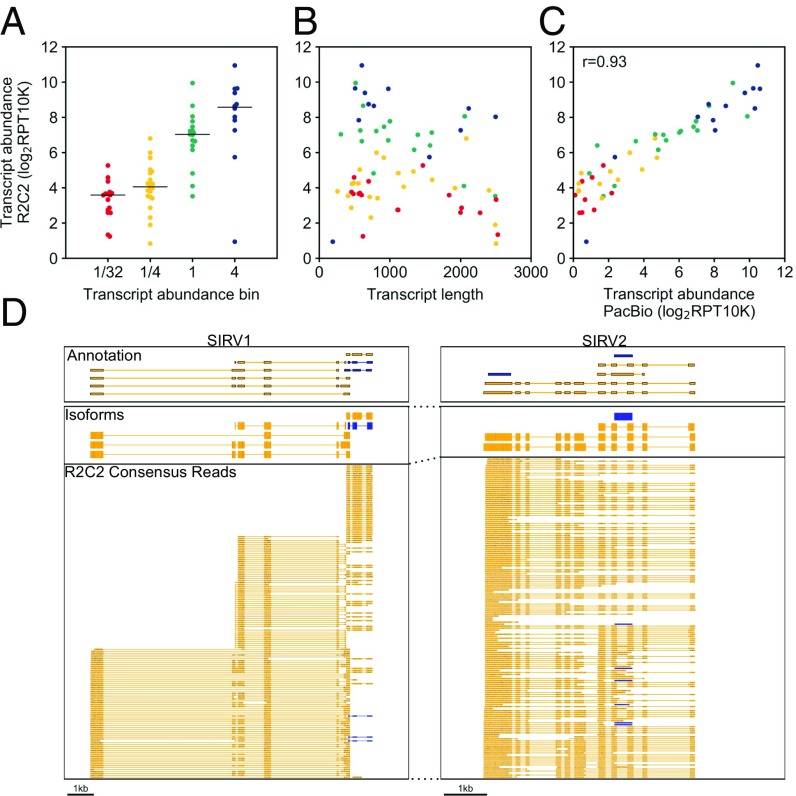
By analyzing the same SIRV E2 cDNA pools using R2C2 and PacBio IsoSeq we found that our R2C2 transcript counts generally matched nominal SIRV concentrations (Fig. 3A). Additionally, there seems to be no clear length bias (Fig. 3B), and our R2C2 transcript counts matched PacBio transcript counts very well with a Pearson correlation coefficient of 0.93 (Fig. 3C). This indicates that the potential variations in transcript quantification seen in Fig. 3A were either rooted in differences in the initial RNA concentration found in the SIRV E2 mix or biases of our modified Smart-seq2–based cDNA amplification step rather than new biases introduced by the sequencing technology.
Fig. 3.
R2C2 reads can quantify SIRV transcripts. R2C2 reads were aligned to SIRV transcripts using minimap2, and expression values’ transcript abundance was determined as Reads Per Transcript Per 10,000 reads (RPT10K). The transcript count ratio was plotted on the y axis against the (A) nominal transcript abundance bin reported by the SIRV transcript manufacturer (Lexogen), (B) transcript length, and (C) transcript count ratio calculated from PacBio Isoseq reads. Pearson correlation coefficient (r) is reported in C. Each point represents a transcript and is colored according to its transcript abundance bin in all panels. (D) Genome browser view of Transcriptome annotation, isoforms identified by Mandalorion, and R2C2 consensus reads is shown of the indicated synthetic SIRV gene loci. Transcript and read direction are shown by colors (blue: +strand; yellow, −strand).
R2C2 Enables Simple and Accurate Isoform Identification.
Next, we tested whether the increased accuracy of R2C2 reads would benefit splice junction and isoform identification. To this end, we aligned PacBio, ONT, and R2C2 reads to the artificial SIRVome sequence provided as a genome reference for their SIRV transcripts (Fig. 3D). The percentage of splice sites detected in sequencing reads that matched annotated splice sites perfectly was 91.26% (95% CI: 91.2176–91.2940%) for R2C2 consensus reads, far exceeding the 80.43% (95% CI: 80.1400–80.7341%) for ONT 1D raw reads and approaching the 96.88% (95% CI: 96.8592–96.9137%) for PacBio CCS reads.
This increased accuracy allowed us to simplify our Mandalorion pipeline for isoform identification (see Methods). To test how this updated version of Mandalorion would perform we subsampled R2C2 consensus read alignments to levels found in highly expressed genes in whole transcriptome analysis (500 read alignments per SIRV gene locus). Some of these subsampled R2C2 consensus reads did not align from end to end to a SIRV transcript (Fig. 2D). We suspect they are products of cDNA synthesis of degraded RNA molecules likely caused by repeated freeze–thaw cycles of the SIRV E2 standards, for they all contained complete 5′ and 3′ priming sites and adapter sequences. This highlighted the importance of RNA integrity for full-length transcriptome sequencing. Indeed, R2C2 reads created from single B cell lysates, which are thawed only once immediately before cDNA synthesis showed evidence of degradation products at much lower levels (SI Appendix, Fig. S2).
Because these degradation products appear to be largely random, they had little effect on the Mandalorion pipeline. The SIRV E2 mix contains 69 transcript isoforms; 28 of these are present at high concentration (concentration bins “1” and “4”), while 40 are present at low concentration (concentration bins “1/4” and “1/32”). Based on subsampled R2C2 reads, Mandalorion identified 34 high-confidence isoforms (Fig. 2D). Twenty-four of these isoforms matched one of the 29 annotated transcripts (83%) from the “1” and “4” concentration bins, while eight isoforms matched one of the 40 annotated transcripts (20%) from the “1/4” and “1/32” concentration bins. Two of 34 (5.6%) high-confidence isoforms represented truncated transcripts, caused by an oligo(dT) mispriming on an A-rich region of the SIRV303 transcript or a premature template switch on the (likely degraded) SIRV602 transcript, respectively. This indicated that R2C2 consensus reads paired with the Mandalorion pipeline can identify complex transcript isoforms. It also highlights the difficulty of correct identification of low-abundance transcript isoforms and the abiding problem of incomplete cDNA amplification.
R2C2 Allows the Demultiplexing of 7- to 8-nt Cellular Indexes.
Next, we tested whether R2C2 reads are accurate enough to demultiplex reads based on short cellular indexes like those employed by 10x, Drop-Seq, or our own Tn5Prime single-cell RNAseq protocols. To this end, the SIRV cDNA we sequenced with the R2C2 method was indexed with eight distinct combinations of 7-nt [template switch oligo (TSO)] and 8-nt (Nextera adapter) indexes. We could confidently assign one 7-nt and one 8-nt index to 74% of R2C2 reads using a custom demultiplexing script based on Levenshtein distance between the observed sequence at the index position and our known input indexes. In 99.8% of these R2C2 reads the combination of assigned indexes matched one of the distinct combinations present in the cDNA pool. Next, we performed the same analysis on 1D reads. We could confidently assign one 7-nt and one 8-nt index to 22% of full-length 1D reads. In 92.9% of these 1D reads the combination of assigned indexes matched one of the distinct combinations present in the cDNA pool. This showed that demultiplexing 1D reads results in the loss of the majority of reads and introduces high levels of index crosstalk that would negatively affect downstream analysis. In contrast, the majority of R2C2 reads covering highly multiplexed cDNA could be accurately demultiplexed.
Analysis of 96 Single B Cell Transcriptomes Using R2C2.
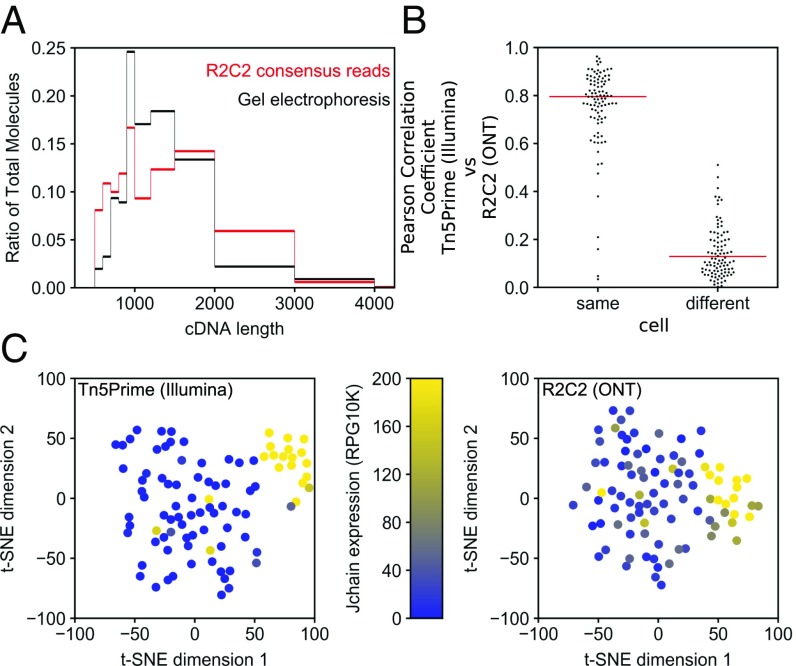
Having established that we could demultiplex our Tn5Prime data using R2C2 reads with very little crosstalk between samples, we sequenced cDNA from 96 single B cells, which we have recently analyzed using Illumina sequencing (13). To streamline the sequencing reaction, we used the ONT RAD4 (RAD004) kit, which has a lower average read output than the ligation-based 1D kit but has a much shorter sequencing library preparation protocol (∼20 min) and, in our hands, more consistent and less error-prone workflow. Using the ONT RAD4 kit we generated 2,064,911 raw reads across four sequencing runs using R9.5 flow cells. C3POa generated 1,132,707 full-length R2C2 consensus reads which matched the length distribution of the sequenced cDNA closely (Fig. 4A). Out of these 1,132,707 R2C2 consensus reads, 975,500 successfully aligned to the human genome, and 730,023 of those aligned reads were assigned to single B cells based on their 7-nt and 8-nt cellular indexes. We found that the vast majority of those reads were complete on the 5′ end by comparing the alignment ends of these reads to TSSs previously identified (13) using Illumina sequencing. Out of the 730,023 aligned and assigned reads, 653,410 (90%) either aligned to within 10 bp of a predicted TSS (604,940 reads) or aligned within a rearranged antibody locus (48,470 reads), which makes accurate read alignment impossible.
Fig. 4.
R2C2 length bias and gene expression quantification. (A) B cell cDNA molecule length distribution as determined by electrophoresis on 2% agarose gel is compared with R2C2 consensus read length distribution. (B) Pearson correlation coefficient (r) is shown for R2C2 and Illumina-based gene expression quantification of the same or different cells. Red lines indicate medians. All 96 correlation coefficients from same cell comparisons and 96 subsampled correlation coefficients from different cell comparisons are shown as a swarm plot to display their distributions. (C) t-SNE dimensional reduction plots of the same 96 B cells whose transcriptomes were quantified with either the Tn5Prime Illumina-based method or the R2C2 ONT-based method. Cells are colored based on the J chain expression, which is strongly associated with plasmablast cell identity.
R2C2 Quantifies Gene Expression in Single Human B Cells.
Individual cells were assigned 7,604 reads on average. We detected an average of 532 genes per cell (at least one R2C2 consensus read overlapping with the gene). Both the number of genes detected as well as gene expression quantification based on these R2C2 consensus reads closely matched RNAseq-based quantification (13). When comparing gene expression of the same cell, RNAseq and R2C2 quantification had a median Pearson correlation coefficient (r) of 0.79 opposed to 0.14 when comparing different cells with one another (Fig. 4B). Using t-distributed stochastic neighbor embedding (t-SNE) clustering on R2C2 and Illumina data resulted in the subclustering of the same J chain-positive cells which we previously identified as plasmablasts (as opposed to memory B cells) (Fig. 4C).
R2C2 Identifies Isoforms in Single Human B Cells.
We used our updated Mandalorion pipeline to identify high-confidence isoforms separately for each of the 96 B cells we analyzed. By grouping R2C2 consensus reads based on their splice sites and alignment starts and ends, Mandalorion identified an average of 163 high-confidence isoforms per cell. We found that identification of high-confidence isoforms was dependent upon R2C2 consensus read coverage. We identified at least one isoform in 3.1%, 64.9%, and 92.2% of genes covered by 1–4 reads, 5–9 reads, or >10 reads, respectively. The vast majority of genes with >10 R2C2 consensus reads contained one (78%) or two (11%) isoforms, highlighting the low complexity of single-cell transcriptomes.
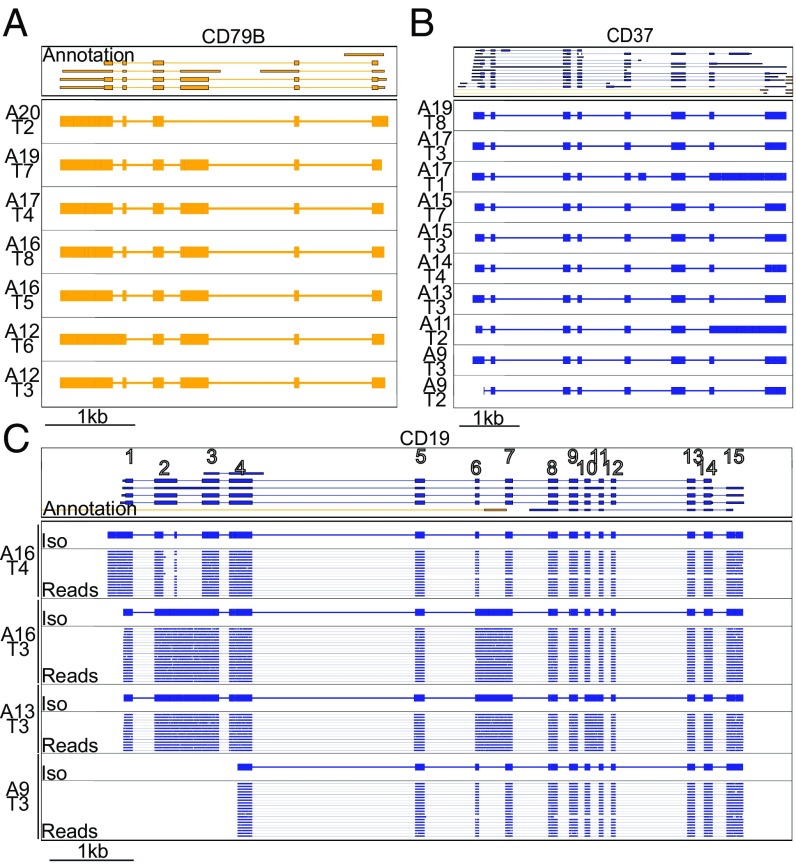
Overall, the isoforms we identified had a 99.1% sequence similarity with the human genome. As previously observed for mouse B1 cells (7), human B cells show a diverse array of isoforms across their surface receptors. CD37 and CD79B, which were expressed in several B cells, showed diverse isoforms. These isoforms were defined by (i) intron retention events (CD79B, Cell A12_T6; CD37, Cells A11_T2 and A17_T1), (ii) variable transcription start sites (TSSs) and alternatively spliced exons (CD79B, Cell A20_T2; CD37, Cell A17_T1), with the alternatively spliced exon being only partially annotated (Fig. 5).
Fig. 5.
R2C2 reads identify isoforms in B cell surface receptor genes. (A–C) Genome browser views of Transcriptome annotation, isoforms (Iso) identified by Mandalorion, and R2C2 consensus reads (Reads) (C only, downsampled to 20 reads) are shown for CD79B (A), CD37 (B), and CD19 (C) gene loci. Transcript and read direction is shown by colors (blue, +strand; yellow, −strand). Cell IDs are indicated by combinations of A and TSO (T) indexes. (C) CD19 exon numbers are indicated on the transcript annotation in white.
Finally, for the B cell-defining CD19 receptors we also observed multiple isoforms across cells, which is of particular interest because CD19 is a target for CAR T cell therapy. Alternative splicing of CD19 has been shown to confer therapy resistance to B cell lymphomas. Interestingly, when we reference-corrected [sqanti-qc (17)] and translated the four isoforms we identified, only one contained the epitope required for FMC63-based CAR T cell therapy (Fig. 5 and SI Appendix, Fig. S3) (10, 11), which has been mapped to exon 4 (12).
Discussion
While RNAseq analysis has fundamentally changed how transcriptional profiling is performed, it is ultimately a stop-gap solution born from the limitations of short-read sequencing technologies. The need to fragment transcripts to fit short-read technologies like Illumina results in often unsurmountable analysis challenges. As a result, RNAseq analysis is often used like gene expression microarrays with the data used for downstream analysis being gene expression values. Single-cell RNAseq has further exacerbated this limitation because it is often restricted to 3′ or 5′ tag counting and generates gene expression values that are sparse due to both biological and technical reasons.
This results in a loss of information because individual genes can express many different isoforms, often with different functions. However, many bulk and single-cell RNAseq methods do generate full-length cDNA as an intermediate product in library preparation. Long-read technology is able to take advantage of this full-length cDNA. While long-read sequencing technologies do not currently match Illumina’s read output and accuracy, their outputs and accuracies are increasing. Here, we produced over 200,000 reads at close to 99% accuracy per run using the PacBio Sequel. Further, in our hands, the standard ONT 1D2 protocol can generate one million 1D cDNA reads at 87% accuracy and 50,000 1D2 reads at 95% accuracy in a single run. The ONT-based R2C2 sequencing method we developed takes advantage of this high throughput and increases ONT read accuracy. The R2C2 method we developed offers a compromise between PacBio and ONT technologies that generates on average 316,000 full-length cDNA reads at 94% accuracy in a single run. Further, because R2C2 relies on high-fidelity polymerases (Phi29 and KAPA HiFi polymerases are both reported to generate an error about 1/1,000,000 nucleotides), the accuracy of R2C2 consensus reads is almost entirely dependent on ONT-sequencing technology. Improvements in ONT MinION library preparation and sequencing kits will therefore directly benefit the R2C2 assay. Increasing read length and single pass base accuracy as well as combining R2C2 with an improved, more efficient 1D2 library protocol will all increase R2C2 consensus read accuracy.
While the per-run cost of flow cells and reagents of PacBio and ONT are roughly comparable, the capital cost of the PacBio Sequel sequencer (∼$300,000) vastly exceeds the cost of the ONT MinION (∼$1,000). This effective lack of capital costs associated with the ONT-based R2C2 method results in much lower total cost of accurate full-length transcriptome analysis compared with the PacBio IsoSeq workflow (SI Appendix, Table S2). Indeed, at its current throughput and accuracy and combined with the low cost of the ONT MinION we believe that R2C2 brings comprehensive full-length transcriptome analysis within reach of most molecular biology laboratories.
In the immediate future, the R2C2 method will be a suitable complement for short-read sequencing. To this end, the R2C2 can be easily adapted to any RNAseq library preparation protocol that produces full-length double-stranded cDNA molecules with known adapter/primer sequences at their ends. This includes standard Smart-seq2 and protocols requiring capped mRNA like the TeloPrime protocol (Lexogen) for bulk sample analysis. For single-cell analysis and in contrast to 1D reads, R2C2 reads will also likely be able to demultiplex reads in highly multiplexed cDNA pools generated by 10x Genomics and Drop-seq protocols.
Adapting R2C2 to these protocols only requires the generation of a compatible DNA splint by modifying the primers used for amplifying the DNA splint. The same cDNA pool can then be sequenced by both Illumina and R2C2 methods. However, cDNA generation protocols may have to be optimized to ensure R2C2 reads cover transcripts end-to-end. In this study, 80% of R2C2 reads generated from cDNA prepped with the Tn5Prime protocol from the (likely somewhat degraded) SIRV E2 mix aligned within 20 nt of both ends of SIRV transcripts and were therefore considered complete. Avoiding sample degradation, optimizing cDNA synthesis, and limiting amplification cycles is likely to increase this percentage.
We believe that R2C2 has the potential to replace short-read RNAseq and its shotgun approach to transcriptome analysis entirely, especially considering the impending wide release of the high-throughput ONT PromethION sequencer. This will be a significant advance, considering the strength of full-length transcriptome sequencing showcased here.
Using R2C2 we generated isoform-level transcriptomes for 96 B cells by analyzing a highly multiplexed pool of cDNA amplified using our Tn5Prime protocol. Generating isoform-level transcriptomes from these cells would have been possible using 1D reads and, to a more limited extent, short Illumina reads as well. However, this would have required each cell to be individually processed to introduce cellular indexes suitable for the respective technology. R2C2 therefore simplifies large-scale single-cell workflows by enabling the analysis of highly multiplexed full-length cDNA generated originally for Illumina-based transcript-end tag counting.
We used Mandalorion to analyze the R2C2 reads of the 96 B cells and identified several surface receptor isoforms of CD79B, CD37, and CD19 expressed by 96 distinct single human B cells. The CD19 RNA isoforms we identified in the single B cells may have implications regarding immunotherapy efficacy because they confirmed that the epitope targeted by FMC63-based CAR T cell therapy and located on exon 4 is not included in all CD19 transcript isoforms. This implies that even healthy individuals contain RNA isoform diversity for CD19, which may ultimately contribute to immunotherapy resistance when undergoing FMC63-based CAR T cell therapy (10, 11). Going forward, full-length cDNA sequencing could be employed to identify truly constitutive exons and epitopes for targeting by immunotherapy.
Materials and Methods
All study protocols were approved by the Institutional Review Board at University of California, Santa Cruz (UCSC). A blood sample of a fully consented healthy adult was collected at UCSC. The sample was deidentified upon collection. B cells were isolated by FACS (13). RNA and cell lysates were amplified using the Tn5Prime (13) method, which represents a modification of the Smart-seq2 (18, 19) method. Briefly, 100 pg of SIRV E2 (Lexogen) RNA or single B cell lysates were reverse-transcribed (RT) using Smartscribe Reverse Transcriptase (Clontech) in a 10-μL reaction including an oligo(dT) primer and a Nextera A TSO containing a 7-nt sample index (SI Appendix, Table S3). RT was performed for 60 min at 42 °C. The resulting cDNA was treated with 1 μL of 1:10 dilutions of RNase A (Thermo) and Lambda Exonuclease New England Biolabs (NEB) for 30 min at 37 °C. The treated cDNA was then amplified using KAPA HiFi Readymix 2x (KAPA) and incubated at 95 °C for 3 min, followed by 15 cycles for SIRV RNA or 27 cycles (single B cells) of 98 °C for 20 s, 67 °C for 15 s, and 72 °C for 4 min, with a final extension at 72 °C for 5 min. cDNA amplification requires both the ISPCR primer and a Nextera A Index primer, which contains another 8-nt sample index.
SIRV RNA.
Eight SIRV E2 RNA aliquots were reverse-transcribed and amplified in separate reactions, adding distinct 7-nt TSO and 8-nt Nextera A indexes to each resulting cDNA aliquot. The separate aliquots were used directly as input into our R2C2 method or amplified using KAPA HiFi Readymix 2x (KAPA) (95 °C for 3 min, followed by 15 cycles; 98 °C for 20 s, 67 °C for 15 s, and 72 °C for 4 min), with a final extension at 72 °C for 5 min with ISPCR and Nextera_A_Universal primers and pooled at equal amounts for input into PacBio Iso-Seq pipeline at the University of Georgia sequencing core.
Single B Cell Lysates.
Single B cells, each cell in a separate well of a 96-well plate, were reverse-transcribed using a distinct 7-nt TSO index for each row. Columns were then pooled and amplified using a distinct 8-nt Nextera A index for each pool. This resulted in the cDNA of all 96 cells carrying a unique combination of TSO and Nextera A index. This cDNA was then pooled for Illumina sequencing (HiSeq4000 2 × 150) (13) or amplified using KAPA HiFi Readymix 2x (KAPA) [95 °C for 3 min, followed by 15 cycles (98 °C for 20 s, 67 °C for 15 s, and 72 °C for 4 min)], with a final extension at 72 °C for 5 min with ISPCR and Nextera_A_Universal primers. The amplified cDNA was used for input into our R2C2 method.
DNA Splint Amplification.
An ∼200-bp DNA splint to enable Gibson Assembly (20) circularization of cDNA was amplified from Lambda DNA using KAPA HiFi Readymix 2x (KAPA) (95 °C for 3 min, followed by 25 cycles; 98 °C for 20 s, 67 °C for 15 s, and 72 °C for 30 s) using primer Lambda_F_ISPCR (RC) and Lambda_R_NextA (RC) (SI Appendix, Table S3). This generated a double-stranded DNA with matching overlaps to full-length cDNA.
R2C2 Sample Preparation.
Circularization of cDNA.
Two hundred nanograms of cDNA was mixed with 200 ng of DNA splint. Volume was adjusted to 6 μL, and 6 μL of 2x NEBuilder HiFi DNA Assembly Master Mix (NEB) was added. The reaction was incubated for 60 min at 55 °C. Volume was adjusted to 20 μL, and noncircularized DNA was digested using 1 μL of 1:10 Exonuclease III and Lambda Exonuclease as well as 1 μL of Exonuclease I (all NEB). Circularized DNA was extracted using Solid Phase Reversible Immobilization (SPRI) beads with a size cutoff to eliminate DNA <500 bp (0.8 beads:1 sample) and eluted in 50 μL of ultrapure water.
Rolling circle amplification.
Circularized DNA was split into five aliquots of 10 μL, and each aliquot was amplified in its own 50-μL reaction containing Phi29 polymerase (NEB) and exonuclease resistant random hexamers (Thermo) [5 μL of 10× Phi29 Buffer, 2.5 μL of 10 uM (each) dNTPs, 2.5 μL random hexamers (10 uM), 10 μL of DNA, 29 μL ultrapure water, 1 μL of Phi29]. Reactions were incubated at 30 °C overnight. All reactions were pooled, and volume was adjusted to 300 μL with ultrapure water. DNA was extracted using SPRI beads with a size cutoff to eliminate DNA <2,000 bp (0.5 beads:1 sample). A mix of 90 μL of ultrapure water, 10 μL NEB buffer 2, and 5 μL T7 Endonuclease was added to the beads to elute and debranch the DNA. Beads were incubated for 2 h on a thermal shaker at 37 °C under constant agitation. The tubes containing the beads were then placed on magnets, and supernatant was recovered. The DNA in the supernatant was then extracted again using SPRI beads with a size cutoff to eliminate DNA <2,000 bp (0.5 beads:1 sample) and eluted in 15 μL of ultrapure water.
ONT sequencing.
SIRV E2 RCA product was sequenced using the ONT 1D sample prep kit and a single 9.5 flow cell according to manufacturer’s instructions with the exception that DNA was not sheared before library preparation. Single B cell RCA product was sequenced using the ONT RAD4 kit and four 9.5 flow cells. The resulting raw data were base-called using the albacore (version 2.1.3) read_fast5_basecaller script.
C3POa Data Processing.
Preprocessing.
Base-called raw reads underwent preprocessing to shorten read names and remove short (<1,000 kb) and low-quality (Q < 9) reads. Raw reads were first analyzed using BLAT (14) to detect DNA splint sequences. If one or more splint sequences were detected in a raw read, the raw read underwent consensus calling.
Consensus calling.
The following steps are performed to generate a consensus sequence:
(i) Tandem repeats in each raw read are detected using a modified EMBOSS WATER (21–23) Smith–Waterman self-to-self alignment. (ii) Raw reads are then split into complete subreads containing full repeats and incomplete subreads containing partial repeats at the read ends. If there is more than one complete subread, a preliminary consensus of these complete subreads is computed using poaV2 (15). If only one complete subread is present in the raw read, its sequence is used as consensus in the following steps. (iii) Complete and incomplete subreads are aligned to the preliminary consensus sequence using minimap2 (24). (iv) These alignments are used as input to the racon (16) algorithm, which error-corrects the consensus. To speed up consensus calling, we divided raw reads into bins of 4,000 and used GNU Parallel (25) to run multiple instances of C3POa.py.
Postprocessing.
ISPCR and Nextera Sequences are identified by BLAT, and the read is trimmed to their positions and reoriented to 5′ → 3′.
Alignment.
Trimmed, full-length R2C2 reads and PacBio reads are aligned to the appropriate genomes and transcripts using minimap2. Percent identity of sequencing reads were calculated from minimap2 alignments. For isoform identification and visualization, SAM files were converted to PSL file format using the jvarkit sam2psl (26) script.
Isoform identification and quantification.
Isoforms were identified and quantified using an updated version of the Mandalorion pipeline (EII)
Availability.
C3POa and Mandalorion will be available at github under https://github.com/rvolden/C3POa and https://github.com/rvolden/Mandalorion-Episode-II, respectively.
Raw read data will be available upon publication at the SRA under PRJNA448331 (SIRV E2) and PRJNA415475 (B cells).
Supplementary Material
Acknowledgments
We thank the Georgia Genomics and Bioinformatics Core (GGBC) for PacBio Sequencing Services. We acknowledge funding from the 2017 Hellman Fellowship (to C.V.) and the National Human Genome Research Institute/National Institute of Health Training Grant 1T32HG008345-01 (to A.B. and C.C.).
Footnotes
Conflict of interest statement: C.V., R.E.G., T.P., and R.V. have filed a provisional patent on the methodology described in the paper. The other authors have nothing to declare.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive database (accession nos. PRJNA448331 and PRJNA415475).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806447115/-/DCSupplemental.
References
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Tilgner H, et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilgner H, et al. Microfluidic isoform sequencing shows widespread splicing coordination in the human transcriptome. Genome Res. 2017;28:231–242. doi: 10.1101/gr.230516.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi L, et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun. 2016;7:12065. doi: 10.1038/ncomms12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo RI, et al. Normalized long read RNA sequencing in chicken reveals transcriptome complexity similar to human. BMC Genomics. 2017;18:323. doi: 10.1186/s12864-017-3691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne A, et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oikonomopoulos S, Wang YC, Djambazian H, Badescu D, Ragoussis J. Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations. Sci Rep. 2016;6:31602. doi: 10.1038/srep31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, et al. INC-seq: Accurate single molecule reads using nanopore sequencing. Gigascience. 2016;5:34. doi: 10.1186/s13742-016-0140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sotillo E, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5:1282–1295. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer J, et al. CD19 isoforms enabling resistance to CART-19 immunotherapy are expressed in B-ALL patients at initial diagnosis. J Immunother. 2017;40:187–195. doi: 10.1097/CJI.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommermeyer D, et al. Fully human CD19-specific chimeric antigen receptors for T-cell therapy. Leukemia. 2017;31:2191–2199. doi: 10.1038/leu.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole C, Byrne A, Beaudin AE, Forsberg EC, Vollmers C. Tn5Prime, a Tn5 based 5′ capture method for single cell RNA-seq. Nucleic Acids Res. 2018;46:e62. doi: 10.1093/nar/gky182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent WJ. BLAT–The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 16.Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tardaguila M, et al. SQANTI: Extensive characterization of long read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 2018;28:396–411. doi: 10.1101/gr.222976.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 19.Picelli S, et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 21.Li W, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McWilliam H, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 24.Li H. 2017. Minimap2: Fast pairwise alignment for long nucleotide sequences. arXiv:1708.01492. Preprint, posted March 16, 2018.
- 25.Tange O, et al. Gnu parallel: The command-line power tool. USENIX Mag. 2011;36:42–47. [Google Scholar]
- 26.Lindenbaum P. 2015. Data from “JVarkit: Java-based utilities for bioinformatics.” Figshare. 10.6084/m9.figshare.1425030.v1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.