Significance
Epigenetic memory for experience-based gene expression has not been well studied in higher organisms. Here we demonstrate that cells previously exposed to interferons exhibit a memory response and mount faster and higher transcription upon restimulation in fibroblasts and macrophages. Genome-wide analysis showed that memory was ascribed to accelerated recruitment of transcription factors to the genes. This process rested upon a distinct chromatin state involving the histone H3.3 and H3K36 modification. Our findings provide a mechanistic framework for the previously proposed idea of “trained innate immunity” representing memory, independent of adaptive immunity. Together, this study highlights learning as a fundamental faculty of mammalian somatic cells.
Keywords: memory, interferons, transcription, innate immunity, histone H3.3
Abstract
Epigenetic memory for signal-dependent transcription has remained elusive. So far, the concept of epigenetic memory has been largely limited to cell-autonomous, preprogrammed processes such as development and metabolism. Here we show that IFNβ stimulation creates transcriptional memory in fibroblasts, conferring faster and greater transcription upon restimulation. The memory was inherited through multiple cell divisions and led to improved antiviral protection. Of ∼2,000 IFNβ-stimulated genes (ISGs), about half exhibited memory, which we define as memory ISGs. The rest, designated nonmemory ISGs, did not show memory. Surprisingly, mechanistic analysis showed that IFN memory was not due to enhanced IFN signaling or retention of transcription factors on the ISGs. We demonstrated that this memory was attributed to accelerated recruitment of RNA polymerase II and transcription/chromatin factors, which coincided with acquisition of the histone H3.3 and H3K36me3 chromatin marks on memory ISGs. Similar memory was observed in bone marrow macrophages after IFNγ stimulation, suggesting that IFN stimulation modifies the shape of the innate immune response. Together, external signals can establish epigenetic memory in mammalian cells that imparts lasting adaptive performance upon various somatic cells.
Lineage-specific gene expression in differentiated cells is somatically inherited through epigenetic mechanisms, conveyed in part by chromatin (1–3). In particular, enhancer DNAs and associated chromatin modifications, such as H3K4me1 and H3K27ac have a large impact on lineage specification and extension (4–7). In addition, the replacement histone, H3.3 contributes to epigenetic memory during transition to embryonic pluripotency (8–10). H3.3 is deposited in the genome by coupling with transcription and distributed across actively expressed genes (11–15).
Epigenetic memory for signal-dependent transcription has not been well studied in multicellular organisms, although transcriptional memory has been reported in yeast following environmental cues (16–19). Nutrient response genes in yeast, such as GAL1 and INO1, are induced more rapidly and at higher levels when cells had previously experienced nutrient signaling. This trait is transmitted across cell generations and may involve histone H2A.Z and other pathways (20–22).
Given the conservation of transcriptional processes in the eukaryotes, one can assume that the mechanism of signal-dependent memory has also withstood evolutionary selection. An impetus for studying transcriptional memory for IFN-stimulated genes (ISGs) came from our previous observation that ISGs accumulate histone H3.3 as they are transcribed after IFN treatment (10, 23, 24). IFNs, both type I IFN (α/β) and type II IFN (IFNγ) stimulate many ISGs in various cell types and enhance antimicrobial activity (25–27). Here we show that IFN stimulation confers transcriptional memory that permits faster and greater ISG transcription in mouse embryonic fibroblasts (MEFs) and bone marrow (BM)-derived macrophages. The memory was attributed to faster and greater recruitment of phospho-STAT1 and RNA polymerase II (Pol II), but not Pol II pausing. Further, memory establishment coincided with acquisition of chromatin marks by H3.3 and H3K36 trimethylation. This study highlights a previously ill-defined epigenetic process through which external signals give rise to transcriptional memory that endows adaptive behavior in response to changing external milieu.
IFNβ Stimulation Generates Heritable Transcriptional Memory
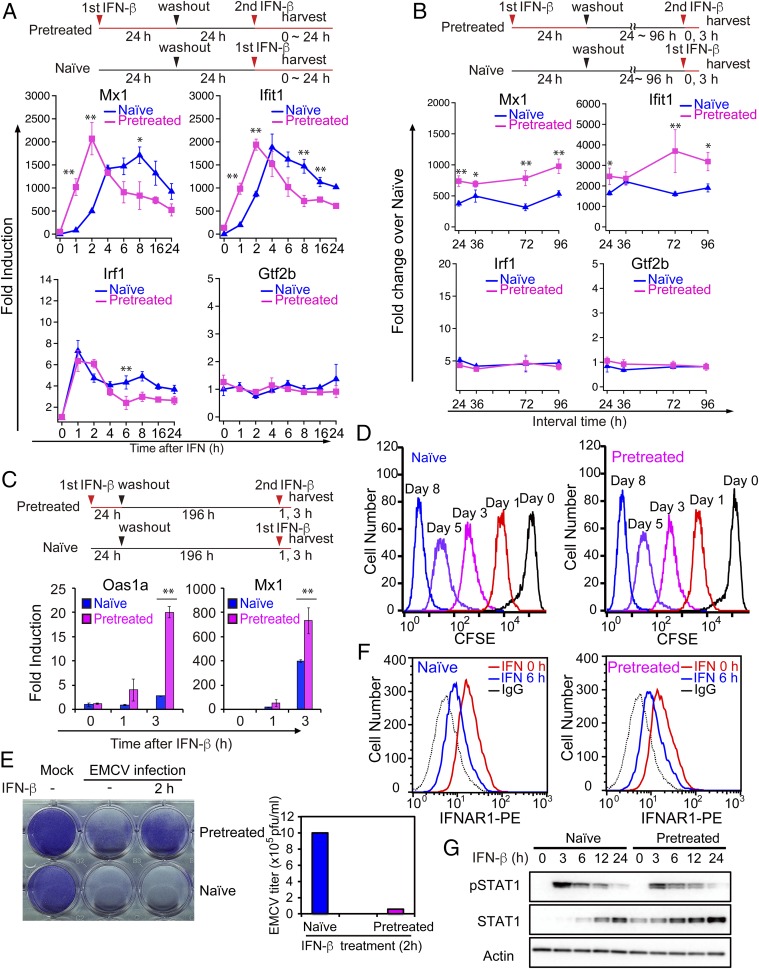
To investigate whether IFN affords epigenetic memory, MEFs were treated with IFNβ (hereafter IFN) for 3 h or 24 h, washed, and left without IFN for 24 h. Pretreated cells were then restimulated with IFN, and ISG induction was compared with naïve cells that were not treated with IFN before (See experimental diagram in Fig. 1A, Top). Time course analysis in Fig. 1A and SI Appendix, Fig. S1A showed that typical ISGs, such as Mx1, Ifit1, Oas1a, and others, were induced faster and at higher levels in pretreated cells than naïve cells. Another ISGs, Irf1 and constitutive Gtf2b were not (see below also). Thus, pretreated cells exhibited a memory response similar to that in yeast. To assess the duration of memory, pretreated cells were left without IFN for up to 96 h and then restimulated to examine memory. Mx1, Ifit1, Oas1a, and other ISGs maintained memory over this period (Fig. 1B and SI Appendix, Fig. S1B). Moreover, Oas1a and Mx1 retained memory farther, for up to 8 d (Fig. 1C). IFN memory was also detected with nascent ISG transcripts, showing that this memory response represents de novo transcription, rather than mRNA carryover (SI Appendix, Fig. S1C). Some ISGs, such as Irf1, did not exhibit memory, in that induction levels were similar in naïve and pretreated cells, indicating variability in memory formation (Fig. 1 A and B). Additionally, the memory response was not detected in constitutively expressed genes that did not respond to IFNs (e.g., Gtf2b in Fig. 1 A and B). Flow cytometry analysis of carboxyfluorescein succinimidyl ester (CFSE)-stained cells showed that both naïve and pretreated cells divided multiple times equally up to 8 d, demonstrating that IFN memory was transmitted across cell divisions (Fig. 1D). Other MEF cell lines and NIH 3T3 fibroblasts displayed similar memory responses after IFN stimulation (SI Appendix, Fig. S1D). Thus, IFN stimulation generates epigenetic memory that confers some ISGs faster and greater transcription upon restimulation.
Fig. 1.
IFNβ stimulation generates transcriptional memory in MEFs. (A, Top) Experimental design. Naïve and pretreated MEFs were first treated with vehicle alone or 100 units/mL of IFNβ (IFN) or for 24 h, respectively, washed, and incubated without IFN for 24 h. These MEFs were then stimulated with IFN (100 units/mL) for indicated times (in hours). (A, Bottom) ISG mRNAs were measured by qRT-PCR, normalized by Gapdh, and expressed as fold induction. Mx1, Ifit1, and Irf1 are examples of memory response and nonmemory response, respectively. Constitutively expressed Gtf2b was run as a control. The data represent the mean of three independent experiments ±SD. Statistically significant differences are indicated (Student’s t test, *P < 0.05, **P < 0.01). (B) Naïve and pretreated cells were left without IFN for indicated times (in hours) and stimulated with IFN for 3 h. ISG mRNAs were measured as above. See SI Appendix, Fig. S1C for nascent ISG mRNA. (C) Naïve and pretreated cells were left without IFN for 196 h (8 d), stimulated with IFN for 1 h or 3 h, and ISG mRNAs were measured as above. The data represent the average of three independent experiments ±SD. Statistically significant differences are indicated (Student’s t test, *P < 0.05, **P < 0.01). (D) Naïve and pretreated cells were labeled with CFSE, treated with or without IFN for 24 h, washed, and then incubated without IFN for indicated days. CFSE staining was detected by flow cytometry. Repeated cell divisions were detected in three separate 96-h washout experiments. (E) Naïve and pretreated cells were treated with IFN for 2 h and infected with EMCV for 24 h (MOI = 10). Cell survival was assessed by crystal violet staining (Top), and EMCV viral titers in the culture supernatants were determined by plaque assay (Bottom). Similar data were obtained in three independent assays. This applies to F and G below. (F) Naïve and pretreated cells were stimulated with IFN for 6 h and IFNAR1 surface expression was detected by flow cytometry using anti-mouse IFNAR1 antibody. (G) Naïve and pretreated cells were stimulated with IFN for indicated times (in hours) and expression of pSTAT1 and STAT1 was detected by immunoblot analysis of whole cell extracts using corresponding antibodies. β-Actin was used as a loading control. Uncropped Western blots are presented in SI Appendix, Fig. S1F.
To assess the functional significance of the memory, we examined IFN’s antiviral activity. Naïve and memory cells were infected with encephalomyocarditis virus (EMCV) with or without brief IFN treatment. Naïve cells underwent extensive cytopathic death after infection, irrespective of IFN treatment, as detected by sparse crystal violet staining (Fig. 1E and SI Appendix, Fig. S1E). In contrast, most of pretreated cells remained viable upon viral infection. Accordingly, the EMCV viral titers in supernatants were >10-fold higher in naïve cells compared with pretreated cells. Thus, IFN memory provides improved antiviral activity, suggesting the ability to create an adaptive response.
IFNβ activates the JAK/STAT signaling pathway to elicit its biological activity (25). To assess whether the memory is ascribed to changes in IFN signaling, we tested expression of IFNAR1 receptor and phosphorylated (p) STAT1, critically required for ISG transcription. Flow cytometry and Western data (Fig. 1 F and G) showed that expression of IFNAR1 and pSTAT1 was virtually identical in naïve and pretreated cells. These data support the view that IFN memory is not attributed to substantial changes in IFN signaling, but to a subsequent event(s).
Memory Response Is Selective: RNA-Seq Analysis
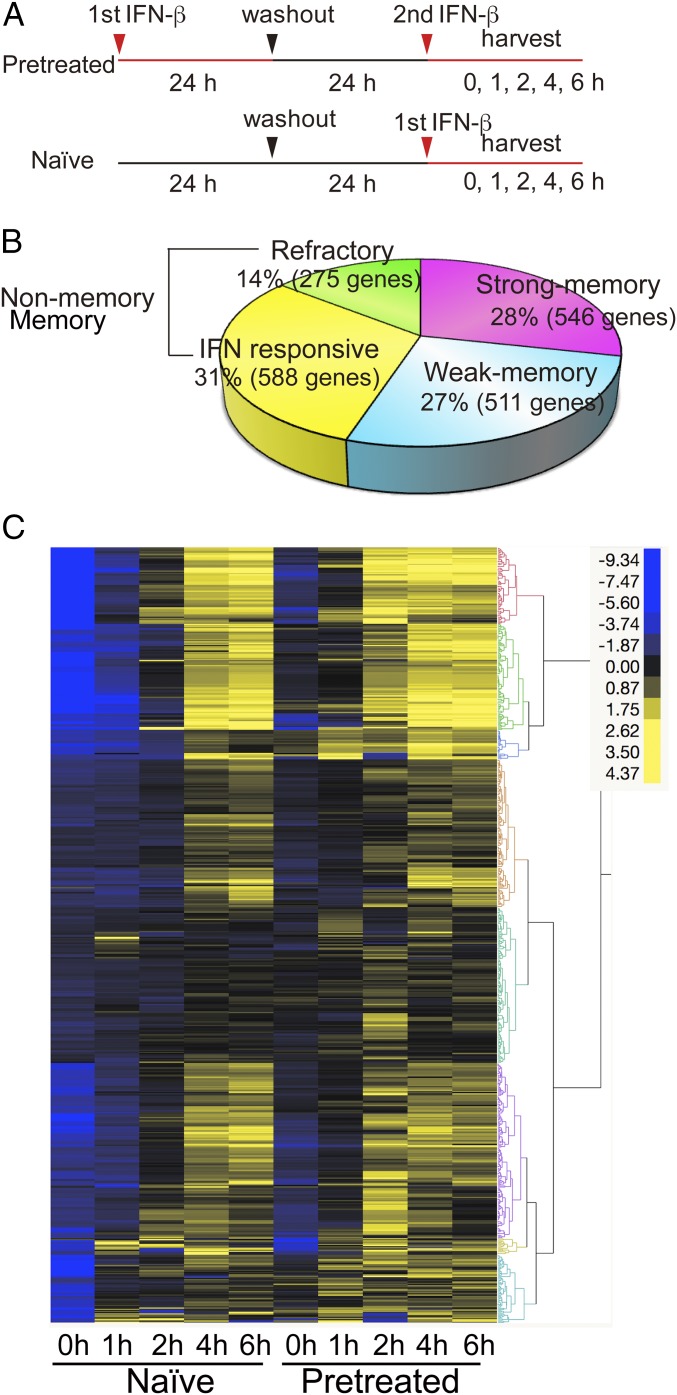
To globally identify ISGs that gain memory after IFN treatment, RNA-seq analysis was performed for naïve and pretreated MEFs that were restimulated by IFN for up to 6 h (Fig. 2A). About 2,000 ISGs were up-regulated or down-regulated by IFN both in naïve and pretreated cells [SI Appendix, Fig. S2 A and B for Gene Ontology (GO) analysis] (28). Of up-regulated ISGs, approximately half (1,057 genes) displayed a typical memory response, showing faster and/or higher expression in pretreated cells at some point during 6 h of IFN stimulation (Fig. 2B). Of these, 546 ISGs showed strong memory [reads per kilobase million (RPKM) levels more than twofold higher in pretreated cells than naïve cells]. The rest of 511 ISGs showed somewhat weaker memory where mRNA levels in pretreated cells were higher than naïve cells, but by less than twofold. On the other hand, about 600 ISGs did not show memory and were classified as nonmemory ISGs, since their RPKM levels were similar in naïve and pretreated cells. This group included Irf1 that did not show memory in the initial qRT-PCR analysis (Fig. 1 A and B). Moreover, an additional 275 genes were designated refractory ISGs, as they were induced in naïve cells but not in pretreated cells. qRT-PCR data in SI Appendix, Fig. S2C confirmed the loss of induction in pretreated cells for these ISGs. This acquired unresponsiveness is analogous to “endotoxin tolerance” caused by bacterial lipopolysaccharide (LPS) (29–31). GO analysis showed that memory and nonmemory ISGs share overlapping and related categories, such as innate immune and defense responses (SI Appendix, Fig. S2D). In contrast, GO categories enriched in refractory ISGs were unrelated to those in memory and nonmemory ISGs, pointing to negative transcriptional regulation, indicating distinct functional traits for this group of ISGs. Kinetic profiles of memory ISG expression in Fig. 2C highlighted faster transcript induction in pretreated cells compared with naïve cells. A fraction of ISGs showed somewhat elevated expression levels at 0 h in pretreated cells, suggestive of carryover transcripts. Nevertheless, their expression greatly exceeded that in naïve cells upon subsequent restimulation, verifying authentic memory response. Thus, IFN stimulation imparts transcriptional memory to more than 1,000 ISGs. Further, this memory response varied in different ISGs, revealing selectivity in memory formation. These data indicate that the second IFN stimulation does not produce a carbon copy of the first response, but it likely imposes reprogramming of gene expression patterns.
Fig. 2.
Memory response in selective RNA-seq analysis. (A) Experimental design for RNA-seq. Naïve and pretreated cells were stimulated with IFN for 0, 1, 2, 4, and 6 h. ISGs were defined by those genes showing >2-fold higher transcript expression (RPKM) at any time point during 6 h of IFN treatment in naïve cells. (B) Total memory ISGs (1,057) were subdivided into two groups. Strong memory: ISG mRNA levels were >2-fold higher in pretreated cells than naïve cells (>2 in RPKM). Weak memory: ISG mRNA levels were >1.5-fold higher (>1.5 in RPKM), but <2-fold (<2 in RPKM). Nonmemory ISGs were those with similar expression levels in naïve and pretreated cells [difference <1.5-fold (<1.5 in RPKM)]. Refractory ISGs showed >1.5-fold lower levels of expression in pretreated cells than naïve cells (>1.5 in RPKM). See qRT-PCR examples in SI Appendix, Fig. S2C, GO analysis in SI Appendix, Fig. S2D. Strong memory ISGs and nonmemory ISGs were subjected to further analysis. List of memory, nonmemory, and refractory ISGs is in Dataset S1. (C) Hierarchical clustering analysis for memory ISGs (strong) was performed by the Ward’s minimum variance method using JMP software (SAS Institute). ISG expression levels are depicted by differential coloring. The clustering analysis was done by using Ward’s minimum variance method for unsupervised hierarchical clustering.
Pol II Is Recruited to Memory ISGs Faster and at Higher Levels Upon Restimulation
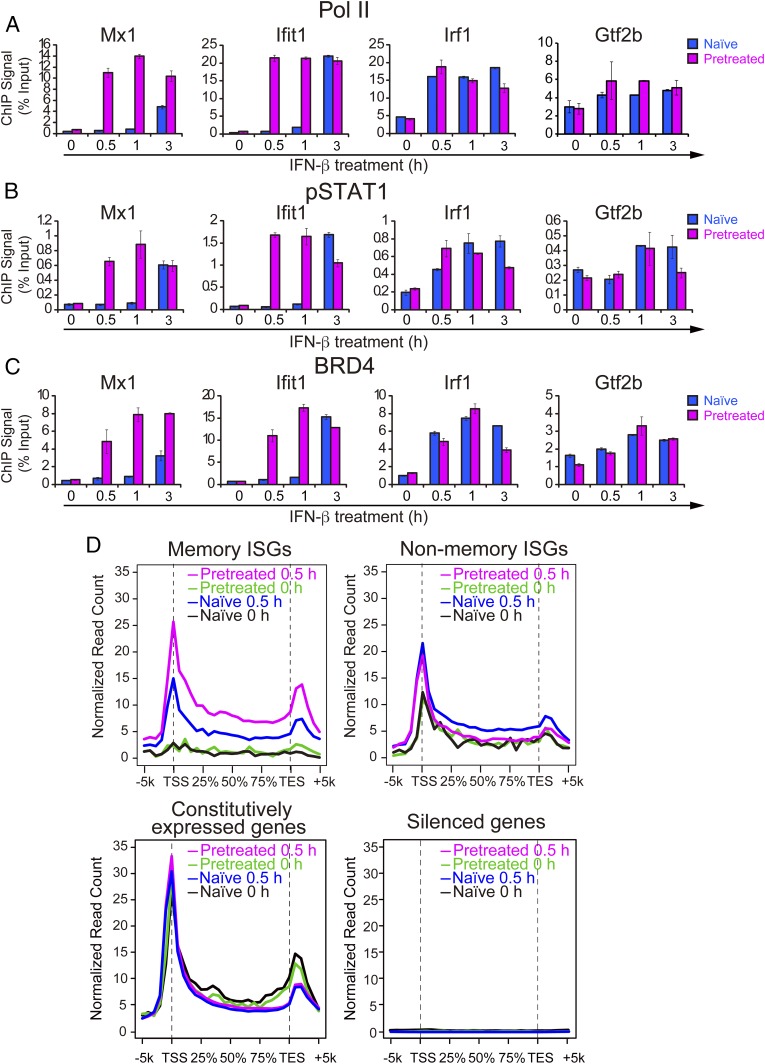
Pol II is paused near the transcription start sites (TSSs) of various genes (32, 33). Some genes with paused Pol II, such as heat shock genes and LPS responsive genes are induced rapidly after stimulation (34–36). However, we previously observed that many ISGs do not have paused Pol II before IFN; rather Pol II is recruited after stimulation along with binding of phosphorylated STAT1 and BRD4 (24, 37). Here we asked whether IFN memory is attributed to a change in the Pol II binding status, from nonpaused to paused state. qChIP analysis of memory ISGs (Mx1, Ifit1, and Oas1) showed virtually no Pol II on the TSS before restimulation both in naïve and pretreated cells (Fig. 3A). However, upon restimulation, Pol II was recruited to the ISGs faster in pretreated cells than naïve cells. In contrast, Irf1, a nonmemory ISG, had sizable amounts of prebound Pol II in both naïve and pretreated cells where Pol II was recruited similarly after restimulation. As expected, Pol II binding on Gtf2b was not affected by IFN treatment. Likewise, memory ISGs had neither prebound pSTAT1 nor prebound BRD4. But these factors were recruited faster in pretreated cells upon restimulation (Fig. 3 B and C). Mirroring the Pol II data, pSTAT1 and BRD4 binding was very similar for the nonmemory ISG, Irf1 in naïve and pretreated cells. These data support the possibility that accelerated Pol II recruitment, rather than Pol II pausing accounts for IFN memory.
Fig. 3.
Pol II is recruited to memory ISGs faster and at higher levels upon restimulation. (A–C) Naïve and pretreated cells (24 h pretreated and 6 h interval time) were stimulated with IFN for indicated times (0, 1, and 3 h) and qChIP assays were performed to detect binding of Pol II (A), pSTAT1 (B), and BRD4 (C) at the TSS/promoter region of memory ISGs (Mx1 and Ifit1), nonmemory ISG (irf1), and Gtf2b (control). Values represent the average of three independent experiments ±SD. (D) ChIP-seq analysis for global distribution of Pol II on the memory ISGs, nonmemory ISGs, constitutively expressed genes or silent genes. See SI Appendix, Fig. S3 A–C for IGV examples.
To further evaluate this possibility, ChIP-seq was performed for global Pol II occupancy on memory and nonmemory ISGs (Fig. 3D and SI Appendix, Fig. S3 A and B). Consistent with the above qChIP data, little to no prebound Pol II was detected on memory ISGs before IFN restimulation, both in naïve and pretreated cells. After 30 min of IFN restimulation, however, Pol II was recruited at higher levels in pretreated cells than in naïve cells. Thus, memory ISGs lose Pol II after IFN washout and are without it until restimulation. In contrast, nonmemory ISGs were already bound by Pol II before IFN stimulation in both naïve and pretreated cells (Fig. 3D and SI Appendix, Fig. S3B). Interestingly, refractory genes also had prebound Pol II (SI Appendix, Fig. S3C). Constitutively expressed genes had constitutive Pol II binding, whereas silent genes were without Pol II, as expected (Fig. 3D). Together, memory and nonmemory ISGs showed contrasting Pol II binding patterns. These results show that memory and nonmemory ISGs differ in the Pol II binding status and that acquisition of IFN memory is not due to conversion of Pol II binding status, but to enhanced factor accessibility.
Memory ISGs Are Marked by Histone H3.3 and H3K36me3
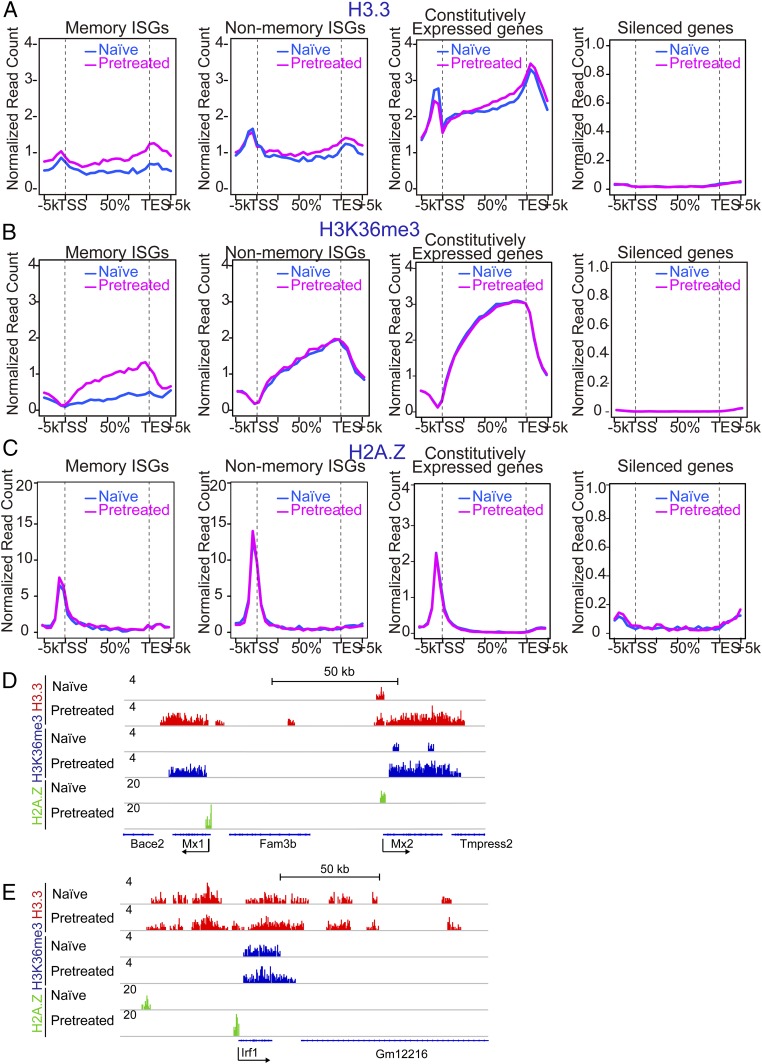
Given that Pol II and other transcription factors do not remain on memory ISGs after IFN washout, but gain improved accessibility, IFN memory information may be stored elsewhere, presumably in the chromatin milieu. To identify chromatin marks that coincide with IFN memory, we investigated distribution of replacement histones, H3.3 and H2A.Z, since they are reported to play a role in epigenetic memory (8, 9, 20). The H3K36me3 mark was also tested, since we previously found that IFN stimulation triggers H3K36me3 marking on ISGs along with H3.3 deposition (23, 24). ChIP-seq analysis was performed for these marks in naïve and pretreated cells without restimulation (Fig. 4 A–C). For H3.3, we analyzed MEFs derived from mice in which the endogenous H3f3b was replaced by the HA-tagged H3.3 cDNA. The HA tag on H3.3 allowed us to detect H3.3 distribution with higher reliability than using anti-H3.3 antibodies. Memory and nonmemory ISGs exhibited disparate H3.3 distribution patterns. Memory ISGs did not have H3.3 in naïve cells, but gained this mark in pretreated cells. Conversely, nonmemory ISGs were already marked with H3.3 in naïve and pretreated cells. To delineate whether the gain of H3.3 mark is important for memory response, we knocked down H3.3 in pretreated cells by siRNA during 48 h of IFN washout (SI Appendix, Fig. S4 A and B). H3.3-specific siRNA, but not control siRNA, significantly reduced IFN memory response, as evidenced by reduced ISG mRNA expression and reduced Pol II recruitment. However, H3.3 siRNA, when added to naïve cells, did not inhibit ISG induction as much under these conditions. These data are in line with the idea that H3.3 deposition contributes to the acquisition of functional IFN memory. We also observed reduced ISG induction in cells stably expressing H3.3 shRNA relative to control shRNA (SI Appendix, Fig. S4C). The H3K36me3 mark displayed the same feature, showing a clear dichotomy between memory and nonmemory ISGs. Thus, memory ISGs acquired H3.3 and H3K36me3 marks only after initial IFN stimulation, while nonmemory ISGs possessed these marks without IFN stimulation. Like Pol II binding, refractory ISGs also possessed both H3.3 and H3K36me3 (SI Appendix, Fig. S4H). As expected, both marks were present in constitutively expressed genes, irrespective of IFN, whereas they were absent in silent genes (Fig. 4 A and B). In contrast, H2AZ did not show an obvious correlation with ISG memory: H2AZ showed a sharp peak at the TSS of all ISGs and constitutively expressed genes (Fig. 4 C–E and SI Appendix, Fig. S4 D–H) (12). Other histone marks representing expressed genes such as H3K4me3 and H4ac did not show a clear correlation with memory, although H3K4me1, an enhancer mark showed a modest correlation (SI Appendix, Fig. S4 I–P).
Fig. 4.
Memory ISGs are marked by the histones H3.3 and H3K36me3. (A–C) ChIP-seq analysis was performed for genome-wide distribution of H3.3 (A), H3K36me3 (B), and H2AZ (C) over memory ISGs, nonmemory ISGs (Left), and constitutively expressed or silent genes (Right) in naïve or pretreated MEFs without restimulation. Pretreated cells were left without IFN for 48 h. See SI Appendix, Fig. S4 D–H for IGV examples. (D) IGV images of H3.3, H3K36me3, and H2AZ peaks over memory ISGs (Mx1 and Mx2) in naïve and pretreated cells. (E) IGV images of H3.3, H3K36me3, and H2AZ peaks over nonmemory ISG (Irf1) in naïve and pretreated cells.
IFNγ Stimulation Generates Transcriptional Memory in Macrophages
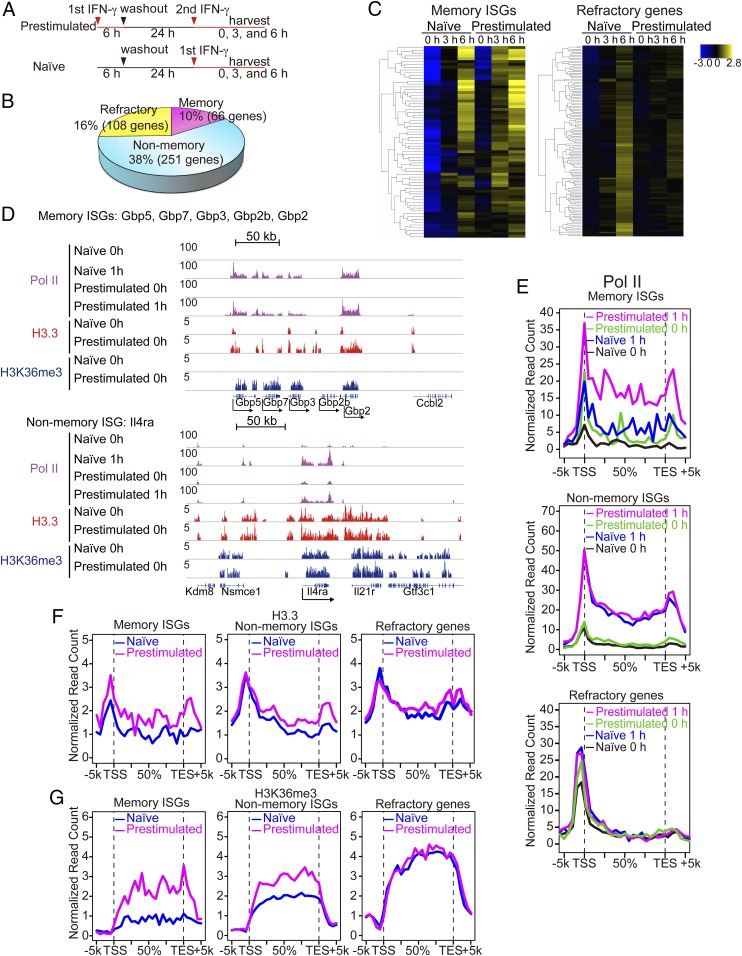
To test whether memory is generated in other cells, we next examined IFNγ-stimulated BM-derived macrophages. IFNγ activates another JAK/STAT pathway distinct from IFNβ to establish antimicrobial activity (25, 26). Macrophages are a major cell type responsible for innate immunity in the body and unlike MEFs, they are postmitotic under normal conditions. Naïve and pretreated macrophages were stimulated with IFNγ and tested for ISG expression (experimental diagram in Fig. 5A). Initial qRT-PCR analysis found that some ISGs were expressed higher in prestimulated macrophages, indicative of IFN memory (SI Appendix, Fig. S5A). Microarray analysis found that naïve macrophages expressed about 425 ISGs (SI Appendix, Fig. S5 B–D for GO analysis). Of these, 66 ISGs were deemed memory ISGs, as their expression was at least >1.5-fold higher in prestimulated macrophages than in naïve ones (Fig. 5B, e.g., Gbp), while 251 ISGs were judged nonmemory ISGs, as their levels were similar in naïve and prestimulated macrophages (e.g., Irf1 in SI Appendix, Fig. S5A). In addition, as many as 108 ISGs were found to be refractory genes, in that their expression was abrogated or markedly lower upon restimulation (e.g., Il12d in SI Appendix, Fig. S5A). Clustering profiles in Fig. 5C illustrate that memory ISGs were expressed earlier and/or higher in prestimulated macrophages than in naïve cells, whereas, refractory ISGs were not induced or reduced in prestimulated macrophages. Some memory and nonmemory ISGs were found both in MEFs and macrophages (e.g., Mx1, Irf7, and Ifi44 for memory; Irf1 for nonmemory ISGs), suggesting a common feature, while others were unique to IFNγ and/or macrophages (Nos2, Il12br, Ciita, and Tlr11). GO analysis revealed that memory and nonmemory ISGs have similar processes and pathways related to immunity and defense (SI Appendix, Fig. S5D). In contrast, refractory ISGs showed distinct categories such as biosynthesis and transcription, a feature similar to MEFs. These results indicate that IFNγ pretreatment alters the overall character of macrophage host defense programs.
Fig. 5.
IFNγ stimulation generates transcriptional memory in macrophages. (A) Experimental design for microarray analysis. (B) ISGs were defined as those genes expressed at least twofold higher in naïve macrophages treated with IFNγ for 3 or 6 h than untreated macrophages (P < 0.05). ISGs were classified into three groups. Memory ISGs: showing at least 1.5-fold higher in expression (>1.5) in prestimulated macrophages than naïve cells. Nonmemory ISGs: showing less than 1.2-fold (<1.2) difference between naïve and prestimulated macrophages. Refractory ISGs: showing at least 1.5-fold less (<1.5) expression in prestimulated macrophages with IFNγ for 3 or 6 h. List of memory, nonmemory, and refractory ISGs is in Dataset S2. (C) Hierarchical clustering of memory (Left) and refractory ISGs (Right). Memory ISGs exhibited faster and/or greater ISG induction in prestimulated macrophages than naïve cells. Refractory ISGs failed to express or showed reduced ISG expression in prestimulated macrophages. (D) IGV images of Pol II, H3.3, and H3K36me3 distribution over the Gbp gene cluster (memory ISGs) and Il4ra (nonmemory ISG). (E) Global Pol II distribution was analyzed for naïve and prestimulated macrophages at 0 h or 1 h after IFNγ stimulation. Distribution of H3.3 and H3K36me3 was tested for untreated naïve and memory macrophages (24 h after IFN washout). (F and G) Global distribution of H3.3 (F) and H3K36me3 (G) over memory ISGs, nonmemory ISGs, or refractory ISGs in naïve and prestimulated macrophages.
To assess whether IFN memory generated in macrophages and fibroblasts share a common mechanism, we tested the status of Pol II recruitment. ChIP-seq analyses were performed for global distribution of Pol II along with H3.3 and K3K36me3 in naïve and pretreated macrophages, without restimulation. For Pol II, binding was tested 1 h after IFNγ stimulation as well. Integrative Genomics Viewer (IGV) examples of memory ISGs (Gbp cluster), and nonmemory ISG (Il4ra) (Fig. 5D, Top and Bottom) showed a remarkable similarity with those in fibroblasts. For Gbp genes, Pol II was absent before stimulation, but vigorous binding was ensured after stimulation.
Global Pol II distribution shown in Fig. 5E largely reproduced the pattern in fibroblasts, in that Pol II was already bound to nonmemory and refractory ISGs even in naïve cells, while memory ISGs were without prebound Pol II. We noted that a fraction of memory ISGs exhibit prebound Pol II, suggesting contribution of paused Pol II to memory or to other aspects (38).
The H3.3 and H3K36me3 marking patterns found in macrophages were also similar to those in MEFs: memory ISGs gained these marks after IFNγ stimulation, whereas these marks were present in nonmemory and refractory ISGs even in naïve cells before IFNγ stimulation (Fig. 5 F and G and SI Appendix, Fig. S5 E–G). Another histone mark, H3K4me1, showed a modest correlation (SI Appendix, Fig. S5 E–H). Together, transcriptional memory, generated by different IFNs in different cell types, exhibited similar molecular features, underscoring a shared mechanism and biological significance.
Discussion
IFN stimulation generated lasting transcriptional memory in MEFs and macrophages, conferring faster and higher ISG induction upon restimulation. This memory was inherited through mitoses in MEFs, while sustained in a postmitotic state in macrophages. The memory led to improved resistance to viral infection, despite the fact that not all ISGs acquired memory. The memory was largely attributed to accelerated Pol II recruitment along with accelerated binding of pSTAT1, rather than Pol II pausing, which were associated with acquisition of H3.3 and H3K36me3 marks.
A salient feature of IFN memory was faster ISG induction both in MEFs and macrophages. Faster induction is also a main characteristic of transcriptional memory in yeasts (16–19, 39). IFN memory we observed here also shares common features with that reported for HeLa cells and mouse macrophages upon IFNγ stimulation (6, 38).
Genome-wide analyses revealed three types of ISGs: memory, nonmemory, and refractory ISGs in both cell types, highlighting nonuniformity among ISGs. Memory and nonmemory ISGs shared a number of GO categories that were mostly related to innate immune responses in both cell types. In contrast, GO categories for refractory ISGs were unrelated to innate immunity. This indicates that the memory response is not a simple repeat of ISG expression, but represents readjustment of gene expression programs in the cell to accommodate changing environments.
Pol II binding status in memory ISGs differed from that in nonmemory and refractory ISGs in both cell types. Pol II occupancy was promptly lost from memory ISGs after IFN washout, while some prebound Pol II was present on nonmemory and refractory genes even without IFN. Likewise, pSTAT1 was recruited to memory ISGs only after IFN stimulation in both naïve and pretreated cells. Thus, Pol II pausing and STAT1 signaling are not likely to be a major mechanism of memory. However, paused Pol II was shown to be associated with memory in some genes in yeast and IFNγ-treated HeLa cells (38). This report and our results are not mutually exclusive, since multiple mechanisms likely contribute to transcriptional memory. Indeed, prebound Pol II was detected in a small fraction of memory ISGs in macrophages in this study as well. Given that Pol II was prebound on nonmemory ISGs and refractory ISGs, it is possible that these ISGs have already been epigenetically engaged before IFN stimulation.
Acquisition of IFN memory correlated with the histone H3.3 and H3K36me3 chromatin marks in both MEFs and macrophages. On memory ISGs, these marks were absent in naïve cells but accorded after IFN stimulation. Our study identifies H3.3 as a chromatin signature that denotes IFN memory. Our data that H3.3 knockdown during IFN washout decreased memory response may support the possibility that this mark has a functional importance. With an additional correlation found between IFN memory and H3K4me1, albeit weaker, IFN memory may be associated with the recently proposed “latent enhancers” (6, 7). While constitutive and poised enhancers already carry Pol II and certain chromatin marks, latent enhancers do not have prebound Pol II, but are associated with cytokine stimulation (6). H3.3 deposition was another memory feature found in our study, suggesting that memory acquisition is linked to histone replacement. The role of H3.3 histone replacement in transcriptional memory was reported during inducible pluripotent stem (iPS) transition in frog embryos (9, 10).
There is growing evidence that innate immune responses have a memory aspect, termed “trained” innate immunity, that is independent of classical immunological memory, which depends on lymphocyte-mediated adaptive immunity (40, 41). For example, earlier nonlethal infection with Candida albicans provides resistance to the subsequent lethal infection in mice (42). This memory depends on cells of the monocyte/macrophage lineage and is associated with changes in chromatin. Trained innate immunity to candida is shown to require intact STAT1, but not STAT3 (30). Given that ISG induction depends on STAT1, we surmise that trained innate immunity may partly include IFN memory described here. Also, bacillus Calmette–Guérin that stimulates IFNγ response is shown to provide protection against heterologous, viral, or bacterial infection (43). In contrast to IFNs which increase innate protection, LPS exposure, after a brief burst of inflammatory responses, leads to a period of profound tolerance and immunosuppression (30). While detailed mechanisms are still elusive, LPS tolerance is shown to depend on the transcription factor ATF7 (44). LPS tolerance likely affects the course of various infectious diseases, particularly sepsis, a prevalent, but hard-to-treat disease. It is interesting to note that since ISGs are induced after LPS through IFNβ-dependent feedback, LPS tolerance may well be modulated by aspects of IFN memory.
In conclusion, this study offers insight into signal-induced epigenetic memory that conveys enhanced adaptive performance upon ordinary cells.
Materials and Methods
Cells and Treatment.
MEFs and BM-derived macrophages were prepared from wild-type and mutant mice in which the H3f3b locus was replaced by a H3.3-HA cDNA. Details of the derivation of the mouse strain will be presented elsewhere. The procedures used in this work were approved by the National Institute of Child Health and Human Development animal care and use committee with the animal study proposal no. 14–044. MEFs were prepared from day 13.5 embryos and maintained in DMEM with 10% FBS and antibiotics, and treated with 100 units/mL of murine recombinant IFNβ (PBL IFN Source) for indicated periods. BM-derived macrophages were cultured in DMEM/F12 medium containing 10% FBS, 20% L929-conditioned medium as a source of macrophage colony-stimulating factor (M-CSF), 10% glutamine, and antibiotics treated with 100 units/mL of murine recombinant IFNγ (Invitrogen) for indicated periods (45).
For EMCV infection, MEFs treated with or without IFNβ for 3 h were infected with EMCV [multiplicity of infection (MOI) = 10] in serum-free and antibiotics-free medium for 1 h at 37 °C, washed, and allowed to proceed in culture for 24 h. Cells were fixed with methanol and incubated with crystal violet solution (1% crystal violet, 10% ethanol) for 20 min at room temperature. For viral titration, plaque assays were performed with L929 cells incubated with serial dilutions of culture supernatants (10−2∼105) followed by incubation with agar overlay for 24 h at 37 °C. The plaques were visualized by staining with a 0.02% neutral red. The viral titers were expressed as plaque forming units (pfu).
Cell proliferation assay was carried out with 1.5 μM CellTrace CFSE (Molecular Probes) according to the manufacturer’s instructions and fluorescent signals were detected by flow cytometry in FACSCalibur (BD) with the FlowJo Software.
ISG mRNA, ISG nascent transcripts detected by qRT-PCR with appropriate primers (24, 37) are listed in Dataset S3. pSTAT1, total STAT1, and surface IFNAR1 were detected by immunoblot assay and flow cytometry, respectively. Antibody for murine pSTAT1 was from Santa Cruz Biotechnology (sc-417), STAT1 Cell Signaling Technology (no. 9172), and PE anti-mouse IFNAR1 from Biolegend (no. 127311).
For siRNA transfection to knockdown H3.3, negative control (si-Ctrl) siRNA and siRNA for H3.3a and H3.3b were transfected into MEF cells with Lipofectamine 2000 (Invitrogen) for 48 h before IFN stimulation. H3f3a and H3fb siRNA oligomers were purchased from Life Technology. siRNA sequences tested here are for H3f3a, sense GAUUGCGAGUGGAAACAUAtt antisense UAUGUUUCCACUCGCAAUCat (5′-4′). siRNA ID no. s234173M; for H3f3b, sense CAGCAUCAUCUAUUACUAAtt antisense UUAGUAAUAGAUGAUGCUGgt; siRNA ID no. s67338; and sense CCCAGGAUUUCAAAACCGAtt antisense UCGGUUUUGAAAUCCUGGGcg, siRNA ID no. s67336.
Knockdown of H3.3 was performed by lentiviral vectors and transduction or siRNA transfection. The shRNA-expressing lentiviral vector to knock down both H3f3a and H3f3b genes was constructed with pLKO.1-puro vector and the same shRNA sequence as described previously (23). pLKO.1-puro control shRNA vector (CAACAAGATGAAGAGCACCAA) was used as a control. The shRNA lentiviruses were produced by transient transfection of HEK293T cells with a pLKO.1-puro vector and the packaging plasmids using polyethylenimine following the standard procedure. Lentiviral supernatants were collected at 24 and 48 h and MEF cells were transduced with lentiviruses with 8 µg/mL Polybrene (Sigma).
RNA-Seq.
Total RNA was obtained from TRIzol (Invitrogen) and purified by the RNeasy mini kit (Qiagen). Oligo(dT) selection was performed by using Dynal magnetic beads (Invitrogen) according to the manufacturer’s protocol. mRNAs were fragmented by heating at 94 °C for 3 min in the fragmentation buffer [40 mM Tris-acetate (pH 8.2), 100 mM potassium acetate, and 30 mM magnesium acetate]. RNA fragments were precipitated with GlycoBlue (Ambion) as a carrier and reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) with random primer and RNasin (Promega). Second-strand cDNAs were synthesized using Escherichia coli DNA polymerase I and RNaseH (Invitrogen). Purification of second-strand cDNAs was performed with ZYMO DNA clean and concentrator-5 kit. Library preparation, including ligating barcode, was conducted using a Mondrian SP (NuGEN Technologies, Inc.) and the Ovation SP Ultralow Library system (NuGEN). Fragments ranging from 250 to 450 bp were selected and subjected to paired-end sequencing on a HiSeq2000 (Illumina). For RNA-Seq analysis, paired-end reads were aligned to the Mus musculus reference genome mm10 using Bowtie/Tophat allowing up to two mismatches. Transcript abundance was quantified using Cufflinks 1.2.1. Genes showing an RPKM of more than a twofold increase in treated samples at least one time point during IFN treatment (1, 2, 4, and 6 h) over untreated samples (0 h) were considered up-regulated genes (ISGs) and were analyzed further. Raw data files are available at the NCBI Gene Expression Omnibus (GEO) server under accession no. 89440.
qChIP and ChIP-Seq.
qChIP was performed essentially as described (24, 37). Chromatin (4 × 105 cells) was incubated with antibodies bound to Dynabeads Protein G (Invitrogen) for 3 h, followed by immunoprecipitation, decrosslinking, DNA purification, and qPCR. The following antibodies were used: antibody for RNA polymerase II (8WG16) were obtained from Covance; antibodies for HA (ab9110), H3 (ab1791), H3K36me3 (ab9050), H3K4me1 (ab8895), and H3H4me3 (ab8580) were from Abcam; and antibody for H4Ac (06-866) was from Millipore. BRD4 was described (37). Primer sequences are listed in Dataset S3.
For ChIP-seq, cells were crosslinked with 1% formaldehyde for 10 min and nuclei were pelleted after sequential wash and resuspended in sonication buffer (10 mM Tris⋅HCl, 1 mM EDTA, 0.1% SDS, protease inhibitor mixture) and sonicated in a Bioruptor (Qsonica) to shear the chromatin into 200- to 300-bp fragments. Chromatin from 2 to 10 × 106 cells was used for each ChIP-seq experiment. Chromatin was incubated with antibody and Protein G beads (Dynabeads) at 4 °C for overnight and washed sequentially with RIPA buffers. Immunoprecipitated chromatin was decrosslinked and DNA purified. ChIP-seq libraries were constructed using a Mondrian SP (NuGEN Technologies, Inc.) and the Ovation SP Ultralow Library system (NuGEN). Fragments ranging from 250 to 450 bp were selected for single-read sequencing on a HiSeq2000 (Illumina). Resulting reads were aligned to the mouse reference genome (mm10) using the Burrows–Wheeler aligner (46) with default parameters. Uniquely mapped reads were retained for downstream analysis. Peak calling was performed using SICER (47). SICER was used with a window size setting of 200 bp, a gap setting of 600 bp, and a false discovery rate setting of 0.001. Data from immunoprecipitated samples were compared with those with input DNA. Raw data files are available at the NCBI Gene Expression Omnibus (GEO) server under accession no. 89440.
Microarray Analysis of BM Macrophages.
Macrophages were stimulated with 100 units/mL IFNγ and total RNA purified with the RNeasy mini kit (Invitrogen) and then subjected to microarray analysis using the Mouse Exon 1.0 ST Array (Affymetrix) essentially as described (48). Quality analysis of total RNA, cDNA synthesis, hybridization, and data extraction were performed at Expression Analysis, Inc. Data were analyzed using GeneSpring software (Silicon Genetics). Values with P ≤0.05 and twofold cutoff were considered significant. ANOVA was used to identify differentially expressed genes. Genes showing more than twofold increase or decrease with P <0.05 either at 3 or 6 h after IFNγ treatment were considered up- or down-regulated genes. Among up-regulated genes (ISGs), those showing >1.5-fold higher expression (P < 0.05) in pretreated macrophages over naïve macrophages were selected as memory ISGs. Genes whose expression levels did not differ (<1.5-fold difference) in pretreated and naïve cells, were classified as nonmemory ISGs. ISGs that did not increase the expression (<1.5 over 0 h) in prestimulated macrophages after 3 or 6 h of IFNγ were classified as refractory ISGs. Raw data files are available at the NCBI Gene Expression Omnibus (GEO) server under accession no. 89440.
Supplementary Material
Acknowledgments
We thank T. McFarlan, K. Ge, D. Levens, D. Singer, K. Pfeifer, H. Levin [National Institute of Child Health and Human Development (NICHD), National Institute of Diabetes and Digestive and Kidney Diseases, National Cancer Institute, NIH] and K. Takaoka and Y. Murakami (Hokkaido University) for suggestions and discussions; and T. Wu, M. Bachu, and C. Chen in the K.O. laboratory for advice on experiments and data interpretations. R.K. and R.O. were supported in part by a Japan Society for the Promotion of Science Kaitoku–NIH Fellowship. This work was supported by the Intramural Program of NICHD, NIH (Grant ZIA HD008815-08) and the National Heart, Lung, and Blood Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.A.F. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. 89440).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720930115/-/DCSupplemental.
References
- 1.Campos EI, Stafford JM, Reinberg D. Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol. 2014;24:664–674. doi: 10.1016/j.tcb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brookes E, Shi Y. Diverse epigenetic mechanisms of human disease. Annu Rev Genet. 2014;48:237–268. doi: 10.1146/annurev-genet-120213-092518. [DOI] [PubMed] [Google Scholar]
- 3.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 10.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 14.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Rai TS, et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014;28:2712–2725. doi: 10.1101/gad.247528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickner JH. Transcriptional memory: Staying in the loop. Curr Biol. 2010;20:R20–R21. doi: 10.1016/j.cub.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brickner DG, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51:118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 23.Tamura T, et al. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem. 2009;284:12217–12225. doi: 10.1074/jbc.M805651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarai N, et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 2013;32:2392–2406. doi: 10.1038/emboj.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 26.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 30.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 31.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat Immunol. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 35.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel MC, et al. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Light WH, et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends Genet. 2014;30:230–236. doi: 10.1016/j.tig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzetto L, et al. Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. J Biol Chem. 2016;291:7961–7972. doi: 10.1074/jbc.M115.699645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinnijenhuis J, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ifrim DC, et al. Defective trained immunity in patients with STAT-1-dependent chronic mucocutaneaous candidiasis. Clin Exp Immunol. 2015;181:434–440. doi: 10.1111/cei.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida K, et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat Immunol. 2015;16:1034–1043. doi: 10.1038/ni.3257. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1951–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta M, et al. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nat Commun. 2015;6:6379. doi: 10.1038/ncomms7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.