Abstract
Metastatic disease remains the primary cause of mortality in cancer patients. Yet the number of available in vitro models to study metastasis is limited by challenges in the recapitulation of the metastatic microenvironment in vitro, and by difficulties in maintaining colonized-tissue specificity in the expansion and maintenance of metastatic cells. Here, we show that decellularized scaffolds that retain tissue-specific extracellular-matrix (ECM) components and bound signaling molecules enable, when seeded with colorectal cancer (CRC) cells, the spontaneous formation of three-dimensional cell colonies that histologically, molecularly and phenotypically resemble in vivo metastases. Lung and liver metastases obtained by culturing CRC cells on, respectively, liver and lung decellularized scaffolds retained their tissue-specific tropism when injected in mice. We also found that the engineered metastases contained signet ring cells, which has not previously been observed ex vivo. A culture system with tissue-specific decellularized scaffolds represents a simple and powerful approach for the study of organ-specific cancer metastases.
Introduction
Metastasis is the main cause of morbidity and mortality in cancer patients. Although it is generally accepted that tissue-specific microenvironments play an important role in modulating the behavior of metastases1, the biology underlying this interaction remains poorly understood, owing to a lack of accessible experimental models that demonstrate organ-specificity2. Conventional in vitro systems are not suitable for the study of metastatic cancer. Although collagen and Matrigel can be used to provide a 3D culture matrix more comparable to the 3D environment encountered in vivo, the composition of these substrata are highly dissimilar from the tissue-specific microenvironments encountered by metastases1,3. Although genetically engineered animal models that develop metastases in vivo can be used to study metastatic cancer in a tissue-specific manner, they are costly and difficult to use. Understanding the biology underlying the interaction between metastases and the tissue microenvironment they inhabit may lead to more effective cancer treatments for metastatic cancer patients, as prior studies have determined that the treatment response of metastases can differ between metastatic sites3,4.
A recent breakthrough in the field of tissue engineering has been the development of tissue decellularization methods especially those done by perfusion protocols5–11. Decellularization is a technique wherein an organ is chemically stripped of its cells, leaving behind an intricately structured extracellular matrix5,12. Importantly, decellularization preserves the complex composition of extracellular matrices found in normal organs, which would be nearly impossible to recreate using synthetic techniques. We hypothesized that we could use decellularized tissues to create a tissue-specific in vitro culture system to engineer cancer “metastases” (Figure 1a). While previous studies have successfully used a variety of methods to decellularize tissues and engineer complex organs, including liver and lung, the degree to which cell signaling molecules are preserved using these methods remains largely unknown6,7,9,10. Consequently, we utilized a unique decellularization technique that retains >98% of the tissue’s decellularized matrix components and preserves physiological levels of matrix-bound growth factors and cytokines11. Decellularized tissues derived using this technique have been termed “biomatrix scaffolds (BMSs)”11. As proof of concept, we use our culture system to study metastatic CRC. Given that liver and lung are the most common sites of metastasis in CRC patients, we aimed to engineer in vitro liver and lung metastases that can be utilized for therapeutic studies.
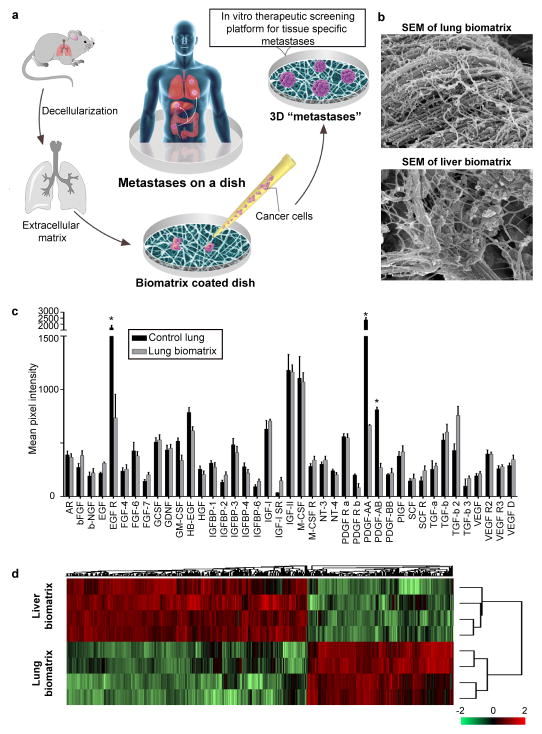
Figure 1.
BMSs recapitulate tissue-specific microenvironments found in vivo. (a) Schematic depicting methodology used for studying metastatic disease in vitro. (b) Scanning electron micrographs of lung and liver BMSs. Scale bars, 100 nm. (c) Analysis of growth factors and cytokines present in lung tissues before and after decellularization (n = 4 biologically independent decellularized samples from four different rats). Differences in the relative abundance of signaling molecules were determined using multiple t-tests, two-sided. Data represent mean ± S.E.M.; statistical significance determined using the Holm-Sidak method, with alpha = 0.05. * p < 0.05 (p = 0.001, 0.00011, and 0.00015 for EGF R, PDGF-AA, and PDGF-AB, respectively); (d) Heat map comparing the composition and relative abundance of proteins present in liver and lung BMSs (n = 4 biologically independent samples). Hierarchical clustering was performed using z-score normalized label-free quantification intensities.
Results
Organ-specific biomatrix scaffolds recapitulate the in vivo biochemical environment
To prepare lung BMSs, we used a perfusion based ECM isolation technique11. The rat’s inferior vena cava (IVC) was cannulated for the infusion of decellularization reagents and the superior vena cava (SVC) was clamped using a vessel clip. An opening was made in the rat’s carotid artery for outflow. The color change of the rat lung (from white to nearly transparent) provided a preliminary indication of successful decellularization (Supplementary Figure 1a). Decellularized liver BMSs was prepared by cannulating the hepatic portal vein for the infusion of decellularization reagents (Supplementary Figure 1a). Complete decellularization was confirmed histologically and by assessing nucleic acid content of the BMSs material (Supplementary Figure 1a,b). Notably, these BMSs naturally polymerized to form a meshwork of fibrous proteins that completely coated tissue culture plates (Figure 1b).
To assess whether lung BMSs contained signaling molecules present within the in vivo lung microenvironment, we evaluated the relative abundance of growth factors and cytokines retained by our liver BMSs following decellularization using semi-quantitative enzyme-linked immunosorbent assay (ELISA). In agreement with previous data demonstrating that extracellular matrix bound signaling molecules are retained following liver decellularization11, lung BMSs retained almost all (93%) of the analyzed growth factors and cytokines at near physiologic levels (Figure 1c). Note that the relative abundance of these signaling molecules varies between liver and lung BMSs, consistent with their tissue-specific nature (Supplementary Figure 1c). To further evaluate molecular differences present between liver and lung BMSs, we performed a mass spectrometric analysis. As with extracellular matrix bound growth factors and cytokines, we found that the relative composition of the extracellular matrix itself also differed between liver and lung BMSs (Figure 1d; Supplementary Figure 2).
CRC cell lines form liver and lung “metastases” in vitro
To engineer tissue-specific CRC cancer metastases, we cultured CRC cancer cell lines (HT-29, CRC119, SW480, and Caco2) on tissue culture dishes coated with liver and lung BMSs13. Excitingly, all four CRC cell lines spontaneously formed 3D spheroid colonies comprised of tumor cells bound together via tight junctions (Figure 2a; Supplementary Figure 3; Supplementary Figure 4a,b). These “metastases” are relatively large in scale, attaining diameters of up to a millimeter. Tumor spheroids that attain a diameter of greater than 500 micrometers contain necrotic cores due to a general lack of oxygen and nutrient availability as well as the internal accumulation of cytotoxic metabolites14. Consistent with this observation, metastases engineered on our BMSs also contain necrotic regions similar to the hypoxic and necrotic regions found in in vivo metastases (Supplementary Figure 4c).
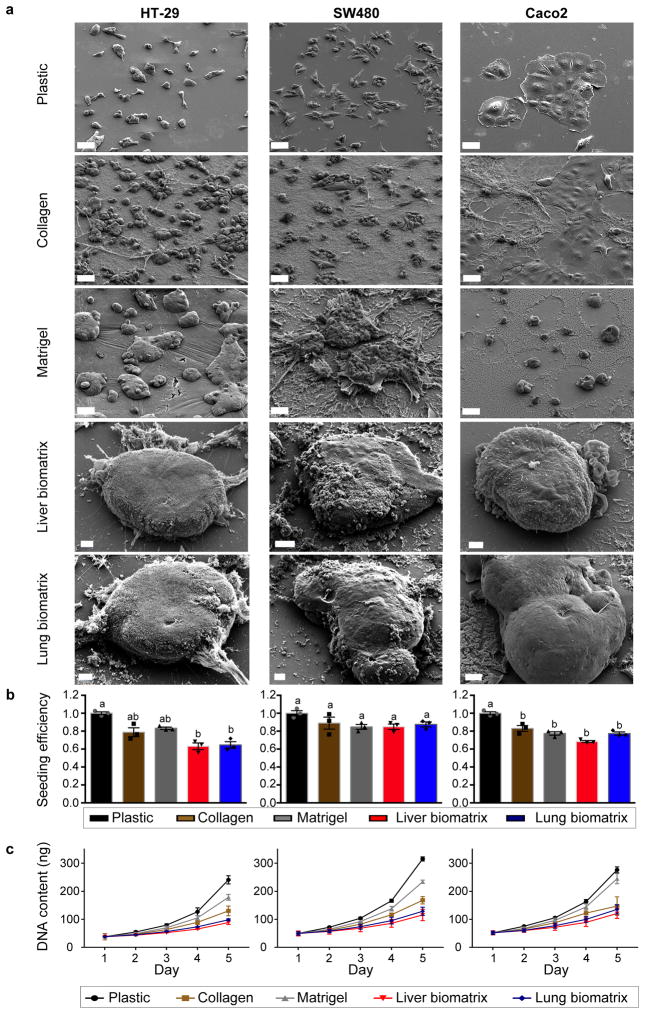
Figure 2.
Colorectal cancer cells spontaneously form 3D engineered metastases when cultured on liver and lung BMSs. (a) Scanning electron micrographs of HT-29 (left), SW480 (middle), and Caco2 (right) cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs. Scale bars, 50 μm. Experiments were repeated three times independently with similar results. (b) Seeding efficiencies and (c) growth rates of HT-29 (left), SW480 (middle), and Caco2 (right) cells seeded on plastic, collagen, Matrigel, liver BMSs, and lung BMSs (n = 3 biologically independent cell samples). Data represent mean ± S.E.M. Differences in seeding efficiency and growth rate were determined using a one-way ANOVA with Tukey’s multiple comparison post-test. Statistical significance is indicated with letters above (p < 0.05). Groups that share the same letter are not significantly different (p = 8.19e-05, 0.1043, and 1.07e-05 for HT-29, SW480, and Caco2, respectively).
To characterize the behavior of our engineered metastases, we compared the seeding efficiencies and growth rates of CRC cells grown on liver and lung BMSs to that of cells grown on plastic, collagen, and Matrigel. We found that CRC cells demonstrate reduced seeding efficiencies on collagen, Matrigel, liver BMSs, and lung BMSs when compared to cells grown on plastic (Figure 2b). Furthermore, we found that cells grown on collagen, liver BMSs, and lung BMSs grow more slowly than cells grown on plastic and Matrigel (Figure 2c). The slower growth rate observed in our engineered metastases is consistent with the behavior of metastatic cancer cells in vivo.
We next sought to determine whether the relatively slow growth rate displayed by liver and lung engineered metastases is due to increased rates of apoptosis or reduced rates of proliferation. To characterize apoptosis in cancer cells grown on different substrata, we assessed the expression of Cleaved Caspase 3, a marker of apoptosis, using flow cytometry. We found that CRC cells in all culture conditions exhibited comparable rates of apoptosis (Supplementary Figure 5a, Supplementary Figure 6). To assess the relative proliferation rates of cancer cells grown on different substrata, we performed a 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation assay. We incubated cultures with EdU, a thymidine analog that is incorporated into deoxyribonucleic acid (DNA) as cells enter S-phase, for four hours. Subsequently, we quantified the number of cells that underwent S-phase (EdU positive) using flow cytometry. We found that tumor cells grown on liver and lung BMSs demonstrated the lowest proliferation rates of all culture conditions tested across all three CRC cell lines (Supplementary Figure 5b, Supplementary Figure 6). Cumulatively, these data demonstrate that CRC cells seeded on BMSs generate relatively large slowly growing cultures that are more comparable to in vivo metastases than cultures produced by other culture systems.
Engineered liver metastases closely resemble metastases found in vivo
To further assess the degree of similarity that cancer cells grown on BMSs and conventional substrata share with in vivo metastases, we conducted a histopathological analysis. The histology of cells grown on plastic, collagen, Matrigel, and liver BMSs was compared with in vivo liver metastases present in nude mice and liver metastases from patients with metastatic CRC cancer. Classic histological features of liver metastases of gastrointestinal origin found in vivo include: (1) signet ring cells, (2) bizarre mitotic figures, (3) necrotic debris (extracellular accumulations of eosinophilic and nuclear debris), (4) pleomorphic cell size and shape, and (5) multinucleated cells. We were able to identify all of these features in engineered liver metastases generated from HT-29, SW480, and Caco2 CRC cells (Figure 3a; Supplementary Figure 7; Supplementary Table 1). In contrast, CRC cells grown on collagen and Matrigel only demonstrated bizarre mitotic figures and multinucleated cells and CRC cells grown on standard plastic culture dishes contained none of these histological features (Figure 3a; Supplementary Figure 7; Supplementary Table 1). Signet ring cells are an in vivo pathologic finding that has not been reported in ex vivo model systems. These data demonstrate that engineer metastases contain the same pathologic features found in in vivo liver metastases.
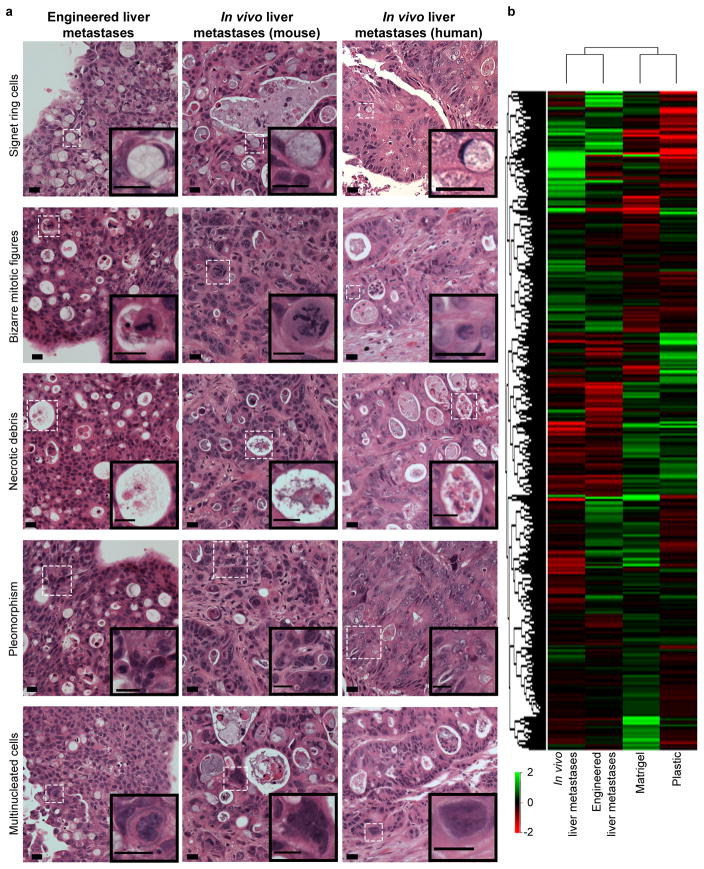
Figure 3.
Engineered liver metastases are comparable to liver metastases found in vivo. (a) Engineered HT-29 liver metastases (left panel), liver metastases formed following intrasplenic injection of HT-29 cells (middle panel), and liver metastases biopsied from late stage human colorectal cancer patients (right panel) all demonstrate classic histologic features of liver metastases of gastrointestinal origin. Experiments were repeated four times independently with similar results. Scale bars, 20 μm. (b) Hierarchal cluster analysis of average global gene expression patterns displayed by HT-29 cells grown on plastic (“Plastic”), Matrigel (“Matrigel”), liver BMSs (“Engineered liver metastases”), and in vivo liver metastases derived from intrasplenic injection of HT-29 cells (“In vivo liver metastases”) (n = 4 biologically independent samples).
In addition to exploring the histologic characteristics shared by cancer cells grown on different substrata and in vivo metastases, we also sought to investigate the degree of similarity between their respective transcriptomes. Specifically, we compared the global gene expression profiles of HT-29 cells cultured on plastic, Matrigel, and liver BMSs to in vivo liver metastases formed via splenic injection of HT-29 cells (Supplementary Figure 8a,b). Hierarchical clustering analysis revealed that the gene expression signature of our engineered liver metastases is more comparable to in vivo liver metastases than to HT-29 cells grown on plastic or Matrigel (Figure 3b). The relatively high degree of similarity shared by engineered metastases and in vivo metastases was further assessed by comparing gene expression profiles of engineered metastases to CRC cells grown on Matrigel and plastic. A total of 619 genes were observed to be discretely up-regulated in engineered metastases and in vivo liver metastases when compared to cells grown on Matrigel and plastic (Supplementary Figure 8c). Many of these commonly up-regulated genes were found to be involved in angiogenesis, cell adhesion, and drug metabolism (Supplementary Figure 8c). We also observed that in vivo and engineered liver CRC metastases express higher levels of tissue inhibitor of metalloproteinases 1 (Timp1) than CRC cells cultured Matrigel and plastic (Supplementary Figure 8d). This finding is consistent with the clinical observation that liver metastases generally exhibit relatively high expression levels of TIMP1 in metastatic CRC patients15,16. Taken together, these data demonstrate that the engineered metastases closely mimic in vivo metastases phenotypically and biologically.
Engineered metastases demonstrate increased metastatic potential in vivo
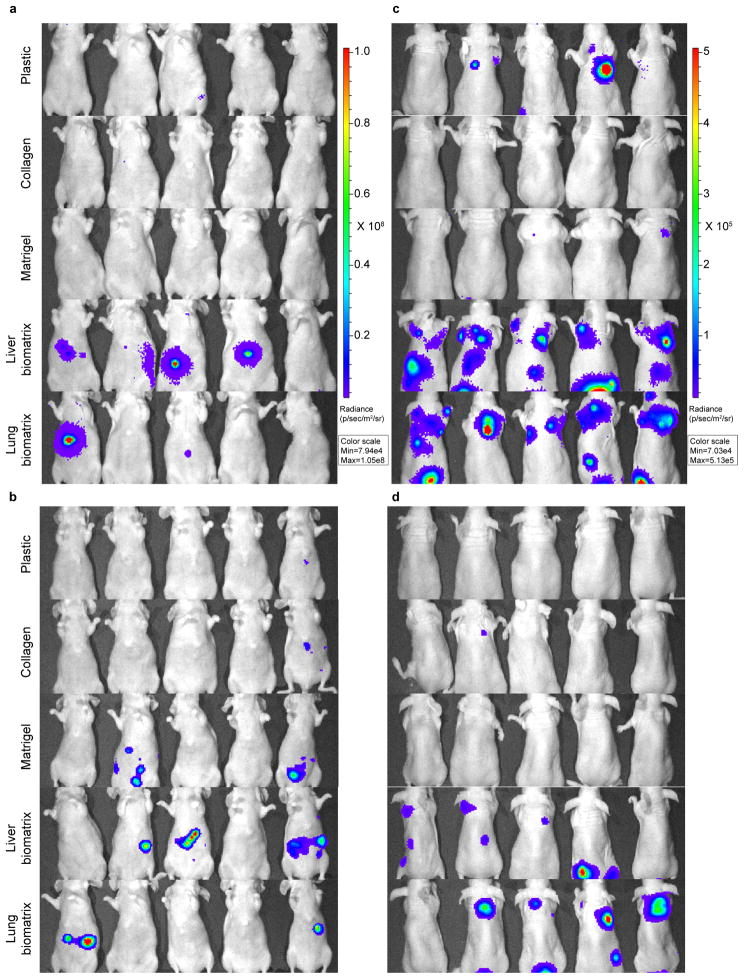
We sought to determine whether the histological and molecular similarities that engineered metastases share with in vivo metastases would functionally translate to an increased ability to grow within respective in vivo tissues. Namely, we hypothesized that if BMSs recapitulate tissue-specific acellular microenvironments, then cells grown on BMSs should be able to survive and grow in their corresponding in vivo microenvironments. For example, cells grown on liver BMSs should be more adapted to grow in liver tissue. To test this hypothesis, we delivered HT-29-luc2, CRC119, SW480 or Caco2 cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs to the liver and lung and subsequently assessed their ability to form metastases in vivo using bioluminescent imaging or gross histology (Figure 4a,b; Supplementary Figure 9a,b; Supplementary Figure 10a; Supplementary Figure 11a; Supplementary Figure 12). To determine the relative ability of engineered liver metastases to grow in liver tissue, we used direct hepatic injections to deliver CRC cells grown in different culture conditions to the liver and found that cells isolated from engineered liver metastases were more capable of forming liver metastases in vivo than cells grown on plastic, collagen, Matrigel, or lung BMSs (Figure 4a,b; Supplementary Figure 9a,b; Supplementary Figure 10a; Supplementary Figure 11a; Supplementary Figure 12).
Figure 4.
Engineered metastases demonstrate increased metastatic potential in vivo. Bioluminescence images of animals 30 days post (a,b) direct hepatic injection or (c,d) tail vein injection with CRC cells isolated from plastic, collagen, Matrigel, liver BMSs, and lung BMSs. (a,c) HT-29-luc2 cells were used. (b,d) CRC119 cells were used. n = 10 biologically independent animals per group. The experiment was repeated twice and the results were pooled.
To assess the relative ability of engineered lung metastases to grow in lung tissue, we delivered HT-29-luc2, CRC119, SW480 or Caco2 cells grown on different substrata to the lung using tail vein injection. We found that cells grown on lung BMSs demonstrated a higher capacity to form lung metastases than cells grown on all conventional culture substrata (Figure 4c,d; Supplementary Figure 9c,d; Supplementary Figure 10b; Supplementary Figure 10b; Supplementary Figure 12). Unexpectedly, we also found that cells grown on liver BMSs can also form lung metastases (Figure 4c,d; Supplementary Figure 9c,d; Supplementary Figure 10b; Supplementary Figure 10b; Supplementary Figure 12). Moreover, we found that cells grown on liver BMSs and to a lesser extent lung BMSs formed in vivo liver metastases following tail vein injection (Figure 4c,d; Supplementary Figure 9c,d; Supplementary Figure 12).
Basement membrane is a thin layer of extracellular matrix enveloping organs and represents “the barrier” to tumor cells. Matrigel, extracted from murine Engelbreth-Holm-Swarm sarcomas, has been served as models of basement membrane invasion. We found that CRC cells grown on biomatrices demonstrated comparable invading potential when compared to cells grown on other culture substratas (Supplementary Figure 13b). To better reproduce the complexity of invasion in living tissues, we instead used liver or lung BMSs as invasion barriers. CRC cells grown on tissue BMSs acquired enhanced invasion potential. In consistent with in vivo observations, CRC cells grown on liver BMSs invaded liver BMS-coated membrane more efficiently than cells grown on other culture substratas (Supplementary Figure 13c). Similarly, cells derived from lung BMSs demonstrated the highest invasion capability on “lung BMS barriers” (Supplementary Figure 13d).
Cancer cells exposed to hypoxic conditions can demonstrate enhanced survival and resistance to undergoing apoptosis17. To determine if the increased lung metastatic potential displayed by engineered metastases is attributable to the presence of hypoxic cells, we delivered HT-29-luc2 cells grown on plastic under hypoxic conditions to the lung using tail vein injection. We found that hypoxic preconditioning of CRC cells grown on plastic did not enhance their capacity to form lung metastases (Supplementary Figure 14). Additionally, cells delivered using tail vein injection must be able to avoid anoikis and colonize lung tissue prior to developing lung metastases. Importantly, we found that the increased lung metastatic potential displayed by HT-29 cells grown on liver and lung BMSs was not attributable to increased resistance to anoikis (Supplementary Figure 13a).
Taken together, these data demonstrate that engineered metastases are more capable of growing in their corresponding in vivo microenvironment than cultures grown on conventional substrata.
Engineered metastases demonstrate organ-specific therapeutic response
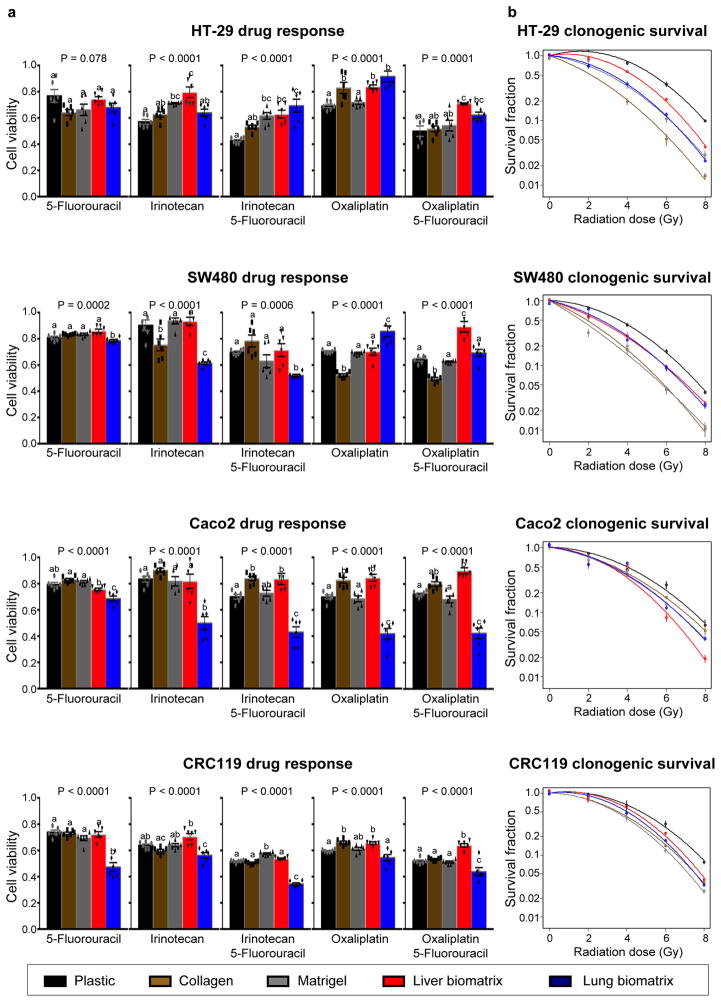
The identification of treatment regimens that are effective in treating metastases in a tissue-specific manner remains an active area of interest, as metastases in different organs of the same patient can respond differently to the same therapeutic regimen18,19. To determine if the substrata upon which CRC cells are grown effects treatment response, we treated CRC cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs to standard CRC chemotherapy regimens as well as radiotherapy. Four commonly used chemotherapy treatment regimens for metastatic CRC were examined: 5-fluorouracil (5-FU), irinotecan alone, irinotecan + 5-FU, oxaliplatin alone, and oxaliplatin + 5-FU. We found that the response of CRC cell lines to chemotherapy and radiotherapy is affected by their in vitro acellular microenvironment (Figure 5a). For example, engineered Caco2 lung metastases are uniformly more sensitive to chemotherapy regimens than engineered Caco2 liver metastases (Figure 5a). Additionally, we found that the response of CRC cell cultures to radiotherapy was dependent upon their culture substrata. (Figure 5b). Importantly, we observed that the treatment response of engineered liver and lung metastases differed. Our results demonstrate that the treatment response of CRC cells is impacted by the organ-specific composition of the BMSs on which they are cultured.
Figure 5.
CRC cells grown on different substrata’s respond differently to chemotherapeutics and radiotherapy. (a) Response of CRC cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs to chemotherapeutics (n = 6 biologically independent cell samples). Data represent mean ± S.E.M. Differences in therapeutic responses were determined using a one-way ANOVA with Tukey’s multiple comparison post-test. Statistical significance is indicated with letters above (P < 0.05). Groups that share the same letter are not significantly different. (b) Response of CRC cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs to radiotherapy (n = 3 biologically independent cell samples). Data represent mean ± S.E.M.
Discussion
The behavior of metastases is strongly influenced by the tissue-specific microenvironments they inhabit20. Recognizing the importance of this interaction, we developed a 3D in vitro culture system using decellularized organs to study metastases in a tissue-specific manner. Our goal is to develop a model that can better recapitulate the development of metastasis and not to model the metastatic process. As proof-of-principle, we engineered liver and lung metastases from a panel of CRC cell lines with a diverse genetic complexity. For instance, HT-29 and Caco2 cells don’t contain Kirsten Rat Sarcoma Viral Proto-Oncogene (KRAS) mutation, while SW480 and CRC119 cells carry each two different KRAS mutations21,22. Genetic alteration provides CRC cells with the ability to seed and colonize different organs, but we don’t think KRAS mutations account for tissue-specific metastases in our culture system. This culture system may not be able to recapitulate CRC cells with KRAS mutation on the organ-specific metastases. While some studies use co-cultures to recreate the tumor microenvironment2, our data demonstrates that we are able to engineer metastases that possess histologic features and gene expression profiles that are similar to those present in metastatic lesions in vivo by recapitulating the acellular biochemical environment. For example, we identified signet ring cells in the engineered liver metastases, an observation that has not been reported in any ex vivo cancer model system to date. Importantly, we show that engineered metastases are more adapted to growth in their respective in vivo acellular tissue-specific microenvironments when compared to cells grown on conventional culture substrata. To provide more specificity to study cell invasion in vitro, we designed a customized invasion chamber coated with liver or lung BMSs. We observed that CRC cells derived from BMSs exhibited significantly enhanced invasion capability in their respective tissue matrices in vitro than cells grown on plastic, collagen, and Matrigel. We also observed that engineered liver metastases were capable of forming metastases in lung. Potential explanations for this observation are either that the lung is a generally more “permissive” environment than the liver23, or that tumor cells grown in a liver microenvironment develop characteristics that promote their ability to grow within the lung.
Organ microenvironment may influence the response of metastases to chemotherapy24. Our data demonstrate that the therapeutic response of CRC cells to standard treatment regimens is dependent upon the tissue-specific acellular microenvironment they are exposed to in vitro. It is interesting that cells grown on lung BMSs are generally more sensitive to treatment than cells grown on liver BMSs. This data is consistent with the clinical observation that liver metastasis is the main cause of morbidity and mortality in metastatic CRC patients25.
Our in vitro culture system has some limitations, including lack of multiple cell populations. It cannot fully recapitulate the tumor microenvironment. However, our culture system, capable of engineering metastases in vitro, represents a powerful tool to better study of metastatic cancer biology in a tissue-specific manner than current available culture systems. Future studies may incorporate resident cell types to more fully recapitulate the tissue microenvironment which allows studying the dynamic and reciprocal dialogue between cells and their extracellular microenvironment. Additionally, future work will involve identifying specific ECM components that affect cancer cell behavior, as such evaluations may reveal new therapeutic targets. Our model is also useful for the study of tissue-specific treatment responses of metastases. Namely, this model can be used for drug-screening assays to test newly developed therapeutics for metastatic disease. Importantly, this is the only system that allows for high-throughput screening assays aimed at identifying therapeutics designed to treat metastases in an organ-specific manner.
Materials and methods
Perfusion-based decellularization of liver and lung
Sprague-Dawley rats (male, 250–300 g) were used to produce liver and lung BMSs. BMSs were prepared by cannulating the portal vein (liver BMSs) or inferior vena cava (lung BMSs) for perfusion of decellularization reagents. The vasculature was perfused with basal medium (e.g. serum-free DMEM/F12) until blood was eliminated and then with 250 mL of 1% sodium deoxycholate (SDC) containing 36 units/L phospholipase. Next, organs were perfused with 3.5 M Sodium Chloride (NaCl) until the perfusate was negative for proteins as assessed by optical density (OD 280.) Finally, tissues were rinsed with basal medium and snap frozen. Frozen decellularized organs were pulverized into a fine powder using a freezer mill (Spex SamplePrep 6770, Metuchen, NJ). Processed BMS powder was stored at −80°C.
Preparation of BMSs coated tissue culture plates
To determine protein concentrations of BMS materials, BMSs were dissolved in a solution composed of 4 M guanidine hydrochloric acid (HCl), 50 mM sodium acetate (pH 5.8), and 25 mM Ethylenediaminetetraacetic acid (EDTA) containing proteinase and phosphatase inhibitor cocktails. Bicinchoninic acid assay (BCA) assays were then performed to determine total protein concentrations. To prepare BMS coated surfaces, BMSs were suspended in Medium (DMEM/F12), added to tissue culture plates or onto Nunc Thermanox coverslips (Thermofisher Scientific), and allowed to dry overnight. Plates were sterilized using 100 Gy of external beam irradiation (Precision X-Ray).
Growth factors antibody array
Tissues and BMSs were sent to RayBiotech (Norcross, GA) where they were processed and submitted for growth factor array analysis. Specifically, the relative levels of growth factors and cytokines were quantified by the RayBio Human Growth Factor Antibody Array G-Series 1 (Cat #AAH-GF-G1-8). Data are expressed as normalized signal intensity.
Mass spectrometry analysis
Decellularized lung (n=4) and liver (n=4) tissue was pulverized and protein was extracted and purified as previously described26. Each sample (50 μg) was reduced with 5 mM dithiothreitol (DTT), alkylated with 15 mM iodoacetamide, and digested with trypsin (Promega) overnight at 37°C. The peptide samples were desalted using C18 spin columns (Pierce).
The peptide samples (1 μg) were analyzed by liquid chromatography/mass spectrometry LC/MS/MS using an Easy nLC 1000 coupled to a QExactive HF mass spectrometer (Thermo Scientific). Samples were injected onto an Easy Spray PepMap C18 column (75 μm id × 25 cm, 2 μm particle size) (Thermo Scientific) and separated over a 2 hr method. The gradient for separation consisted of 5–32% mobile phase B at a 250 nl/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in acetonitrile. The QExactive HF was operated in data-dependent mode where the 15 most intense precursors were selected for subsequent fragmentation. Resolution for the precursor scan (m/z 400–1600) was set to 120,000 with a target value of 3 × 106 ions. MS/MS scans resolution was set to 15,000 with a target value of 5 × 104 ions. The normalized collision energy was set to 27% for higher energy collision dissociation (HCD). Peptide match was set to preferred, and precursors with unknown charge or a charge state of 1 and ≥ 7 were excluded.
Raw data files were processed using MaxQuant version 1.5.3.17 and searched against a Uniprot rat database (downloaded Dec 2016, containing 29,795 entries), using Andromeda within MaxQuant. Enzyme specificity was set to trypsin, up to two missed cleavage sites were allowed, carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. A 1% false discovery rate (FDR) was used to filter all data. Label-free quantification using razor + unique peptides and match between runs (1 min time window) were enabled. A minimum of 3 unique peptides per protein was required for quantification. Proteins with >60% missing values were removed. Statistical analysis was performed in Perseus version 1.5.6.0 using analysis of variance (ANOVA) with p<0.05 considered significant. Hierarchical clustering using the z-score normalized label-free quantification (LFQ) intensities of the significant proteins was performed.
Cell culture
Human colorectal cancer cell lines (HT-29, Caco2 and SW480) were acquired from the Tissue Culture Facility at UNC Lineberger Comprehensive Cancer Center. The luciferase-expressing cell line, HT-29-luc2, was purchased from Caliper Life Sciences (Hopkinton, MA). Another luciferase-expressing cell line, CRC119, was derived from human colorectal cancer liver metastases and obtained from Dr. David Hsu from Duke University. Cell lines were authenticated using short tandem repeat and were tested for mycoplasma contamination. HT-29, HT-29-luc2, and SW480 cells were cultured in DMEM/F12 (Gibco) supplemented with 10% fetal bovine serum (Gibco) and penicillin/streptomycin (Mediatech). Caco2 cells were cultured in DMEM/F12 (Gibco) supplemented with 20% fetal bovine serum (Gibco) and penicillin/streptomycin (Mediatech). CRC119 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and penicillin/streptomycin (Mediatech). Cells were passaged on normal tissue culture plates or tissue culture plates coated with collagen, Matrigel, liver BMSs, and lung BMSs (100 ug/cm2).
Cell seeding efficacy
CRC cells were seeded at plates coated with collagen, Matrigel, liver BMSs, and lung BMSs (100 ug/cm2). After 24 h, cultures were washed with phosphate buffered saline (PBS) and lysed in 500 μL of DNA lysis solution. DNA concentrations were assessed using a Qubit dsDNA BR Assay Kit (Thermofisher Scientific).
Cellular growth rates
CRC cells were grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs (100 ug/cm2) in 6-well plates. Cells were collected at various time points post seeding and placed in DNA lysis solution. DNA concentrations were assessed using a Qubit dsDNA BR Assay Kit (Thermofisher Scientific). Growth rate over time was standardized based on seeding efficiencies.
Assessment of proliferation and apoptosis in vitro
For proliferation assays, CRC cells grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs (100 ug/cm2) were incubated with 10μM EdU for 4 hours. Cells were then washed with PBS, processed into single cells using TrypLE, and stained for EdU using a Click-iT Plus EdU Assay for Flow Cytometry kit (Thermofisher Scientific) according to the manufacturer’s instructions. Cells were then washed in PBS containing 10% FBS three times and submitted for flow cytometric analysis.
For apoptosis assays, CRC cells grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs (100 ug/cm2) were collected, processed into single cells, and fixed in 4% paraformaldehyde for 10 minutes at room temperature. Cell suspensions were blocked over night in Dako block (Agilent Technologies). Cells were then resuspended and stained with primary conjugated Cleaved Caspase 3 (Cell Signaling Technologies) (1:100) for 2 hours at room temperature. Cells were then washed with PBS containing 10% FBS three times and submitted for flow cytometric analysis. All flow cytometric analysis was done using a Beckman Coulter CyAn ADP and analyzed using software Summit 5.2.
Anoikis assays
CRC cells grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs were seeded at 1 x 104 /well onto the Anchorage Resistance Plate (Cell Biolabs) or a control 96-well cell culture plate. Cells were allowed to culture for 48 h. Live cells were detected with Calcein AM and the fluorescence was read using a micro-plate reader.
Invasion assay
CRC cells grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs in serum free media were placed into the upper chamber of an insert coated with Matrigel (Corning), liver BMSs, or lung BMSs. For BMS coating, liver or lung BMSs were diluted with serum free media to coat the transwell inserts. 50 μL of BMS solution were added to each insert and dried overnight under laminar air flow. The lower chamber contained media with 10% fetal bovine serum. After 16 h (Matrigel-coated) or 72h (BMS-coated) culture, non-invading cells were removed by a cotton swab and the cells on the lower surface of the membrane were fixed with 100% methanol and stained with 1% Toluidine Blue. Cells were counted in five fields using an inverted microscope.
Scanning electron microscopy (SEM)
Cover slips were coated with collagen, Matrigel, liver BMSs, or lung BMSs. Cultures grown on these substrata were fixed in a 0.15 M sodium phosphate buffer (pH 7.4) solution containing 3% glutaraldehyde overnight at 4°C. Samples were rinsed with PBS three times and then dehydrated using serial incubations in increasingly concentrated ethanol solutions (30%, 50%, 75% to 100%) for 10 min each. Cover slips were transferred to a Samdri-795 critical point dryer and dried using CO2 as the transitional solvent (Tousimis Research Corporation). The coverslips were then mounted on 13mm aluminum planchets with double sided carbon adhesive tabs and sputter-coated with 10nm of gold palladium alloy (60Au: 40Pd, Hummer X Sputter Coater). Images were acquired using a Zeiss Supra 25 FESEM operating at 5 kV, with working distance of 5 mm, and 10 um aperture (Carl Zeiss Microscopy).
Transmission electron microscopy (TEM)
Cell colonies were fixed in 0.15M sodium phosphate buffer (pH 7.4) solution containing 3% glutaraldehyde for 1h at room temperature. Samples were then rinsed with PBS and post-fixed with 1% osmium tetroxide/1.25% potassium ferrocyanide/0.15M PBS for 1h. Following post-fix, samples were rinsed with deionized water and dehydrated using serial incubations in increasingly concentrated ethanol solutions (30%, 50%, 75% to 100%) for 10 min each (30%, 50%, 75%, to 100%). Samples were then incubated in a 1:1 mixture of propylene oxide/Polybed 812 epoxy resin (Polysciences, Inc.) overnight followed by 100% resin for 24 hours. Samples were then placed in fresh Polybed 812 epoxy resin and sectioned transversely at 70 nm using a diamond knife and a Leica Ultracut UCT microtome (Leica Microsystems). Sections were mounted on mesh copper grids and stained with 4% aqueous uranyl acetate and Reynold’s lead citrate. The grids were observed at 80 kV using a LEO EM910 transmission electron microscope (Carl Zeiss SMT, LLC). Images were acquired using a Gatan Orius SC 1000 CCD Camera with DigitalMicrograph 3.11.0 software.
Histology
Cell colonies were fixed in 4% paraformaldehyde for 1 h at room temperature, paraffin embedded, and cut into 4 μm sections. Sections were stained with hematoxylin and eosin (H&E).
Intrasplenic injection
Nu/Nu mice (male, 8–10 weeks old) were obtained from the animal colony at UNC Animal Studies Core. Mice were anesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and dexdomitor (1 mg/kg). An incision was made on the left side of the abdomen to expose the spleen. 5 x 106 HT-29 or HT-29-luc2 cells in 50 ul PBS suspension per mouse were intrasplenically injected. All animal experiments were approved by UNC Institutional Animal Care and Use Committee.
Global gene expression analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions from HT-29 cells grown on plastic, Matrigel, and liver BMSs. RNA was also isolated from liver metastases generated following intrasplenic injection of HT-29 cells. Four biological replicates of cells grown under each condition were used. RNA quality was evaluated by Agilent Bioanalyzer 2100. RNA samples were sent to the UNC Genomics Core Facility. Samples were analyzed using Agilent SurePrint G3 Unrestricted Gene Expression 8x60K Microarray (human G4858A). cDNA was labeled with Cy3-CTP prior to hybridization.
Microarray data analyses were performed using GeneSpring 12.6 GX software. Raw signal values were quantile normalized and probe sets were filtered based on flag values. Normalized intensity values were performed hierarchical clustering analysis using Euclidean distance to sort both entities and samples. Differently expressed genes were selected based on statistical (one way ANOVA p<0.05) and fold change (>=4) thresholds. Genes that were upregulated in both engineered liver metastasis and in vivo metastasis were analyzed using gene ontology. Data have been submitted to the Gene Expression Omnibus database (accession number GSE76180).
Real-time polymerase chain reaction (qPCR)
Total RNA was isolated from CRC cells grown on plastic, collagen, Matrigel, and liver BMSs using the RNeasy mini kit (Qiagen). Total RNA was reverse transcribed into cDNA using the Quantitek cDNA synthesis kit (Qiagen). Quantification of Timp1 transcript was conducted by the 2−ΔΔCt method using β-Actin as a house keeping gene. The sequence of primers used for Timp1 gene expression were: forward primer 5′-AGACCTACACTGTTGGCTGTGAG-3′;reverse primer 5′-GACTGGAAGCCCTTTTCAGAG-3′.
Drug response assays
CRC cells were seeded at 2 x 104 cells/well in 96-well plates coated with collagen, Matrigel, liver BMSs, and lung BMSs (100 ug/cm2). One day post seeding, cells were treated with chemotherapeutics for 24 hours. Chemotherapeutics were then removed and cells remained in culture for another 24 hours in standard culture media. Cell viability was determined by MTS [(3-(4,5-dimethylthiazol-2-yl)-5-(3-car-boxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)] cell proliferation assays using the CellTiter 96® Aqueous One Solution Cell Proliferation assay kit (Promega). Treatment response for each culture condition was standardized to untreated cultures.
Clonogenic assays
Plating efficiency (PE) of each cell line was determined. Cells grown on plastic, collagen, Matrigel, liver BMSs, and lung BMSs were irradiated with 0, 2, 4, 6, and 8 Gy. Following irradiation, cells were plated into 25mL flasks at densities ranging from 100 to 250,000. Cells were incubated for 14 days, fixed, and then stained a solution composed of 4% formaldehyde, 80% methanol, and 0.25% crystal violet. Only colonies containing 30 or more cells were counted. The surviving fraction (SF) was calculated using the formula: (# of colonies formed)/(# of plated cells) (Plating Efficiency). The SF was plotted against the radiation dose on a log scale. Linear-quadratic formula SF=e^(−αD-βD2) was used to generate survival curves using the R package “CFAssay”.
In vivo assessment of metastatic potential
HT-29-luc2 or CRC119 cells grown on plastic, collagen, Matrigel, liver BMSs, or lung BMSs were trypsinized, harvested and processed into single cell suspensions. SW480 or Caco2 cells grown on plastic, liver BMSs, or lung BMSs were trypsinized, harvested and processed into single cell suspensions. For hypoxic cell culture, HT-29-luc2 cells were cultured in a hypoxia incubator chamber with 1% O2 and 5% CO2 with 37°C for 24 hours. Mice were randomly assigned to groups (10 mice per group). Cells were suspended 1xPBS and administered into Nu/Nu mice (8–10 weeks old) via tail vein injection (total volume 200μl, 1 x 106 cells/mouse) or direct hepatic injection (total volume 20μl, 2 x 103 cells/mouse). Cell injection and bioluminescence imaging were not blinded to investigators. For mice injected with HT-29-luc2 or CRC119 cells, bioluminescence was measured every week using an IVIS in vivo imaging system (Perkin Elmer, previously Caliper) in mice anaesthetized with 2% isoflurane. 100 μl Firefly Luciferin was administered to each mouse intraperitoneally and luminescence data was acquired 10 minutes after each injection with an exposure time of 1 minute. Luciferase intensity for each time point was normalized to the respective intensity value at day 0. Lung and liver from the sacrificed mice were then removed and examined by ex vivo bioluminescence. For mice injected with SW480 or Caco2 cells, lung and liver from sacrificed mice were removed and imaged.
All animal experiments were conducted in accordance with guidelines provided by University of North Carolina at Chapel Hill (UNC) Institutional Animal Care and Use Committee.
Statistical analysis
Data analysis was performed using either GraphPad Prism 6 (GraphPad) or R statistical software. Results from cell growth rate, seeding efficiency, MTS, flow cytometry, anoikis assay, invasion assay and real-time PCR, were analyzed by one-way ANOVA followed by Tukey’s Honestly Significant Difference post-hoc test. For tumor incidence in mice injected with cells from different substrata, statistical analysis was conducted using the Fisher’s exact test. For the clonogenic assay, linear-quadratic cell survival curves were analyzed using R package “CFAssay”. No statistical methods were used to predetermine sample size. F-test was used to test the equality of two variances. P-values less than 0.05 were considered significant.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
Acknowledgments
We would like to thank Microscopy Service Laboratory, Animal Studies Core, Genomics Core Facility, and Animal Histopathology Core Facility at the University of North Carolina at Chapel Hill. This work was supported by the University Cancer Research Fund from the University of North Carolina and R21 CA182322 from the National Institutes of Health/National Cancer Institute. This work was also supported by a generous gift from Mr. And Mrs. Edward Barkley.
Abbreviations
- bFGF
basic fibroblast growth factor
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FGF-4
fibroblast growth factor-4
- FGF-6
fibroblast growth factor-6
- FGF-7
fibroblast growth factor-7
- G-CSF
granulocyte-colony stimulating factor
- GDNF
glial-derived neurotrophic factor
- GM-CSF
granulocyte macrophage-colony stimulating factor
- HB-EGF
heparin-binding epidermal growth factor
- IGFBP-1
insulin-like growth factor binding protein 1
- IGFBP-3
insulin-like growth factor binding protein 3
- IGFBP-4
insulin-like growth factor binding protein 4
- IGFBP-6
insulin-like growth factor binding protein 6
- IGF-I
insulin-like growth factor-I
- IGF-1 SR
insulin-like growth factor-I receptor
- IGF-II
insulin-like growth factor-II
- M-CSF
macrophage-colony stimulating factor
- M-CSF R
macrophage-colony stimulating factor receptor
- NT-3
neurotrophin-3
- NT-4
neurotrophin-4
- PDGF-R α
platelet-derived growth factor receptor alpha
- PDGF-R β
platelet-derived growth factor receptor beta
- PDGF-AA
platelet-derived growth factor AA
- PDGF-AB
platelet-derived growth factor AB
- PDGF-BB
platelet-derived growth factor BB
- PIGF
phosphatidylinositol glycan anchor biosynthesis class F
- SCF
stromal cell-derived factor-1
- SCF R
stromal cell-derived factor receptor
- TGF-α
transforming growth factor alpha
- TGF-β
transforming growth factor beta
- TGF-β 2
transforming growth factor beta 2
- TGF-β 3
transforming growth factor beta 3
- VEGF
vascular endothelial growth factor
- VEGF R2
vascular endothelial growth factor receptor 2
- VEGF R3
vascular endothelial growth factor receptor 3
- VEGF-D
vascular endothelial growth factor receptor D
Footnotes
Competing interests
The authors declare no competing financial interests.
Contributions
X.T., M.E.W., K.C.R., H.P.F., S.K.Y., J.A.C., H.L., and L.Z. performed the experiments. X.T., M.E.W., H.P.F., and L.E.H. analyzed data. A.D.H., S.B.W., and E.L.W., D.S.H. provided essential protocols and technical support. X.T., M.E.W., K.C.R., T.Z., and A.Z.W. designed the experiments. X.T., M.E.W., K.C.R., H.P.F., and A.Z.W. wrote the manuscript. X.T., A.H., K.C.R., J.E.T., J.M.C., T.Z., L.M.R., and A.Z.W. edited the manuscript. A.Z.W. directed the study.
References
- 1.Fidler IJ. The organ microenvironment and cancer metastasis. Differentiation. 2002;70:498–505. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- 2.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higashiyama M, et al. Differences in chemosensitivity between primary and paired metastatic lung cancer tissues: In vitro analysis based on the collagen gel droplet embedded culture drug test (CD-DST) J Thorac Dis. 2012;4:40–47. doi: 10.3978/j.issn.2072-1439.2011.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay A, et al. Inhibition of pulmonary and skeletal metastasis by a transforming growth factor-beta type I receptor kinase inhibitor. Cancer Res. 2006;66:6714–6721. doi: 10.1158/0008-5472.CAN-05-3565. [DOI] [PubMed] [Google Scholar]
- 5.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott HC, et al. Perfusion-decellularize matrix using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 7.Baptista PM, et al. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–617. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 8.Uygun B, et al. Organ re-engineering through develpment of a transplantatble recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soto-Gutierrez A, et al. Engineering of an hepatic organoid to develop liver assist devices. Cell Transplant. 2010;19:815–822. doi: 10.3727/096368910X508933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen TH, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Lineage restriction of hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2011;53:293–305. doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 12.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flatmark K, Maelandsmo GM, Martinsen M, Rasmussen H, Fodstad O. Twelve colorectal cancer cell lines exhibit highly variable growth and metastatic capacities in an orthotopic model in nude mice. Eur J Cancer. 2004;40:1593–1598. doi: 10.1016/j.ejca.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Vinci M, et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schelter F, et al. Tumor cell-derived Timp-1 is necessary for maintaining metastasis-promoting Met-signaling via inhibition of Adam-10. Clin Exp Metastasis. 2011;28:793–802. doi: 10.1007/s10585-011-9410-z. [DOI] [PubMed] [Google Scholar]
- 16.Weidle UH, Birzele F, Kruger A. Molecular targets and pathways involved in liver metastasis of colorectal cancer. Clin Exp Metastasis. 2015;32:623–635. doi: 10.1007/s10585-015-9732-3. [DOI] [PubMed] [Google Scholar]
- 17.Yao K, et al. In vitro hypoxia-conditioned colon cancer cell lines derived from HCT116 and HT29 exhibit altered apoptosis susceptibility and a more angiogenic profile in vivo. Br J Cancer. 2005;93:1356–1363. doi: 10.1038/sj.bjc.6602864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 19.Ring BZ, Ross DT. Predicting the sites of metastases. Genome Biol. 2005;6:241. doi: 10.1186/gb-2005-6-12-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed D, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uronis JM, et al. Histological and molecular evaluation of patient-derived colorectal cancer explants. Plos One. 2012;7:e38422. doi: 10.1371/journal.pone.0038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna C, Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26:513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 24.Fidler IJ, et al. Modulation of tumor cell response to chemotherapy by the organ environment. Cancer Metastasis Rev. 1994;13:209–222. doi: 10.1007/BF00689637. [DOI] [PubMed] [Google Scholar]
- 25.Helling TS, Martin M. Cause of death from liver metastases in colorectal cancer. Ann Surg Oncol. 2014;21:501–506. doi: 10.1245/s10434-013-3297-7. [DOI] [PubMed] [Google Scholar]
- 26.Klaas M, et al. The alterations in the extracellular matrix composition guide the repair of damaged liver tissue. Sci Rep. 2016;6:27398. doi: 10.1038/srep27398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.