Significance
This paper addresses how the structures of voltage-sensing domains within voltage-gated ion channels respond to physiological changes in electric potential across membranes. We report three new structures of TPC1 by cryo-EM and X-ray crystallography, two with voltage-sensing domains in intermediate activated conformations and a third structure representing the deactivated state under low cytoplasmic Ca2+. The intermediate states of the voltage sensor undergo significant rotation in the membrane plane and a twisting motion, with respect to our previously determined resting state, that alters access of gating charges from the inside to the outside of the membrane, with little vertical movement across the membrane. Together these structures suggest a general mechanism for the activation of voltage-gated ion channels.
Keywords: cryo-EM, X-ray crystallography, ion channel, voltage sensor, two pore channnel
Abstract
Voltage-sensing domains (VSDs) couple changes in transmembrane electrical potential to conformational changes that regulate ion conductance through a central channel. Positively charged amino acids inside each sensor cooperatively respond to changes in voltage. Our previous structure of a TPC1 channel captured an example of a resting-state VSD in an intact ion channel. To generate an activated-state VSD in the same channel we removed the luminal inhibitory Ca2+-binding site (Cai2+), which shifts voltage-dependent opening to more negative voltage and activation at 0 mV. Cryo-EM reveals two coexisting structures of the VSD, an intermediate state 1 that partially closes access to the cytoplasmic side but remains occluded on the luminal side and an intermediate activated state 2 in which the cytoplasmic solvent access to the gating charges closes, while luminal access partially opens. Activation can be thought of as moving a hydrophobic insulating region of the VSD from the external side to an alternate grouping on the internal side. This effectively moves the gating charges from the inside potential to that of the outside. Activation also requires binding of Ca2+ to a cytoplasmic site (Caa2+). An X-ray structure with Caa2+ removed and a near-atomic resolution cryo-EM structure with Cai2+ removed define how dramatic conformational changes in the cytoplasmic domains may communicate with the VSD during activation. Together four structures provide a basis for understanding the voltage-dependent transition from resting to activated state, the tuning of VSD by thermodynamic stability, and this channel’s requirement of cytoplasmic Ca2+ ions for activation.
Voltage-sensing domains (VSDs) are four-helical bundle domains, termed S1–S4, that respond to changes in membrane potential by allowing “gating” charges, generally positively charged arginine or occasionally lysine side chains in the fourth transmembrane helix S4 (charges referred to as R1–R5), to move relative to a charge-transfer center (CT) (1, 2) that contains countercharges in the surrounding helices S1–S3 and an aromatic residue (Y, F) that seals the VSD to solvent passage.
The number of gating charges in each VSD that move across the membrane from connection to the cytoplasmic side to the extracellular (or luminal) side during activation is typically measured as two to three and up to five positive charges. The basis for structural and electrical changes in S4 that give rise to voltage dependence is key to understanding the response of voltage-gated ion channels to changes in membrane potential.
In voltage-gated ion channels, the movement of S4 is connected via an S4–S5 linker helix on the cytoplasmic side to the pore helices S5–S6 of the ion channel, and also through Van der Waals hydrophobic contacts between S4 and the pore-forming domains. In excitable cells or organelles, activation of the VSD in response to membrane depolarization greatly increases the probability of channel opening (3, 4).
Several models have been proposed for voltage-dependent activation. The “sliding helix” (5), “rotating helix” (6, 7), and “paddle” (8) models suggest that S4 moves substantially (vertically or in rotation) in the membrane to translate the gating charges across the membrane (SI Appendix, Fig. S1). In sliding helix and rotating helix models, the gating charges interact with counteranions or aqueous environments to avoid the energetic penalty of placing a charge in a hydrophobic environment. Lanthanide-based resonance energy transfer measurements suggest that gating charges do not move extensively during activation, but rather achieve alternating exposure to the internal and external milieu through conformational changes in the VSD helices S1–S4 (9). A “ratchet” model also includes the possibility of multiple intermediate states of S4 (10). Electrophysiology, EPR spectroscopy, X-ray structures, disulfide cross-linking, and simulations support a combination of translation and rotation of S4 during activation (11–18). No structural information until now had existed for voltage-gated channels captured in multiple activation states, precluding an atomic-scale evaluation of the mechanism of voltage-dependent changes, or how they translate to channel activation. Our previous crystal structure of wild-type Arabidopsis thaliana two-pore channel 1 (AtTPC1WT) provided the first resting-closed state in an intact ion channel (19, 20), the electrophysiological state that forms under high luminal 1 mM (>100 µM) Ca2+-ion concentration. Here we sought to determine structures for the activated state of AtTPC1 that is formed in low luminal (<100 µM) Ca2+ ions by removing the inhibitory luminal Ca2+-binding site in VSD2 (Cai2+), while keeping the cytoplasmic activation site (Caa2+) occupied with >300 µM cytoplasmic Ca2+ ions (21). Second, we wanted to determine the mechanism for the channel’s requirement of cytoplasmic Ca2+ ions for activation by removing the Caa2+ site.
TPCs are a family of ion channels that regulate ion conductance across endolysosomal membranes (21–23). Located in endosomes that endocytose from the plasma membrane, initially with ∼1 mM extracellular Ca2+ concentration, they regulate the conductance of Na+ and/or Ca2+ ions out of the endolysosome, intravesicular pH (24), trafficking (25), and membrane excitability (26). Cytoplasmic Ca2+ ions (>300 µM) are required for any activation of AtTPC1 (27), whereas luminal Ca2+ ions (>100 µM) suppress voltage-dependent activation (20, 28). TPCs encode two pore-forming domains on a single chain with two nonequivalent VSDs (S1–S4 and S7–S10) and pore helices (S5–S6 and S11–S12). In AtTPC1, only VSD2s (S7–S10) respond to changes in voltage (20). Three arginines on S10 of each VSD2 (equivalent to S4 in the VSD of tetrameric ion channels) in AtTPC1 are required for voltage-dependent activation. A homodimer of two TPCs forms the central functional channel surrounded by four pore-forming domains.
The dependence of AtTPC1 on external and internal Ca2+ offers the opportunity to visualize the resting state of a VSD of an intact channel and the activated state and to ask how voltage changes are detected and relayed.
Luminal Ca2+ ions suppress activation of AtTPC1 via binding to Cai2+ located in the active voltage sensor VSD2 with EC50 ∼0.1 mM. This previously enabled trapping of the resting-state VSD2 of wild-type AtTPC1 by including 1 mM Ca2+ ions (19, 20). Replacement of the Ca2+-chelating amino acids by mutagenesis (D240N, D454N, and E528Q; termed AtTPC1DDE) shifts voltage-dependent activation by −50 mV, such that the channel is open at 0 mV (20, 28) and VSD2 is in an activated conformation. Using AtTPC1DDE we sought to determine the structure of an activated state of the same intact channel where we had previously determined a resting state.
Channel opening requires Ca2+-ion binding to Caa2+ mediated by D376 of cytoplasmic EF-hand domain helix 3–4 loop (EF3-EF4). Removal of cytoplasmic Ca2+ ions or the mutation D376A (AtTPC1DA) yields permanently closed channels (29). The absolute requirement of the Caa2+ site for voltage-dependent activation led us to hypothesize that channel activation depends on communication between Caa2+ and VSD2 (21). Using the D376A mutation we sought to determine how cytoplasmic Ca2+ evokes activation.
Results and Discussion
Cryo-EM Structures of AtTPC1DDE.
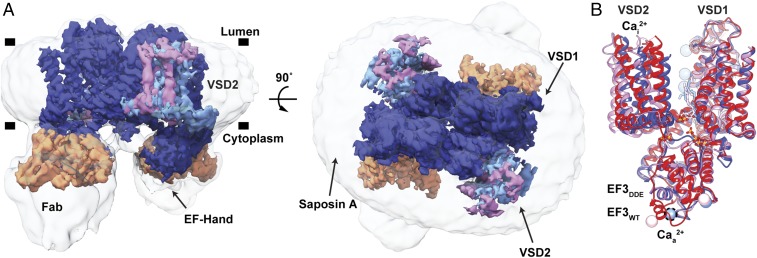
As a basis for understanding the voltage-dependent activation mechanism and its modulation by Ca2+ ions, we determined the structure of AtTPC1DDE by cryo-EM (Fig. 1A and SI Appendix, Figs. S2–S4 and Table S1). We employed saposin A nanoparticles (30) to reconstitute AtTPC1DDE into a membrane environment and an antibody Fab (antigen-binding fragment) made against AtTPC1 (CAT06/H12) (31) to facilitate particle alignment (32) (SI Appendix, Supplementary Discussion and Figs. S5 and S6).
Fig. 1.
Cryo-EM Structure of the AtTPC1DDE–saposin A–Fab complex. (A) Side (Left) and top-down (Right) views of cryo-EM density. The composite map is colored to highlight high-resolution features of TPC1 (blue, EMDB entry no. 8957), VSD2 in state 1 (cyan, EMDB entry no. 8958) and state 2 (pink, EMDB entry no. 8960), and the Fab variable domains (orange, EMDB entry no. 8956). Unsharpened density is shown at low contour (transparent gray). Membrane boundaries (black bars) defined by the nanodisc are marked. (B) Overlay of AtTPC1WT (red, PDB ID code 5DQQ), state 1 (blue), and state 2 (pink). Cai2+ and Caa2+ denote sites of luminal inhibition and cytoplasmic activation by Ca2+ ions. Dash lines around Caa2+ indicate hypothetical position in cryo-EM structure. Ca2+ ions shown as colored balls.
With a nominal resolution of 3.3 Å, the density map of AtTPC1DDE is of high quality (SI Appendix, Fig. S4A), allowing additional de novo interpretation of the AtTPC1 N-terminal domain (NTD), the S1–S2 linker, EF3-EF4 with an intact Caa2+ site, the upper vestibule of the pore, and the C-terminal domain (CTD). Three ions lie in the selectivity filter, consistent with previously defined Ca2+/Na+-binding sites (19, 20, 33). Fourteen lipid molecules surround the channel in the luminal leaflet with two on the cytoplasmic side (SI Appendix, Fig. S7). S1–S6 are well defined and remain stationary between all of the AtTPC1 structures. In AtTPC1DDE, major rearrangements are observed in the VSD2, the upper vestibule of the pore, and EF3-EF4 on the cytoplasmic side, relative to AtTPC1WT (Fig. 1B). The positions of three residues known to be phosphorylated (S22, T26, and T29) are observed in previously unresolved portions of the NTD (SI Appendix, Supplementary Discussion and Fig. S4A) (19).
The Fab binds to the EF1–EF2 loop, EF4, and S6 on the cytoplasmic side. The Fab binding affinity is the same with and without Ca2+ ions, in other molecular conditions tested (1 mM EGTA, 1 µM trans-NED19, and 1 µM nicotinic acid adenine dinucleotide phosphate), and in three different constructs (AtTPC1WT, AtTPC1DDE, and AtTPC1DA), indicating that the Fab binding does not distinguish between nor is it likely to influence the activation state (SI Appendix, Fig. S5 C and D). Most of the variable domain of the Fab is visible, allowing interpretation of the AtTPC1DDE–Fab interface (SI Appendix, Fig. S6 E and F). The constant domain of the Fab is flexible and not resolved to high resolution.
Activation of the Voltage Sensor.
Luminal Ca2+ ions inhibit AtTPC1 channel activation half-maximally at ∼0.1 mM concentration (20) via binding to Cai2+ (21) between VSD2 (D454 in the S7–S8 loop and E528 in S10) and the pore (D240). D454N, also named fou2 (fatty acid oxygenation up-regulated 2) (28), abolishes inhibition by luminal Ca2+ ions, increasing channel open probability by shifting the voltage-dependent channel opening toward more hyperpolarizing potentials (20, 28). Ca2+ ions do not inhibit AtTPC1DDE at 1 mM and up to 10 mM (33). Thus, the Cai2+ site is functionally abolished in AtTPC1DDE under cryo-EM conditions and thereby mimics the activated state under voltage and low luminal Ca2+-ion conditions (20, 33).
While the overall resolution of AtTPC1DDE is excellent, density for S7–S10 of VSD2 is significantly weaker, indicating conformational heterogeneity. Focused classification identified two states of VSD2, each with an overall resolution of 3.7 Å, but with distinct and different conformations of the S7–S10 domain structure (state 1 and 2) (Methods and SI Appendix, Fig. S3 and Table S1). We therefore conclude that these two conformations represent different functional states of the AtTPC1DDE channel. They suggest a model for the voltage activation of AtTPC1 and a mechanism for dependence on luminal Ca2+ ions.
To confirm that the structural rearrangements in VSD2 were not induced by saposin A we determined a ∼7-Å reconstruction of AtTPC1WT in both detergent and in saposin A. In these the overall channel architecture is comparable to the AtTPC1WT crystal structure (SI Appendix, Fig. S8).
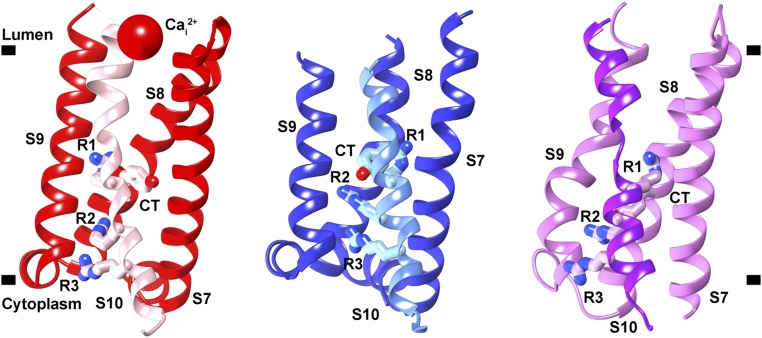
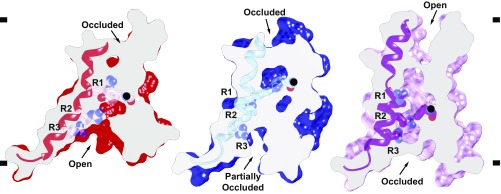
The local resolution of VSD2 (S7–S10) ranges from 4 to 6 Å in both states, making it possible to model the intermediate active state of VSD2 based on the 2.8-Å X-ray structure of AtTPC1WT (Structure Determination and Refinement and SI Appendix, Fig. S4 B and C) (19). Atomic structures for states 1 and 2 were determined by real-space and B-factor refinement against the cryo-EM densities. Changes in solvent accessibility at the luminal and cytoplasmic boundaries of VSD2 were apparent from the comparison of these new structures of AtTPC1 and the previous resting state. Taking the resting-state structure of AtTPC1WT as a reference [Protein Data Bank (PDB) ID code 5DQQ; ref. 19], transmembrane helices S7–S10 in state 1 rotate in a counterclockwise manner with respect to AtTPC1WT (Fig. 2). The helices twist to partially close the cytoplasmic solvent access to the gating charges of VSD2 (Fig. 3), while the luminal face remains occluded. Among all helices of VSD2, the key arginine-rich S10 has the least movement, and R537 (R1) remains interacting with the CT (Y475), albeit the changes in S8 conformation place R1 on the opposite horizontal face of the CT compared with AtTPC1WT. S8 moves upward in the membrane plane by nearly one helical turn, thus moving the CT into place to interact with R1 in manner similar to AtTPC1WT. State 1 probably represents a resting-state structure present in low luminal (<0.1 mM) Ca2+-ion concentrations.
Fig. 2.
Three states of VSD2. View of VSD2 in the membrane plane from the center of the channel looking outward that illustrates the rotation and twisting of VSD2 helices S7, S8, S9, and S10. Connections to the pore domains are omitted for clarity. S10 is highlighted in each state with a different color than the other helices. Gating charges R1–R3 (R537, R540, and R543) and CT residue Y475 are shown. (Left) Resting-state AtTPC1WT (red, PDB ID code 5DQQ), (Center) AtTPC1DDE state 1 (blue), and (Right) AtTPC1DDE state 2 (pink).
Fig. 3.
Activation of the voltage sensor. Side views of a common slice perpendicular to the membrane surface of VSD2 in AtTPC1WT resting state (red, PDB ID code 5DQQ) and AtTPC1DDE state 1 (blue) and state 2 (pink). S10 shown with gating charges R1–R3. The CT Cα position (black ball) is marked. Based on a structural alignment with respect to the pore helices S6–S7 of each structure.
In state 2, VSD2 rotates ∼20° clockwise in the plane of the membrane with respect to AtTPC1WT (Fig. 2). Helices S7–S10 reorient dramatically leading to an opening of the luminal face of VSD2 (Fig. 3). Tilting of S10 and rotation of S8 around S10 moves the CT downward, placing R1 in an activated conformation. The cytoplasmic face is fully closed in state 2 while the luminal side is partially open. VSD2 has effectively alternated solvent access, or electrical contact from the cytoplasmic to the luminal side of the membrane.
Clockwise rotation was proposed to connect resting- and active-state conformations on the basis of comparing previous voltage-gated channel structures (13, 19, 34). During review of this paper, activated-state structures of mouse TPC1 were determined (35). Comparing the activated-state mouse TPC1 to resting- and intermediate-state AtTPC1 supports the proposed role of VSD2 rotation during channel activation. The observation of clockwise rotation in multiple activated-state structures supports our conclusion that state 2 represents an intermediate-activated state of AtTPC1.
These two states of VSD2 in AtTPC1 are likely to represent structures that VSD2 adopts in low luminal Ca2+ during activation. The overall movement of VSD2 during activation serves to dilate the luminal face of VSD2, close off the cytoplasmic leaflet to solvent, and move the CT below a gating charge in the membrane. This would reconnect the positive gating charges from the potential of the cytoplasmic side to the potential of the luminal side upon activation, mediated by tilt and twisting of S7, S8, and S9 around S10. This is all that is necessary to transport the gating-charge arginine residues from cytoplasmic electrical connection, to the external potential. A hydrophobic sealed region between all four helices forms a thin outer insulating layer in the resting state, while in the activated state this becomes open to solvent and the bundle twists to form a hydrophobic insulating region closer to the cytoplasmic side.
Drug and Lipid Binding Sites.
The high-resolution map enabled refinement of a total of 14 lipids on the luminal and 2 on the cytoplasmic leaflet of the membrane. The luminal lipids are modeled as the 16-carbon-containing palmitic acid, the predominant lipid length in soy polar lipids, whereas the cytoplasmic lipids are modeled as 18-carbon phosphatidic acid (PA). Twelve lipids bind to the long axis of AtTPC1, occupying the binding site for Ned19 (refs. 19 and 21 and SI Appendix, Fig. S7). Ned19 binding may disrupt these structured lipids, acting as a steric block to prevent S7 and pore movements during gating. One lipid binds to a buried site along the short axis, sandwiched in between the pore domains. Based on homology to Cav channels in this region and recent structures of bacterial Cav channels (CavAb) bound to dihydropyridines (DHP) (amlodipine and nimodipine) and phenylalkylamine (verapamil) inhibitors (36), the short axis in transmembrane segments S6 and S12 is a likely site for binding of tetrandrine, a bisbenzylisoquinoline alkaloid isolated from the Chinese herb Stephania tetrandra (37) and approved medications of the DHP class of L-type Cav antagonists—all TPC channel blockers.
The luminal PA lipids bind along the short axis in a pocket formed by S1 of VSD1, the S10–S11 linker, S11, and the S8–S9 linker of VSD2. The alkyl chains make Van der Waals interactions, whereas the phosphoglycerol headgroup makes hydrogen bonds to the backbone of W492 and to the sidechain of R498 and S8–S9 of VSD2. The S8–S9 linker moves 4 Å inward in the AtTPC1DDE structure to form the PA binding site, which would not be intact in the AtTPC1WT resting state. Therefore, this lipid binding site is likely specific for AtTPC1DDE. Certain lipids may occupy this site during activation of AtTPC1. Polyunsaturated fatty acids inhibit plant TPC1 activation, but the binding site has not been determined (38). In principle, polyunsaturated fatty acids could mimic the observed site in the resting state and prevent activation.
Charge-Transfer Mechanism.
State 2 represents an intermediate activation state of VSD2 where the gating charge R1 is transferred across the membrane (Fig. 3). In this state, the CT moves downward, R1 moves slightly upward, and overall the VSD2 bundle rotates by ∼20° in the membrane plane with respect to the adjacent pore domains. In this the voltage-sensing helix S10 pivots only 2–3° inward toward the pore. Importantly, the three observed states of the VSD thus far are not related by rigid-body rotations. These changes in VSD2 make R1 more accessible to solvent on the luminal side while charges R2 and R3 are shielded from access to the cytoplasmic side; there is a net transfer of charge from cytoplasmic side (in the resting state) to the outside (in the activated state).
Pore Conformational Changes.
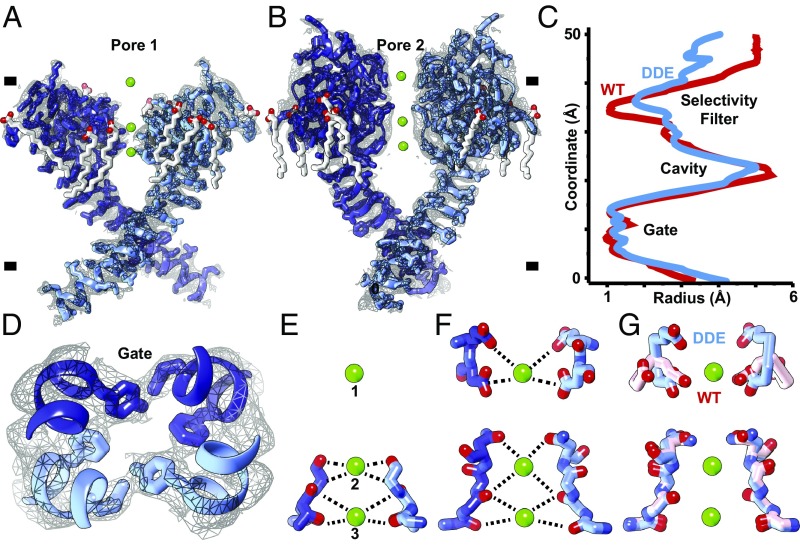
High-resolution density for the pore defines the consequences of activation by removal of the external Cai2+ site on VSD2 on the central channel (Fig. 4 A and B). As a consequence of the rearrangement of VSD2 the upper vestibule and the upper selectivity filter of the channel open along the ion permeation pathway (Fig. 4C), while the lower gate remains closed (Fig. 4D). Full channel opening may be evoked by passage of ions or an additional energy barrier, because electrophysiological studies show the channel is maximally open at 0 mV with removal of the Cai2+ site.
Fig. 4.
Ion permeation pathway. (A and B) Orthogonal side views through pore helices (A) S5–S6 (pore 1) and (B) S11–S12 (pore 2) of the channel homodimer overlaid with high-resolution cryo-EM density (gray mesh). (C) HOLE plot of pore radii along central channel coordinate of AtTPC1DDE (red) and AtTPC1WT (blue). (D) Top-down view through central pore. Gate residues Y305, L673, and F676 are shown. (E–G) Side views through the selectivity filter in (E) pore 1, (F) pore 2, and (G) an overlay of pore 2 of AtTPC1DDE (blue) and AtTPC1WT (pink). (Upper) E605 and D606 and (Lower) S265, T263, T264, V628, M629, and N631 selectivity filter residues are shown. Density for lipids (SI Appendix, Fig. S7) and ions is omitted for clarity.
In the selectivity filter, three ions are observed (Fig. 4 E and F). Sites 1 and 3 were occupied by Ca2+ mimetics, Yb3+ or Ba2+, in AtTPC1WT structures (19, 20). Site 2 was seen to be occupied by Na+ in the Na+-selective chimera of AtTPC1 (33). Since AtTPC1DDE contains 1 mM Ca2+ and the channel is Ca2+-selective, these are probably hydrated Ca2+ ions (SI Appendix, Supplementary Discussion). Side-chain interactions from the upper vestibule of S11–S12 exclusively coordinate site 1. The side chains of D606 move inward to coordinate site 1 with E605 (Fig. 4G). E605–Ca2+ (5.5 Å) and D606–Ca2+ (4.6 Å) distances are consistent with the radius of a hydrated Ca2+ ion. There are readjustments throughout the channel, suggesting that VSD2 activation by removal of the luminal Cai2+ site can indeed potentiate channel opening, as observed by electrophysiology.
The observation of upper and lower selectivity filter and activation gate operating independently in our structures suggests a multistep gating mechanism that resembles that proposed for TRPV1 channel activation (39). Activation of the VSDs could lead to opening of the selectivity filter and activation gate in two steps. First, the upper selectivity filter opens in response to rotational rearrangement and partial activation of the VSDs, then the lower selectivity filter and activation gate open when the VSD achieves maximal activation.
Cytoplasmic Activation by Ca2+ Ions.
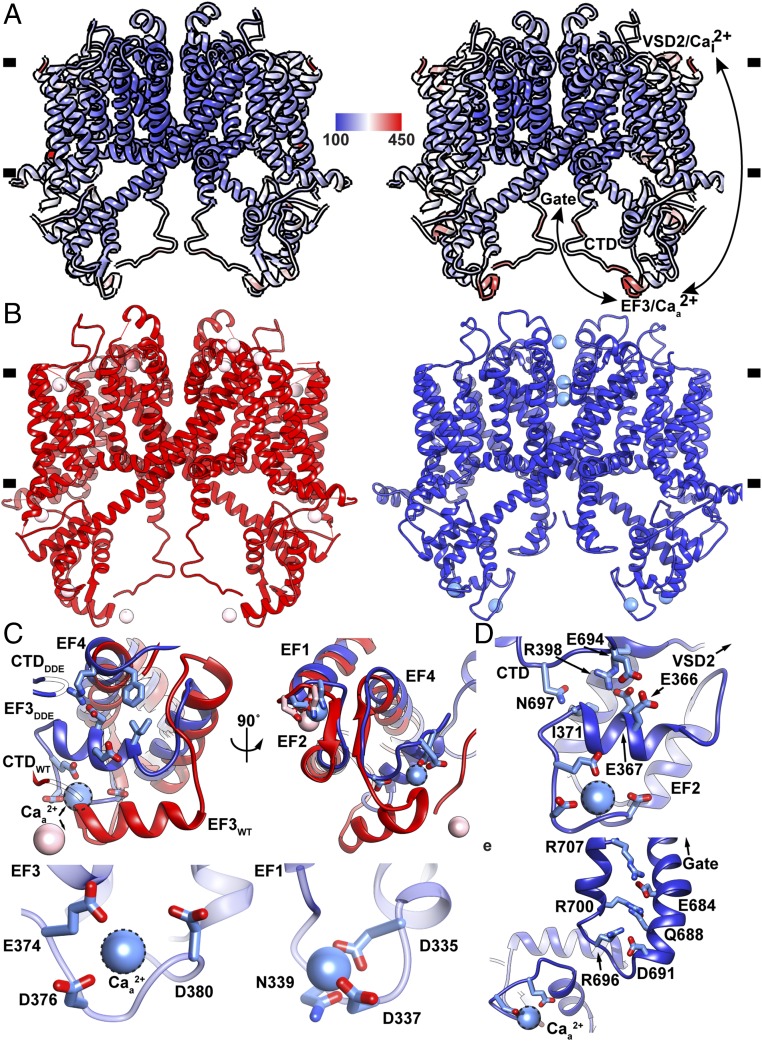
Full voltage-dependent activation of AtTPC1 requires ∼0.3 mM cytoplasmic Ca2+ evoked by Ca2+ binding to the EF3-EF4 loop (20, 29). D376 in EF3 is critical; when it is substituted with D376A in AtTPC1DA the channel remains closed and is no longer responsive to membrane potential (29). We determined the crystal structure of AtTPC1DA (Fig. 5A and SI Appendix, Table S2) to 3.5-Å resolution by X-ray crystallography. Compared with the AtTPC1WT crystal structure, removing the activating cytoplasmic Caa2+ binding site leads to higher dynamic motion (B-factors) not only in the EF-hand and the CTD, but also in the VSDs and upper vestibule of the pore, showing that the activating cytoplasmic site may act through effects on the VSDs (SI Appendix, Supplementary Discussion).
Fig. 5.
Dynamics of cytoplasmic domains. (A) Overlay of (Left) AtTPC1WT and (Right) AtTPC1DA crystal structures colored by B-factor value (100–450 Å2) (see Methods and SI Appendix, Supplementary Discussion). (B) Side views of AtTPC1WT (red) and AtTPC1DDE (blue). (C) Views from (Top Left) side and (Top Right) bottom of EF3 for overlaid AtTPC1DDE (blue) and AtTPC1WT crystal structures (red, PDB ID code 5DQQ). CTD interactions are omitted for clarity. Ca2+-ion binding sites in (Bottom Right) EF1–EF2 and (Bottom Left) EF3–EF4. (D) View of EF3-CTD and EF3-EF4 interactions and connection to VSD2. (E) Structure of the CTD in AtTPC1DDE and a possible coupling pathway to the pore gate and Caa2+. Dashed lines indicate the hypothetical Caa2+ ion position.
AtTPC1DA is in a resting state as expected. However, there is an overall effect of increasing the dynamical motion of VSD1, VSD2, and the pore (in the context of otherwise identical overall B-factors), showing that Caa2+ has an allosteric effect in stabilization that could impact voltage dependence (VSDs) and conductance (the pore). The regions of increased motion in AtTPC1DA correspond to the regions that undergo conformational change in AtTPC1DDE, suggesting that there is a pathway for coupling cytoplasmic activation at Caa2+ to voltage dependence of activation in VSD2 and transmission to the pore.
The Caa2+ site in EF3 is fully formed in the cryo-EM structure of AtTPC1DDE, whereas it was partially occupied in AtTPC1WT and was in a more extended conformation (19, 20) (Fig. 5B). In AtTPC1DDE, EF3 alone lies 7 Å closer to the transmembrane domain and rotates 20° to make close contacts with EF4 (Fig. 5C). The Caa2+ chelating residues E374, D376, and D380 order the EF3-EF4 loop around the activating Caa2+ site (Fig. 5C). Movement of EF3 leaves the EF1–EF2 Ca2+ site unaltered. Therefore, EF3 is capable of undergoing large-scale movements that change the structure of the cytoplasmic domains and their connection to VSD2, suggesting that activation on the cytoplasmic side can act reciprocally through VSD2.
The conformation of the cytoplasmic domains seen in AtTPC1DDE probably reflects the predominant structure in AtTPC1WT (rather than the crystal structure) because the conformation of EF3 is conserved in cryo-EM structures from several conditions: detergent and saposin A nanoparticles of AtTPC1WT, and AtTPC1DDE, and when bound to either of two different Fab molecules (SI Appendix, Fig. S8C).
The CTD of AtTPC1 is indispensable for channel activation (40). Removal or truncation by 29 residues abolishes channel function. In the crystal structure of AtTPC1WT, the CTD forms an intramolecular complex with the EF3 via salt-bridge interaction D376–R700, leading to the hypothesis that the EF3–CTD complex could undergo conformational changes upon activating cytoplasmic Ca2+ binding (19). In the cryo-EM structures the CTD moves toward the membrane vertically by 13 Å to accommodate the upward movement of EF3 upon forming the cytoplasmic activating Caa2+ site (Fig. 5B). The salt bridge (D376–R700) that previously linked the CTD to EF3 in AtTPC1WT breaks to allow D376 to chelate the Caa2+ site. This can explain why only substitution of D376 and not the other chelating residues abolishes Ca2+ activation (29). In AtTPC1DDE, the CTD now forms a hydrogen bond (E366–E694) with EF3 and hydrogen bonds (N697–I371) via the backbone (Fig. 5D). Following the interaction with EF3 and a β-turn the CTD to folds back on itself to form a charged zippered interaction (R696–D691, R700–Q688, and R707–E684) between the signal poly-R and poly-E motifs of the CTD (19). The poly-E motif connects directly to the pore gate (Fig. 5E). The role of the CTD on channel activation may be to stabilize the EF3 helix in both apo- and Ca2+-bound conformations to allow communication of EF3 movement to VSD2 and the pore gate. Upon removal of Ca2+, the CTD could adopt an extended conformation to reform the salt-bridge interaction D376–R700 with EF3. Without the CTD, EF3 may not be able to reform the Caa2+ site and would become trapped in an inactive state.
To further investigate the conformation of EF3 in solution, we performed continuous wave (CW) EPR experiments using spin-labeled full-length AtTPC1 lacking cysteines (AtTPC1cysless; see Methods) in detergent micelles. Ten positions in the CTD, gate, and EF-hand domains were examined by spin-probe mobility and responsiveness to Ca2+ ions. Labeling at a site on EF3 (R379) indicates a conformational shift to higher probe mobility compared with EGTA, indicating that the probe changes environment in the presence of Ca2+ ions (SI Appendix, Fig. S9). Mutation of the Caa2+ site, but not the Ca2+ site, in EF1–EF2 abolishes the Ca2+-dependent increase in probe mobility, suggesting that EF3/Caa2+ changes conformation upon increasing cytoplasmic Ca2+-ion concentrations. The data suggest that the AtTPC1WT crystal structure likely represents an apo-state of EF3, whereas AtTPC1DDE represents a Ca2+-bound conformation present along the pathway of activation.
Alternate Access Mechanism of Activation.
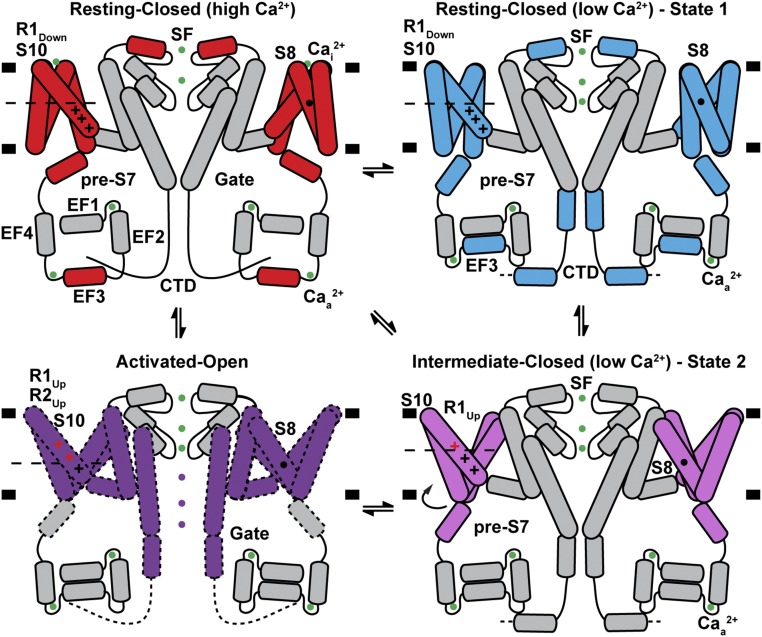
As the electric potential changes across a membrane, functional voltage sensors find a new free energy minimum. Under hyperpolarizing conditions, the favorable electrostatic free energy component of positive gating charges’ attraction to internal negative potential is balanced against a structural “distortion” free energy cost. Depolarization therefore releases both of these components to populate a new overall equilibrium state that includes a component that regulates the open probability of the pore. Release of the electrostatic free energy is achieved by alternating access of the positively charged gating charges on S10 to solvent from the cytoplasmic side in the hyperpolarized resting state, to a state in which they are insulated from the cytoplasmic side and become accessible to solvent from the luminal side, without any necessity for vertical movement of the gating charges themselves. The structures of AtTPC1WT and AtTPC1DDE suggest that this “alternating access” mechanism plays the key role. The movement of the VSD2 helices S7, S8, and S9 with respect to the gating charges on S10 achieve this (Fig. 6).
Fig. 6.
Mechanism of TPC1 channel activation. Schematic summarizing the conformations of VSD2, EF3, CTD, and selectivity filter (SF) observed in crystal and cryo-EM structures under high and low effective luminal Ca2+-ion concentrations. A potential model for coupling between luminal inhibition Cai2+, cytoplasmic activation Caa2+, voltage sensing, and channel gating is shown. Helices and ions that move between the AtTPC1WT resting state (red), AtTPC1DDE state 1 (cyan), AtTPC1DDE state 2 (pink), and hypothetical activated-open state (purple) are colored. Ca2+ ions are shown as green balls. Gating charges that change position with respect to the CT (dashed line) between states are colored red.
Hence, the potential gradient is focused across a thin hydrophobic region close to the luminal (external) side in the resting state and alternately across one on the cytoplasmic side in the activated state. Thus, the gating charges do not require any vertical movement across the bilayer as the electrically insulating region, which includes the CT and across which the voltage gradient is formed, moves from outside of the gating charges in the resting-state voltage sensor to the cytoplasmic side of gating charges in the activated state. Thus, the electric component of free energy is released.
The mechanism is unique versus most previously proposed models in that S7–S9, not only S10 that is closest to the pore, are crucial during voltage activation to move gating charges from the electric potential on one side of the membrane to the potential of the other side without any necessary vertical translation of S10. Even after this major driving effect, gating-charge side chains will also readjust in context of their different environments and some vertical movement may find more favorable energetics in the shallow energy gradient around them.
There is a teleologically favorable aspect to this alternate access mechanism in that it does not need to remove any hydrophobic residues out of the bilayer, as would be required of a model involving large vertical movement of S10. It also does not need to rebuild the hydrophobic interface between S10 of the VSD and the walls of the pore with which it forms the primary contact that may be important in changes that alter open probability of the channel.
Simulations of VSDs that assume a linear gradient of electric potential across the bilayer that changes only in slope on depolarization, but not position of start and end points across the VSD, may not represent the true physiological state.
The Gibbs free energy change on moving one charge across a transmembrane potential difference of −70 mV is −1.6 kcal/mol. The measured gating charge transfer for activation of AtTPC1 is 3.9 charges (20) or −6.3 kcal/mol in free energy difference. Ca2+ binding to the luminal Cai2+ site effectively stabilizes the resting state. The EC50 for the Cai2+ site is ∼0.1 mM. Therefore, assuming a Kd of ∼0.1 mM, the free energy released by removing each Cai2+ site is ∼5.6 kcal/mol per site (or ∼11.2 kcal/mol per AtTPC1 channel). It is not unreasonable that this difference, in releasing the restraint at Cai2+, could allow the VSD to reach its active state that would normally be evoked by depolarizing the transmembrane potential at low luminal Ca2+. The fact that we observe intermediate states of TPC1 upon removing Cai2+ rather than the fully activated state suggests that the membrane voltage or the passage of ions may contribute additional energy to maximize activation of VSD2 and channel open probability.
The mechanism suggested for AtTPC1 voltage-dependent activation provides atomic structures that have been determined for intermediates on transition from resting state to activated state in an intact voltage-dependent channel. The structures also capture large-scale rotation of VSDs that could play a role in activation. It could be a general way in which gating charge can move energetically downhill in response to a change in voltage, without the energetic cost of removing any hydrophobic lipophilic regions out of the membrane.
Methods
Protein Production.
AtTPC1.
X-ray and cryo-EM trials used AtTPC1 with deletion of residues 2–11 (19). Antibody generation employed full-length AtTPC1. AtTPC1 constructs were expressed and purified by nickel affinity chromatography (NiNTA), thrombin cleavage, and size exclusion, as described previously (19). For reconstitution into saposin A lipoprotein nanoparticles and MSP nanodiscs, the size-exclusion step was omitted after NiNTA but performed after reconstitution. To introduce single cysteine labels for CW-EPR, a cysteine-less variant of AtTPC1, AtTPC1cysless, was constructed to replace all native cysteines with serines (C93S, C101S, C159S, C347S, C392S, C574S, C577S, C580S, C687S, and C728S). Cysteine substitutions for EPR spectroscopy were purified in the presence of 1 mM TCEP during solubilization and binding to nickel beads but then removed during wash and elution steps to prevent interference with MTSL labeling. AtTPC1cysless has identical biochemical behavior to wild-type AtTPC1.
Saposin A.
Human saposin A (plasmid was a kind gift from J. Frauenfeld, Salipro Biotech AB) was expressed and purified from Escherichia coli Rosetta-gami 2(DE3) cells essentially as described (30). Several colonies from transformed cells were used to inoculate overnight 300 mL LB cultures containing 25 µg/mL chloramphenicol, 10 µg/mL tetracycline, and 15 µg/mL kanamycin at 37 °C. Overnight cultures were used to seed 6 L of Terrific Broth, grown to an absorbance value at 600 nm of 1. The temperature was adjusted to 20 °C 30 min before induction by 0.7 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 15 h at 20 °C. Cells were harvested by centrifugation at 4,000 × g for 10 min and resuspended in 200 mL 50 mM Tris, pH 7.4, and 150 mM NaCl (lysis buffer) and frozen at −80 °C before further use. Bacterial pellets were lysed in the presence of 1 mM PMSF by sonication for 5 min then clarified by centrifugation at 26,000 × g for 20 min. The supernatant was then incubated at 70 °C for 10 min followed by centrifugation at 26,000 × g for 20 min to remove contaminant proteins. Imidazole (pH 7.4) was then added to 20 mM and passed through two HisTrap FF crude 5-mL columns equilibrated in lysis buffer with 20 mM imidazole (pH 7.4) using a peristaltic pump. Following 50-mL washes with lysis buffer with 20 mM imidazole (pH 7.4) and lysis buffer with 35 mM imidazole (pH 7.4), pure human saposin A was eluted in 25 mL lysis buffer with 400 mM imidazole (pH 7.4) in 1.5-mL fractions. Fractions containing human saposin A were dialyzed against 1 L of lysis buffer with 4 mg of tobacco etch virus (TEV) protease. The following morning an additional 2 mg of TEV was added with an exchange of the dialysis buffer and dialyzed at room temperature for 3 h. Uncleaved saposin A and free TEV were removed by passing the protein through a single HisTrap FF crude 5-mL column equilibrated in lysis buffer with 20 mM imidazole (pH 7.4) and washed with 30 mL of lysis buffer with 20 mM imidazole (pH 7.4). The flow-through and wash fractions were pooled and concentrated in 5-kDa molecular weight cutoff concentrators and serially injected in 20-mg aliquots over TSKgel G3000SW 7 mm × 60 cm or Superdex S200 10/300 columns equilibrated in lysis buffer with 1 mM CaCl2. High- and low-molecular-weight peaks were observed during size exclusion. The low molecular weight, most consistent with the size of the saposin A monomers and dimers, was concentrated to 2 mg/mL for use in reconstitution of AtTPC1 and frozen at −80 °C. Final yields of human saposin A were 5–10 milligrams of human saposin A per liter of culture.
Membrane scaffold protein.
Plasmid harboring His-tagged E3D1 variant of membrane scaffold protein (MSP) of the nanodiscs was obtained from Addgene (20066). MSP was expressed, purified, and cleaved with TEV protease as described (41) with minor modifications. Colonies of transformed BL21 (DE3) Gold cells (Stratagene) were used to inoculate overnight 30 mL LB cultures containing 30 μg/mL kanamycin. Overnight cultures were used to inoculate 2 L of Terrific Broth, grown at 37 °C until OD600 of 1.3 and then induced with 1 mM IPTG for 3 h. Cultures were harvested by centrifugation at 4,000 × g for 10 min, resuspended in 50 mM Tris (pH 8.0) and 300 mM NaCl (MSP lysis buffer) supplemented with 1% Triton X-100, protease inhibitor tablet, and 1 mM PMSF. A microtip sonicator (Branson) was used to lyse the cells for three cycles at 1 s on, 1 s off, and at 60% amplitude for 3 min each. For each 1 L culture 1 mL of NiNTA resin equilibrated with MSP lysis buffer was used in batch-binding for 1 h at 4 °C. Resin was then washed with MSP lysis buffer supplemented with (i) 1% Triton X-100, 10 column volumes (CVs), (ii) 50 mM sodium cholate and 20 mM imidazole, 5 CVs, and (iii) 50 mM imidazole, 7 CVs. MSP was eluted with 4 CVs of MSP lysis buffer supplemented with 400 mM imidazole. TEV cleavage was performed overnight in dialysis against 4 L of 50 mM Hepes (pH 7.5) and 200 mM NaCl. Uncut protein was removed with subtractive step on NiNTA resin and cleaved MSP was concentrated to 5 mg/mL using Millipore concentrators with 10-kDa molecular weight cutoff and frozen in aliquots at −80 °C. Typical yield of MSP E3D1 from 1 L culture was 15 mg.
Antibodies.
Monoclonal antibodies were expressed and purified by Dan Cawley from hybridoma culture supernatants using standard methods in the Monoclonal Antibody Core Facility at Oregon Health & Science University (OHSU). The 4B8 Fab fragment was generated by papain digestion 1/20 papain:4B8 (wt/wt) at 30 °C for 1 h in the presence of 5 mM l-cysteine and quenched with 20 mM iodoacetamide. Full-length IgG and Fc regions were removed via protein A chromatography and the flow-through containing 4B8 Fab was concentrated and purified by TSK3000/Superdex200 size-exclusion chromatography in 20 mM Hepes (pH 7.4), 0.2 M NaCl, and 5% glycerol.
Antibody fragments (Fabs) generated by phage display were expressed and purified essentially as described (42) from constructs subcloned into the expression vectors pRH2.2 or pSFV4, a gift from S. Sidhu, Karolinska Institutet, Stockholm. DNA was transformed into BL21-Gold(DE3) (Agilent) and used directly to set up 15-mL overnight starter cultures in 2xYT media supplemented with 50 μg/mL ampicillin. Cultures were then inoculated into 1 L of 2xYT media, grown until OD600 of ∼0.8–1.0, induced with 1 mM IPTG for 4 h at 37 °C. Cells were harvested by centrifugation and disrupted in lysis buffer containing 20 mM sodium phosphate dibasic (pH 7.4), 500 mM NaCl, 1 mM PMSF, and 2 mM DNase, using a sonicator (Branson). Lysate was heated to 55 °C for 30 min, cleared by centrifugation, and incubated with 1 mL of MabSelect ProteinA resin (GE Healthcare) in batch for 1 h at 4 °C. Resin was washed with 15 column volumes of 20 mM sodium phosphate dibasic (pH 7.4) and 500 mM NaCl and then eluted with 10 mL 0.1 M acetic acid into tubes containing 1 mL of 1 M Hepes (pH 7.5) to immediately neutralize elutions containing Fab fragments. Fabs were then dialyzed against 4 L of 50 mM Hepes (pH 7.5) and 200 mM NaCl, concentrated with Millipore concentrators with 30-kDa molecular weight cutoff and stored in aliquots at −80 °C. The typical yield from 1 L culture varied depending on Fab fragment and was between 0.5 and 3 mg. For cryo-EM studies Fab CAT06 was modified with H12 elbow variant as described previously (43), resulting in the CAT06/H12 antibody fragment. Briefly, the H12 elbow variant exchanges heavy chain residues (112)SSASTK(117) [numbering according to Kabat et al. (44)] to (112)FNQI-K(117) with one position deleted from the sequence.
Saposin A and Nanodisc Reconstitution.
Lipids.
All lipids were prepared from thin films. Lipids were dissolved in chloroform, aliquoted into glass vials, dried under nitrogen, and stored at −20 °C dry. Unless otherwise noted, resuspending lipids was accomplished by adding buffer with or without detergent and sonicating 10–30 min until clear.
Saposin A nanoparticles.
Saposin A nanoparticles containing AtTPC1 were formed by mixing AtTPC1 alone or AtTPC1–Cat06/H12 Fab complexes with soy polar lipids and saposin A in a 1:8:5–10 ratio by mass. A typical preparation consisted of 2.4 mg (8 mg/mL) of NiNTA-purified AtTPC1 incubated with 2.4 mg (9 mg/mL) of protein A-purified CAT06/H12 [adjusted to 0.05% n-dodecyl β-d-maltoside (DDM) and 1 mM CaCl2], incubated at room temperature for 5–10 min. AtTPC1-Cat06/H12 Fab complexes were then mixed with 4.8 mL of 5 mg/mL soy polar lipids (diluted from a 25 mg/mL stock in 1% DDM) dissolved in size-exclusion buffer (20 mM Hepes, pH 7.3, 0.2 M NaCl, 5% glycerol, 1 mM CaCl2, and 0.05% DDM) and incubated at room temperature for 30 min. Then, 9.6 mL (2 mg/mL) saposin A was added and incubated 1 h at room temperature,then diluted to 40 mL with size-exclusion buffer. Detergent was removed by adding 1 g of activated, washed, Bio-Beads SM2 (Bio-Rad) resin overnight at 4 °C. Bio-Beads were activated by incubation in methanol and successively washed with copious amounts of water. The following morning, beads were removed by filtration through 5-µm syringe filter. Then, 0.5 molar equivalent of Fab CAT06/H12 was added, incubated 30 min at 4 °C, and then concentrated to ∼0.3 mL in a 100-kDa MWCO concentrator and filtered before injection on a TSK4000 or Superose 6 column equilibrated in nanodisc size-exclusion buffer (20 mM Hepes, pH 7.3, 0.2 M NaCl, and 1 mM CaCl2). A single fraction was selected for cryo-EM analysis by screening for homogeneous particles by negative-stain EM (SI Appendix, Fig. S6 B–D). Cryo-EM grids were frozen following ultracentrifugation at 100,000 × g (TLA-55) for 5 min at 4 °C without further concentration. AtTPC1-CAT06/H12–saposin A complexes typically eluted from size exclusion at 0.5–1 mg/mL; 16 lipids were observed bound to AtTPC1DDE (SI Appendix, Fig. S7).
MSP nanodiscs.
Reconstitution of AtTPC1 into E3D1 nanodiscs was performed using standard protocols (45). For the purpose of antibody generation by phage display, full-length AtTPC1WT was reconstituted into nanodiscs using biotinylated version of MSP E3D1, and size-exclusion fractions corresponding to AtTPC1 in biotinylated nanodiscs were used without the removal of empty nanodiscs. Final mixed lipid–detergent micelles used in reconstitution contained 10 mM soy polar extract, 1 mM cholesterol hemisuccinate (CHS), and 30 mM DDM. Nanodisc assembly mix was prepared using purified TPC1, mixed micelles, and MSP at 2:10:1,000 ratio. All components were mixed together in nanodisc size-exclusion buffer and incubated on ice for 1 h. Next, 400–500 mg activated Bio-Beads were added per each milliliter of assembly mix, and the reconstitution reaction was left shaking overnight at 4 °C. As a control, empty nanodiscs were prepared using the same phospholipids. All nanodisc samples for phage display experiments were concentrated, supplemented with 5% wt/vol sucrose, aliquoted, flash-frozen in liquid nitrogen, and kept at −80 °C.
Proteoliposomes.
Proteoliposomes of full-length wild-type AtTPC1 were formed by resuspending 20 mg of soy polar lipids dried from chloroform resuspended in 4 mL nanodisc size-exclusion buffer, subjected to 10× alternating liquid nitrogen and warm water bath freeze–thaw cycles, then sonicated for 10 min. The opaque mixture was then passed through an Avestin hand extruder with a 400-nm pore size 10 times until mildly translucent. Vesicles were disrupted by addition of 69.8 µL 10% DDM added (4 mM final), 12.1 µL 2% CHS in 10% DDM added (0.1 mM final), and 1 mM CaCl2 and stirred 30 min at room temperature. Two milligrams of AtTPC1 was added to disrupted liposomes (1:10 protein:lipid ratio) and stirred for 1 h at room temperature using a magnetic “flea” stir bar. To form proteoliposomes, methanol-washed Bio-Beads were added in increments of 300 mg, 300 mg, 500 mg, and 1 g in 1-h intervals at room temperature. The last incubation was done at 4 °C overnight. The following morning the samples were centrifuged at 100,000 × g for 1 h in a table-top ultracentrifuge (TLA-55). The proteoliposome pellet was resuspended in 0.4 mL of nanodisc size-exclusion buffer. The concentration was estimated by gel and frozen in liquid nitrogen and stored at −80 °C. Empty liposomes were made in a similar fashion by substituting AtTPC1 protein with size-exclusion buffer.
Antibody Generation.
Hybridoma.
Hybridoma cell lines were generated by standard methods at Vector and Gene Therapy Institute, OHSU, by Daniel Cawley. Animals were handled under protocols approved by OHSU Institutional Animal Care and Use Committee. Briefly, four BALB/c mice were injected with 25 µg of AtTPC1 proteoliposomes at 0 and 14 d. On day 26, serum was prepared and tested for antibody titer in ELISA. The mice with the highest titers were used to derive hybridomas. The two best-responding mice were injected on day 45–55 with 10 µg Ag as a final boost. Four days later, spleen cells are fused with P3X mouse myeloma cells. Hybridomas were assayed 12 d later in ELISA. All candidate cell lines were expanded and given a secondary ELISA screen 12–14 d later. All antigen-specific antibodies were made available as culture supernates for screening. All candidate cell lines are frozen in liquid nitrogen. Fourteen hybridoma antibodies were identified that specifically bind to immobilized biotinylated AtTPC1 in DDM detergent by ELISA. To select for antibodies that recognize 3D structural epitopes a counterselection against antibodies that bind linear epitopes on denatured protein in 8 M urea was performed, yielding five antibodies that bind only under native conditions by ELISA. Selection of high-affinity antibodies that were capable of complex formation was conducted by fluorescence size-exclusion chromatography (46) (FSEC) using FITC-labeled AtTPC1 and hybridoma supernatants containing 10–50 µg/mL whole IgG. FSEC samples were prepared by mixing 1 µg FITC-labeled AtTPC1 with 0.4 mL hybridoma supernatants (supplemented with components to make size-exclusion buffer + 0.03 mg/mL soy polar lipids) in a total volume of 490 µL of size-exclusion buffer. Samples were incubated at 25 °C for 1 h or 4 °C for 4–5 h and injected on a Superose 6 column, 0.4 mL/min, equilibrated in size-exclusion buffer at 4 °C, using an autosampler and inline fluorescence detector set at excitation = 495 nm and emission = 518 nm. The fluorescence detector (Shimadzu RF-10AXL) was calibrated before use by injecting varying amounts of FITC-AtTPC1 (0.1 ng–1 µg) with known labeling efficiency.
Phage display.
To generate Fabs, the efficiency of biotinylation of AtTPC1WT-E3D1 nanodiscs was evaluated by pull-down on streptavidin-coated magnetic beads (Promega). Library sorting steps were performed using Fab Library E [DNA kindly provided by S. Koide, New York University School of Medicine, New York (47)] based on previous protocols (31, 48). Six independent phage library sorting experiments were performed against biotinylated AtTPC1WT -E3D1 nanodiscs in two buffers containing 20 mM Hepes, pH 7.3, 200 mM NaCl, 5% glycerol, 1% BSA, and either 1 mM CaCl2 or 1 mM EGTA. Additionally, for each CaCl2 or EGTA condition, sorting was carried out in the presence of following ligands: either 1 μM NAADP, 1 μM trans-NED19, or no ligand control to maximize the number of multiple states of TPC1 channel during sorting (sorting buffer). A total of five rounds of phage library sorting were performed for each sample with decreasing concentration of immobilized biotinylated TPC1-E3D1 nanodiscs between subsequent rounds in the following order: 1,000, 600, 200, 200, and 100 nM. Round 1 of sorting was performed manually using nanodisc bound to streptavidin-coated magnetic beads, and upon washing with respective sorting buffer whole beads were used to infect log phase E. coli XL-1 strain (Stratagene) in the 2xYT media (Fisher) supplemented with 50 µg/mL ampicillin and 109 pfu/mL of KO7 helper phage (NEB) overnight at 37 °C and 280 rpm to amplify the phage particles. The amplified phage particles after round 1 were used as an input for four additional rounds of library sorting performed semiautomatically with a KingFisher magnetic bead handler (Thermo) according to described protocols (49). Starting from round 2, in every step except elution, the buffer was supplemented with 10-fold molar excess of nonbiotinylated empty E3D1 nanodiscs to counterselect for MSP, lipid, and nonspecific phage particles. Finally, in each of rounds 2–5, phage particles were eluted from magnetic beads by 15-min incubation with 1% Fos-Choline-12 in respective sorting buffer.
Initial validation of selection clones was performed by single-point direct phage ELISA using clones from rounds 3, 4, and 5. Amplified phage particles at 10-fold dilution were assayed against 50 nM biotinylated nanodiscs (either empty or with AtTPC1) using HRP-conjugated anti-M13 monoclonal antibody (27-9421-01; GE Healthcare). Assays were performed in respective sorting buffer supplemented with 2% BSA. Each Fab clone with A450 signal above 0.2 (four times the average background level of the assay) was sequenced; unique Fabs were subcloned into pSVF4 or pRH2.2 vectors, a kind gift of S. Sidhu, and purified as described above. In total 16 unique Fab sequences were obtained from total of 192 single colonies screened: nine Fabs from library sorting with CaCl2 and seven Fabs from library sorting with EGTA.
Cryo-EM Structure Determination.
Sample preparation.
Samples were analyzed by negative-stain EM to determine suitability for cryo-EM. To prepare grids, 3 μL of sample at 10–50 μg⋅mL−1 was applied to a glow-discharged continuous carbon grid, which was then treated with 0.75% (wt/vol) uranyl formate. Grids were imaged on an FEI Tecnai T12 microscope operated at 120 kV at a nominal magnification of 52,000× using an UltraScan 4000 camera (Gatan) or an FEI Tecnai T20 microscope operated at 200 kV and at a nominal magnification of 80,000× using a TemCam-F816 8k × 8k complementary metal-oxide-semiconductor camera (TVIPS), corresponding to pixel sizes of 2.21 Å and 0.95 Å, respectively, on the specimen.
Grids for cryo-EM were prepared with FEI Mark IV vitrobot. Quantafoil R1.2/1.3 400 mesh holey carbon grids (EMS) were glow-discharged for 30 s. Then, 2.5 μL protein sample at a concentration of 0.6–1.5 mg⋅mL−1 was applied onto the carbon face of the grids. The grids were blotted with Whatman no. 1 filter paper for 2–4 s at 100% humidity and plunge-frozen in liquid ethane. The grids were loaded onto a 300-kV Polara (FEI) with a K2 Summit direct electron detector (Gatan). Data were collected at nominal magnification of 31,000×, corresponding to a physical pixel size of 1.2156 Å (0.6078 Å superresolution pixel size) on the specimen with a dose rate of 7.6 electrons per physical pixel per second. Images were recorded with SerialEM in superresolution counting mode and a defocus range of −0.8 to −2.0 μm. A total exposure of 12 s was used, with 0.2 s subframes (60 total frames) to give a total dose of 60 electrons per Å2 (1.35 electrons per Å2 per subframe). Data for AtTPC1WT in saposin A were also collected at the Janelia Cryo-EM facility FEI Titan Krios microscope operated at 300 kV with a K2 camera with a physical pixel size of 1.02 Å.
Image processing.
For negative-stain data, GCTF (50), Gautomatch (Kai Zhang, https://www.mrc-lmb.cam.ac.uk/kzhang/), and RELION-2 (51) were used for CTF estimation, particle picking, and 2D classification, respectively. For cryo-EM data, dose-fractionated superresolution image stacks were drift-corrected and binned 2 × 2 by Fourier cropping using MotionCor2 (52) (after discarding the first frame). CTF determination and particle picking was performed on motion-corrected sums without dose-weighting using GCTF and Gautomatch. To generate particle picking templates and initial models a Gaussian reference was used to pick particles and 2D classification was performed in RELION-2 followed by ab initio reconstruction in cryoSPARC. Particles were then picked using six 2D classes as templates. Two-dimensional classification, 3D classification, and refinement procedures were carried out using and RELION-2 and cryoSPARC (53) (SI Appendix, Fig. S3). After 3D classification the reconstruction was filtered to 30-Å resolution and used as an initial reference model for 3D refinement in cryoSPARC v2 using the beta version of nonuniform refinement yielding a reconstruction with a gold-standard Fourier shell correlation of 3.5 Å with a 0.143 cutoff criterion. The map from cryoSPARC was used for modeling the Fab variable domain. The final 3.3-Å high-resolution reconstruction was performed in RELION-2 using 3D autorefinement by masking out the saposin A membrane belt and the Fab using a mask created from a 30-Å low-pass-filtered model of AtTPC1WT. Local resolution estimates were performed using RELION-2. The final maps were sharpened in RELION-2 using a B-factor of −104 and −117 Å2 for the TPC1-Fab-saposin and TPC1-only reconstructions, respectively.
States 1 and 2 were identified by focused classification of VSD2 without image alignment using the angles determined from the high-resolution reconstruction (SI Appendix, Fig. S3). Focused classification benefitted from applying C2 symmetry. After 3D classification each particle set was refined in RELION-2 with global angular searches using a 30-Å low–pass-filtered reconstruction of the combined dataset. The maps for states 1 and 2 extend to 3.7 Å as determined by the gold-standard FSC. These maps were used to build atomic models for VSD2 with local resolution in this region ranging from 4 to 6 Å as estimated by RELION-2 (SI Appendix, Table S1). The final maps were sharpened in RELION-2 using a B-factor of −95 and −86 Å2 for the state 1 and state 2 reconstructions, respectively.
Structure determination and refinement.
The entire structure of AtTPC1DDE excluding VSD2 was built into the highest-resolution map (High-res, SI Appendix, Table S1) manually after initial real-space flexible fitting and refinement of the AtTPC1WT structure into the map using Rosetta (54) with the electron scattering table. Manual fitting in Coot (55), followed by real-space coordinate and B-factor/atomic displacement factor refinement in PHENIX (56), was used in de novo building of the NTD, EF-hand, CTD, Ca2+ ions, lipid molecules (palmitic acid and phosphatidic acid), and the Fab–AtTPC1DDE interface. This map was calculated with a mask around TPC1, excluding the Fab. A structure of the Fab complex (Fab-bound, SI Appendix, Table S1) was determined from the same dataset by extending the mask to include the Fab variable domains (VH and VL). The Fab was built using a homologous structure of a Fab from the same library (PDB ID code 3PGF) with the variable CDR loops deleted.
Building of the state 1 (State 1, SI Appendix, Table S1) was accomplished by rigid body alignment of the AtTPC1WT structure to the cryo-EM map using CHIMERA (57) followed by flexible fitting and refinement in Rosetta, and extensive manual fitting in Coot. State 2 (State 2, SI Appendix, Table S1) was built by first rotating VSD2 of AtTPC1WT to place S10. Movement of S7–S9 around the S10 axis was done manually in real space. Gating charges R1–R3 were placed in favored rotamer positions using the rotamer library in Coot and PHENIX, while applying the additional restraint that R1–R3 must contact solvent, a polar side chain, or countercharge in the membrane. One position satisfied these criteria for each gating charge.
Model validation employed MolProbity (58) and EMRinger (59). Low-resolution maps of AtTPC1WT-4B8 were interpreted by flexible fitting of the AtTPC1WT crystal structure and a homology model of 4B8 in real space using Rosetta.
Crystal structure determination.
AtTPC1 D376A (AtTPC1DA) crystals were obtained in the presence of CaCl2, CHS, soy polar lipids, and 1 mM trans-NED19 (NED19), using HiLiDe (60) as described previously (19). Average diffraction was 4 Å, with 10% diffracting to 3.5- to 3.7-Å resolution, the best being 3.5 Å. Anisotropic resolution was determined using the CCP4 program Truncate (61) and the UCLA Anisotropy Server (62). Crystals were partially dehydrated by incubation with additional 15% (vol/vol) of polyethylene glycol 300 before freezing in liquid nitrogen.
X-ray diffraction datasets were collected at the Advanced Light Source (ALS) beamlines 8.3.1 and 5.0.2 and at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2. Data were reduced using XDS (63). The best native dataset extends to overall resolution 3.5 × 6.0 × 4.5 Å (62). Phases were calculated by molecular replacement using PHASER (64) and AtTPC1 wild-type (PDB ID code 5DQQ) as a search model. Structure interpretation was using Coot (55), with refinement in PHENIX (56). The structure of AtTPC1DA was refined to 3.5-Å resolution with final Rwork/Rfree of 31.84% and 35.19%. A sharpening B-factor of −142.05 Å2 was used for refinement, as described previously (19). Analysis by Molprobity shows Ramachandran geometries of 93.09, 6.25, and 0.66% for favored, allowed, and outliers. The structure contains 11.31% rotamer outliers. For comparison of B-factors between AtTPC1WT (mean Wilson B-factor 108 Å2; PDB ID code 5DQQ) (19) and AtTPC1DA (mean Wilson B-factor 109.86 Å2) structures, B-factors were first scaled by adding the difference in mean wilson B-factor (1.86 Å2) to the mean wilson B-factor of the wild-type AtTPC1 crystal structure (PDB ID code 5DQQ) (19).
Sample preparation for CW-EPR.
Concentrated (7–12 mg/mL) NiNTA-purified AtTPC1cysless mutations were incubated with 10-fold molar excess MTSL (Toronto Research Chemicals, Inc) dissolved at 10 mg/mL in DMSO for 2 h at room temperature. The samples were then split in half and treated with either 1 mM CaCl2 or 5 mM EGTA, pH 7.4, before filtration and purification by size-exclusion chromatography in 1 mM CaCl2 or 5 mM EGTA, pH 7.4, respectively. AtTPC1cysless has biochemical behavior identical to AtTPC1WT and maintains structural integrity as evidenced by negative-stain EM analysis. Fractions containing labeled AtTPC1cysless were pooled and concentrated to 5 mg/mL for EPR measurements.
EPR data collection.
For CW-EPR experiments, X-band spectra were collected on a Varian E-102 Century series spectrometer fitted with a loop-gap resonator (Medical Advances). Samples (10 μL) were contained in a 0.64/0.80 mm (i.d./o.d.) quartz capillary (Vitrocom) and spectra were recorded at room temperature over 100 G with an incident microwave power of 2 mW, modulation amplitude of 1 G, and modulation frequency of 100 kHz; typical total scan times were 5 min. CW spectra were normalized and corrected for minute deviations in baseline before comparison and analysis. All data were plotted using Origin 6 after normalizing the area under integrated CW spectrum.
Cryo-EM structural data of AtTPC1DDE have been deposited to the Protein Data Bank under ID codes 6E1K, 6E1M, 6E1N, and 6E1P and the Electron Microscopy Data Bank under entry nos. 8956, 8957, 8958, and 8960. X-ray structural data of AtTPC1DA have been deposited to the Protein Data Bank under ID code 6CX0.
Supplementary Material
Acknowledgments
We thank J. Finer-Moore for aiding with manual building, refinement, and critical reading of the manuscript; A. Brilot, K. Verba, E. Palovcak, and D. Asarnow for help with data processing and refinement; R. Wang for help using Rosetta; D. Bulkley and M. Braunfeld (University of California, San Francisco) and Z. Yu and his colleagues (HHMI Janelia Cryo-EM Facility) for aiding with data collection; and Ali Punjani for a prerelease version of cryoSPARC v2. This work was supported by NIH Grant GM24485 (to R.M.S.); a postdoctoral independent research grant (to A.F.K.) from the University of California, San Francisco Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation; NIH Grants R01GM098672, S10OD020054, and S10OD021741 (to Y.C.), GM117372 and GM087519 (to A.A.K.), and R01EY05216 and P30EY00331; the Jules Stein Professor Endowment; and the Bruce Ford and Anne Smith Bundy Foundation (W.H.). E.M.G. was supported by National Science Foundation Graduate Research Fellowship NSF 1144247. Beamline 8.3.1 at the ALS is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599. The Berkeley Center for Structural Biology is supported in part by NIH, National Institute of General Medical Sciences (NIGMS), and HHMI. The ALS is supported by Contract DE-AC02-05CH11231and the SSRL by Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the US Department of Energy and Grant P41GM103393. Chimera was supported by NIGMS Grant P41-GM103311.
Footnotes
The authors declare no conflict of interest.
Data deposition: Final AtTPC1DDE coordinates and maps have been deposited in the Electron Microscopy Data Bank (EMDB) [entry nos. 8957 (high-resolution), 8956 (Fab), 8958 (state 1), and 8960 (state 2)], and the Protein Data Bank, www.wwpdb.org (PDB ID codes 6E1M, 6E1K, 6E1N, and 6E1P). X-ray structural data of AtTPC1DA have been deposited to the Protein Data Bank under ID code 6CX0.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805651115/-/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 2.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 4.Seoh S-A, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 5.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 6.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandhi CS, Isacoff EY. Molecular models of voltage sensing. J Gen Physiol. 2002;120:455–463. doi: 10.1085/jgp.20028678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 9.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 10.Bezanilla F, Perozo E, Stefani E. Gating of Shaker K+ channels: II. The components of gating currents and a model of channel activation. Biophys J. 1994;66:1011–1021. doi: 10.1016/S0006-3495(94)80882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 13.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 17.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc Natl Acad Sci USA. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kintzer AF, Stroud RM. Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:258–262. doi: 10.1038/nature17194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J, et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:196–201. doi: 10.1038/nature16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kintzer AF, Stroud RM. On the structure and mechanism of two-pore channels. FEBS J. 2018;285:233–243. doi: 10.1111/febs.14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedrich R, Marten I. TPC1-SV channels gain shape. Mol Plant. 2011;4:428–441. doi: 10.1093/mp/ssr017. [DOI] [PubMed] [Google Scholar]
- 23.Patel S. Function and dysfunction of two-pore channels. Sci Signal. 2015;8:re7. doi: 10.1126/scisignal.aab3314. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakurai Y, et al. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cang C, Bekele B, Ren D. The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat Chem Biol. 2014;10:463–469. doi: 10.1038/nchembio.1522. [DOI] [PubMed] [Google Scholar]
- 27.Hedrich R, Neher E. Cytoplasmic calcium regulates voltage-dependent ion channels in plant vacuoles. Nature. 1987;329:833–836. [Google Scholar]
- 28.Beyhl D, et al. The fou2 mutation in the major vacuolar cation channel TPC1 confers tolerance to inhibitory luminal calcium. Plant J. 2009;58:715–723. doi: 10.1111/j.1365-313X.2009.03820.x. [DOI] [PubMed] [Google Scholar]
- 29.Schulze C, Sticht H, Meyerhoff P, Dietrich P. Differential contribution of EF-hands to the Ca2+-dependent activation in the plant two-pore channel TPC1. Plant J. 2011;68:424–432. doi: 10.1111/j.1365-313X.2011.04697.x. [DOI] [PubMed] [Google Scholar]
- 30.Frauenfeld J, et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominik PK, et al. Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure. 2016;24:300–309. doi: 10.1016/j.str.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S, et al. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J, Zeng W, Jiang Y. Tuning the ion selectivity of two-pore channels. Proc Natl Acad Sci USA. 2017;114:1009–1014. doi: 10.1073/pnas.1616191114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catterall WA, Wisedchaisri G, Zheng N. The chemical basis for electrical signaling. Nat Chem Biol. 2017;13:455–463. doi: 10.1038/nchembio.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.She J, et al. Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature. 2018;556:130–134. doi: 10.1038/nature26139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang L, et al. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature. 2016;537:117–121. doi: 10.1038/nature19102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, et al. Ionic liquid-based ultrasound-assisted extraction of fangchinoline and tetrandrine from Stephaniae tetrandrae. J Sep Sci. 2009;32:3550–3554. doi: 10.1002/jssc.200900413. [DOI] [PubMed] [Google Scholar]
- 38.Gutla PVK, Boccaccio A, De Angeli A, Gambale F, Carpaneto A. Modulation of plant TPC channels by polyunsaturated fatty acids. J Exp Bot. 2012;63:6187–6197. doi: 10.1093/jxb/ers272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larisch N, et al. The function of the two-pore channel TPC1 depends on dimerization of its carboxy-terminal helix. Cell Mol Life Sci. 2016;73:2565–2581. doi: 10.1007/s00018-016-2131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez FJD, Orelle C, Davidson AL. Functional reconstitution of an ABC transporter in nanodiscs for use in electron paramagnetic resonance spectroscopy. J Am Chem Soc. 2010;132:9513–9515. doi: 10.1021/ja104047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borowska MT, Dominik PK, Anghel SA, Kossiakoff AA, Keenan RJ. A YidC-like protein in the archaeal plasma membrane. Structure. 2015;23:1715–1724. doi: 10.1016/j.str.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey LJ, et al. Locking the elbow: Improved antibody Fab fragments as chaperones for structure determination. J Mol Biol. 2018;430:337–347. doi: 10.1016/j.jmb.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabat EA, Wu TT, Foeller C, Perry HM, Gottesman KS. Sequences of Proteins of Immunological Interest. Diane Publishing; Collingdale, PA: 1992. [Google Scholar]
- 45.Ritchie TK, et al. 2009. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Liposomes, Part F, Methods in Enzymology, ed Düzgünes N (Academic, New York), pp 211–231.
- 46.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Miller KR, et al. T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS One. 2012;7:e43746. doi: 10.1371/journal.pone.0043746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominik PK, Kossiakoff AA. Phage display selections for affinity reagents to membrane proteins in nanodiscs. Methods Enzymol. 2015;557:219–245. doi: 10.1016/bs.mie.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 49.Fellouse FA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 54.Wang RY-R, et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife. 2016;5:e17219. doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barad BA, et al. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gourdon P, et al. HiLiDe—Systematic approach to membrane protein crystallization in lipid and detergent. Cryst Growth Des. 2011;11:2098–2106. [Google Scholar]
- 61.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 64.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.