Significance
Tuberculosis results in over 1.7 million deaths annually; this problem is worsened by the emergence of drug-resistant strains. New drugs and a better understanding of the mechanisms of existing interventions are needed to prevent the development of drug resistance. Reactive oxygen species (ROS)-mediated lethality by bactericidal drugs was observed in Mycobacterium tuberculosis (Mtb). Our studies demonstrate that arginine deprivation sterilizes for Mtb in vitro and in the host. Using several complementary approaches, we show that arginine deprivation leads to the depletion of thiols, accumulation of ROS, DNA damage, and rapid Mtb death. We propose enzymes in the de novo arginine biosynthesis pathway represent promising drug target candidates to be inhibited to treat Mtb and its drug-resistant strains.
Keywords: l-arginine, reactive oxygen species, oxidative damage, Mycobacterium tuberculosis, bactericidal auxotrophy
Abstract
Reactive oxygen species (ROS)-mediated oxidative stress and DNA damage have recently been recognized as contributing to the efficacy of most bactericidal antibiotics, irrespective of their primary macromolecular targets. Inhibitors of targets involved in both combating oxidative stress as well as being required for in vivo survival may exhibit powerful synergistic action. This study demonstrates that the de novo arginine biosynthetic pathway in Mycobacterium tuberculosis (Mtb) is up-regulated in the early response to the oxidative stress-elevating agent isoniazid or vitamin C. Arginine deprivation rapidly sterilizes the Mtb de novo arginine biosynthesis pathway mutants ΔargB and ΔargF without the emergence of suppressor mutants in vitro as well as in vivo. Transcriptomic and flow cytometry studies of arginine-deprived Mtb have indicated accumulation of ROS and extensive DNA damage. Metabolomics studies following arginine deprivation have revealed that these cells experienced depletion of antioxidant thiols and accumulation of the upstream metabolite substrate of ArgB or ArgF enzymes. ΔargB and ΔargF were unable to scavenge host arginine and were quickly cleared from both immunocompetent and immunocompromised mice. In summary, our investigation revealed in vivo essentiality of the de novo arginine biosynthesis pathway for Mtb and a promising drug target space for combating tuberculosis.
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is a global health problem that causes 1.7 million deaths each year. Long chemotherapy, misdiagnosis, and emergence of multidrug-resistant strains mandate the need to identify novel vulnerable Mtb metabolic pathways to develop more efficient drugs to shorten chemotherapy (1). Moreover, systematic understanding of the adaptive mechanisms used by Mtb to develop resistance to the existing mycobactericidals will help to identify novel therapeutic approaches to prevent sensitive bacteria from developing resistance during treatment (2).
Presently, in both Gram-positive and Gram-negative bacteria, the role of reactive oxygen species (ROS) generated by metabolic perturbations leading to oxidative damage is well understood. Most of the bactericidal antibiotics that target Escherichia coli induce lethality by a common mechanism despite having different primary targets (3–7). This common response involves changes in iron and redox metabolism and accumulation of ROS causing oxidative damage of DNA, RNA, and proteins, ending in cell death (7). ROS-mediated oxidative damage is also a recognized rapid killing mechanism for Mtb (8–13). The mycobactericidals isoniazid (INH) and vitamin C have been shown to induce rapid death of Mtb by ROS generation, leading to oxidative damage (8, 9, 13). Nonetheless, early adaptive responses of Mtb to INH- and vitamin C-induced ROS have yet to be characterized.
Through transcriptional profiling, this study identifies the de novo arginine biosynthesis pathway as one of the early adaptive responses of Mtb to oxidative stress promoting mycobactericidals vitamin C and INH. Since other studies in cancer cells, Burkholderia cenocepacia, and Leishmania donovani have shown that arginine deprivation leads to ROS generation, oxidative damage, and cell death (14–17), we have further investigated the effect of arginine deprivation in Mtb. Using an integrated approach of genetics, transcriptomics, flow cytometry-based analysis, metabolomics, and animal models, our study demonstrates the absolute essentiality of the de novo arginine biosynthesis pathway for Mtb in the absence of pathway intermediates or exogenous arginine. Disruption of the de novo arginine biosynthesis pathway leads to depletion of antioxidant thiols, accumulation of upstream metabolite substrates of the deleted enzymes (ArgB or ArgF) in the arginine biosynthetic pathway, followed by ROS-mediated oxidative damage and rapid sterilization of Mtb populations both in vitro and in vivo.
Results
Arginine Deprivation of Mtb Leads to Rapid Sterilization.
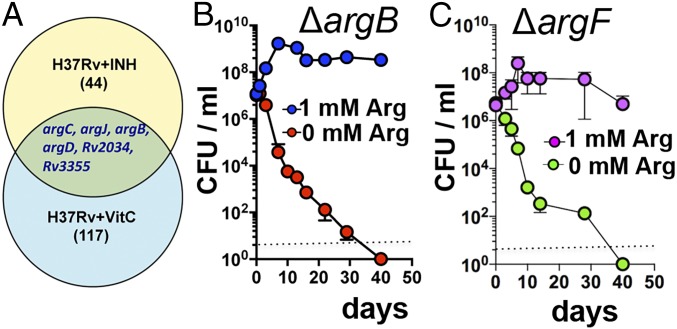
Previous studies have shown that INH and vitamin C kill Mtb in part by inducing oxidative damage (8, 9). Therefore, to identify Mtb’s initial transcriptional signatures as an adaptive response to these mycobactericidals, we performed RNA-Seq on Mtb cultures both untreated and treated with INH (1 µg/mL) or vitamin C (4 mM) for 4 h. Of the six genes up-regulated in response to both of the treatments, four (argB, argC, argD, argJ) are involved in the de novo arginine biosynthesis (Fig. 1A). This result suggests a role for the arginine biosynthesis pathway in tolerating oxidative stress generated by these bactericidal agents. This prompted us to investigate the role of this pathway in Mtb survival. The biosynthetic pathway for de novo biosynthesis of l-arginine in Mtb consists of eight catalytic steps performed by eight enzymes encoded by the genes argA, argB–D, argF–H, and argJ. Using bioinformatics approaches and the results of genetic screens (18), we determined that the gene argB, which encodes an acetylglutamate kinase and the second enzyme in the pathway (SI Appendix, Fig. S1A), is essential to Mtb and lacks a human homolog, thus fulfilling key drug-target criteria. Ornithine carbamoyltransferase, encoded by argF, was additionally chosen for study as it produces the important nonproteinogenic amino acid citrulline (SI Appendix, Fig. S1A). ΔargB and ΔargF mutants, generated using specialized transduction in the clinical isolate CDC1551 and in the laboratory strain H37Rv of Mtb (SI Appendix, Tables S1–S3), were confirmed by PCR (SI Appendix, Fig. S1B). Neither mutant grew in unsupplemented media (SI Appendix, Fig. S1 C and D), but growth was restored completely by the addition of 0.5–1 mM l-arginine or by genetic complementation (SI Appendix, Fig. S1 C–E and Tables S1 and S2). Supplementation with the arginine pathway intermediates l-ornithine or l-citrulline restored ∆argB growth in the absence of l-arginine (SI Appendix, Fig. S2A). We further tested whether arginine starvation is bactericidal or bacteriostatic for Mtb CDC1551. Arginine deprivation remarkably caused a greater than 4-log decrease in ΔargB and ΔargF colony-forming units (cfu) within day 10, whereas when other auxotrophs were deprived of their respective nutrients, the effect was bacteriostasis (∆leuCD, leucine; ∆panCD, pantothenate) or a more limited ∼3-log decrease for a bactericidal auxotrophy (∆metA, methionine) (SI Appendix, Fig. S2B). The rapid killing of ΔargB and ΔargF was independent of strain background, as we observed the same rapid death with ΔargB and ΔargF mutants of H37Rv; no cfu were isolated for ∆argB, indicating complete sterilization by day 20 or 40, depending on the initial number of Mtb cells used, 106 or 107 cfu/mL, respectively (Fig. 1B and SI Appendix, Fig. S2C). Similarly, no cfu were isolated for ∆argF after day 40 of the arginine starvation (Fig. 1C). Addition of exogenous l-arginine to the arginine-deprived cultures at day 40 was unable to rescue the ΔargB mutant (SI Appendix, Fig. S2D), suggesting a lack of persistent viable cell populations to resume growth. No suppressor mutants were isolated after plating 109 cells of ∆argB or ∆argF in three independent experiments. Thus, using two independent mutants in the de novo arginine biosynthetic pathway, we found that arginine deprivation leads to rapid sterilization of ΔargB and ΔargF in vitro without the emergence of suppressor mutants.
Fig. 1.
Deprivation of l-arginine sterilizes pathogenic Mtb. (A) Venn diagram showing up-regulation of de novo arginine biosynthesis in Mtb cells treated with 1 µg/mL INH or 4 mM vitamin C (Vit C) for 4 h. Starvation-death response of Mtb ∆argB (B) and ∆argF (C) ±1 mM l-arginine. Aliquots were plated on l-arginine–containing plates for cfu determination. The dotted line represents the limit of detection. Data are representative of one of three independent experiments. Error bars show mean ± SD (n = 3).
Transcriptional Profiling and Flow Cytometry Analysis of Arginine-Deprived Mtb Reveal Envelope and Oxidative Stresses, ROS Accumulation, and DNA Fragmentation.
We have demonstrated early up-regulation of the de novo arginine biosynthesis pathway by Mtb as an adaptive response to oxidative stress elevating the mycobactericidals INH or vitamin C. Furthermore, argB or argF deletion leads to sterilization of Mtb under arginine deprivation. Therefore, we conducted time-course transcriptomics and flow cytometry studies on arginine-deprived ∆argB and ∆argF mutants to investigate the mechanisms underlying their rapid death.
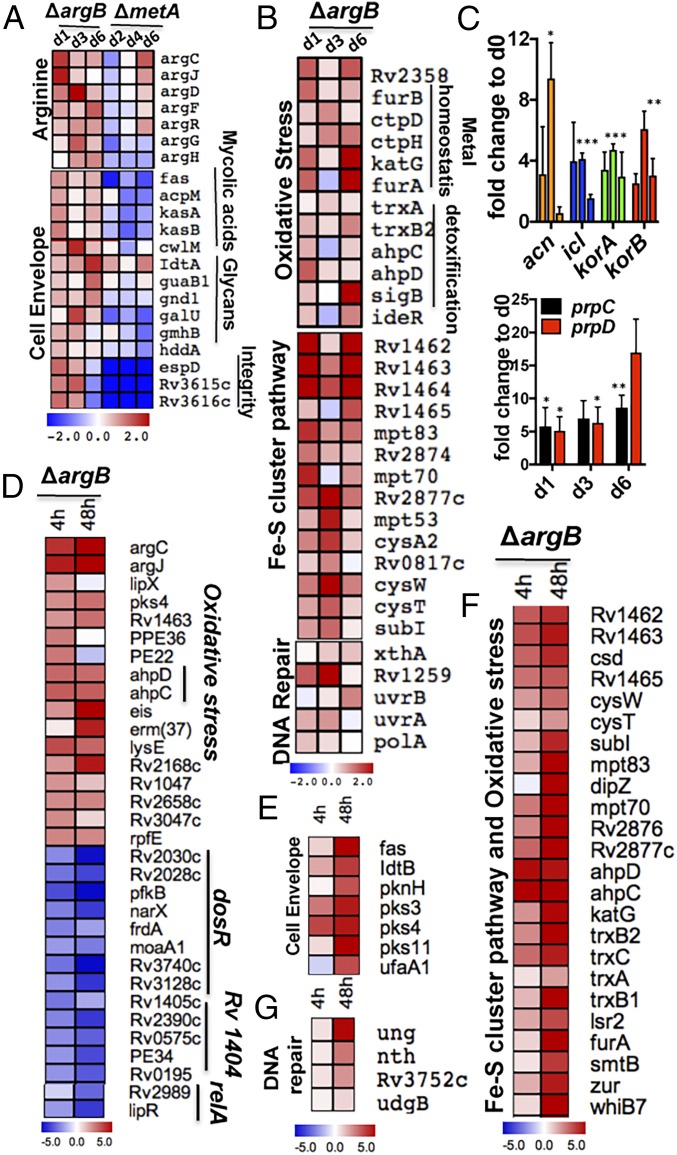
Transcriptomic microarrays analysis of arginine-deprived ∆argB (SI Appendix, Fig. S3 A and B) indicated a more than twofold increase in expression of genes for arginine biosynthesis (Fig. 2A), mycothiol, and the ergothioneine precursor histidine (SI Appendix, Fig. S3C). In contrast to the response of methionine-starved ∆metA, the only reported Mtb bactericidal amino acid auxotrophy, arginine-starved ∆argB at day 1, displayed induction of genes involved in cell-envelope stress and remodeling (including genes of the FAS II pathway); synthesis of mycolic acids, phospholipids, and glycans; and maintenance of membrane integrity as seen during INH treatment of Mtb (Fig. 2A) (19). Mtb genes involved in oxidative damage response, Fe-S cluster synthesis, as well as assembly (Fig. 2B) and Fe-S cluster-dependent essential enzymes (SI Appendix, Fig. S3D), were highly up-regulated (20). The bioenergetically less efficient microaerobic respiration pathway (SI Appendix, Fig. S3E) and the anaplerotic genes involved in an antioxidant defense to antibiotic stress (Fig. 2C) were also highly expressed (12, 21, 22). Increased ROS causes DNA damage, and there was a consequent up-regulation of genes involved in DNA repair (Fig. 2B) (3, 19). Increased expression of genes for arginine biosynthesis, antioxidant defense, and DNA repair genes was also observed in ∆argF (SI Appendix, Fig. S4 A–C), whereas the respiratory genes nuoA–N were down-regulated in both ∆argB and ∆argF (SI Appendix, Figs. S3F and S4D).
Fig. 2.
Transcriptomic analysis of arginine-deprived Mtb ∆argB revealed signatures of cell-envelope stress and oxidative damage. To compare time-course changes in transcriptomic profiles of l-arginine–deprived H37Rv and ΔargB, RNA was isolated from samples harvested at 0, 1, 3, and 6 d for microarrays (A–C) and 0, 4, and 48 h for RNA-Seq (D–G). (A) Enhanced expression of genes involved in arginine biosynthesis and cell-envelope maintenance. (B) Up-regulation of genes associated with antioxidant defense, Fe-S cluster biosynthesis, and DNA repair. (C) Enhanced expression of anaplerotic genes (containing Fe-S) involved in antioxidant defense during antibiotic treatment (12). (D–G) RNA-Seq data of arginine-deprived Mtb ∆argB. (D) At 4 h of arginine starvation, genes involved in arginine biosynthesis, cell-envelope remodeling, antioxidative response, and heme and sulfur import were highly up-regulated. Conversely, dosR-, Rv1401-, and relA- regulated genes were down-regulated. (E–G) Up-regulation of genes involved in cell-envelope remodeling (E), Fe-S cluster biogenesis assembly and antioxidative defense (F), and DNA repair (G) at 48 h. Raw data for microarrays were deposited in the GEO database (accession no. GSE98821). All transcriptomic heat maps show log twofold changes. Mean with SD is plotted; error bars represent the SD of three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t test.
To extend our study of arginine-deprived ∆argB, we also performed RNA-Seq analysis following 4 and 48 h of arginine starvation. Consistent with the microarrays, genes involved in arginine biosynthesis were four- to sixfold up-regulated within 4 h of starvation. Significantly expressed genes (two- to fivefold) at 4 h included genes in cell-envelope remodeling (lipX, pks4) (23), sulfur import (Rv1463), heme binding at the cell surface for iron import PE22-PPE36 (24), and antioxidative response (ahpC and ahpD; Fig. 2D) (25). This was followed by a more than twofold enhanced expression of genes involved in the biosynthesis of thiols (mycothiol and the precursor of ergothioneine, histidine) and anaplerotic genes required for antioxidant defense against antibiotic stress at 48 h (SI Appendix, Fig. S3G). Genes involved in cell-envelope remodeling, antioxidant stress response, and DNA damage (ung) were also highly up-regulated at 48 h (Fig. 2 E–G). In contrast, at 4 and 48 h, dosR-regulated genes involved in hypoxia, Rv1404-regulated genes for a response against acid stress, and the relA regulon genes were three- to fourfold down-regulated (Fig. 2D) (25).
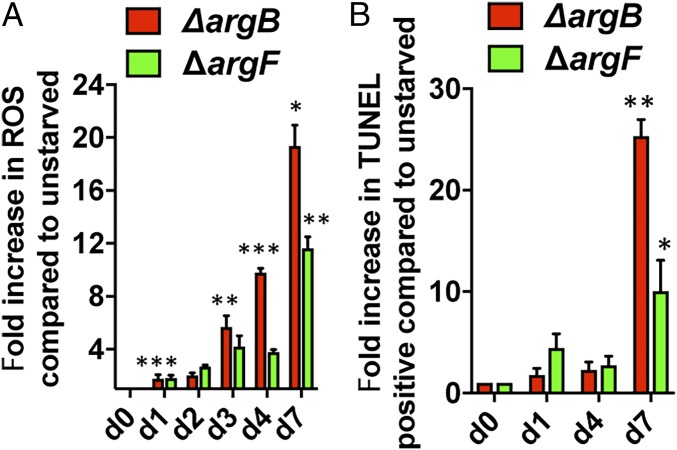
Our transcriptomics data on arginine-deprived ΔargB and ΔargF revealed signatures of oxidative stress, as seen in INH or vitamin C treatment of Mtb (9, 19). Therefore, we measured the accumulation of ROS in l-arginine–starved ΔargB and ΔargF by flow cytometry using fluorescent dihydroethidium dye and DNA damage using the terminal deoxynucleotidyl transferase-mediated dUTP-fluorescein nick-end labeling assay (TUNEL). The gating strategy used for both the assays is depicted in SI Appendix, Fig. S5. At day 1, we observed an approximately twofold increase in ROS in arginine-deprived ΔargB and ΔargF cultures compared with the arginine-supplemented cultures. In arginine-deprived ΔargB, from days 3–7, the ROS signal increased from approximately 6- to 20-fold over the nondeprived (Fig. 3A). In ΔargF, a slower increase of up to 12-fold at day 7 was observed (Fig. 3A). DNA damage in arginine-deprived cells correlated well with ROS accumulation and increased ∼25-fold (ΔargB) or ∼12-fold (ΔargF) compared with nondeprived cultures at day 7 (Fig. 3B).
Fig. 3.
Arginine deprivation leads to ROS accumulation and DNA damage. ∆argB and ∆argF were grown in l-arginine–supplemented 7H9 medium, washed, and transferred to l-arginine–deprived 7H9 medium. At the indicated time points, cell pellets were collected and stained with (A) dihydroethidium for ROS measurement or (B) TUNEL for measuring DNA double-stranded breaks using flow cytometry. Results were compared with nondeprived or untreated control samples. Mean ± SD is plotted (n = 3). ***P < 0.001, **P < 0.01, *P < 0.05; Student’s t test (two-tailed).
To determine whether arginine deprivation enhances sensitivity to external ROS-mediated oxidative stress and DNA damage, we exposed day 1 arginine-starved ΔargB cells to 16 mM H2O2 for 4 h. An ΔmshA mutant deficient in mycothiol biosynthesis and hypersensitive to oxidative stress was used as a control (26). Within 4 h, we found approximately fivefold more ROS accumulation in ΔargB and ΔmshA than in H37Rv, although a decrease in ROS signal at later time points (days 2–3) was possibly due to nonviability of cells (SI Appendix, Fig. S6A). ROS production correlated well with increased DNA double-strand breaks. Within 4 h, H2O2 caused approximately twofold more DNA damage to ΔargB than to H37Rv (SI Appendix, Fig. S6B). To assess whether arginine-deprived Mtb is more sensitive to external ROS-mediated cell death, arginine-deprived ΔargB and ΔargF were treated at day 1 with 16 mM H2O2 for 4 h and plated to determine cfu on arginine-containing plates. The arginine-deprived mutants were highly sensitive and vulnerable to oxidative damage, as indicated by the approximately fivefold decreases in cfu within 4 h, but genetic complementation restored H2O2 sensitivity to H37Rv levels (SI Appendix, Fig. S6C).
To further test whether ROS has a role in the rapid killing of arginine-deprived ΔargB, cultures were starved for arginine in an hypoxic chamber containing 1% O2 and 5% CO2. The killing kinetics of arginine-starved ΔargB was reduced under hypoxic conditions (SI Appendix, Fig. S6D), suggesting a role for oxygen in the sterilization mechanism of arginine deprivation. Overall, our transcriptomics and flow cytometry data showed that arginine-deprived Mtb induces pleiotropic stress responses involving cell-envelope stress and ROS accumulation leading to DNA fragmentation and cell death.
Metabolomic Studies of Arginine-Deprived Mtb Show the Accumulation of Upstream Metabolites and Depletion of Antioxidant Thiols.
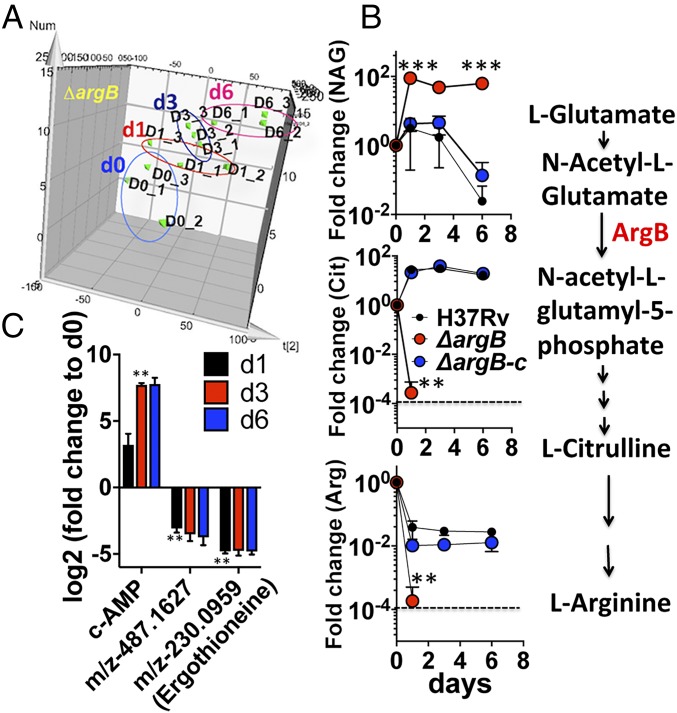
To investigate the upstream molecular events underlying the ROS-mediated oxidative damage and cell death of arginine-deprived ∆argB and ∆argF, we conducted time-course metabolomics during arginine deprivation. Mutants and their complemented strains were grown in l-arginine–supplemented broth, washed, and then transferred to the arginine-free broth. Cells were harvested at days 0, 1, 3, and 6, and extracted metabolites were subjected to ultra-performance liquid chromatography mass spectrometry (UPLC-MS). Principal components analysis of m/z retention time pairs showed changes in the ∆argB metabolome over time (Fig. 4A), with substantial accumulation of the ArgB substrate N-acetyl-l-glutamate (∼100-fold, Fig. 4B). The downstream metabolites, l-citrulline and l-arginine, decreased ∼1,000-fold at day 1 and were later undetectable (Fig. 4B). At day 3, levels of ergothioneine and a compound with m/z 487.16 (consistent with mycothiol), which have roles in redox homeostasis (27), were decreased ∼20-fold, whereas the secondary messenger cAMP accumulated by ∼150-fold (Fig. 4C). In ∆argF, N-acetyl ornithine levels increased, and l-citrulline increased transiently before returning to initial levels while l-arginosuccinate and l-arginine were depleted below the detection limit after day 1 (SI Appendix, Fig. S4E). These results confirmed that arginine deprivation of Mtb leads to accumulation of upstream metabolite substrates of the deleted enzymes ArgB or ArgF and depletion of l-arginine, l-citrulline, and antioxidant thiols.
Fig. 4.
Metabolomic analysis of arginine-deprived Mtb ∆argB. Time-course changes in metabolic profiles of H37Rv, ΔargB, and ΔargB-c were compared during 6 d of l-arginine deprivation. Samples were harvested at 0, 1, 3, and 6 d from three biologically independent replicates for extraction of metabolites. Aqueous-phase metabolites were measured by UPLC-MS, and fold changes in metabolite abundance were calculated relative to time 0. (A) Principal component analysis showed changes in the overall metabolomic profile of ∆argB over time during arginine deprivation. (B) Upstream metabolite N-acetyl glutamate (NAG), the substrate for ArgB, accumulated while the downstream metabolites l-citrulline and l-arginine were depleted. (C) Accumulation of cAMP and depletion of antioxidant thiols (ergothioneine and a compound with m/z 487.16, consistent with mycothiol). Error bars represent SD of three biological replicates. Dashed lines represent the limit of detection.
Arginine Deprivation Sterilizes Mtb in Immunocompetent and Immunocompromised Mice.
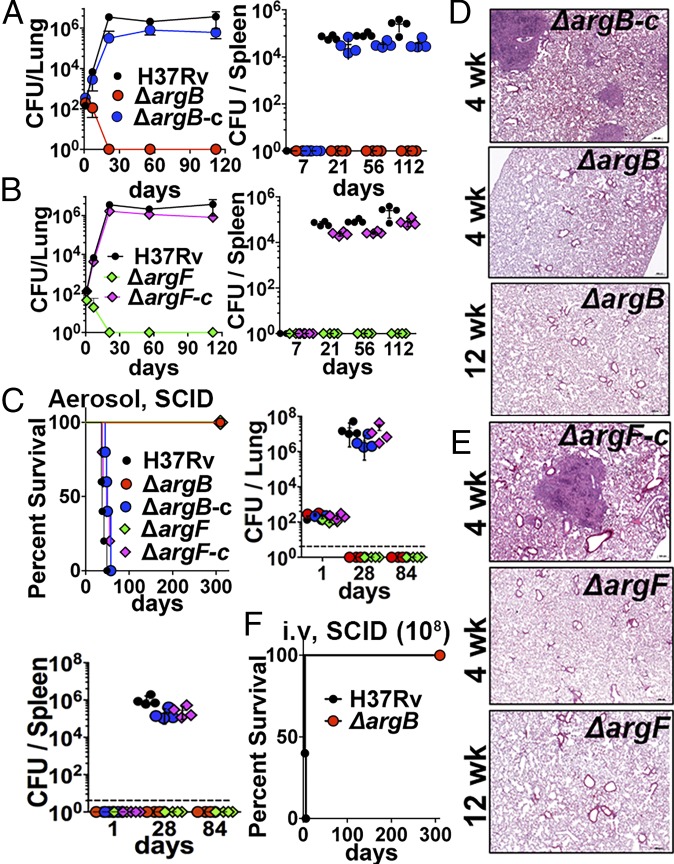
Our previous results demonstrated that arginine deprivation leads to the rapid death of Mtb in vitro. Arginine concentration in mouse plasma is ∼200 μM (28). Therefore, to determine whether Mtb can acquire arginine or pathway intermediates (ornithine and citrulline) in vivo, we tested the growth of the arginine auxotrophs ∆argB and ∆argF in immunocompetent C57BL/6 mice and in Severe Combined Immune-Deficient (SCID) mice. Following low-dose aerosol infection, both ∆argB and ∆argF were sterilized within 21 d in lungs of C57BL/6 mice (Fig. 5 A and B, Left) and were unable to disseminate to the spleen (Fig. 5 A and B, Right). Additionally, there was no sign of inflammation in the lungs at 4 wk postinfection (SI Appendix, Fig. S7A). In comparison, H37Rv and complemented strains (∆argB-c and ∆argF-c) had increased burdens by 3 wk postinfection, with 104–106 cfu detected in lungs and spleen and gross pathology showing granulomatous lung disease (Fig. 5 A and B and SI Appendix, Fig. S7A). The rapid death phenotype of ∆argB and ∆argF was also observed in the lungs (Fig. 5C, Upper Right) and spleens (Fig. 5C, Lower) of aerosol-infected SCID mice, whereas H37Rv and complemented strains grew unimpeded, causing 100% mortality by 7–8 wk postinfection (Fig. 5C). There was no sign of disease pathology or acid-fast bacilli (AFB) in the lungs of SCID mice infected with ∆argB (Fig. 5D and SI Appendix, Fig. S7B and Table S4) or ∆argF (Fig. 5E and SI Appendix, Fig. S7C and Table S4). Conversely, mouse lungs infected with H37Rv or complemented Mtb strains exhibited severe lesions and tissue inflammatory infiltrates, primarily granulomas that obscured normal pulmonary architecture and that were often admixed with necrosis and associated with large numbers of AFB (Fig. 5 D and E, Upper; SI Appendix, Fig. S7 B and C, Left; and SI Appendix, Table S4).
Fig. 5.
Mtb arginine auxotrophic mutants are rapidly sterilized in immunocompetent (C57BL/6) and immunocompromised (SCID) mice. Mice were infected with low-dose aerosol (100 bacilli per animal) of strains ∆argB, ∆argB-c ∆argF, ∆argF-c, and H37Rv. Lung and spleen burdens were determined at days 1, 7, 21, 56, and 112 postinfection for C57BL/6 or at days 1, 28, and 84 postinfection for SCID. (A and B) The cfu in the lung (Left) and spleen (Right) of C57BL/6 mice. (C) Survival of SCID mice (n = 6) after low-dose aerosol infection (Left); cfu burden in lung and spleen (Upper Right and Lower, n = 4). (D and E) Lung histopathology of SCID mice. (F) Survival of SCID mice (n = 6) after high-dose (108 bacilli per animal) i.v. infection. One of two independent experiments is shown. Data are mean ± SD for n = 4 biological replicates. Dashed lines represent the limit of detection.
Interestingly, ∆argB is so severely attenuated that even after i.v. infection with ∼106 or 108 cfu, 100% of SCID mice were alive after 300 d, whereas those infected with H37Rv (108 cfu) died within 6 d (Fig. 5F and SI Appendix, Fig. S7D). Moreover, no mutant colonies or suppressors were recovered from lungs or spleens of SCID mice infected with ∆argB at 106 cfu (i.v.) even at 57 wk postinfection (SI Appendix, Fig. S7D). Together, these results establish that arginine deprivation causes rapid sterilization of Mtb cells in vivo without the emergence of any suppressor mutants and independently of the immune status of the host. These results suggest that Mtb is restricted to host niches where it cannot scavenge arginine or intermediates in the pathway, potentially due to restricted availability or defective recruitment of arginine transport systems to the phagosome, thus leading to its rapid death.
Discussion
Shortening TB chemotherapy requires novel drugs that can rapidly sterilize Mtb. Our study provides compelling evidence that the arginine biosynthetic pathway is an attractive target for antitubercular drug development. Arginine deprivation leads to ROS accumulation, DNA damage, and rapid sterilization of Mtb.
ROS-mediated oxidative damage is a potent killing mechanism by bactericidal antibiotics for Gram-positive and Gram-negative bacteria, as well as Mtb (3–12). Our RNA-Seq data from Mtb treated with INH or vitamin C at 4 h indicated early up-regulation of de novo arginine biosynthesis. An increase in metabolites in the arginine biosynthesis pathway of Mtb at 24 h in monotherapy with the mycobactericidals INH, rifampicin, or streptomycin has been reported (12). Therefore, we investigated the role of the de novo arginine biosynthesis pathway in Mtb.
Arginine deprivation leads to a rapid sterilization of Mtb both in vitro and in vivo. Furthermore, no suppressor mutants were obtained when mice were challenged with ΔargB (108 cfu) or in three independent experiments on Middlebrook 7H9 media supplemented with oleic acid–albumin–dextrose–catalase OADC and glycerol up to 6 mo. While suppressors of complete gene deletions are very rare but could occur by biochemical bypass as has been observed for diaminopimelate auxotroph (29), these two sets of data suggest a multifactorial mechanism of death. Several potential mechanisms are possible such as futile cycling to generate precursors for arginine biosynthesis leading to enhanced ROS generation as observed in arginine-deprived cancer cells (14, 15). Our metabolomics studies showed an accumulation of N-acetyl glutamate and N-acetyl ornithine, an upstream metabolite substrate for ArgB or ArgF (Fig. 4 and SI Appendix, Fig. S4E). These intermediates are derived from l-glutamate, a precursor for l-arginine biosynthesis. This indicates that arginine-deprived Mtb is undergoing futile cycles to generate either l-glutamate from α-ketoglutarate or by glutamine. Both conditions are accompanied by increased flux into the tricarboxylic acid cycle and oxidative phosphorylation followed by ROS generation, DNA damage, and cell death. Another potential explanation is altered protein synthesis and secretion due to the indispensability of arginine for the twin-arginine transport (TAT) secretion signal motif “RR.” TAT effectors are involved in respiration, nutrition uptake, and envelope maintenance (30–32). Defects in the TAT system are associated with cell death and envelope and oxidative stress susceptibility in many bacterial pathogens including Mtb (30, 31). Transcriptomics reveals that arginine-deprived Mtb exhibited up-regulation of signatures of envelope stress as seen during INH exposure (19) (Fig. 2A). Arginine is also the source of polyamines involved in scavenging ROS and protects membranes from lipid peroxidation in plants, bacteria, and parasites (16, 17, 33); similar mechanisms may be involved in Mtb. At day 3 of arginine deprivation, an ∼20-fold decrease was observed in the antioxidant thiols, ergothioneine, and a compound with an m/z 487.16, which corresponds to mycothiol (Fig. 4C). However, we have not detected the polyamines putrescine and agmatine in any sample possibly due to their lower abundance. We observed an ∼150-fold increase in accumulation of cAMP in arginine-deprived Mtb (Fig. 4C) in contrast to the observed depletion of cAMP in wild-type cells starved in PBS (34). The possibility of the accumulation of toxic metabolites or loss of an arginine-dependent essential metabolite contributing to killing was not ruled out, but we were unable to identify such metabolites. We propose a model in which futile cycles for generation of precursors for arginine biosynthesis, envelope stress, defects in protein synthesis and secretion, depleted thiols, or other undetermined factors initiate immediate and extreme oxidative stress, causing oxidative damage leading to multitargeted rapid Mtb death (SI Appendix, Fig. S8).
Much of Mtb’s success may stem from being self-sufficient as it has conserved biosynthesis pathways for most of the essential nutrients. While several Mtb amino acid auxotrophs are attenuated in vivo, only methionine deprivation has been reported to be bactericidal (35–40). In our study, high doses of ΔargB (106–108 cells) or ΔargF (106 cells) delivered via the i.v. route revealed that these arginine biosynthesis mutants were unable to replicate in SCID mice and were rapidly sterilized (Fig. 5). These results were somewhat unexpected, as Mtb has two arginine transporters (41) and serum concentrations of arginine are substantial (28). The inability of host-derived arginine to compensate for arginine auxotrophy reflects either inaccessibility of arginine in Mtb niches or inefficiency in expression, function, or translocation of import systems in vivo. Additionally, our results contrast with those using an Mtb argF mutant, which could acquire arginine, replicate in, and kill SCID mice (42). However, the above-mentioned study lacked an argF-complemented control strain, and the possibility of contamination with the parental or single cross-over strain during mouse infection cannot be ruled out.
This study underlines the critical sterilization role of the de novo arginine biosynthesis pathway for the survival of Mtb under arginine deprivation. This work suggests that the inhibitors against enzymes in this pathway have potential to sterilize Mtb and therefore can prevent or kill persistent populations. An absence of human homologs for many enzymes in the arginine biosynthetic pathway and lack of detection of arginine or its metabolites in NMR studies of lung granulomas of Mtb-infected guinea pigs (43) further reinforce the de novo arginine biosynthesis pathway as an excellent anti-TB drug target. These strains will provide a useful tool for screening novel inhibitors against enzymes in the arginine biosynthesis pathway using target-based approaches as whole-cell screening or new approaches as fragment-based drug discovery (44). Promising candidates that cocrystalize with enzymes in the de novo arginine biosynthesis pathway and inhibit its enzymatic activity could be tested for their potency and specificity in vitro and in vivo against Mtb. These strains are also a valuable tool to screen for compounds that act in synergy with arginine deprivation to enhance ROS-mediated Mtb cell death. Additionally, further insights into the common death mechanisms involved in arginine deprivation and other Mtb sterilization phenotypes will open avenues for drug screening to develop antibacterial strategies and interventions against Mtb.
Methods
See SI Appendix, SI Methods, for method details.
Bacterial Strains and Culture Conditions.
All of the primers, plasmids, cosmids, and bacterial strains used in the study are listed in SI Appendix, Tables S1–S3. Mtb strains were grown at 37 °C in Middlebrook 7H9 liquid medium or 7H10 solid media supplemented with 10% OADC, 0.5% glycerol, and 0.05% tyloxapol. For Mtb strains bearing antibiotic cassettes, hygromycin (75 μg/mL) and/or kanamycin (20 μg/mL) were added wherever required. The l-arginine (1 mM) was added to liquid or solid media as described above. For the time-course starvation experiments, samples were grown in the presence of 1 mM l-arginine to 0.5 OD600, washed with PBS tyloxapol (PBST), and switched to arginine-free medium. Mtb mutants were constructed by specialized transduction, unmarked and complemented as mentioned in SI Appendix.
RNA Isolation, Microarrays, and RNA-Seq.
Samples were prepared in triplicate from cultures of H37Rv, ΔargB, and ΔargF for microarray analysis at days 0, 1, 3, and 6 and for RNA-Seq at 0, 4, and 48 h after starvation. RNA isolation, cDNA preparation, and microarray analysis were performed as described before (9). All of the data have been deposited at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE98821. For RNA-seq, libraries were prepared and checked for quality on a 2100 Bioanalyzer. Sequencing of 75-bp paired-end reads was performed on the Illumina HiSEq. 4000 platform followed by differential expression analyses, normalization of raw counts, and generation of heat maps. Details can be found in SI Appendix.
Flow Cytometry-Based ROS Detection and DNA Fragmentation Assays.
Mtb strains were grown in 7H9 media with or without l-arginine to OD600 ∼ 0.8. Cultures were pelleted and washed five times with PBST and diluted to OD600 ∼ 0.2 in fresh media with or without l-arginine or H2O2. Aliquots were removed at indicated time points, and cells were washed twice in PBS and stained with dihydroethidium for 30 min at 37 °C. Cells were analyzed immediately on a BD FACS Calibur (BD Biosciences) with the following instrument settings: forward scatter (FSC), EO2 log gain; side scatter, 550V log; fluorescence (FL1), 674V log; fluorescence (FL2), 628V log and threshold set on SSC (236V). For each sample, 10,000 events were acquired. The analysis was performed by using unlabeled and untreated/treated cells as negative controls and gating intact cells. DNA strand breaks in Mtb strains undergoing starvation or H2O2 treatment were measured by TUNEL assay using the in situ cell death detection kit (Roche). Additional details are given in SI Appendix.
Metabolite Extraction and Mass Spectroscopy.
Bacterial strains were grown in l-arginine media to OD600 (∼0.5–0.6), washed, and transferred to starvation media. The 5-mL samples (OD600 of 0.5) were quickly quenched in 10 mL methanol at −20 °C. Samples were centrifuged at 3,300 × g/10 min/−9 °C. Cell pellets were suspended in 1 mL extraction solvent [40% acetonitrile (vol/vol), 40% methanol (vol/vol), and 20% water (vol/vol)] and transferred to screw-cap tubes containing silica beads. Cells were lysed and processed, and samples collected at various time points (days 0, 1, 3, and 6) were analyzed using an Acquity UPLC system (Waters) coupled with a Synapt G2 quadrupole-time-of-flight hybrid mass spectrometer. Method details, metabolite abundances, retention time, and m/z values can be found in SI Appendix, SI Methods and Dataset S1.
Animal Experiments and Pathology.
All animal protocols used were approved by the Animal Use and Care Committee of the Albert Einstein College of Medicine (Bronx, NY). The 6- to 8-wk-old C57BL/6 and SCID mice (Jackson Laboratories) were infected with Mtb and various mutants via the aerosol route using a 1 × 107 cfu/mL mycobacterial suspension in PBST and 0.004% antifoam. Infection yielded ∼100 bacilli per lung. To determine bacterial burden, four mice were killed at various mentioned time points. Lungs and spleens were harvested and plated to determine cfu. For SCID i.v. infections, strains grown to midlog phase were washed and injected via tail vein (106 or 108 cfu/mouse). Lung tissues were fixed in 10% (vol/vol) formalin for 48–72 h, followed by paraffin-embedded sectioning and H&E or acid-fast staining. Five or six mice per group were kept for survival experiments. All mice infected with Mtb were maintained under appropriate conditions in an animal biosafety level 3 laboratory. All experiments were repeated to confirm the reproducibility.
Supplementary Material
Acknowledgments
We thank Regy Lukose, Annie Zhi Dai and Laura Cole for technical support, Core Facilities at Albert Einstein College of Medicine; Dr. Amanda Beck for histopathology; Dr. Paras Jain and Dr. Lydia Tesfa (Grant P30CA013330) for flow cytometry. We thank students Lara Kleinfelter, Karen Giraldo and Humira Ferdush for their help in different aspects of paper. This work was supported by a Human Challenge grant from the Bill and Melinda Gates Foundation, a grant from the National Institutes of Health (R01AI026170) and Wellcome Trust Grant 098051.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray raw data have been deposited in the Gene Expression Omnibus (GEO) database at https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98821).
See Commentary on page 9658.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808874115/-/DCSupplemental.
References
- 1.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 2.Levin-Reisman I, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 3.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belenky P, et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasouly A, Nudler E. Antibiotic killing through oxidized nucleotides. Proc Natl Acad Sci USA. 2018;115:1967–1969. doi: 10.1073/pnas.1800255115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhandayuthapani S, Zhang Y, Mudd MH, Deretic V. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: Characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol. 1996;178:3641–3649. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilchèze C, Hartman T, Weinrick B, Jacobs WR., Jr Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun. 2013;4:1881–1891. doi: 10.1038/ncomms2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilchèze C, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114:4495–4500. doi: 10.1073/pnas.1704376114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan XY, et al. Oxidation of dCTP contributes to antibiotic lethality in stationary-phase mycobacteria. Proc Natl Acad Sci USA. 2018;115:2210–2215. doi: 10.1073/pnas.1719627115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandakumar M, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 13.Middlebrook G, Cohn ML, Schaefer WB. Studies on isoniazid and tubercle bacilli. III. The isolation, drug-susceptibility, and catalase-testing of tubercle bacilli from isoniazid-treated patients. Am Rev Tuberc. 1954;70:852–872. doi: 10.1164/art.1954.70.5.852. [DOI] [PubMed] [Google Scholar]
- 14.Qiu F, et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci Signal. 2014;7:ra31. doi: 10.1126/scisignal.2004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer JC, et al. Arginine deprivation inhibits the Warburg effect and upregulates glutamine anaplerosis and serine biosynthesis in ASS1-deficient cancers. Cell Rep. 2017;18:991–1004. doi: 10.1016/j.celrep.2016.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Halfawy OM, Valvano MA. Putrescine reduces antibiotic-induced oxidative stress as a mechanism of modulation of antibiotic resistance in Burkholderia cenocepacia. Antimicrob Agents Chemother. 2014;58:4162–4171. doi: 10.1128/AAC.02649-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal A, et al. Deprivation of L-arginine induces oxidative stress mediated apoptosis in Leishmania donovani promastigotes: Contribution of the polyamine pathway. PLoS Negl Trop Dis. 2016;10:e0004373. doi: 10.1371/journal.pntd.0004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 19.Boshoff HI, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 20.Huet G, Daffé M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: Evidence for its implication in the pathogen’s survival. J Bacteriol. 2005;187:6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsoso LG, et al. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J Bacteriol. 2005;187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubey VS, Sirakova TD, Kolattukudy PE. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol. 2002;45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- 24.Mitra A, Speer A, Lin K, Ehrt S, Niederweis M. PPE surface proteins are required for heme utilization by Mycobacterium tuberculosis. MBio. 2017;8:1–14. doi: 10.1128/mBio.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cumming BM, et al. The physiology and genetics of oxidative stress in mycobacteria. Microbiol Spectr. 2014;2:1–22. doi: 10.1128/microbiolspec.MGM2-0019-2013. [DOI] [PubMed] [Google Scholar]
- 26.Vilchèze C, et al. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol. 2008;69:1316–1329. doi: 10.1111/j.1365-2958.2008.06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini V, et al. Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep. 2016;14:572–585. doi: 10.1016/j.celrep.2015.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gobert AP, et al. L-Arginine availability modulates local nitric oxide production and parasite killing in experimental trypanosomiasis. Infect Immun. 2000;68:4653–4657. doi: 10.1128/iai.68.8.4653-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consaul SA, Jacobs WR, Jr, Pavelka MS., Jr Extragenic suppression of the requirement for diaminopimelate in diaminopimelate auxotrophs of Mycobacterium smegmatis. FEMS Microbiol Lett. 2003;225:131–135. doi: 10.1016/S0378-1097(03)00502-0. [DOI] [PubMed] [Google Scholar]
- 30.Sargent F, Berks BC, Palmer T. Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch Microbiol. 2002;178:77–84. doi: 10.1007/s00203-002-0434-2. [DOI] [PubMed] [Google Scholar]
- 31.McDonough JA, et al. Identification of functional Tat signal sequences in Mycobacterium tuberculosis proteins. J Bacteriol. 2008;190:6428–6438. doi: 10.1128/JB.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oertel D, Schmitz S, Freudl R. A TatABC-type Tat translocase is required for unimpaired aerobic growth of Corynebacterium glutamicum ATCC13032. PLoS One. 2015;10:e0123413. doi: 10.1371/journal.pone.0123413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha J, et al. Polyamines as redox homeostasis regulators during salt stress in plants. Front Environ Sci. 2015;3:21. [Google Scholar]
- 34.Kahramanoglou C, et al. Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: Relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42:8320–8329. doi: 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YJ, Rubin EJ. Feast or famine: The host-pathogen battle over amino acids. Cell Microbiol. 2013;15:1079–1087. doi: 10.1111/cmi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouzy A, Poquet Y, Neyrolles O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat Rev Microbiol. 2014;12:729–737. doi: 10.1038/nrmicro3349. [DOI] [PubMed] [Google Scholar]
- 37.Hondalus MK, et al. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect Immun. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, et al. Protection elicited by two glutamine auxotrophs of Mycobacterium tuberculosis and in vivo growth phenotypes of the four unique glutamine synthetase mutants in a murine model. Infect Immun. 2006;74:6491–6495. doi: 10.1128/IAI.00531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith DA, Parish T, Stoker NG, Bancroft GJ. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect Immun. 2001;69:1142–1150. doi: 10.1128/IAI.69.2.1142-1150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berney M, et al. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2015;112:10008–10013. doi: 10.1073/pnas.1513033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peteroy-Kelly MA, Venketaraman V, Talaue M, Seth A, Connell ND. Modulation of J774.1 macrophage L-arginine metabolism by intracellular Mycobacterium bovis BCG. Infect Immun. 2003;71:1011–1015. doi: 10.1128/IAI.71.2.1011-1015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordhan BG, et al. Construction and phenotypic characterization of an auxotrophic mutant of Mycobacterium tuberculosis defective in L-arginine biosynthesis. Infect Immun. 2002;70:3080–3084. doi: 10.1128/IAI.70.6.3080-3084.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somashekar BS, et al. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: Ex vivo 1H magic angle spinning NMR studies. J Proteome Res. 2011;10:4186–4195. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 44.Mendes V, Blundell TL. Targeting tuberculosis using structure-guided fragment-based drug design. Drug Discov Today. 2017;22:546–554. doi: 10.1016/j.drudis.2016.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.