Significance
The RNA delivery field is mostly focused on lipid nanoparticles (LNPs). Although promising, LNPs have several limitations with respect to pharmacokinetics, biodistribution, and toxicity. The mechanism of RNA charge-altering releasable transporters (CART) delivery and release is unique. It proceeds dynamically with a controllable change in physical properties. Differing from all mRNA delivery systems, a key attribute of CARTs is a charge-altering degradation mechanism, which transforms the initial polycationic CART into neutral byproducts, thereby enabling endosomal escape, release, and subsequent translation of the polyanionic mRNA cargo. With this study, we introduce a potentially general approach to therapeutic vaccination enabled by a dynamic drug-delivery system (mRNA-CART) and demonstrate its utility in suppressing tumor formation and in eliminating established tumors.
Keywords: cancer vaccination, mRNA, nanoparticles, T cell responses, immunotherapy
Abstract
In vivo delivery of antigen-encoding mRNA is a promising approach to personalized cancer treatment. The therapeutic efficacy of mRNA vaccines is contingent on safe and efficient gene delivery, biological stability of the mRNA, and the immunological properties of the vaccine. Here we describe the development and evaluation of a versatile and highly efficient mRNA vaccine-delivery system that employs charge-altering releasable transporters (CARTs) to deliver antigen-coding mRNA to antigen-presenting cells (APCs). We demonstrate in human peripheral blood mononuclear cells that CART vaccines can activate a robust antigen-specific immune response against mRNA-encoded viral epitopes. In an established mouse model, we demonstrate that CARTs preferentially target professional APCs in secondary lymphoid organs upon i.v. injections and target local APCs upon s.c. injection. Finally, we show that CARTs coformulated with mRNA and a Toll-like receptor ligand simultaneously transfect and activate target cells to generate an immune response that can treat and cure mice with large, established tumors.
Immunotherapy has come of age and with it has come an increased understanding of the complex interactions between cancers and the immune system (1). Emerging therapies rely on fundamental insights into the mechanisms that suppress immune cell function and enable tumor cells to evade recognition and death. To these ends, vast resources have been invested in high-throughput screening for novel targets, genetic alterations, immune cell characterization, and optimized drug-delivery platforms (2–6). From these efforts, personalized medicine is emerging as the promising strategy for cancer treatment.
Vaccination can provide long-term protection against external pathogens and initiate the clearance of resident diseased cells (7). For that reason, experimental approaches of cancer vaccination began with prophylactic vaccination against cancer-associated antigens. Now, new developments have shifted the focus to the more clinically relevant therapeutic use of cancer vaccination (3, 7–9). These include vaccination platforms that target antigen delivery to specialized immune cells, the rapid identification of immunogenic tumor-specific antigens, and alternative molecular compositions of the antigen (2, 3, 10).
To this end, mRNA-based cancer vaccination has emerged as a promising approach. The concept of using mRNA as the basis for a vaccine was first proposed in 1972 (11) and subsequently was reported in 1993 for antigen-specific influenza vaccination (12) and then in 1999 for vaccination against cancer-specific antigens (13). In this strategy, an mRNA coding for specific antigen epitopes is introduced into the immune system, effectively “training” the body’s natural defenses to recognize and destroy antigen-presenting cancerous tissue. Antigen presentation is the fundamental starting point in all adaptive immune responses. Antigen-specific T cells are unable to react without proper antigen presentation, and B cell responses are often inferior without T cell help. mRNA delivery provides an exquisite strategy to induce intracellular synthesis of any desired protein or peptide antigen. Notably, because mRNA produces protein catalytically, a relatively small dose of mRNA is needed to produce many copies of protein, an advantage over costly whole-protein vaccination (14). Additionally, mRNA therapy does not carry with it the same risks of insertional mutagenesis associated with plasmid DNA-based gene therapy (15). Moreover, the delivery of specific/customized protein encoding mRNA can be leveraged to induce the transient expression of exogenous proteins in antigen-presenting cells (APCs) (3), a flexibility ideal for effective cancer vaccination against rapidly mutating cancers (due to the large number of potential mutated antigens).
Notwithstanding this promise, mRNA is a large, polyanionic molecule that does not readily cross cell membrane barriers and is generally unstable in circulation due to scission by native nucleases (16). Therefore, a key to enabling this methodology is development of safe and effective agents that package, protect, deliver, and intracellularly release mRNA (7, 14, 17, 18). Despite these challenges, early reports showed promise for mRNA-based vaccination for cancer immunotherapy. However, safe and effective delivery remains a major challenge to enabling the broader implementation of this approach (19–22). Due to the critical role of mRNA delivery in therapeutic vaccinations, the introduction and evaluation of safe, effective, and tunable mRNA delivery vehicles are required for broader clinical implementation.
We recently reported the robust in vivo delivery and expression of mRNA using charge-altering releasable transporters (CARTs) (10, 23). The mechanism of mRNA release for CARTs differs from other delivery vehicles as it proceeds dynamically with a tunable change in physical properties. Unique among other mRNA delivery systems such as lipid nanoparticles (LNPs), a key attribute of CARTs is a charge-altering degradation mechanism which transforms the initial polycationic backbone into neutral small-molecule byproducts, thereby assisting endosomal escape by allowing the electrostatic release and subsequent translation of mRNA. CARTs represent a completely new class of transfection agents, and because these CARTs degrade to well-tolerated neutral small molecules, they avoid the known toxicity concerns associated with cationic lipids and materials (24). Herein, we report that mRNA-CART vaccines using whole-protein–encoding mRNA not only induce antigen-specific activation of T cells in human peripheral blood mononuclear cells (PBMCs) but do so more effectively than the conventional synthetic viral peptide mixtures. Moreover, our data demonstrate the potency of these vectors in the therapeutic treatment of tumor-bearing animals, highlighting that this strategy protects mice against inoculation with cancer cells and cures mice with established cancers.
Results
Human CMV pp65 mRNA-CARTs Transfect PBMCs and Activate Antigen-Specific T Cells.
We have previously reported that in immortalized cell lines and in living animals mRNA-CART complexes are taken up by endocytosis, escape the endosomes, and release their mRNA cargo for cytoplasmic translation (Fig. 1A) (10). To test if CARTs are suitable for gene delivery to primary human cells, we used a GFP reporter readout in PBMCs from healthy donors. PBMCs were thawed and rested before treatment with GFP mRNA/CART complexes. The cells were then stained for surface lineage markers and analyzed for GFP expression by flow cytometry. Notably, we observed that GFP mRNA was delivered and translated in ∼50% of the dendritic cell (DC) population, ∼75% of the monocyte population, and ∼25% of the B cell population (SI Appendix, Fig. S1 A and B). With DCs, monocytes, and B cells representing the three major professional APC populations in humans, our data indicate the translational promise of using CARTs as vaccine-delivery agents in a clinical setting. Moreover, the observation that CARTs efficiently transfected ∼10% of the CD4+ T cells, 20% of the CD8+ T cells, and 20% of lineage-negative cells establishes the high transfection efficiency of CARTs across the wide phenotypic range found in PBMCs.
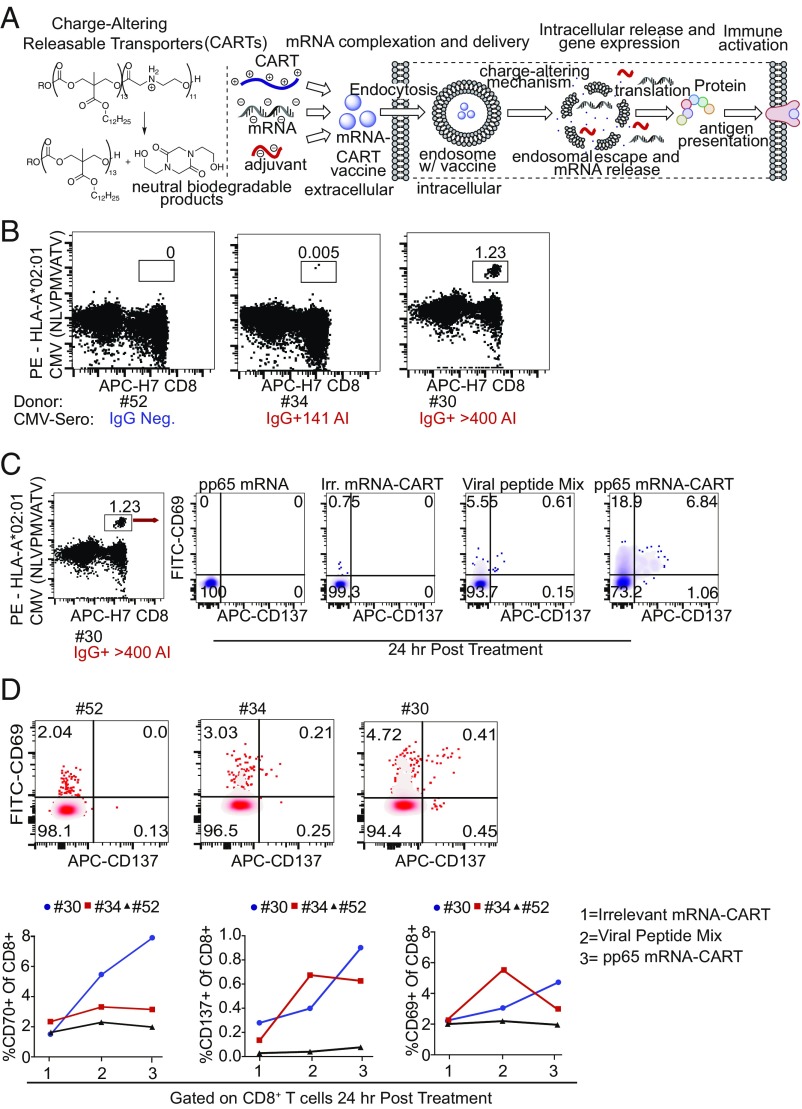
Fig. 1.
hCMV pp65 mRNA CARTs transfect PBMCs and activate antigen-specific T cells. (A, Left) Charge-altering mechanism responsible for release of mRNA for expression. (Center) CART + mRNA + CpG coformulation strategy. (Right) Mechanism of CART-mediated cellular uptake, endosomal escape, and release of mRNA leading to protein synthesis. (B) PBMCs from three HLA-A2+ donors were tested for the presence of CMV pp65 NLVPMVAATV-specific CD8 T cells by tetramer staining. Donors no. 30 and no. 34 were CMV+ by serological IgG test; donor no. 54 was CMV−. (C) PBMCs from these three donors were then treated with unformulated hCMV pp65 mRNA, alkaline phosphatase mRNA-CARTs [an irrelevant (Irr.) mRNA-CART], a viral peptide mix that includes several CMV epitopes, or hCMV pp65 mRNA-CARTs. (D) Expression of early activation markers CD137 and CD69 on tet+ CD8 T cells from donor no. 30 24 h post treatment. (Upper) CD137 and CD69 expression on all CD8 T cells from donors no. 52, no. 34, and no. 30 24 h post treatment with hCMV pp65 mRNA-CARTs. (Lower) Percent CD69+, CD70+, and CD137+ CD8 T cells of all CD8 T cells from donors no. 52, 34, and 30 24 h post treatment with irrelevant mRNA-CART, viral peptide mix, or hCMV pp65 mRNA-CART.
To investigate if the mRNA delivery capabilities of CARTs could be leveraged as an effective personalized vaccination strategy, we first sought to test the ability of mRNA-CARTs to evoke an antigen-specific T cell response in human PBMC cultures. To that end we tested T cell activation against epitopes within the human cytomegalovirus (hCMV) pp65 protein. The pp65 protein sequence contains several HLA-binding epitopes (25). In our experiments we chose two donors that we knew were CMV-positive and one that was negative by serological testing. All three donors were HLA*A02:01-positive, enabling us to test for CD8+ T cells specific to the HLA*A2-restricted pp65 epitope NLVPMVATV using an HLA-A*02:01 CMV pp65 (NLVPMVATV) tetramer. By flow cytometry we could detect a substantial tetramer-positive (tet+) CD8 T cell population in PBMCs from donor no. 30 and a small tet+ CD8 T cell population from donor no. 34 but no tet+ CD8 T cell population from the CMV-negative donor no. 52 (Fig. 1B). PBMCs from these three donors were then treated with either naked hCMV pp65 mRNA (unformulated), alkaline phosphatase mRNA-CART (an irrelevant mRNA-CART), a viral peptide mix that includes several CMV epitopes, or hCMV pp65 mRNA-CART. CD8 T cell activation was assayed by flow cytometry. For donor no. 30, we found that tet+ CD8 T cells from the hCMV pp65 mRNA-CART–treated PBMCs expressed high levels of early activation markers CD69 and CD137 (Fig. 1C) in striking contrast to treatment with naked hCMV pp65 mRNA, irrelevant mRNA-CART, or the viral peptide mix.
Previous studies have shown that the pp65 protein contains several immunogenic epitopes that are restricted to specific HLA alleles (25). All three donors had more than one HLA allele known to bind different epitopes from the pp65 protein (SI Appendix, Fig. S1C). Thus, we next assayed treatment-induced responses in whole T cell populations. Comparing CD8 and CD4 T cell expression levels of CD69, CD70, and CD137 in the different treatment groups, we observed robust CD8 and CD4 T cell activation in hCMV pp65 mRNA-CART–treated PBMCs from donors no. 30 and no. 34 but not from donor no. 52 (Fig. 1D and SI Appendix, Fig. S1E). Notably, CD8 T cell activation was increased by approximately one order of magnitude if assayed 72 h after treatment (SI Appendix, Fig. S1D).
Encouraged by the induction of a robust antigen-specific immune response in human PBMCs, we turned to animal models both to evaluate the therapeutic efficacy of mRNA-CART vaccination in vivo and to correlate these effects with specific immune responses in living animals. In an in vitro optical reporter assay, we previously demonstrated superior transfection efficiency and protein expression using the mRNA-CART delivery system benchmarked against the commercial transfection reagent Lipofectamine 2000 (∼98% versus ∼50%, respectively) (SI Appendix, Fig. S1F and ref. 10). To demonstrate the relevance of this superior transfection efficiency to an induced immunotherapeutic response, we first tested if antigenic epitopes presented on MHC I molecules could be detected on the surface of murine DC line DC2.4 cells following mRNA transfection. DC2.4 cells were transfected with ovalbumin (OVA) mRNA using Lipofectamine or OVA mRNA/CART complexes, and the presence of the well-known OVA-derived SIINFEKL peptide on MHC class I molecules was measured. Positive staining was observed only for OVA mRNA-CART–treated DC2.4 cells (SI Appendix, Fig. S1F). In an in vivo experiment, we performed s.c. vaccination of an OVA-specific CD8 T cell receptor transgenic mouse with 5 μg of either unformulated OVA mRNA or OVA mRNA-CART. Six days post vaccination we measured CD8 T cell activation by means of CD69, IFNγ, and the proliferation marker Ki-67 expression. Although activated CD8 T cells were observed in both groups, significantly more CD8 T cells were activated in mice vaccinated with OVA mRNA-CART (SI Appendix, Fig. S1G). Prompted by these findings, we decided to characterize the in vivo biodistribution of mRNA-CARTs and assay the optimal routes of administration in a murine model, with the eventual goal being vaccination against OVA-expressing lymphoma cells in wild-type BALB/c mice.
In Vivo Gene Delivery with CARTs.
To demonstrate that the mRNA delivery by CARTs translates to an effective vaccination strategy, the in vivo biodistribution following delivery of mRNA encoding the firefly luciferase (Fluc) reporter gene via two common vaccinations routes—i.v., and s.c.—was quantified by bioluminescence imaging (BLI). CARTs successfully delivered mRNA in vivo through both routes of administration (Fig. 2A). Signal intensity increased over the first hours after injection and peaked after 4 and 8 h for s.c.- and i.v.-injected mice, respectively (Fig. 2A). Notably, we observed an almost exclusive expression of Fluc mRNA in the spleen and lymph nodes of i.v.-injected mice (SI Appendix, Fig. S2A) while the signal remained localized at the site of s.c. injection (Fig. 2A). To observe in vivo transfection at very early time points after i.v. injection, blood samples and spleens were taken at 2, 5, and 10 min and were incubated for 2 h to allow protein translation. Even the earliest sample (2 min) showed detectable expression, consistent with literature reports that in mice the blood takes ∼30 s to circulate (26). This signal increased over the course of the first 10 min after injection with a preferential expression in circulating cells over spleen cells (SI Appendix, Fig. S2B).
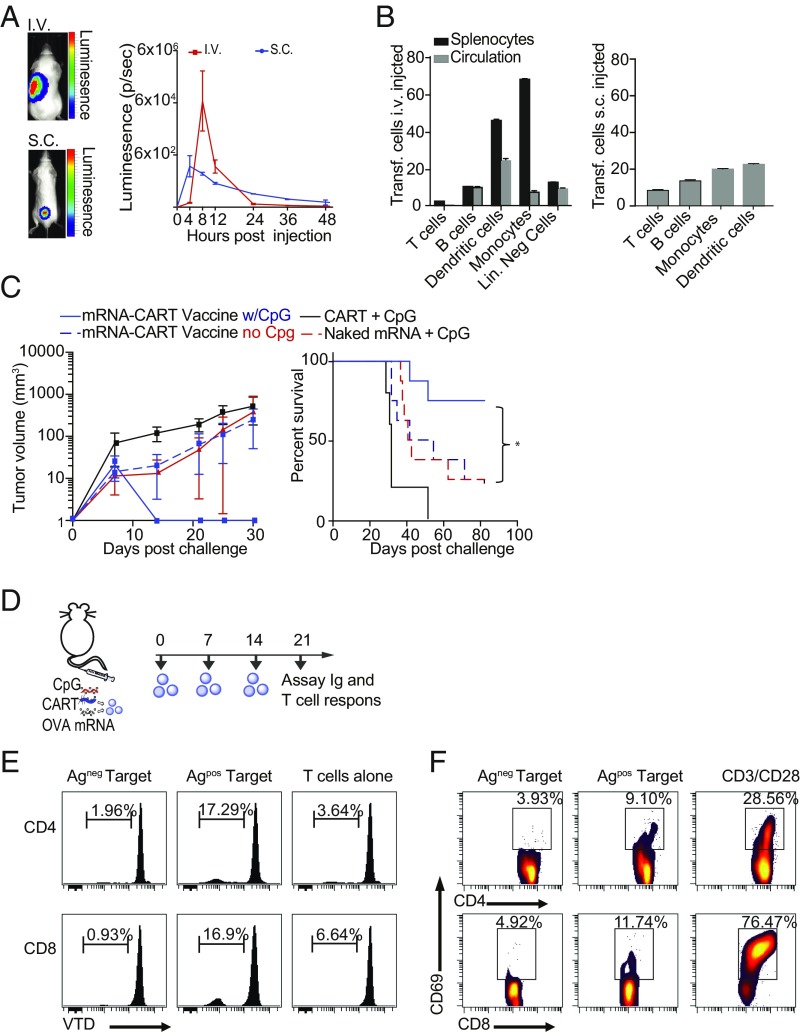
Fig. 2.
CARTs target professional APCs in secondary lymphoid organs and trigger therapeutic antigen- specific T cell responses. (A, Left) Representative bioluminescence in mice after s.c. (gated on injected site) and i.v. (gated on whole animal) injections of 7.5 μg of Fluc mRNA with CART. (Right) Kinetics of BLI in animals injected s.c. or i.v. with Fluc mRNA-CART. Data are the mean of three separate experiments ± SD. (B) Identification of in vivo transfected cells upon s.c. and i.v. injections of mRNA BDK-CART following injection of 10 μg mRNA complexed with BDK-CART. Skin and spleens of injected animals were collected 1 h post injection, and cells were stained for phenotypic markers. Graphs represent the fraction of BDK-CART–positive cells among the different cell phenotypes. The i.v. and s.c. results are each the mean of three separate BALB/c mice ± SD. Lin. Neg, lineage negative. (C) Tumor volume and percent survival of BALB/c mice inoculated with 107 A20-OVA cells and concurrently treated with mRNA-CART vaccine candidates. Variations of vaccine components including mRNA-CART vaccine (CART + 7.5 μg OVA-mRNA + 5 μg CpG; solid blue trace; n = 8), mRNA-CART vaccine without CpG (dashed blue trace; n = 8), CART alone (black trace; n = 5), CART + 5 μg CpG (solid red trace; n = 8), or 7.5 μg mRNA + 5 μg CpG (no CART; dashed red trace; n = 5). Data are representative of three individual experiments; n = 5–8 for all conditions. Mice were treated on day 0 only. Tumor size was measured every 2–4 d for the first 25–40 d after inoculation. Survival was monitored for >80 d. Mice that developed tumors that exceeded 15 mm in the largest diameter were killed. (D) Experiment setup. BALB/c mice were immunized i.v. through tail-vein injections with 10 μg OVA mRNA CART + CpG (mRNA CART vaccine) on days 0, +7, and +14. On day 21, sera were collected, and splenic CD4+ and CD8+ T cells were isolated. As control, we included serum from a naive mouse. (E) T cell proliferation in response to coculture with mitomycin C-treated OVA-expressing or -nonexpressing target cells. CellTrace Violet dye (VTD)-labeled CD4+ and CD8+ T cells were cocultured in a 1:1 ratio with mitomycin C-treated A20-OVA cells (Agpos), or wild-type A20 (Agneg) cells for 5 d. T cell proliferation was measured by dilution of VTD signal. T cells cocultured with OVA-expressing target cells divided substantially more than T cells left alone or cocultured with the Agneg target cell. (F) Antigen-specific ex vivo restimulation was measured by CD4 and CD8 T cells’ ability to up-regulate the activation marker CD69 when cocultured with Agpos or Agneg target cells in a 1:1 ratio for 24 h. As positive control, cells were activated in the presence of CD3/CD28 monoclonal antibodies to mimic T cell receptor engagement.
Prior work has demonstrated that in vivo expression levels using reporter genes such as GFP or mCherry are below detection limits for flow cytometric analysis (9). Therefore, to further evaluate the cellular distribution of CART/mRNA complexes within the transfected organs, we developed a CART functionalized with a difluoroboron-β-diketonate fluorophore (BDK-CART) (23, 27). This fluorophore has a high fluorescence quantum yield (28) and was easily installed on every CART molecule by using it as an initiator for ring-opening polymerization (SI Appendix, Fig. S2C), resulting in ∼1,000 fluorophores per mRNA molecule in the CART complex based on formulation stoichiometry. The BDK fluorophore shares excitation and emission properties with the well-known Pacific Blue fluorochrome and is easily detected by flow cytometry. In vitro flow cytometry analysis of HeLa cells transfected with complexes of a BDK-CART with a labeled Cy5-mRNA demonstrates a linear correlation between BDK-CART fluorescence and both the uptake of a Cy5-labeled mRNA and the resulting expressed GFP fluorescence, confirming this as a robust method for determining CART/mRNA complex biodistribution in vivo (SI Appendix, Fig. S2 C and D).
Using this BDK-CART/mRNA complex, we were able to identify transfected cells in the circulating blood, spleen, and skin isolated 1 h after injection. Flow cytometric analysis of isolated cells from the spleen, circulation, and skin of i.v.- and s.c.-injected animals revealed that BDK-CARTs are primarily taken up by monocytes and DCs but also are taken up to a substantial degree by B cells (Fig. 2B and SI Appendix, Fig. S2E). Transfected T cells were observed only in the spleen and skin and not in the circulation, which suggests that BDK-CART uptake happens both in the circulation and in the secondary lymphoid organs simultaneously (Fig. 2B and SI Appendix, Fig. S2 B and E). Notably, we were able to detect eGFP-transfected monocytes and DCs in the lymph nodes draining the site of s.c. injection 24 h after administration (SI Appendix, Fig. S2F), indicating either that these cells have migrated to the lymph nodes or that the mRNA/BDK-CART complex has entered circulation to transfect APCs at a different anatomical site. Thus, for the rest of the study s.c. injection/vaccination was the preferred route of administration due to the ease of injections and the control of potential local side effects. In short, the striking preferential mRNA delivery to professional APCs bodes well for the use of CARTs to deliver antigen mRNA for cancer vaccination.
Therapeutic Anticancer mRNA Vaccination Is CART Dependent and Augmented by Adjuvant Coformulation.
The clinical relevance of cancer vaccination strategies depends on the vaccine’s ability to induce therapeutic effects against established tumors. This requires the induction of robust antigen-specific immune responses. The induction of such responses often requires the use of an adjuvant (29). Based on our electrostatic complexation strategy, we envisioned being able to coformulate the polyanionic mRNA and short oligoanionic nucleic acid adjuvant CpG into the same CART nanoparticles (referred to as “mRNA-CART vaccine”). This strategy ensures that each cell is transfected with both the antigen mRNA and the adjuvant (SI Appendix, Fig. S2G). Importantly, formulating CpG into CARTs ensured that significantly more CpG was delivered to the transfected cells and was retained there compared with cells treated with FITC-CpG alone (SI Appendix, Fig. S2G). This coformulation maneuver allows the use of much lower CpG doses than would be required if relying on unassisted uptake alone.
In a series of same-day vaccination and tumor inoculation experiments, we tested whether CARTs are needed for efficient antigen delivery and the need for and effectiveness of different adjuvants in eliciting therapeutic immune responses. To test the need for CARTs and adjuvant in these studies, we compared our mRNA-CART(+CpG) vaccine formulation to mRNA-CART without added CpG and to naked mRNA+CpG. To control for the possible anticancer effect of CARTs alone, we excluded mRNA from two groups, one with CARTs alone and one with CARTs+CpG (Fig. 2C). Furthermore, to explore the therapeutic efficacy of adjuvants other than CpG, we compared the mRNA-CART vaccine (CpG as adjuvant) with mRNA-CART vaccine in which CpG was replaced with the TLR7 agonist resiquimod, the STING agonist DMXAA, or an agonistic CD40 antibody. Notably, these alternative adjuvants do not harbor the oligoanionic properties of CpG and thus are not complexed into CARTs. Instead, they were administered immediately following injection of the mRNA-CART complexes. As an additional comparison, we included complexed mRNAs encoding the costimulatory molecules CD80 and CD86 with OVA antigen mRNA into our CART formulation to mimic APC costimulation (SI Appendix, Fig. S2 I and J). As a positive control we compared the efficacy of the mRNA-CART vaccine to the established OVA protein + LPS vaccine strategy (SI Appendix, Fig. S2J). BALB/c mice were inoculated with 107 A20-OVA cells and were vaccinated the same day s.c. at a distant anatomical site with the indicated vaccine formulations. The efficacy of the different treatments was measured by means of tumor growth and overall survival. Compared with the mRNA-CART vaccine, we observed inferior responses in all other treatment groups (Fig. 2C and SI Appendix, Fig. S2 I and J). These data establish that CARTs are needed for efficient antigen mRNA delivery and that an adjuvant is needed for the induction of robust anticancer immunity (Fig. 2C). Moreover, CpG was superior to the other adjuvants tested, with agonistic CD40 antibody as the best alternative (Fig. 2C and SI Appendix, Fig. S2 I and J). The efficacy of CpG was not limited to one specific class of CpGs. In a side-by-side test we observed no significant difference in efficacy between class B and class C CpGs (SI Appendix, Fig. S2K). Importantly, induction of antitumor immunity was significantly better in mice vaccinated with the mRNA-CART vaccine than in mice vaccinated with the OVA protein+LPS. In addition, we were able to activate human DCs from PBMCs via high expression of HLA-DR when we coformulated CpG into the GFP mRNA-containing CARTs (SI Appendix, Fig. S2H). Thus, our strategy of using gene/antigen delivery with adjuvant (CpG) in the same particles is effective on human as well as mouse DCs.
mRNA-CART Vaccines Primarily Induce Antigen-Specific T Cell Responses.
Vaccination could induce antigen-specific B and/or T cell responses. To assess the functional response to our mRNA-CART vaccine, we immunized normal, non–tumor-bearing mice three times over the course of 14 d with 3 μg of OVA mRNA and analyzed antigen-specific T cell and Ig responses 7 d after the last immunization (Fig. 2 D–F). As a positive control for antigen-specific Ig responses, we vaccinated mice with OVA protein + LPS. Consistent with existing literature (30), we observed high titers of antigen-specific Ig in the sera of mice immunized with OVA protein + LPS (SI Appendix, Fig. S2L). However, only trace amounts of OVA-specific Ig were detected in the sera of mice immunized with mRNA-CART vaccine. Notwithstanding, we were able to detect robust OVA-specific T cell responses from these mice immunized with mRNA-CART vaccine. Both CD4+ and CD8+ T cells responded by proliferation and up-regulation of the activation marker CD69 when cocultured with mitomycin C-fixed antigen-expressing target cells (AgPos) but not with antigen-negative (AgNeg) target cells (Fig. 2 D–F). Hence, the mRNA-CART vaccine primarily induces antigen-specific T cell responses.
mRNA-CART Vaccines Induce Long-Lasting, Cytotoxic, Antigen-Specific T Cell Responses.
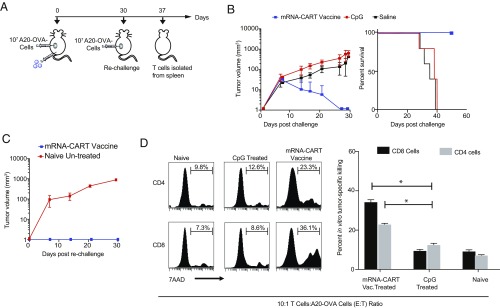
We tested the antigen-specific reactivation and cytotoxic activity of explanted T cells from vaccinated mice. Mice were vaccinated and challenged with 107 A20-OVA cells on the same day. Tumor growth and survival were monitored, and mice that had successfully rejected the tumors were rechallenged on day +30 to test the protective recall responses against a second challenge with the same tumor (Fig. 3 A–C). The rechallenged group was then divided in two. The first group was monitored for tumor growth over the next 40 d. As control for establishing new tumors, we included a group of naive mice that received the same amount of tumor cells but no prior treatment. The rechallenged mice completely rejected the second challenge, while the previously unchallenged and untreated mice developed tumors. T cells were isolated from the spleen of a parallel set of mice 7 d after tumor challenge and were tested in vitro for cytotoxicity and antigen-specific reactivation. We included T cells isolated from the spleens of tumor-bearing CpG-treated mice and naive mice as negative controls and supraphysiological activation with CD3 and CD28 antibodies as a positive control. Isolated CD4 and CD8 T cells were either cocultured in a 1:1 ratio with mitomycin-fixed AgPos target cells to assay antigen-specific reactivation or were cocultured with AgNeg cells or an irrelevant target (CT26 colon carcinoma) in a 10:1 effector:target ratio to test antigen-specific cell cytotoxicity. Reactivation was measured by means of IFNγ up-regulation (SI Appendix, Fig. S3C). Cytotoxicity was also measured in the target population. By means of IFNγ production, we observed a profound up-regulation in both CD4 and CD8 T cells from vaccinated mice when cocultured with AgPos target cells but not in T cells from CpG-treated mice (SI Appendix, Fig. S3C). Moreover, both CD4+ and CD8+ T cells explanted from vaccinated and protected mice were able to kill target cells efficiently in vitro (Fig. 3D). CD4+ T cell cytotoxicity was antigen-restricted (Fig. 3D and SI Appendix, Fig. S3B). Significantly, CD8+ cells killed both A20-OVA cells and wild-type A20 cells with similar efficiency but did not kill unrelated CT26 colon carcinoma cells (Fig. 3D and SI Appendix, Fig. S3B). This striking result indicates preferential CD8+ recognition of antigens shared between wild-type A20 and A20-OVA. Notably, T cells from naive mice and mice treated only with CpG were not able to kill any target efficiently, regardless of antigen expression (Fig. 3D). The OVA-independent cytotoxicity of CD8+ T cells and the lack of cytotoxicity of CD8+ T cells from the unprotected control indicates that a robust OVA-specific CD4+ T cell response is needed to bolster the CD8+ T cell response. In sum, these data show that the therapeutic mRNA-CART vaccine primarily induces cytotoxic T cell responses.
Fig. 3.
mRNA-CART vaccine induces long-lasting cytotoxic antigen-specific T cell responses. In vitro cytotoxic activity of T cells explanted from vaccinated and protected mice against antigen-expressing tumor cells. (A) BALB/c mice were inoculated with 107 A20-OVA cells. On day 0, mice were treated with mRNA-CART vaccine (CART + 7.5 μg OVA mRNA + 5 μg CpG; blue; n = 10), 5 μg CpG alone (n = 10), or saline (n = 5). On day +30, all mice in the mRNA-CART vaccine group had completely rejected the tumor; these mice were then rechallenged with 107 A20-OVA cells s.c. On day 37, T cells were isolated from the spleens of 5 out of 10 of the rechallenged mice. (B) Tumor growth (Left) and percent survival (Right) were monitored every 2–4 d for the first 30 d after tumor inoculation. (C) On day +30, mice in the mRNA-CART vaccine group (red trace; n = 5) were rechallenged with 107 A20-OVA cells s.c. and compared with naive mice (blue trace; n = 5). (D, Left) Isolated CD4+ and CD8+ T cells from naive and CpG- and mRNA-CART–treated mice (n = 2 each) show in vitro cytotoxicity against A20-OVA cells. (Right) CD4+ and CD8+ T cells from mRNA-CART–vaccinated, protected, and rechallenged mice, CpG-treated mice, and naive mice were cocultured for 18 h with OVA-expressing A20 cells in a 10:1 effector-to-target ratio. Cytotoxicity was measured by the percentage of 7-AAD+ cells in the target population. Data represent the mean ± SD of two or three individual experiments. *P > 0.05, nonparametric Mann–Whitney U test.
mRNA-CART Vaccines Cure Established Tumors.
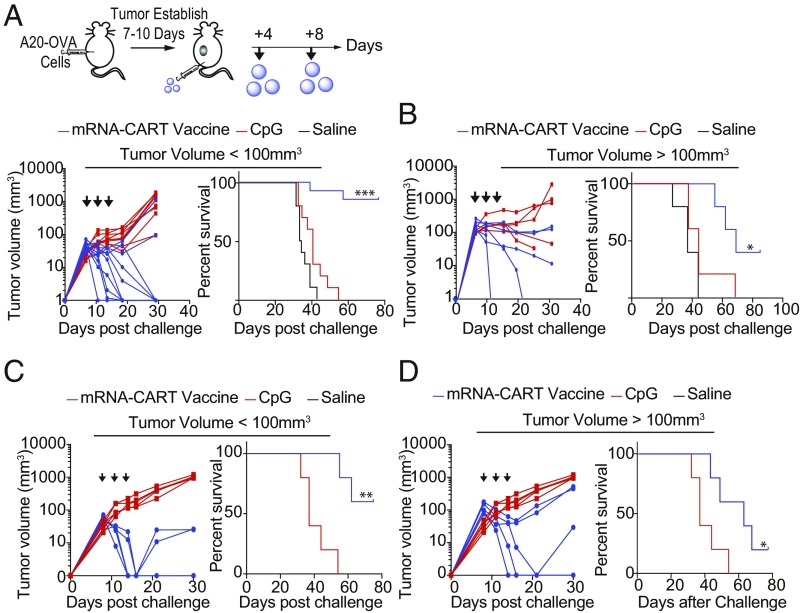
We tested the efficacy of the mRNA-CART vaccine on established tumors in mice that had either medium (<100 mm3) or large (>100 mm3) tumor volume at the start of the treatment. BALB/c mice were inoculated s.c. with 107 A20-OVA lymphoma cells. After tumors were established, we started our treatment regimen with s.c. injections of either CART-formulated mRNA-CART vaccine (3 μg OVA-mRNA+5 μg CpG) or 5 μg of CpG alone. Mice were treated every 4 d for a total of three treatments. Tumor growth and overall survival were monitored over the course of 30 and 75 d, respectively (Fig. 4 A and B). In mice with medium tumor volume, we observed complete regression of tumors in 8 of 10 mice and a significant prolonged survival exceeding 75 d compared with no survival in the animals treated only with CpG. In the second group with established tumor volume >100 mm3, we observed a significant delay in tumor growth and the complete cure of 40% of the treated animals (Fig. 4 A and B). Notably, this profound therapeutic effect of the mRNA-CART vaccine on established tumors was not limited to the s.c. route of administration but could be reproduced when treatments were given i.v. (Fig. 4 C and D).
Fig. 4.
Low-dose mRNA-CART vaccines cure large, established experimental tumors. (A) Established tumor vaccination schedule and outcomes for BALB/c mice containing either medium-sized or large A20-OVA tumors. (Upper) Experiment setup for s.c. treatments. Mice received a total of three treatments on day 7 (day 10 for large tumors), 11 (day 13), and 14 (day 17) after tumor inoculation. Tumor size was measured every 2–4 d for the first 35 d after inoculation. Survival was monitored for >30 d. Mice that developed tumors >15 mm in the largest diameter were killed. (Lower) Tumor volumes and survival curves for mice that developed medium-sized tumors (50–100 mm3) and were treated with either 3 μg OVA mRNA CART coformulated with 5 μg CpG (mRNA-CART vaccine) (blue traces; n = 10), received 5 μg CpG only (red traces; n = 10), or a saline solution (black traces; n = 5). Results are representative of two individual experiments. (B) Tumor volumes and Kaplan–Meier survival curves for mice that developed tumors <100 mm3 (Left) or large tumors >100 mm3 (Right) and were treated with mRNA-CART vaccine (blue traces; n = 5) or received 5 μg CpG only (red traces; n = 5) or a saline solution (black traces; n = 5). All treatments were administered s.c. on the anatomical site opposite the tumors (on the back near the root of the tail). (C) Tumor volumes (Left) and survival curves (Right) for mice that developed medium-sized tumors (50–100 mm3) and were treated with either 3 μg OVA mRNA CART coformulated with 5 μg CpG (mRNA-CART vaccine) (blue traces; n = 5) or received 5 μg CpG only (red traces; n = 5). (D) Tumor volumes (Left) and Kaplan–Meier survival curves (Right) for mice that developed large tumors (>100 mm3) and were treated with either mRNA-CART vaccine (blue traces; n = 5) or received 5 μg CpG only (red traces; n = 5). All treatments were administered i.v. *P > 0.05, **P > 0.005, ***P > 0.0005, Kaplan–Meier log-rank test. Data are shown as mean ± SD. All data are representative of two or three individual experiments.
Discussion
The efficient delivery of mRNA to APCs in vivo will enable the clinical advancement of personalized therapeutic cancer vaccination. An essential and unsolved obstacle to achieving this goal is safe and effective mRNA delivery. While the majority of work in the field has focused on a single delivery strategy using LNPs (14, 17, 18), we have shown that CARTs are remarkably effective in delivering customized functional mRNAs to APCs of the immune system. These studies have resulted in the striking demonstration of an mRNA-CART vaccine for the induction of polyclonal antigen-specific human T cells in vitro and the safe and therapeutic eradication of large, established tumors in vivo.
In our studies, we have investigated the mechanisms underlying this curative vaccination strategy. Using fluorescently labeled CARTs and reporter systems, we have demonstrated that CARTs are able to package both mRNA molecules and the synthetic TLR9 agonist CpG into a nanoparticle complex which can be administered through both i.v. and s.c. routes. mRNA-CART vaccines efficiently transfect human APCs in vitro and mouse APCs in the spleen and lymphatic system, providing the enabling foundation for mRNA vaccination. Upon delivery, mRNA is efficiently translated, processed, and presented by MHCs. Codelivery of a strong TLR9 agonist ensures the bona fide activation of the APCs and the concurrent up-regulation of costimulatory molecules. The versatility of the CART platform to deliver multiple components to achieve a powerful synergistic effect grants the ability to fine-tune and amplify the immunogenicity of the vaccine.
In a lymphoma model, we have investigated the power of therapeutic vaccination for both large (>100 mm3) and medium-sized (50–100 mm3) tumors using only three doses of 3 μg of OVA-mRNA coformulated with 5 μg of CpG. Others have shown therapeutic efficacy with mRNA-based vaccines, but few are able to cure mice with medium to large established tumors (8, 9). In addition, compared with existing mRNA vaccination platforms, we have developed an efficient strategy that uses relatively small amounts of mRNA and few treatments. We provide evidence that our delivery vehicle combines high transfection efficacy, biocompatibility, selectivity, and specificity with minimal toxicity (10). We believe that our delivery system and vaccination methodology represent an attractive alternative to existing LNP vaccination platforms (1, 14, 17, 18).
We have sought to establish a fundamental understanding of the immunological mechanisms behind these mRNA-CART vaccines. Notably, we have demonstrated a robust and long-lasting antigen-specific T cell response both in vitro and in vivo as a result of vaccination. Our CART construct on its own has relatively low systemic immunogenicity, and its coformulation with CpG enhances the maturation and priming capability of professional APCs (29). Thus, the ability to codeliver antigen and adjuvant represents a strategy to manipulate and optimize the immune response.
We have shown that mRNA-CART vaccines are effective through multiple routes of administration. We have shown that CARTs are primarily taken up by professional APCs and that the majority of transfected cells reside in the secondary lymphoid tissues upon i.v. injection. Moreover, i.v. injection of antigen-mRNA CART+CpG mediates robust induction of long-lived antigen-specific T cells. However, s.c. or intracutaneous injections are still the preferred route of administration for most vaccines. The availability of APCs in skin is limited compared with the secondary lymphoid organs and circulation. Vaccinations at these sites must attract APCs to the site of injection (31), enter circulation, or deliver antigen to skin-resident APCs able transport the injected antigen to the secondary lymphoid organs (32). As mRNA is short lived in vivo, the time from the delivery of antigen to its presentation on MHC class I and II molecules to antigen-specific T cells is of essence. We have shown that CARTs are taken up locally by APCs that are either recruited or resident in the skin and that APCs that have taken up the CART can be found in the draining lymph nodes as early as 24 h after injection. Thus, we have indications that CARTs could be suitable for both s.c./intradermal and i.v. vaccination, making CARTs extremely versatile.
Treating “cold tumors,” i.e., tumors with limited immune infiltrate, is emerging as one of the largest challenges in immunotherapy. As these tumors do not respond to conventional checkpoint inhibitors, there is an expressed need for innovative approaches. Combined with emerging advances in sequencing, mRNA modifications and synthesis, and antigen identification, the potential of mRNA drugs has come to the forefront of novel therapeutics. These next-generation applications will be aided by the development of new delivery vehicle platforms that may enable preferential targeting to DCs, different expression profiles, and altered immunogenicity and cytotoxicity. CARTs are a unique class of tunable, nontoxic, and dynamic delivery agents for mRNA vaccines that address the limitations of existing approaches and have clinical promise for the treatment of established tumors.
Materials and Methods
CpG.
FITC-conjugated CpG 1826 with sequence 5′-TCCATGACGTTCCTGACGTT was purchased from InvivoGen. Type C CpG-C SD-101 was provided by Dynavax Technologies Corporation. For in vivo applications, CpG was administered alone or was coformulated with CARTs at a final concentration of 5 μg per dose.
CART.
CART D13:A11 was prepared as previously described using benzyl alcohol as an initiator and matched reported characterization. Fluorescent BDK-CART was also prepared as previously described using the difluoroboron dibenzoylmethane initiator prepared according to a procedure described by Fraser and coworkers (33). End group analysis of the protected polymer showed block lengths of 12 dodecyl carbonate monomers and 14 cationic aminoester monomers.
Flow Cytometry.
The following fluorochrome-conjugated rat anti-mouse mAbs were used for flow cytometry: CD4-PE, B220-FITC, CD8-FITC, CD11c-PerCP-Cy5.5, CD11b-APC, IFNγ-PE, CD3 Alexa Fluor 700, 7AAD-APC, and rat isotype controls for the listed fluorochromes. The fluorochrome-conjugated hamster anti-mouse mAb CD69-PE was used. The rat anti-mouse unconjugated antibody anti-CD16/32 was used. The following mouse anti-human mAbs were used: CD11c-PE, CD14-APC, CD19-BV711, HLA-DR-V450, CD4-BV605, CD8-PErCP-Cy5.5, CD8-APC-H7, CD69-FITC, CD137-APC, and CD70-PE. Tetramer staining was done with HLA-A*02:01 CMV pp65 (NLVPMVATV) (MBL International). Staining using two Brilliant Violet fluorochromes was performed in Brilliant stain buffer according to the manufacturer’s protocol (BD Biosciences). For intracellular staining, cells were treated with GolgiStop (BD Biosciences) for 5 h before staining. Cells were fixed and permeabilized according to the manufacturer’s protocol (BD biosciences). Cell proliferation was measured by dilution of CellTrace Violet dye (VTD). Cells were labeled with VTD according to the manufacturer’s protocol (Thermo Fisher Scientific). Antibodies were purchased from BD Biosciences, Invitrogen, or eBioscience. All surface marker staining was done for 20 min at room temperature in PBS (Gibco) containing 0.01% sodium azide (Sigma Aldrich) and 0.5% BSA (Sigma). Stained cells either were fixed in 2% paraformaldehyde or were run fresh and were analyzed by flow cytometry on a FACSCalibur or LSR II system (BD Biosciences). Data were analyzed using either Cytobank (Cytobank Inc.) or FlowJo version 10.0 (FlowJo).
Mice and Cell Lines.
Eight- to twelve-week-old female BALB/c mice and C57BL/6-Tg(TcraTcrb)1100Mjb/J mice were purchased from The Jackson Laboratory and housed in the Laboratory Animal Facility of the Stanford University Medical Center. All experiments were approved by the Stanford Administrative Panel on Laboratory Animal Care and were conducted in accordance with Stanford University Animal Facility and NIH guidelines. The A20 cell line was obtained from ATCC (ATCC no. TIB-208). It is a BALB/c B cell lymphoma line derived from a spontaneous neoplasm found in an old BALB/cAnN mouse, expressing MHC class I and class II H-2d molecules. A20 wild-type tumor was transfected with an expression plasmid DNA encoding a fusion of chicken OVA with the trans membrane and cytoplasmic domains of the transferrin receptor (“membrane OVA”) under hygromycin selection. Both MHC I and II OVA epitopes are presented by the transfectant (34, 35). It was grown in complete medium containing 200 μg/mL hygromycin (Invitrogen). CT26 colon carcinoma cells, the DC2.4 murine DC line, and human cervix carcinoma HeLa cells were purchased from ATCC. Tumor cells were cultured at 37 °C in 5% CO2 in RPMI medium 1640 with l-glutamine (cellgro; Corning) supplemented with 10% heat-inactivated FCS (HyClone), 100 international units/mL penicillin (Gibco), 100 μg/mL streptomycin (Gibco), and 50 μM 2-mercaptoethanol (Gibco), as complete medium.
In Vivo Bioluminescence.
For bioluminescence assessment, mice were anesthetized with isoflurane gas (2% isoflurane in oxygen, 1 L/min) during injection and imaging procedures. The i.p. injections of d-Luciferin (Biosynth AG) were done at a dose of 150 mg/kg, providing a saturating substrate concentration for the Fluc enzyme (luciferin crosses the blood–brain barrier). Mice were imaged in a light-tight chamber using an in vivo optical imaging system (IVIS 100; Xenogen Corp.) equipped with a cooled charge-coupled device camera. During image recording, mice inhaled isofluorane delivered via a nose cone, and their body temperature was maintained at 37 °C in the dark box of the camera system. Bioluminescence images were acquired between 10 and 20 min after luciferin administration. Mice usually recovered from anesthesia within 2 min of imaging.
Tumor Transplantations and Assessment.
A20-OVA B lymphoma cells (107) were implanted into mice while in the exponential growth phase (<1.5 × 106 cells/mL). The s.c. injections were done with tumor cells washed twice in PBS medium and resuspended in 100 μL PBS. The s.c. tumors were implanted on the left flanks. Tumor size was monitored with a digital caliper (Mitutoyo) every 2–3 d and was expressed as volume (length × width × height). Mice were killed when tumor size reached 1.5 cm in the largest diameter, per guidelines.
General CART Formulation Methods.
For all experiments, CARTs were formulated with mRNA at a 10:1 cation:anion ratio assuming full protonation of the CART and full deprotonation of the oligonucleotide. Formulations were carried out in acidic PBS (pH adjusted to 5.5 by the addition of 0.1 M HCl) before injection (in vivo) or to treatment wells (in vitro). In experiments in which CpG was used, CpG and mRNA were first premixed in acidic PBS, and the amount of CART used was adjusted to maintain a 10:1 cation:anion ratio considering the additional charges on the CpG. For all formulations, cargo (mRNA and/or CpG) was first premixed in PBS (pH 5.5), and then CART was added, followed by 20 s of mixing and immediate injection. In experiments in which Lipofectamine 2000 was used, it was formulated in Opti-MEM (Gibco) according to the manufacturer’s suggested protocol.
CART in Vitro Transfection.
For in vitro transfections, HeLa cells were seeded at 40,000 cells per well in 24-well plates or PBMCs were seeded at 50,000 cells per well in 96-well plates. Immediately before treatment, cells were washed and resuspended in serum-free medium. CART formulations were made using the general method described above. In a standard in vitro experiment, 0.42 μg of mRNA (2.1 μL of a 0.2-mg/mL stock) was added to 5.71 μL of PBS (pH 5.5). To this was added 0.59 μL of CART (from a 2-mM stock), and this was mixed for 20 s. Then 2.5 μL of this formulation was added to each of three wells, resulting in a final mRNA dose of 125 ng per well. This was incubated for 8 h, after which gene expression was determined by antibody staining or direct flow cytometric analysis.
Vaccinations/Immunizations.
CART formulations were made according to the general procedure described above. For a standard vaccination, 7.5 μg of OVA-mRNA (37.5 μL of a 0.2-mg/mL stock) and 5 μg CpG (6.25 μL of a 0.8-mg/mL stock) were added to 38.7 μL of PBS (pH 5.5). To this was added CART (17.56 μL of a 2-mM stock), and the mixture was mixed for 20 s before injection. Four hundred micrograms of the STING agonist DMXAA (Sigma Aldrich) was given in a separate syringe. Five micrograms of the TLR7 agonist resiquimod (Invivogen) was given in a separate syringe. Thirty micrograms of agonistic α-CD40 Ab (FGK45; BioXcell) was given i.p. CD80 and CD86 mRNA was coformulated with OVA-mRNA into CARTs and administered as one dose. Fifty micrograms of VacciGrade Ova protein (Sigma Aldrich) was combined with 25 μg of VacciGrade LPS (Sigma Aldrich) and was given as one dose. All antigen (OVA) immunizations were administered s.c. near the root of the tail. The i.v. injections were done in the tail vein after mice had been warmed for 2 min under a heating lamp.
mRNA.
Fluc, OVA, eGFP, and secreted alkaline phosphatase (SEAP) mRNA were purchased from TriLink BioTechnologies. All purchased mRNAs were capped and polyadenylated and contained 5-methoxyuridine modification.
hCMV pp65, CD80, and CD86 mRNA were produced in-house by cloning the CD80 and CD86 genes into pcDNA3.1(+) plasmids. Plasmids were linearized using the DraIII restriction enzyme (New England BioLabs) and extracted using phenol:chloroform extraction with subsequent alcohol precipitation. mRNA was then transcribed from the linearized plasmids using the mMESSAGE mMACHINE T7 Transcription Kit (Thermo Scientific).
CART Transfection of Human PBMCs.
PBMCs from healthy donors were isolated by density-gradient centrifugation using the Ficoll-Hypaque technique (Amersham Biosciences). Cells were rested for 1 h after thawing and were transfected with 125 ng of CART formulated with eGFP ± CpG. All specimens were obtained with informed consent in accordance with the Declaration of Helsinki, and this study was approved by Stanford University’s Administrative Panels on Human Subjects in Medical Research. Samples were collected from patients by peripheral blood leukapheresis and were cryopreserved.
Mitomycin C Treatment.
Cells (1 million/mL) were treated with 50 μg of mitomycin C for 30 min in a rotator at 37 °C in the dark and then were washed out three times with HBSS supplemented with 5% FCS.
Immune Cell Processing, T Cell Isolation, and Effector Function.
Lymph nodes and spleens were turned into single-cell suspensions by processing them through a 70-μm cell strainer (BD Biosciences). Skin samples were prepared by isolating and treating the mouse dermis with 1 mg/mL collagenase IV (Sigma Aldrich) and 0.3 mg/mL DNase I (Sigma Aldrich) for 30 min at 37 °C before single-cell suspensions were prepared as described above. Untouched CD4+ and CD8+ T cells were obtained by negative selection from splenocytes (Miltenyi Biotech). T cells were mixed with autologous A20-OVA lymphoma cells, WT A20 lymphoma cells, or CT26 colon carcinoma cells at an effector-to-target ratio of 10:1. Autologous tumor cells were labeled with CellTrace Violet (Thermo Fisher). After 18 h, cells were stained with 7-aminoactinomycin D (7-AAD) to determine cell death. Target tumor cells were gated by CellTrace Violet labeling.
ELISA.
Plasma levels of αOVA antibodies were measured by ELISA. Plasma was collected from immunized mice on day 21 after completion of three immunizations with mRNA-CART vaccine with CpG, mRNA-CART vaccine without CpG, or 50 μg OVA protein with 25 μg LPS and was added to 96-well MaxiSorp plates (Nunc) coated with OVA protein (Sigma Aldrich). Bound antibodies were detected using HRP-conjugated goat anti-mouse IgG1, IgG2a, IgG2b, or IgG2c (IgG3) (R&D Systems). Samples were diluted from 1:10 to 1:10,000, and absorbance was determined at 650 nm using a VMax Microplate Reader (Molecular Devices).
Statistical Analysis.
Prism software (GraphPad) was used to analyze tumor growth and to determine the statistical significance of differences between groups by applying a nonparametric Mann–Whitney U test. P values <0.05 were considered significant. The Kaplan–Meier method was employed for survival analysis.
Supplementary Material
Acknowledgments
We thank Debra Czerwinski for technical help, guidance, and flow cytometry assistance; Kayvon Pedram for synthetic work; and the Dynavax Corporation for the gift of the CpG formulation SD101. This work was supported by NIH Grant R35CA197353 (to R.L.); National Science Foundation Grant CHE-1607092 (to R.M.W.); NIH Grants CA031841 and CA031845 (to P.A.W.); the Norwegian Cancer Society and the Norwegian Health Authorities (O.A.W.H); Stanford Cancer Translational Nanotechnology Training T32 Training Grant CA196585 funded by the National Cancer Institute (to T.R.B.); and the Stanford Center for Molecular Analysis and Design (C.J.M.). Flow cytometry data were collected on an instrument in the Stanford Shared FACS Facility obtained using NIH S10 Shared Instrument Grant S10RR027431-01.
Footnotes
Conflict of interest statement: Reviewers P.M.S. and M.C. and author R.L. are coauthors of a 2016 review article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810002115/-/DCSupplemental.
References
- 1.van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief CJ. Vaccines for established cancer: Overcoming the challenges posed by immune evasion. Nat Rev Cancer. 2016;16:219–233. doi: 10.1038/nrc.2016.16. [DOI] [PubMed] [Google Scholar]
- 2.Khodadoust MS, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543:723–727. doi: 10.1038/nature21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 4.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kauffman KJ, et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 7.Sahin U, Kariko K, Tureci O. mRNA-based therapeutics–Developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 8.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 9.Oberli MA, et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinlay CJ, et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc Natl Acad Sci USA. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theodore F, Roblin R. Gene therapy for human genetic disease? Science. 1972;175:949–956. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W-Z. RNA melanoma vaccine: Induction of antitumor immunity by human glycoprotein 100 mRNA immunization. Hum Gene Ther. 1999;10:2719–2724. doi: 10.1089/10430349950016762. [DOI] [PubMed] [Google Scholar]
- 14.Hajj KA, Whitehead KA. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat Rev Mater. 2017;2:17056. [Google Scholar]
- 15.Ramamoorth M, Narvekar A. Non viral vectors in gene therapy–An overview. J Clin Diagn Res. 2015;9:GE01-06. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Ortin JE, Alepuz P, Chavez S, Choder M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 17.Quader S, Kataoka K. Nanomaterial-enabled cancer therapy. Mol Ther. 2017;25:1501–1513. doi: 10.1016/j.ymthe.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Lint S, et al. The ReNAissanCe of mRNA-based cancer therapy. Expert Rev Vaccines. 2015;14:235–251. doi: 10.1586/14760584.2015.957685. [DOI] [PubMed] [Google Scholar]
- 19.Chahal JS, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci USA. 2016;113:E4133–E4142, and erratum (2016) 113:E5250. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JB, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramaswamy S, et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc Natl Acad Sci USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramunas J, et al. Transient delivery of modified mRNA encoding TERT rapidly extends telomeres in human cells. FASEB J. 2015;29:1930–1939. doi: 10.1096/fj.14-259531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinlay CJ, Benner NL, Haabeth OA, Waymouth RM, Wender PA. Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters. Proc Natl Acad Sci USA. 2018;115:E5859–E5866. doi: 10.1073/pnas.1805358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Kondo E, et al. Identification of novel CTL epitopes of CMV-pp65 presented by a variety of HLA alleles. Blood. 2004;103:630–638. doi: 10.1182/blood-2003-03-0824. [DOI] [PubMed] [Google Scholar]
- 26.Welsher K, Sherlock SP, Dai H. Deep-tissue anatomical imaging of mice using carbon nanotube fluorophores in the second near-infrared window. Proc Natl Acad Sci USA. 2011;108:8943–8948. doi: 10.1073/pnas.1014501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benner NL, et al. Functional DNA delivery enabled by lipid-modified charge-altering releasable transporters (CARTs) Biomacromolecules. 2018;19:2812–2824. doi: 10.1021/acs.biomac.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, et al. Multi-emissive difluoroboron dibenzoylmethane polylactide exhibiting intense fluorescence and oxygen-sensitive room-temperature phosphorescence. J Am Chem Soc. 2007;129:8942–8943, and erratum (2007) 129:15728. doi: 10.1021/ja0720255. [DOI] [PubMed] [Google Scholar]
- 29.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: Putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharjee AK, Izadjoo MJ, Zollinger WD, Nikolich MP, Hoover DL. Comparison of protective efficacy of subcutaneous versus intranasal immunization of mice with a Brucella melitensis lipopolysaccharide subunit vaccine. Infect Immun. 2006;74:5820–5825. doi: 10.1128/IAI.00331-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludewiga B, et al. In vivo antigen loading and activation of dendritic cells via a liposomal peptide vaccine mediates protective antiviral and anti-tumour immunity. Vaccine. 2000;19:23–32. doi: 10.1016/s0264-410x(00)00163-8. [DOI] [PubMed] [Google Scholar]
- 32.Lin K, Roosinovich E, Ma B, Hung CF, Wu TC. Therapeutic HPV DNA vaccines. Immunol Res. 2010;47:86–112. doi: 10.1007/s12026-009-8141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G, et al. Multi-emissive difluoroboron dibenzoylmethane polylactide exhibiting intense fluorescence and oxygen-sensitive room-temperature phosphorescence. J Am Chem Soc. 2007;129:8942–8943. doi: 10.1021/ja0720255. [DOI] [PubMed] [Google Scholar]
- 34.Ding ZC, Blazar BR, Mellor AL, Munn DH, Zhou G. Chemotherapy rescues tumor-driven aberrant CD4+ T-cell differentiation and restores an activated polyfunctional helper phenotype. Blood. 2010;115:2397–2406. doi: 10.1182/blood-2009-11-253336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marabelle A, et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J Clin Invest. 2013;123:2447–2463. doi: 10.1172/JCI64859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.