Abstract
The structural modeling and representation of cells is a complex task as different microscopic, spectroscopic and other information resources have to be combined to achieve a three-dimensional representation with high accuracy. Moreover, to provide an appropriate spatial representation of the cell, a stereoscopic 3D (S3D) visualization is favorable. In this work, a structural cell model is created by combining information from various light microscopic and electron microscopic images as well as from publication-related data. At the mesoscopic level each cell component is presented with special structural and visual properties; at the molecular level a cell membrane composition and the underlying modeling method are discussed; and structural information is correlated with those at the functional level (represented by simplified energy-producing metabolic pathways).
The organism used as an example is the unicellular Chlamydomonas reinhardtii, which might be important in future alternative energy production processes. Based on the 3D model, an educative S3D animation was created which was shown at conferences. The complete workflow was accomplished by using the open source 3D modeling software Blender.
The discussed project including the animation is available from: http://Cm5.CELLmicrocosmos.org
Keywords: Cell Modeling and Visualization, Microscopic Imaging, Computational Biology
1. Introduction
Cell visualization plays an important role in communicating, analyzing and understanding intra- and extracellular processes. Cytology-related school/university books and standard teaching material, such as Campbell, Alberts or Cooper, use different visualization approaches to communicate internal cellular structures to school as well as undergraduate students [1], [2], [3]. In the past, the learning material usually was limited to printed illustrations and microscopic images in combination with explanatory text. However, in recent years videos illustrating the three-dimensional (3D) properties of cells are becoming increasingly popular. Cell visualization experts such as Drew Berry, David Goodsell or Gaël McGill are specialized in creating spatial illustrations communicating cellular functions to broader audiences which are both visually demanding and reflecting current knowledge of a specific research topic [4], [5], [6]. For this purpose 3D modeling tools – such as Autodesk 3ds Max®, Maya®, or Maxon Cinema4D® – are utilized. These software packages were originally focusing on the creation of 3D designs, animations for entertaining movies or games, and just in the recent years were applied to biomedical visualizations [7]. A famous example is “The Inner Life of the Cell”, released in 2006 by Harvard University, which was an inspiration for many recent cell animations [8], [9].
A decade ago only 3D modeling design experts were able to create this kind of animations. Nowadays, open source 3D modeling tools like Blender are becoming very popular, lowering the entry level for 3D animation beginners [10]. Cell visualization experts such as Christopher Hammang or Christian Grove use Blender; they presented their award-winning visualizations at conferences such as VizBi (Visualizing Biological Data) 2010 and 2014 in Heidelberg [11], [12]. A number of cell visualizations were created by Monica Zoppè et al. [13], other researchers use Blender for visualization of, e.g. quantum mechanical properties [14] or molecular dynamic simulations [15].
However, the process of achieving visually convincing results remains hidden from the audience and is usually not communicated. To address this aspect, this paper will provide a short introduction and guide to cell visualization based on the recently created animation “Chlamydomonas reinhardtii 3D – From Biological Cells to Biofuels”. The green algal cell Chlamydomonas reinhardtii (C. reinhardtii) has great potential to be used as an alternative energy source. Its biochemical aspects are currently being studied with the target to increase its energy production rate and/or sustainability. However, there is still quite fragmentary knowledge concerning the cell’s precise structural properties. Therefore, structural cell modeling plays a central role for elaborating new theories concerning this organism. For this purpose, we used the aforementioned 3D modeling software Blender for the manual creation of highly-detailed 3D models, especially in the context of university education [10].
An important aspect of 3D animation still underdeveloped in biology-related research is the application of Stereoscopic 3D (S3D) visualization methods. This is in contrast to the popularity of S3D technology in cinemas (e. g., in 2016 the number of 3D screens in Asia exceeded non-3D screens [16]) and existing S3D visualizations of cells in the context of animations and interactive applications [17]. In addition, Virtual Reality (VR) cardboards such as Google Cardboard® or Samsung Gear VR® are found in many households nowadays, providing easy ways to watch S3D videos including side-by-side format [18], [19], [20].
Therefore, this work takes advantage of S3D, combining it with an heuristic approach to create a semi-realistic 3D model of C. reinhardtii. Integrative cell visualization was applied [21], combining the mesoscopic (see Section 3.3), molecular (see Section 3.6) and functional level (see Section 3.5) to provide information about the 3D structure, texture and biochemical functionality of this cell. This and similar approaches are relevant for communicating science, as they reduce the complexity of different biochemical aspects to a visually understandable form accessible to non-experts.
In summary, the intention of this article is three-fold:
It provides an overview of the basic workflow and the scientific base for the decision process of creating cells and their components with 3D modeling tools, whereas the focus here is on the open source tool Blender.
It discusses how to integratively combine the molecular, mesoscopic and functional levels.
It shows the basic aspects of stereoscopic 3D visualization in the context of cytology.
2. Background
Before discussing the methods and workflow of our approach in detail, we like to provide some biological and microscopy-related background information required to understand why this heuristic cell visualization approach is valuable.
2.1. Limits of Microscopy
The modeling and visualization of cells requires the integration of different microscopic technologies. Traditional optical/light microscopes are limited to a resolution of approx. 200 nm, a resolution where larger cell components are already distinguishable [22]. New approaches can surpass this limitation, for example, by relying on fluorescent molecules and precisely controlling their excitation [23], or by using statistical algorithms on otherwise noise-afflicted images [24]. Other approaches, such as scanning probe microscopy allow for resolutions at subatomic level [25], [26]. Transmission Electron Microscopy (TEM) and Scanning Electron Microscope (SEM) are technologies which scan the surface of a specimen with an accumulated electron beam, enabling the visualization of small cellular entities, such as ribosomes. Moreover, it is possible to combine different TEM images of the same specimen to create a 3D tomography representation which usually requires a very time-intensive manual segmentation (Section 3.1). Central repositories, such as the Cell-centered database, provide access to a number of microscopic images and including some segmentations [27].
Another relevant topic in this context is the coloring of the cell and its components. In light microscopy the proportions of some cellular compartments are so small that there is insufficient interaction with light to make the tissue visually distinguishable from water/cytosol, as the absorption depends i. a. on traveled distance and chemical composition [28]. Also, proteins and amino acids show very little intrinsic light absorption in the visible part of the spectrum. For example, high spectral activity is shown by the aromatic amino acids tryptophan, tyrosine and phenylalanine, due to the accumulation of multiple-electron bonds, which absorption maxima lie roughly in the ultraviolet part of the spectrum [29]. This is often circumvented by staining tissue samples with dyes like haematoxylin and eosin, which allow for different types of tissues to be distinguished, but in turn cause non-natural colors.
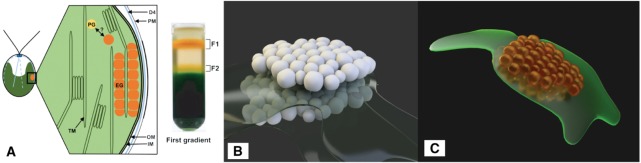
To mimic visual cell component properties, this work applies materials to the cell model based on the colors known from thick layers where the absorption would be strong enough to produce a visible effect (Section 3.1). These properties can, for example, be established by performing a fractioning of cell compartments by chemical lysis and subsequent fractioning [30] (Figure 14A).
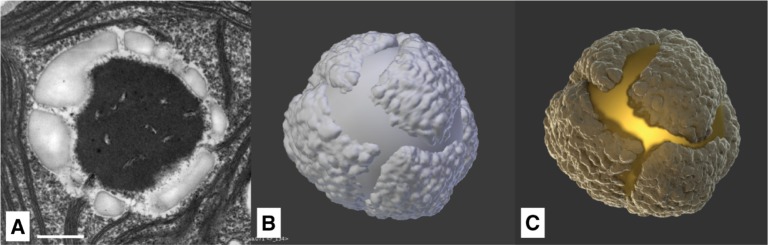
Figure 14:
(A) Left: schematic longitudinal section through the eyespot in the region of the four stranded microtubular root (D4) consisting of local specializations of different compartments: plasma membrane (PM) overlying the eyespot globules (EG), which is close to the inner and outer chloroplast envelope (IM/OM) as well as in contact with the thylakoid membrane (TM) and arranged in layers. The possible link between plastoglobules (PG) and EG is indicated, Right: separation of the cell homogenate in the first gradient using the Schmidt et al. eyespot isolation method, clearly showing the carotenoid spectrum (©2015 Eitzinger, Wagner, Weisheit, Geimer, Boness, Kreimer and Mittag [70]) (B) Eye segments rendered with neutral material. (C) Rendered with final material.
All these different visual information regarding the structure, texture and coloring of the specimen at different magnifications and levels of detail need to be combined to create an appropriate visual cell model.
2.2. Heuristic Cell Modeling and Integrative Cell Visualization
Although there are already low resolution 3D microscopy-based models available for C. reinhardtii [31], the structure at a higher resolution is still not well-described. This work follows an interpretative approach in which we combine heterogeneous information of different sources [21], such as light microscopic images for coloring and the overall structure of cell components at a lower resolution (Figure 7C), detailed electron microscopic images for high resolution images (Figure 7A) and 3D tomography-based models for the spatial distribution of cell components (Figure 11A). These different information sources can be used to model the mesoscopic level, ranging in the area of a few thousands down to tenths of nanometers [17]. At this level small entities like ribosomes are visible, which have an average size of approximately 25 nanometers in diameter (in eukaryotes) [32], [33]. The mesoscopic modeling is discussed in Section 3.3.
Figure 7:
Chloroplast of Chlamydomonas: (A) TEM slice (scale bar 500 nm) (©2018 Trustees of Dartmouth College/Public Domain [64]). (B) Freeze fracture SEM image (×35.000, for scale bar see c) (©2014 Courtesy of Ursula Goodenough/John Heuser). (C) Light microscopy image (scale bar 5 μm) (©2016 Courtesy of Wolfgang Bettighofer). (D) Chloroplast plates rendered with final material, partly truncated, with highlighted cleaved surfaces to be compared with (A).
Figure 11:
Lipid deposits: (A) X-ray tomography model (scale bar 1 μm) (©2012 Hummel et al., CC BY 4.0 [31]). (B) Lipid droplets modeled in Blender. (C) Rendered Lipid droplets with final material.
The molecular level, roughly ranging from tenths of nanometers down to Ångström level, enables the visualization of molecular assemblies in high resolution. Here, X-ray crystallography (besides Nuclear Magnetic Resonance spectroscopy) plays a pivotal role, enabling the computer-based 3D reconstruction of molecular structures. For this purpose, the electron scattering distribution pattern is computationally transformed to 3D coordinates representing the different atom positions. The molecular modeling of a C. reinhardtii membrane with Blender is introduced in Section 3.6.
In addition to the structural features of the cell also the functional ones are relevant. To illustrate functionalities and the communication among different cellular components and their molecular sub-structures, different processes such as metabolic processes and protein–protein interactions have to be analyzed. Here we will focus on the combination of a metabolic pathway with the spatial structure of the C. reinhardtii cell. The functional modeling approach is discussed in Section 3.5.
By integrating the three previously-mentioned cytological levels, a holistic model of the cell is created. Here we propose a structural model which can be used as a base for further research. We argue with this article that cell modeling – although in the bioinformatics community often understood as building dynamic models [34], [35], [36] – is also a relevant topic in terms of geometrical modeling as also shown by other examples: established mathematical cell simulation environments, such as VCell, nowadays also support the import of 3D geometries and the integration of those into mathematical models based on differential equations [37].
2.3. “Chlamydomonas reinhardtii 3D – From Biological Cells to Biofuels”
The genus Chlamydomonas (Gr. chlamys, mantle; monas, solitary) has about 500 species around the world and is very ubiquitous in nature, most commonly found in fresh lakes water, saline water, brackish water and ponds. Depending on the species, whole areas occupied with these organisms are colored, e. g., in red or yellowish green [38], [39], [40], [41].
Chlamydomonas is a unicellular and motile organism which lives in many environments and normally derives its energy through photosynthesis. However, Chlamydomonas can also metabolize carbon sources in order to survive in the dark. Towards its anterior region, it is equipped with two flagella relevant for cell movement [39], [40]. The different cell components will be discussed in Section 3.3.
The development of alternative, environmentally sustainable energy technologies is an important research field, including the synthesis of biofuels, biogas and biohydrogen. Green micro-algae (including C. reinhardtii) and cyanobacteria are two types of photosynthetic organisms that are capable of photobiological hydrogen production [42]. Photosynthetic micro-algae use light energy for growth and can exhibit comparably high biomass yields.
In this work we discuss the creation of the scientific animation “Chlamydomonas reinhardtii 3D – From Biological Cells to Biofuels”. The initial purpose of the video was to present the work of the Algae Biotechnology & Bioenergy group of the University of Bielefeld during the conference “Prospects and challenges for the development of algal biotechnology” at the ZIFF Bielefeld, 2014 [43]. The stereoscopic full HD version of the video won the Best of Show (Animation/CG) award in 2016 at the “Stereoscopic Displays and Applications XXVII.” [44]. However, the first version of the molecular membrane model was very simplistic. During a poster session at VizBi 2016, where we presented the new animation, this membrane structure was discussed and the visual appearance and the wave-like motion of the thylakoid membrane was criticized, which led to a revision of the thylakoid membrane, see Section 3.6.
2.4. Cell Visualization with Blender
Blender is a 3D modeling and animation suite compatible to Linux, Mac OS X®, and MS Windows® systems. It is a free open-source computer graphics program that allows users to produce high quality still images as well as animations primarily using 3D geometry [45]. Blender supports the whole 3D modeling and visualization pipeline, from creating basic and advanced 3D models, adding modifiers, texturing, (bone-based) animations, stereoscopic rendering, etc. A plugin interface enables developers to create scripts which can be used to extend the functionality of Blender [46].
In recent years a number of Blender-based extensions for bio-visualization were developed. BioBlender is a project which visualizes molecular structures with Blender [13]. ePMV especially provides extended import capabilities for atomic coordinates enabling the animation of single proteins [47]. In addition it is the base for cellPACK which can be used to create so-called filled cells based on the work of David Goodsell, well-known from the “Molecule of the Month” section of the PDB database [48]. Although there seems to be no up-to-date cellPACK version for Blender available, parts of cellPACK are still under active development. It is the base for a new instant cell visualization tool which is able to interactively create filled cells [49]. Another alternative is CellBlender which is based on the MCell simulation package which uses the Monte Carlo algorithm to create cellular simulations [50], [51]. As previously mentioned, there are also a number of novel cell visualization experts which make use of Blender’s capabilities [11], [12].
This work is based on the CELLmicrocosmos project for scientific and educational purposes as well as on student projects [17]. The modeling was done with Blender version 2.71, rendering was performed with the Cycles engine included in Blender.
3. Methods
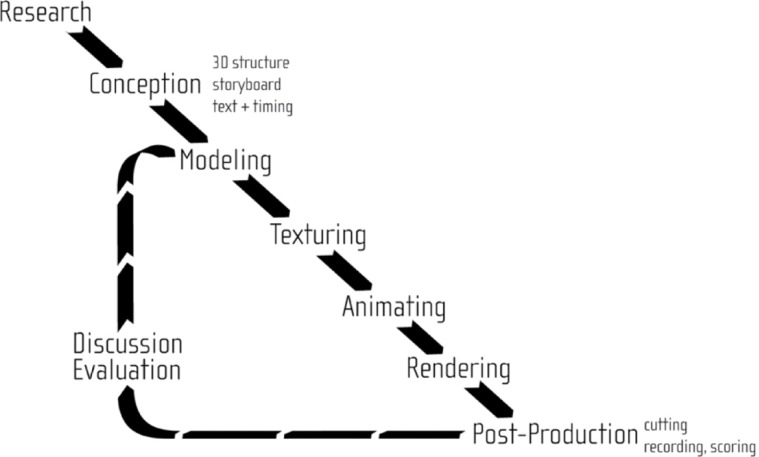
Here the workflow of the heuristic cell modeling and integrative cell visualization processes will be discussed, see also Figure 1. The workflow to create 3D cell models from heterogeneous microscopy data will be described based on a single cell component: the endoplasmic reticulum (ER). While the workflow for each component contains similar elements, it differs in available material.
Figure 1:
Basic workflow of the heuristic modeling and visualization approach.
Therefore, Section 3.3 will focus on the specific characteristics in the modeling of each particular cell component. Section 3.6 will then discuss the modeling of a membrane at the molecular scale, whereas Section 3.5 will explain the combination with functional information represented by metabolic pathways. The chapter will be concluded by an introduction of S3D principles relevant for this work in Section 3.2.
3.1. Workflow
3.1.1. Research and Conception
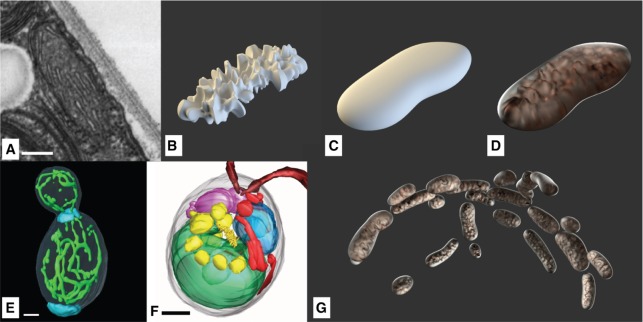
The first step in the cell compartment modeling process is the literature research, in order to find adequate source material. A number of TEM images describing the Golgi apparatus and the adjacent endoplasmic reticulum in Chlamydomonas species were found in the literature [52], [53]. However, as there was no high-resolution 3D structure for Chlamydomonas available, we made use of a segmented set of approximately 500 TEM images, encompassing a single neuron of the mouse primary visual cortex [54]. The comparison of single images of this dataset with TEM images from Chlamydomonas showed comparable structures of the endoplasmic reticulum.
3.1.2. Modeling
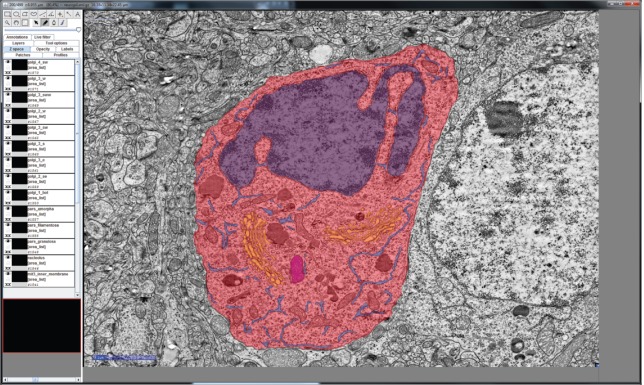
This single neuron image set was manually converted to a 3D structure with the open source software Fiji (Is Just ImageJ) [55]. For this purpose, the integrated TrakEM2 plugin was used to color the cell components of interest at each single slice using a drawing tablet, as shown in Figure 2 [56]. The generated 3D model was transferred from Fiji to Blender. After applying textures to the model, the model reveals a wavy, branch-like structure, as can be seen in Figure 3A. This quite complex 3D structure was then adapted and simplified to serve as an archetype for manual remodeling of the ER of the Chlamydomonas cell. Figure 3B shows the simplified reconstituted mesh and Figure 3C shows the resulting rendering after applying textures to its surface.
Figure 2:
A TEM slide of a mouse neuron cell with manually-colored compartments by using Fiji: red – cell membrane and cytosol, purple – nucleus, blue – endoplasmic reticulum, orange – Golgi apparatus, magenta – mitochondrion. Scale Bar: 2000 nm.
Figure 3:
Model of the endoplasmic reticulum: (A) the ER model of a mouse neuron derived from electron tomography via Fiji, (B) the basic mesh of the simplified ER model via Blender, showing a fragment of the polygonal mesh, and (C) the same ER model rendered with applied materials using the Cycles renderer.
Alternatively, the classical interpretative cell modeling approach can be used to manually model cell components in Blender, based on the manual combination of different microscopic imaging sources (see Section 2.2).
3.1.3. Texturing
After creating a cell component 3D model with the corresponding mesh, the model needs to be associated with a visible surface (a procedure called texturing). The material properties will define the way the Blender Cycles engine will render the model. The Blender Cycles render engine is a ray tracing method that utilizes Bidirectional Scattering Distribution Functions (BSDF) [57]. For every pixel of an image to be rendered, several rays have to be calculated and the way they are scattered by the different surfaces – i. e., the combination of a 3D model and its material – in their paths. Each material features individual BSDF settings. There are several BSDF shaders implemented in Blender which can be freely combined. In this context, shaders are small computer graphics-related scripts generating different visual properties for assigned materials. The four most important shaders for this project are presented in Figure 4.
Figure 4:
The four most relevant BSDF-shader properties in the context of this work: (A) Diffuse. (B) Reflective (“Glossy”). (C) Refractive. (D) Translucent.
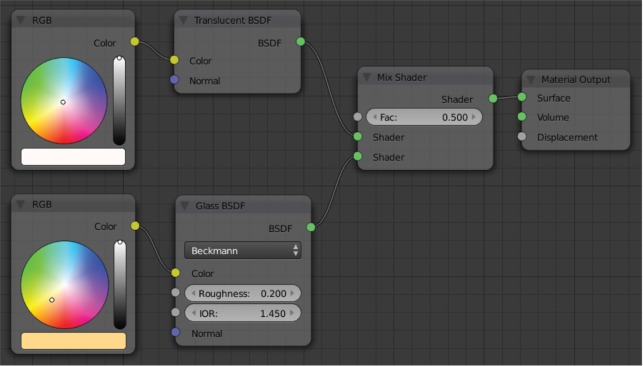
The Diffuse shader is a standard shader simulating diffuse (environment) lighting which is used for many basic materials, and is often used in combination with other shaders. Figure 5 describes a different approach which is especially interesting in the context of cell visualization. Since both, the cell components as well as the surrounding medium, are fluid substances, there are only minor differences in the refractive index at the interfaces. For this purpose, a combination of two predefined material shaders was chosen: (1) a Translucent shader, simulating the properties of a diffuse, light-absorbing liquid (Figure 4D), and (2) a Glass shader with an increased surface roughness of 0.2 to mimic a higher refractive index and determine reflective properties of the surface, as displayed in Figure 5 [46]. The Glass shader is a predefined combination of the Glossy (Figure 4B) and Refraction (Figure 4C) shaders [46]. These settings are a common basis for all cell components that are not transparent and only differ in color, creating an organic impression without being too CPU-intensive (see Section 3.3).
Figure 5:
The basic shader setup for most non-transparent materials.
In Figure 3C the color for the endoplasmic reticulum was defined by a gradient from the dark brown tone of the nucleus to the brighter brown hue of the Golgi apparatus, as clear partitioning is not easy to define. The overlaying ER structure is curved and encompasses around half of the nucleus.
3.1.4. Animating
The animation of the cell model includes parts with changing geometry, like the movement cycle of the flagella (see Section 3.4) or growing starch deposits, and also the movement of the camera, as previously sketched in a storyboard.
3.1.5. Rendering and Post Production
High definition rendering of all separate scenes results in separate images for the left and the right perspective (see also next section). During post processing the images were color-corrected and other visual effects were added and finally all scenes were combined to a final movie. Additionally, a narrating audio track was recorded and added, together with a musical score created with Steinberg Cubase Studio 6®. The complete post-production as well as movie editing can be done in Blender, but during the later process we decided to switch to EditShare Lightworks Pro 12® which provides an improved stereoscopic post production in combination with comfortable movie editing.
3.2. Stereoscopic 3D Visualization
3.2.1. Basics of 3D Stereoscopy
S3D technology is used to compute the two different perspectives simulating the image for the left and right eye enabling the spatial visual perception of a 3D scenario. There exist a number of technologies which can be used to transport the image to the corresponding eyes: for example by anaglyph color-coding, circular or linear polarization, shutter glasses, or two separate displays right in front of the observer’s eyes, like VR cardboard in combination with a smartphone.
We already showed in [17] that S3D visualization drastically improves the communication and understanding of cellular structures, as the cellular structure is a three-dimensional structure which often is not correctly spatially perceived in case S3D is not used. Therefore, it was decided from the start of the herein-discussed project to make use of S3D technology.
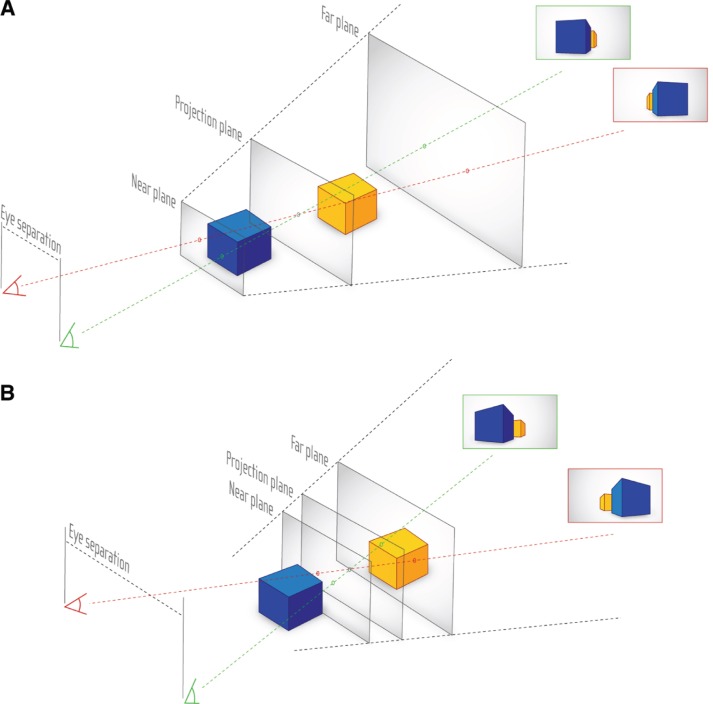
The viewing frustum created by the two eye positions leads to the definition of the three S3D planes shown in Figure 6. These planes can be used to optimize the positioning of the object(s) to be visualized.
Figure 6:
Principles of stereoscopic viewing. (A) Basic parallax of different viewing positions. (B) Shift of Near and Far plane in dependence of eye separation.
The Projection plane represents the distance where the projection of a virtual object onto the canvas matches in both, left and right, perspectives. A flat mesh placed exactly at the location of the Projection plane will not provide any stereoscopic effect.
The distance, at which the optical disparity between the left and right perspective image becomes too strong towards the viewer to be comfortably watched, is marked by the Near plane.
Any objects between Projection and Near plane will appear to be in front of the canvas. This should be avoided if the object in the front is larger than then screen, as the cutting on the edge of the canvas produces contradictory information and thus causes discomfort for the viewer. However, if the object in front of the Projection plane does not exceed the screen boundaries, it will hover outside the screen, providing an intense spatial impression to the viewer. In case of cell visualization, this might be a single cell component, or a molecule which should be highlighted.
Lastly, the Far plane is the maximum position where objects should appear behind the Projection plane, as the disparity becomes again too strong, but this time directed towards the opposite direction in comparison to the Near plane. As objects appear usually smaller in the background, the crosstalk effect is not so strong in comparison to the one occurring towards the Near plane. Therefore, the Projection and Far plane span a viewing frustum in which most events should take place. Moreover, animated objects may leave the Far plane-Projection plane frustum, as this space lies behind the screen.
Another factor for consideration is an effect called crosstalk, as defined by Woods as “the incomplete isolation of the left and right image channels so that one image leaks into the other” [58]. This depends on the separation method of the applied S3D technology as well as the used contrast space.
3.2.2. 3D Stereoscopy in Blender
Based on the plugin mechanism of Blender, we used the Stereoscopic Rendering plugin developed by Sebastian Schneider [59], [60]. Nowadays, Blender natively supports S3D visualization, but for older versions of Blender (such as 2.6X to 2.74) Schneider’s plugin is still relevant for stereoscopic rendering. Another application case using this plugin in context of S3D visualization in combination with cell visualization was previously discussed by us [17].
3.3. Mesoscopic Modeling of C. reinhardtii
As previously mentioned, this work follows an interpretative cell modeling approach. Different information sources were combined: Published articles and books, as well as graphics. Important information sources used as base for the following section were for example [38], [39], [40], [41]. But the most relevant information source was microscopic imaging, as discussed in Section 2.1. In the following sections the mesoscopic modeling process as well as the definition of the used material will be discussed. The accompanying figures will show original microscopic images in combination with the resulting 3D renderings.
3.3.1. Cell Wall and Overall Structure
Chlamydomonas cells have usually spherical, oval and/or oblong shapes (see Figure 7B and C), but also other shapes exist in nature. It has a thin, smooth and firm cell wall. The cell length ranges from 20 to 30 μm.
3.3.2. Chloroplast/Thylakoid
The most prominent feature in many plant cells is the chloroplast, with its internal thylakoid stacks in which the photosynthesis takes place. Its morphology can vary widely over various species, ranging from smooth shapes to heavily convoluted lobes [61]. In Chlamydomonas, the lower part of the cell is the generally cup-shaped chloroplast.
In case of C. reinhardtii, most TEM images show the thylakoids as thin lines, as can be seen in Figure 7A. As the TEM images represent slices through a spatial structure, one can deduce that the thylakoids are formed as extensive plates, which can be straight in shape or slightly curved. This is also affirmed by freeze fracture SEM images (Figure 7B) showing the curved shape of the plates. The thylakoid plates were modeled in stacks – only slightly varying the including plates’ shape – as the thylakoids often appear as multiple lines on the TEM: the grana (Figure 7A and D) (see also, e.g. [62]).
To obtain the optical properties of the thylakoid, light microscopic images were used as a base. Figure 7C shows that large parts of the cell are indeed of greenish color, as the chlorophyll variants (both, a and b) in the thylakoid membranes are strong intense green dyes and give most plant cells their characteristic green color. In addition to the Translucent and Glass mixture basis of the other non-transparent materials (see also Section 3.1), this material consists to a majority of a Diffuse (Figure 4A) shader, to avoid unwanted artefacts of the Translucent (Figure 4D) shader, where sometimes single pixels on a surface appear as bright outliers (known to Blender users as “Fireflies”). Therefore, a special method was applied for optically filtering these “Firefly” outliers [63].
3.3.3. Nucleus, ER, Golgi Apparatus
A dark brownish hue was assigned to the nucleus (Figure 8B) as along its surface (and the one of the subsequent rough endoplasmic reticulum) the ribosomes are located which are high-density molecules largely consisting of RNA. The size difference is approx. 1:100 (Ribosome:Nucleus). Also, a rough texture was assigned to the nucleus, mimicking its pores (Figure 8A). There is also an alternative version of the nucleus, with the darker ribosomes attached externally, created by a randomized particle system (Figure 8C).
Figure 8:
Model of the nucleus: (A) Freeze fracture SEM image, detail of/for scale see Figure 7B (©2014 Courtesy of Ursula Goodenough/John Heuser). (B) Rendered with final material. (C) Alternative version with less dark material and attached ribosomes.
For the ER, a model based on the dataset [54] was taken into account, as already discussed in Section 3.1. This model shows wave-like branch structures which are usually omitted by current visualizations. Though not the same type of cell, it served as reference for the shape of the ER in our cell, whereas the complexity of the outer shape’s structure was reduced. For coloring, again the brown tone of the nucleus was chosen, due to the ER’s proximity.
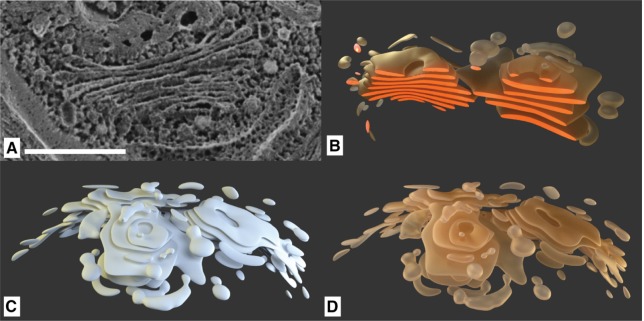
The further branches show a gradient from the darker ER color to the brighter color of the Golgi apparatus, as the compartments’ shapes are transitioning into each other and the outer membranes cannot always be determined. The Golgi apparatus itself was modeled based on several images, exemplary shown in Figure 9A, in accordance to several known educational illustrations of this compartment, with its cup-shaped plates, which can be especially seen in the cleaved model in Figure 9B.
Figure 9:
The Golgi apparatus: (A) Freeze fracture SEM image (scale bar 500 nm) (©2014 Courtesy of Ursula Goodenough/John Heuser). (B) Golgi model cut, with highlighted cleaved surfaces for comparison with SEM image. (C) Golgi model. (D) Golgi model rendered with final material.
3.3.4. Pyrenoid and Starch
Another prominent feature of most C. reinhardtii TEM images is the pyrenoid. Figure 10A and B show its spherical structure. Several starch-plates are attached to the pyrenoid. For practical reasons regarding the resulting animation, the pyrenoid model itself was made translucent with a light source located inside to provide adequate contrast against the starch plates which enclose the whole volume of the compartment (Figure 10C). Additional accumulations of energy-storing starch plates are located between the thylakoid stacks. The material of the starch was chosen in a light grey color, taking a flour-like appearance as a visual metaphor. The texture was applied by a dynamic displacement modifier.
Figure 10:
Model of pyrenoid and starch attachments: (A) TEM slice (scale bar 500 nm) (©2018 Trustees of Dartmouth College/Public Domain [64]). (B) Rendered solid with default OpenGL lights. (C) Rendered with applied final materials using the Cycles renderer.
3.3.5. Lipid Bodies
The lipid bodies are free-floating droplets mostly located in the outer regions of the cell. Size and position, along with the overall composition of the cell, can be extracted from an X-ray tomographic image of Chlamydomonas cells (Figure 11A) [31]. Although the rendered 3D models of Hummel et al. are of lower resolution, the model is of great value to the model proposed here, as it reveals the spatial arrangement and structure of this relatively large compartments and other distinctive parts of the cell.
Due to the fact that the lipid bodies play a vital role in the cellular metabolism and serve as energy storage, we decided that they have to provide a high visual contrast in the context of this cell model. Therefore, their material and color is the typical yellow hue which is associated with oily substances (Figure 11C).
In scenes where the lipid deposits can be seen in close-up, their appearance is additionally altered by a wave modifier. In this way, their model is deformed by a traversing sinusoidal wave. The time-dependence of the deformation results in a wave-like motion that is associated with the fluidic behavior of a liquid.
3.3.6. Mitochondria
Mitochondria are known to create networks [31], [65] which reside in the outer layers of the cell (Figure 12E and F). They are often visualized in the literature as individual entities like here shown in Figure 12D. The structure was duplicated while varying the form factor and aligned in a way creating a network-similar structure (Figure 12G). They consist of a smooth outer membrane, and a rough inner membrane surrounding the mitochondrial matrix. Figure 12B shows the internal structure symbolizing the mitochondrial matrix, resulting in the typical crest-like visual appearance in Figure 12D.
Figure 12:
Mitochondria: (A) TEM-slice (scale bar 200 nm) (©2018 Trustees of Dartmouth College/Public Domain [64]). (B) Model of the inner structure. (C) Model of the outer structure. (D) Combinded model with final materials. (Ee) Yeast cells show elongated mitochondria networks (scale bar 1 μm) (©2002 National Academy of Sciences [65]). (F) Mitochondria structures spanning through C. reinhardtii (red, scale bar 1 μm) (©2012 Hummel et al., CC BY 4.0 [31]). (G) Extended mitochondrial structures positioned following the curvature of the cell membrane.
To be able to visually recognize this characteristic feature, its color was chosen as a very transparent, almost glass-like white with slight roughness. Leighton et al. observed a pink color for purified mitochondria and a light tan color of mitochondrial fractions [66], which was accounted for in our model.
Each mitochondrial compartment model consists of two objects, the outer and inner membrane. The inner models are textured by a displacement modifier, each of which geometry are linked to its respective outer parts, to enable their easy network-like repositioning (Figure 12G).
3.3.7. Cytoskeleton
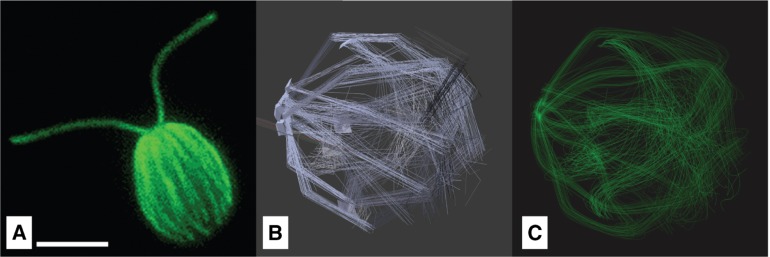
The cytoskeleton consists i.a. of a network of microtubules, originating in four rootlets at the flagellar basal bodies and extending around the cell, located directly beneath the cell membrane [67] in order to distribute the mechanical stress of the movement over the surface of the cell. Aside from that, the microtubules are responsible for intracellular transport [68]. For making the flagella in combination with the cytoskeleton visible, immunofluorescence labeling is often used. Figure 13A shows the cytoskeleton in combination with the flagella made visible by being fluorescently labeled with an antibody to tyrosinated tubulin [69]). Similar to this, in Mittelmeier et al. the microtubules were made visible using indirect immunofluorescence labeling [68].
Figure 13:
Microtubules filaments of the cytoskeleton: A) Chlamydomonas reinhardtii fluorescently labeled with an antibody to tyrosinated tubulin (©2018 Courtesy of Karl Johnson, procedurally as described in [69]) B) Hair Emitter with emitting planes. A) Rendered tubule model.
In our model the cytoskeleton was realized with a Hair Emitter, a particle system where the complete path of a moving particle is rendered at once (Figure 13B). A particle system is a mathematical model in which randomly generated particles follow defined physical influences [46]. It is an adequate option for displaying linear structures with no directly observable surface, which is appropriate for delicate filaments like microtubules. The parameters of the particle system and the green material were optimized until visually being similar to figures found in [68] and Figure 13A.
3.3.8. Eyespot
C. reinhardtii cells possess a light-sensitive eyespot which is used to determine its position in relation to a potential light source (phototaxis) [30].
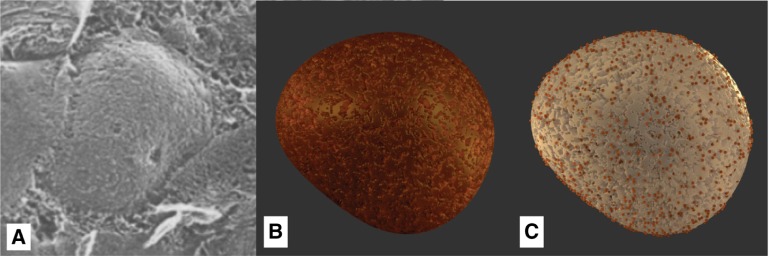
The size and shape of the eyespot can be modeled with high accuracy, as this cell compartment is very well-explored. It consists of one or two layers of small spherical carotenoid-filled lipid granules, approx. 80–130 nm in size, as depicted in Figure 14A. These two layers are embedded between the thylakoid membranes. Its location depends on the asymmetric microtubule cytoskeleton, by which it is placed in relation to the flagella [68].
The diameter of the whole spot is around 1 μm. Analyses of separated eye apparatuses show their carotenoid-rich chemical composition (Figure 14A). This aspect explains their orange color, which is visible in light-microscopic images, despite their low thickness of around 300 nm [30]. Figure 14C shows the final rendering performed with Blender, clearly showing the orange-colored eye spot on top of thylakoid membranes.
3.4. Flagella and the Movement Cycle of C. reinhardtii
The initial scene of the animation (Chapter 4) shows the flagella movement of C. reinhardtii. The animation starts at the macroscopic level, showing a number of C. reinhardtii cells as prepared on a sample slide. In vivo they are able to move with speeds of 50 μm/s to 200 μm/s [71], [72], [73], [74]. Then, the animation slows down to show the exact series of movements the flagella undergo in order to propel the cell forward, which causes a saltatory forward-backward motion, that ultimately results in a forward movement. This flagella beating occurs with a frequency of 40–70 Hz [72], [73].
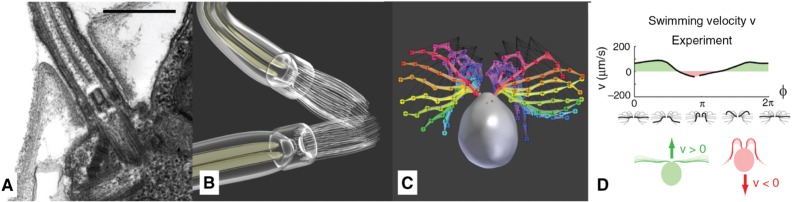
The flagella were modeled with an almost transparent material, which seems reasonable as they are comparably thin with a diameter of 300 nm [64], featuring a very low light absorption. The mechanism of their fixation in the cell body was modeled with respect to TEM slices similar to the one in Figure 15A.
Figure 15:
Modeling of the flagella: (A) TEM slice of the cellular mounting (scale bar 500 nm) (©2018 Trustees of Dartmouth College/Public Domain [64]). (B) Basal bodies and Flagella rendered. (C) Overlay of joint-bone model of several keyframes of the beating cycle, redrawn based on [72]. (D) Swimming velocity profile during one beating cycle [71].
The animation of the flagella was realized via rigging, where the vertices of the model were attached to a simple skeleton consisting of bones and joints. The bones were then positioned after a sequence of micrographs that were captured with high-speed phase-contrast microscopy by Rüffer and Nultsch [72] (Figure 15C). For the movement of the entire cell, a velocity profile of Geyer et al. [71] was adapted (Figure 15D). Similar velocity profiles can be found in [72], [73], [74].
3.5. Functional Modeling of C. reinhardtii Energy-Producing Pathways
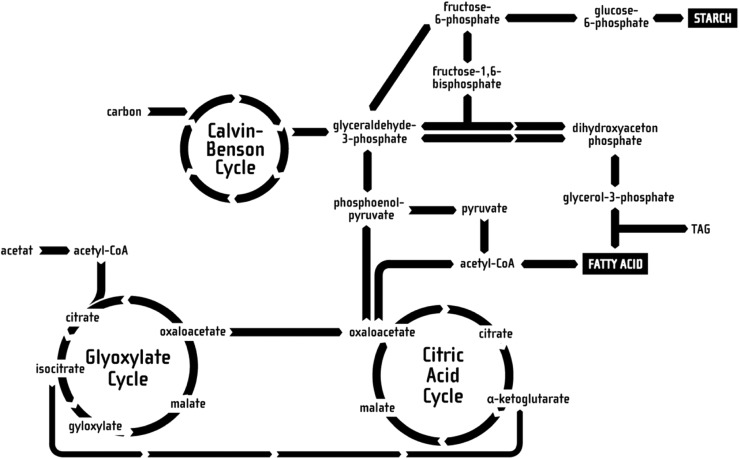
The objective of the educational animation is to illustrate the process of metabolizing nutrients and storing them in energy carriers, such as starch or lipids. Aside from the respective location in the cell, not every molecular step in the whole process of consuming nutrients and transforming them to starch or lipids on a chemical basis can be displayed, as this would be far too time-consuming and overwhelming for the common viewer to process, thus being counterproductive to the educational value. To achieve an optimal educational effect, the complex chemical transitions were simplified to the relevant pathways, as well as geometrically streamlined. This was done based on a pathway diagram from [75], illustrating the complete energy-generating pathway in C. reinhardtii. This pathway illustration was then reduced to three important cycles, namely the Glyoxylate Cycle, the Citric Acid Cycle and the Calvin-Benson Cycle, as well as the corresponding side products, as these are the ones participating in the energy production and transport processes. These pathways are also linked to – as discussed later – the energy storage in form of starch and/or lipids.
The resulting diagram is shown in Figure 16 and is presented in form of a two-dimensional illustration in front of a non-animated S3D background, locating the cycles at their respective compartments while preserving the simplistic pathway diagram illustration as well as the spatial structure of the cell components in the background. Whereas the cell components in the background were located between the Far and Projection plane, the diagram was laid onto the Projection plane (see also Section 3.2).
Figure 16:
The reduced pathways used in the animation, relevant for starch and lipid metabolization.
Only the momentarily relevant parts of the 3D diagram are highlighted and slightly animated, for instance through turning wheels or changes in brightness. Aside from the designations of the most important cycles the names of important intermediate products are displayed.
3.6. Molecular Modeling of a C. reinhardtii membrane
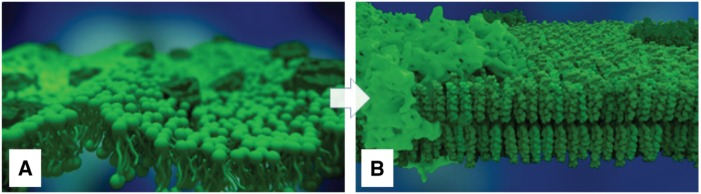
Some processes involved in the previously-discussed metabolic pathway take place inside the thylakoid membrane. The initial membrane model was realized with a simplistic conceptual representation of phospholipid molecules in form of simple ball-stick-models and a generic form of membrane proteins, see Figure 17A. Due to time constraints, this representation was lacking realism and was not up to the standard of the other scenes. This was due to the fact that the scale on which individual molecules can be observed falls into the molecular level – i.e. nanoscopic regime – rather than the mesoscopic one.
Figure 17:
The rendered thylakoid membrane: (A) The old ball-stick-lipid molecules. (B) The enhanced more realistic molecular representation.
The revised thylakoid membrane representation comprises of several phospholipids types and protein complexes. Based on literature research, seven different types of phospholipids namely, MGDG, monogalactosyldiglyceride; DGDG, digalactosyldiglyceride; SQDG, sulfoquinovosyldiacylglycerol; DGTS, diacylglyceryl-(N,N,N-trimethyl)-homoserine; PE, phoshatidylethanolamine; PG, phosphatidylinositol; PI, phospahtidylglycerol; DPG, diphosphatidylglycerol were used. The lipid distribution was based on the publication [76]. In addition, two protein complexes were integrated. Although there are more than 200 protein complexes that are discovered to be part of the thylakoid membrane, we are focusing here for this rather small membrane patch on two photosynthesis-relevant complexes: the Light harvesting II [77] and Photosystem I [78] one. The required 3D models of these lipids and proteins were downloaded from Avanti® Polar Lipids and OPM databases respectively and were imported via Jmol into Blender [79], [80], [81]. A python-scripted panel was used to generate a grid-based membrane model containing only lipids at the moment. This lipid packing approach places lipids along a grid with equal distances between the different lipids [82]. The membrane patch is packed with seven different lipids. The script provides a user-defined seed-based random generation of distribution patterns.
Once the modeling is done, proteins are manually inserted into the membrane patch. In the revised membrane, the problematic wave-like motion of the bilayer membrane was replaced by a more realistic movement. The new movement has been implemented by using different modifiers from Blender, this mainly includes shrink wrap, wave and armature modifiers. The shrink wrap modifier gives the global movement effects such as tilting the lipid models. While the other two modifiers give local movement effect such as the wobbling effect as to mimic the molecules’ movement. This creates an appropriately looking movement effect closer to realistic movement. The resulting membrane is shown in Figure 17B.
The detailed discussion of this approach will be published in a future manuscript and lies out of scope of this work [83].
3.7. Multiscale Stereoscopic 3D Visualization
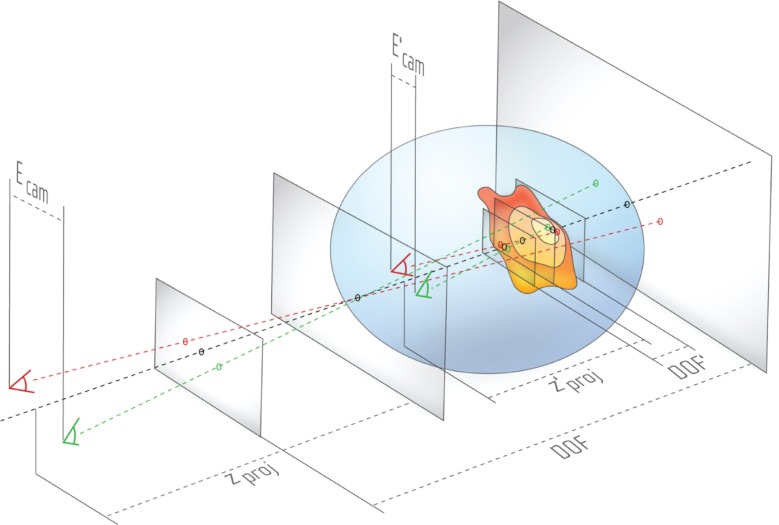
We previously discussed in detail a problem specific for cell visualization [17]: while showing different objects in a cell environment, the viewer is presented with objects at different scales. For example, the video discussed in this work starts at the mesoscopic scale: the viewer watches a moving Chlamydomonas cell with a length of approx. 20 to 30 μm. Then, the camera moves into the cell towards smaller scales, finally ending at the molecular scale, showing lipids featuring a size of a few Ångström. To enable a smooth transition between mesoscopic and the molecular scale, the eye/camera separation Ecam shown in Figure 18 has to be dynamically adjusted.
Figure 18:
While moving into the cell, with decreasing eye separation Ecam and projection distance zproj, the DOF also decreases. Because the smaller viewing frustum is still mapped to the same perceived depth, the longitudinal magnification increases, thus small structures appear bigger. The three planes are illustrated in Figure 6.
By manipulating Ecam as well as the projection distance zproj, the mapping under constant viewing conditions causes an impression of visual shrinking or enlarging, as depicted in Figure 18. Ecam, zproj and Depth of Field DOF for example show the initial setting of the viewing frustum, and then the blue circular space containing , and DOF′ illustrate the state after the cameras have moved towards the cell membrane whereas simultaneously decreasing all aforementioned parameters.
In the example of Figure 18 S3D visualization is used to create an effect of passing through the outer cell membrane into the inner cell’s cytoplasm. Inside the blue circular space, the Projection plane (see also Figure 6) lies beyond the cell membrane. Based on the rule discussed in Section 3.2 – an object after passing the Projection plane towards the Near plane should not be visible anymore – the cell membrane has to disappear in this state, e.g. using frontface culling.
For using a similar approach in combination with interactive cell visualization environments, we previously introduced an approach called the interactive dynamic projection plane S3D method [17].
Instead of creating a smooth transition between different scales by adjusting the stereo camera parameters, it is also possible to use visual effects to change between two scenarios. In the beginning of the video, first, multiple cells are shown, and then the camera shows only a single one whereas simultaneously changing the magnification.
4. Results and Discussion
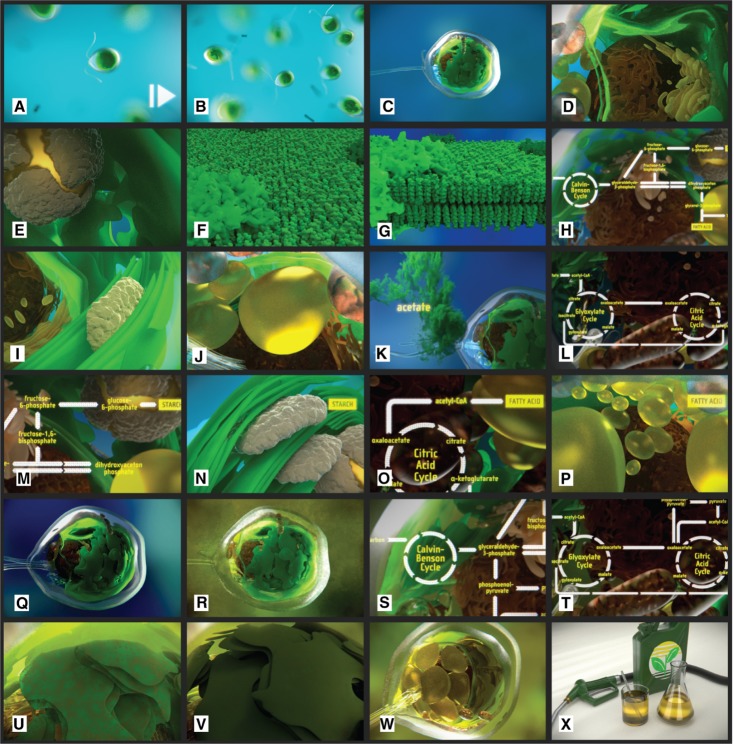
Figure 19 shows an overview of all scenes of the video “Chlamydomonas reinhardtii 3D – From Biological Cells to Biofuels”.
Figure 19:
An overview of all scenes as shown in the animation.
The previously discussed model was used to create an animation to visualize the energy-relevant pathways in the context of the C. reinhardtii cell. Different aspects of this species are illustrated, for example: the flagella movement of single cells (Figure 19A), the assembly in small colonies (Figure 19B), the cellular structure visited from outside towards the interior of the cell (Figure 19C–E), the thylakoid membrane and its role in energy-relevant processes (Figure 19F–G), the intracellular energy production processes and the involved cell components as well as the way how they can be manipulated to increase their output (Figure 19H–T), resulting structural changes and the way to synthesize biological fuel from this cell (Figure 19U–X).
The video unites visualizations at the mesoscopic, molecular and functional level.
At the mesoscopic level, cell components were modeled and visualized based on different information sources and images. We also discussed different coloring methods applied to the cell components, trying to mimic the realistic appearance of the cell. Of course, there exist alternative coloring approaches, like the one we introduced providing a contrasting color set which is ignoring the natural coloring but comes with the advantage that it is also applicable to larger numbers of differentiable cell component types [21], [84]. However, as the cell components here were modeled with different shapes and textures they are easily distinguishable. Moreover, the use of S3D techniques enables that the spatial structure and textures of the cell components can be well-perceived and differentiated. In addition, we provided an animation of the flagella-based movement of C. reinhardtii based on published material. Naturally, the shape and size of the cell component models used in this work are based on our selection of microscopic images and cannot be interpret as standardized representations of a specific component type. For this purpose, specific studies would have to be conducted for C. reinhardtii measuring the sizes of, e.g. fluorescently dyed cell components using image analysis in combination with high resolution image stacks.
The discussed pathways at the functional level were based on complex intracellular reactions found in published material. They were broken down to their most basic parts to enable a comprehensible illustration of the reactions relevant in this context. The time required to explain the complexity of the complete pathway would exceed by far the time available in a scientific animation as well as the regular recipient’s attention span. Here again S3D techniques were used to show the pathways in front of the corresponding cell component structures where the underlying reactions take place.
Also at the molecular level we were following an interpretative approach by introducing the modeling of the thylakoid membrane following a grid-based method. This method was developed based on the plugin functionality of Blender. The detailed discussion of the membrane modeling lies beyond the scope of this publication and is subject of a future manuscript [83]. The modeling and animation of the membrane with Blender in terms of fluidity and membrane dynamics still falls back behind those membrane models simulated by dedicated software. An alternative approach to extend the provided membrane model is to use external packages for membrane modeling – such as the MembraneEditor [85] – as well as molecular simulations. For this purpose, tools like GROMACS can be used to simulate membrane models based on Molecular Dynamics which provides a higher degree of realism [86]. Just recently a new plugin for Blender was released which can be used to visualize underlying GROMACS simulations with Blender [15]. This approach enables visualization of the membrane components at the molecular level with an increased grade of realism.
Finally, we want to give some additional hints regarding lessons learned during the production of this video.
First of all, we want to highlight that this video was not produced by a professional animation studio which was collaborating with a university, like in case of videos like “The Inner Life of the Cell”. This video is the result of a student project starting with a hands-on Blender workshop conducted by B.S., whereas the major part was then realized by N.B. during the student project and finished as a student job, and then extended by M.G. during his Master thesis, providing an improved membrane model. Therefore, the discussed video is not intended to be a final version. Instead of this, we are using versioning for the issued videos. An exemplary Changelog:
Version 1.0 HD-ready Stereo Version (720p), male voice, 2014
Version 1.1 Full HD Stereo Version (1080p), native speaker female voice, 2016
Version 1.2 Improved Membrane Model (1080p), 2017
The big advantage is that in this way future students or researchers can extend the video, or extract scenes to use them for other purposes, discussing for example a specific aspect like the membrane in more detail. Here, Blender is a very good choice as all of its software versions can be directly downloaded from its website and can be used side-by-side on the same computer. In this way, also animations created with older versions – in our case version 2.71 – can be easily reproduced.
An important aspect which applies to most similar animations is the fact that is often not possible to grasp the complete complexity of the discussed topic by watching the video a single time. The recipient should be animated to repeatedly watch the animation and do some additional research in case the topic is able to grasp her/his interest.
It is also important to note that the rendering time is a bottleneck during the video production process. As two perspectives have to be rendered, also the rendering time nearly doubles in comparison to a regular video rendering. The rendering of version 1.1 for example was largely rendered on four server computers with dedicated NVIDIA graphics and took (with administrative breaks) ca. one month. The rendering was executed by using a bash script.
In terms of S3D visualization it is important to plan the visualization of the video right from the start. The obligatory storyboard has to be extended by a concrete design of the stereo camera settings and handling, especially in case camera movement is involved. Then, a test S3D animation has to be rendered without the use of expensive materials and textures in order to check the stereoscopic effect. Especially for the initial S3D process and planning, the use of analgyph red-cyan glasses is fully sufficient, because recent Blender versions directly support this mode in the 3D viewer and the glasses are cheap and can be used everywhere. Then, in the second step, it is of course important to use dedicated 3D screens providing adequate quality. Another important aspect is the wide-spread distribution of VR cardboards which can be used to watch, for example, YouTube® 3D videos with a common smartphone.
5. Conclusions and Outlook
We showed, how computational systems biology in combination with different published information and microscopic image resources can be used to create a consistent holistic 3D model of a C. reinhardtii cell by using the Open Source tool Blender.
For this purpose, we were following an interpretative approach: To create the 3D shapes of cell components and to place them inside the context of a cellular environment at the mesoscopic level, various visual details provided by light as well as electron microscopic image resources were integrated. By doing so, we combined expectations of biology experts regarding the structural appearance and coloring of a cell with practical design choices enabling a clear orientation and differentiation of the different components.
Of course, we do not want to proclaim that the presented images do present the visual reality of the cellular structure. Technology is used to make the invisible visible, and we interpret these images to convey a specific image of a cell, creating its own virtual reality, meeting cultural expectations and empirical observations.
Additional future work includes the improvement of the model by using advanced microscopy techniques. Other extensions of the provided 3D cell include the modeling of additional cell components, such as the spindle complex – important for replication – or the ribosomes – important for the transcription process.
At the moment, still many cell components appear of quite static nature in the animation. Their dynamic appearance could be improved by introducing fluidity effects to all cell components. However, before doing so, additional research would have to be conducted to achieve a visually realistic movement of the cell components. Alternatively, live microscopy imaging data could be included in the video to provide a first glance at realistic data of the cell.
A more-advanced vision is to substitute the interpretive cell models by those ones completely based on 3D microscopic image sets. For this purpose, a very time-consuming segmentation process would be required, resulting in a very complex cell model which then would have to be reduced in the context of the video.
Regarding the S3D visualization, we are currently working on different ways to present the underlying model with Virtual Reality-related interactive technologies, as an important alternative to an animation is an interactive representation of the model. In this context we are also planning a contrasting evaluation of the different visualization approaches. Also external tools could be used to explore the underlying model exported from Blender. For example, the model was already used with Unity 3D® enabling interactive exploration. Similar approaches are already known at the molecular level which are partly also compatible to large atomistic models [87], [88]. We are currently working on interactive visualization methods compatible to web browsers, enabling to conveniently explore the cell and its components in combination with the underlying textual information.
The video as well as additional information regarding the CELLmicrocosmos Cell Visualization project can be found at: http://Cm5.CELLmicrocosmos.org
Acknowledgements
First, we would like to thank all researchers who kindly allowed us to use their microscopic images for this work. Please see the corresponding figure captions for the copyright information. Then, many thanks go to the anonymous reviewers whose critics and suggestions drastically helped us to improve this manuscript. Then, we have to thank the Blender community and in particular Sebastian Schneider who once developed the stereoscopic rendering script for older Blender versions and also kindly allowed us to maintain his script.
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Reece JB, Urry LA, Cain ML, Campbell biology. Boston: Pearson; 2014. [Google Scholar]
- [2].Cooper GM, Hausman RE. The cell: a molecular approach. 7th ed. USA: Sinauer Associates Inc.; 2015. [Google Scholar]
- [3].Alberts B, Johnson A, Lewis J, Molecular biology of the cell. 2017. [Google Scholar]
- [4].Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol. 2012;198:961–71. doi: 10.1083/jcb.201206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Goodsell DS. Inside a living cell. Trends Biochem Sci. 1991;16:203–6. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- [6].McGill G. Molecular movies… coming to a lecture near you. Cell. 2008;133:1127–32. doi: 10.1016/j.cell.2008.06.013. [DOI] [PubMed] [Google Scholar]
- [7].Lok C. Biomedical illustration: from monsters to molecules. Nature. 2011;477:359–61. doi: 10.1038/nj7364-359a. [DOI] [PubMed] [Google Scholar]
- [8].Of Harvard College TPaF. BioVisions: the inner life of the cell. 2007 Available from: http://web.archive.org/web/20081215222914/http://multimedia.mcb.harvard.edu/
- [9].The inner life of the cell. 2017 Page Version ID: 769770989. Available from: https://en.wikipedia.org/w/index.php?title=The_Inner_Life_of_the_Cell&oldid=769770989.
- [10].Blender Online Community. Blender – a 3D modelling and rendering package. Amsterdam: Blender Institute; 2018. [Google Scholar]
- [11].Grove C. The virtual worm: 3D renderings of Caenorhabditis elegans (VizBi2010 Poster) [Poster] Heidelberg, Germany: 2010. Available from: https://vizbi.org/Posters/2010/W23. [Google Scholar]
- [12].Hammang C, O’Donoghue S, Stolte C, Drew B, Sabir K, Patterson K. et al. The Hungry Microbiome: a biomedical animation (VizBi2014 Poster) [Poster] 2014 Available from: https://vizbi.org/Posters/2014/A17.
- [13].Andrei RM, Callieri M, Zini MF, Loni T, Maraziti G, Pan MC. et al. Intuitive representation of surface properties of biomolecules using BioBlender. BMC Bioinformatics. 2012;13(Suppl 4):S16. doi: 10.1186/1471-2105-13-S4-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haschka T, Dauchez M, Henon E. Visualization of molecular properties at the qantum mechanical level using blender; Virtual and Augmented Reality for Molecular Science (VARMS@IEEEVR), 2015 IEEE 1st International Workshop on. IEEE; 2015. pp. 7–13. [Google Scholar]
- [15].Rajendiran N, Durrant JD. Pyrite: a blender plugin for visualizing molecular dynamics simulations using industry-standard rendering techniques. J Comput Chem. 2018;39:748–755. doi: 10.1002/jcc.25155. [DOI] [PubMed] [Google Scholar]
- [16].Motion Picture Association of America I. MPAA’s 2016 Theatrical Market Statistics report. 2017. Available from: http://www.filmjournal.com/mpaas-2016-theatrical-market-statistics-report-showcases-worldwide-box-office-growth. [Google Scholar]
- [17].Sommer B, Bender C, Hoppe T, Gamroth C, Jelonek L. Stereoscopic cell visualization: From mesoscopic to molecular scale. J Electron Imaging. 2014;23:011007. [Google Scholar]
- [18].Google Cardboard – Google VR. 2017 Available from: https://vr.google.com/cardboard/
- [19].Samsung Gear VR with Controller. 2017 Available from: http://www.samsung.com/global/galaxy/gear-vr/
- [20].YouTube. Upload virtual reality videos – YouTube Help. 2018 Available from: https://support.google.com/youtube/answer/6316263?hl=en. [Google Scholar]
- [21].Sommer B. CELLmicrocosmos – integrative cell modeling at the molecular, mesoscopic and functional level [Doctorate Thesis] Bielefeld, Germany: Bielefeld University; 2012. Available from: http://pub.uni-bielefeld.de/download/2557380/2557411. [Google Scholar]
- [22].Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch Mikrosk Anat. 1873;9:413–8. [Google Scholar]
- [23].Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21:1347–55. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- [24].Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–3. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- [26].Giessibl FJ, Bielefeldt H, Hembacher S, Mannhart J. Imaging of atomic orbitals with the Atomic Force Microscope — experiments and simulations. Ann Phys. 2001;10:887–910. [Google Scholar]
- [27].Martone ME, Gupta A, Wong M, Qian X, Sosinsky G, Ludäscher B. et al. A cell-centered database for electron tomographic data. J Struct Biol. 2002;138:145–55. doi: 10.1016/s1047-8477(02)00006-0. [DOI] [PubMed] [Google Scholar]
- [28].Lottspeich F. Bioanalytik. 2nd ed. Heidelberg: Spektrum, Akademischer Verlag; 2006. [Google Scholar]
- [29].Haken H, Wolf HC. Molekülphysik und Quantenchemie: Einführung in die experimentellen und theoretischen Grundlagen. 5th ed. Springer-Lehrbuch. Berlin [u.a.]: Springer; 2006. [Google Scholar]
- [30].Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M. et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18:1908–30. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hummel E, Guttmann P, Werner S, Tarek B, Schneider G, Kunz M. et al. 3D Ultrastructural organization of whole Chlamydomonas reinhardtii cells studied by nanoscale soft X-ray tomography. PLoS One. 2012;7:e53293. doi: 10.1371/journal.pone.0053293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Plattner H, Hentschel J. Zellbiologie. Stuttgart: Georg Thieme Verlag; 2006. [Google Scholar]
- [33].Vournakis J, Rich A. Size changes in eukaryotic ribosomes. Proc Natl Acad Sci. 1971;68:3021–5. doi: 10.1073/pnas.68.12.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. TRENDS Biotechnol. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- [35].Takahashi K, Ishikawa N, Sadamoto Y, Sasamoto H, Ohta S, Shiozawa A. et al. E-Cell 2: multi-platform E-Cell simulation system. Bioinformatics. 2003;19:1727–9. doi: 10.1093/bioinformatics/btg221. [DOI] [PubMed] [Google Scholar]
- [36].Izaguirre JA, Chaturvedi R, Huang C, Cickovski T, Coffland J, Thomas G. et al. CompuCell, a multi-model framework for simulation of morphogenesis. Bioinformatics. 2004;20:1129–37. doi: 10.1093/bioinformatics/bth050. [DOI] [PubMed] [Google Scholar]
- [37].Blinov ML, Schaff JC, Vasilescu D, Moraru II, Bloom JE, Loew LM. Compartmental and spatial rule-based modeling with Virtual Cell. Biophys J. 2017;113:1365–72. doi: 10.1016/j.bpj.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wehr JD, Sheath RG, Kociolek JP. Freshwater algae of North America: ecology and classification. New York: Elsevier; 2015. [Google Scholar]
- [39].Hoshaw RW, Ettl H. Chlamydomonas Smithii sp. nov. – a chlamydonad interfertile with Chlamydononas rheinhardtii. J Phycol. 1966;2:93–6. doi: 10.1111/j.1529-8817.1966.tb04600.x. [DOI] [PubMed] [Google Scholar]
- [40].Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB. et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–50. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Harris EH. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. 1st ed. San Diego [u.a.]: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- [42].Ghirardi ML, Posewitz MC, Maness PC, Dubini A, Yu J, Seibert M. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu Rev Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- [43].2014-09-21 | 4th Int. CeBiTec Research Conference Bielefeld. 2014 Available from: http://www.cebitec.uni-bielefeld.de/index.php/events/conferences/392-2014-09-21-int-bielefeld-cebitec-research-conference.
- [44].Stereoscopic Displays and Applications conference – 3D Theatre Session. 2017 Available from: http://stereoscopic.org/3dcinema/index.html.
- [45].Van Gumster J. Blender for dummies. Hoboken: John Wiley & Sons; 2015. [Google Scholar]
- [46].Blender Online Community. Blender Documentation. Amsterdam: Blender Institute; 2018. Available from: http://docs.blender.org. [Google Scholar]
- [47].Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure (London, England: 1993) 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Johnson GT, Autin L, Al-Alusi M, Goodsell DS, Sanner MF, Olson AJ. cellPACK: A virtual mesoscope to model and visualize structural systems biology. Nat Methods. 2015;12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Klein T, Autin L, Kozlíková B, Goodsell DS, Olson A, Gröller ME. et al. Instant construction and visualization of crowded biological environments. IEEE Trans Vis Comput Graph. 2018;24:862–872. doi: 10.1109/TVCG.2017.2744258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kerr RA, Bartol TM, Kaminsky B, Dittrich M, Chang JCJ, Baden SB. et al. Fast Monte Carlo simulation methods for biological reaction-diffusion systems in solution and on surfaces. SIAM J Sci Comput. 2008;30:3126–49. doi: 10.1137/070692017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].MCell Home. 2017. Available from: http://www.mcell.org/ [Google Scholar]
- [52].Robinson DG, Brandizzi F, Hawes C, Nakano A. Vesicles versus tubes: is endoplasmic reticulum-Golgi transport in plants fundamentally different from other eukaryotes? Plant Physiol. 2015;168:393–406. doi: 10.1104/pp.15.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Engel BD, Schaffer M, Albert S, Asano S, Plitzko JM, Baumeister W. In situ structural analysis of Golgi intracisternal protein arrays. Proc Natl Acad Sci. 2015;112:11264–9. doi: 10.1073/pnas.1515337112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bock DD, Lee WCA, Kerlin AM, Andermann ML, Yurgenson S, Kim HS. et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M. et al. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Asmail C. Bidirectional scattering distribution function (BSDF): a systematized bibliography. J Res Natl Inst Standards Technol. 1991;96:215. doi: 10.6028/jres.096.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Woods AJ. How are crosstalk and ghosting defined in the stereoscopic literature?; IS&T/SPIE electronic imaging. International Society for Optics and Photonics; 2011. pp. 78630Z–12. [Google Scholar]
- [59].Schneider S. Stereoscopic rendering in Blender 2.6. 2013 Available from: http://www.webcitation.org/query?url=http%3A%2F%2Fwww.noeol.de%2Fs3d%2F&date=2013-08-23.
- [60].Schneider S, Sommer B. CELLmicrocosmos.org forum – Stereoscopic Camera Plugin for Blender 2.6X to 2.7X. 2015 Available from: http://www.cellmicrocosmos.org/Cmforum/viewtopic.php?f=62&t=776.
- [61].Škaloud P, Peksa O. Comparative study of chloroplast morphology and ontogeny in Asterochloris (Trebouxiophyceae, Chlorophyta) Biologia. 2008;63:250. [Google Scholar]
- [62].Engel BD, Schaffer M, Cuellar LK, Villa E, Plitzko JM, Baumeister W. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. Elife. 2015;4:e04889. doi: 10.7554/eLife.04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].D’Oliveiro L. Blender 3D: Noob to Pro/Fireflies in Cycles, Continued – Wikibooks, open books for an open world. 2014 Available from: https://en.wikibooks.org/wiki/Blender_3D:_Noob_to_Pro/Advanced_Tutorials/Cycles_Fireflies_2.
- [64].Dartmouth Electron Microscope Facility DC. Dartmouth College Electron Microscope Facility. 2018 Available from: http://www.dartmouth.edu/ emlab/
- [65].Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci USA. 2002;99:3370–5. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Leighton F, Poole B, Beaufay H, Baudhuin P, Coffey JW, Fowler S. et al. The large-scale separation of peroxisomes, mitochondria, and lysosomes from the livers of rats injected with Triton WR-1339: improved isolation procedures, automated analysis, biochemical and morphological properties of fractions. J Cell Biol. 1968;37:482–513. doi: 10.1083/jcb.37.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ringo L. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J Cell Biol. 2011;193:741–53. doi: 10.1083/jcb.201009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Johnson KA. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J Cell Sci. 1998;111:313–20. doi: 10.1242/jcs.111.3.313. [DOI] [PubMed] [Google Scholar]
- [70].Eitzinger N, Wagner V, Weisheit W, Geimer S, Boness D, Kreimer G. et al. Proteomic analysis of a fraction with intact eyespots of Chlamydomonas reinhardtii and assignment of protein methylation. Front Plant Sci. 2015;6:1085. doi: 10.3389/fpls.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Geyer VF, Jülicher F, Howard J, Friedrich BM. Cell-body rocking is a dominant mechanism for flagellar synchronization in a swimming alga. Proc Natl Acad Sci USA. 2013;110:18058–63. doi: 10.1073/pnas.1300895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rüffer U, Nultsch W. High-speed cinematographic analysis of the movement of Chlamydomonas. Cell Motil. 1985;5:251–63. [Google Scholar]
- [73].Fujita S, Matsuo T, Ishiura M, Kikkawa M. High-throughput phenotyping of chlamydomonas swimming mutants based on nanoscale video analysis. Biophys J. 2014;107:336–45. doi: 10.1016/j.bpj.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Racey TJ, Hallett R, Nickel B. A quasi-elastic light scattering and cinematographic investigation of motile Chlamydomonas reinhardtii. Biophys J. 1981;35:557–71. doi: 10.1016/S0006-3495(81)84812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S. et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell. 2014;13:591–613. doi: 10.1128/EC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mendiola-Morgenthaler L, Eichenberger W, Boschetti A. Isolation of chloroplast envelopes from Chlamydomonas. Lipid and polypeptide composition. Plant Sci. 1985;41:97–104. [Google Scholar]
- [77].1rwt – Light-Harvesting Complex II – orientations of proteins in membranes (OPM) database. 2004 Available from: http://opm.phar.umich.edu/protein.php?search=1rwt.
- [78].1jb0 – Photosystem I of cyanobacteria – orientations of proteins in membranes (OPM) database. 2001 Available from: http://opm.phar.umich.edu/protein.php?search=1jb0.
- [79].Avanti Polar Lipids, Inc. 2017 Available from: https://avantilipids.com. [Google Scholar]
- [80].Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–6. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jmol: an open-source Java viewer for chemical structures in 3D. 2018 Available from: http://jmol.sourceforge.net/
- [82].Sommer B. Membrane packing problems: a short review on computational membrane modeling methods and tools. Comput Struct Biotechnol J. 2013;5:e201302014. doi: 10.5936/csbj.201302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ghaffar M, Biere N, Jäger D, Klein K, Schreiber F, Kruse O. et al. 3D modelling and visualisation of heterogeneous cell membranes in Blender. In review. 2018 submitted.
- [84].Sommer B, Kormeier B, Demenkov PS, Arrigo P, Hippe K, Ates O. et al. Subcellular localization charts: a new visual methodology for the semi-automatic localization of protein-related data sets. J Bioinformatics Comput Biol. 2013;11:1340005. doi: 10.1142/S0219720013400052. [DOI] [PubMed] [Google Scholar]
- [85].Sommer B, Dingersen T, Gamroth C, Schneider SE, Rubert S, Krüger J. et al. CELLmicrocosmos 2.2 MembraneEditor: a modular interactive shape-based software approach to solve heterogeneous Membrane Packing Problems. J Chem Inf Model. 2011;5:1165–82. doi: 10.1021/ci1003619. [DOI] [PubMed] [Google Scholar]
- [86].Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- [87].Doutreligne S, Cragnolini T, Pasquali S, Derreumaux P, Baaden M. UnityMol: interactive scientific visualization for integrative biology; Large Data Analysis and Visualization (LDAV), 2014 IEEE 4th Symposium on. IEEE.2014. pp. 109–10. [Google Scholar]
- [88].Grottel S, Krone M, Müller C, Reina G, Ertl T. MegaMol—a prototyping framework for particle-based visualization. IEEE Trans Vis Comput Graph. 2015;21:201–14. doi: 10.1109/TVCG.2014.2350479. [DOI] [PubMed] [Google Scholar]