Abstract
A dissimilatory nitrate reduction to ammonium (DNRA) microbial community was developed under a high organic carbon to nitrate (C/NO3−) ratio in an anoxic semi-continuous sequencing batch reactor (SBR) fed with glucose as the source of carbon and NO3− as the electron acceptor. Activated sludge collected from a municipal wastewater treatment plant with good denitrification efficiency was used as the inoculum to start the system. The aim of this study was to examine the microbial populations in a high C/NO3− ecosystem for potential DNRA microorganisms, which are the microbial group with the ability to reduce NO3− to ammonium (NH4+). A low C/NO3− reactor was operated in parallel for direct comparisons of the microbial communities that developed under different C/NO3− values. The occurrence of DNRA in the high C/NO3− SBR was evidenced by stable isotope-labeled nitrate and nitrite (15NO3− and 15NO2−), which proved the formation of NH4+ from dissimilatory NO3−/NO2− reduction, in which both nitrogen oxides induced DNRA activity in a similar manner. An analysis of sludge samples with Illumina MiSeq 16S rRNA sequencing showed that the predominant microorganisms in the high C/NO3− SBR were related to Sulfurospirillum and the family Lachnospiraceae, which were barely present in the low C/NO3− system. A comparison of the populations and activities of the two reactors indicated that these major taxa play important roles as DNRA microorganisms under the high C/NO3− condition. Additionally, a beta-diversity analysis revealed distinct microbial compositions between the low and high C/NO3− SBRs, which reflected the activities observed in the two systems.
Keywords: dissimilatory nitrate reduction to ammonium (DNRA), semi-continuous sequencing batch reactor, C/NO3− ratio, stable-isotope tracer, Illumina MiSeq 16S rRNA sequencing
Dissimilatory nitrate (NO3−) reduction is part of the nitrogen cycle driven by microorganisms with the ability to use NO3− as their electron acceptor. Known dissimilatory NO3−-reducing pathways include denitrification, which generates nitrogenous gases as its products, and dissimilatory nitrate reduction to ammonium (DNRA), which produces ammonium (NH4+). Both of these processes occur under anoxic environments and utilize the same types of electron donors, such as organic carbon, sulfur, and iron (6, 12, 28). Therefore, denitrifiers and DNRA microorganisms are competitors for electron donors and electron acceptors (NO3− and nitrite [NO2−]), as well as for the habitats in which they grow. Although the microorganisms responsible for denitrification have been extensively examined, limited information is currently available on the DNRA microbial group.
Although different nomenclatures have been termed for this dissimilatory NH4+-forming process, including NO3− or NO2− ammonification, it is mainly known as DNRA. This microbial group may be further classified into respiratory and fermentative types (7, 22), which use different metabolic pathways to catalyze the formation of NH4+. DNRA microorganisms have been hypothesized and observed to occur in a high electron donor, limited NO3− environment, which is normally referred to as a high carbon to NO3− (C/NO3−) condition, whereas denitrifiers appear to prefer a low C/NO3− condition (7, 34). Thermodynamically, the energy gained per mole electron donor and per mole NO3− differs for denitrification and DNRA, with the former obtaining a higher amount of energy per glucose molecule and the latter gaining more energy per NO3− molecule (32). These findings suggest how DNRA microorganisms compete with denitrifiers in a NO3−- limiting ecosystem. The influence of the C/NO3− ratio on NO3−-reducing communities or a pure culture was recently reported (23, 36–38, 44), with selective pressure of the ratio on the NO3− reduction pathways being observed.
In agriculture, DNRA is considered to be a beneficial process because it preserves nitrogen fertilizers within soil (29). However, this pathway is undesirable in a biological wastewater treatment system due to its production of NH4+, a waste product that is generally removed by nitrification-denitrification. The growth and activity of DNRA microorganisms decrease denitrification efficiency, and increase the waste load on nitrification as well as total nitrogen discharged with the effluent. DNRA activity was previously detected in anaerobic digester systems (1, 2, 13, 21), a pilot plant treating sulfate (SO42−)- and NO3−-containing wastewater (14), a lab-scale denitrifying reactor (4), and aquaculture nitrogen removal systems (10). The extent of DNRA measured among these studies suggests its competitive potential in high organic-loaded wastewaters; however, the microorganisms responsible for the process have rarely been identified in these environments.
In order to control the occurrence of microorganisms with the capacity for DNRA within a wastewater treatment system, it is essential to recognize their identity and diversity as the first step towards maneuvering these populations to the required process. Therefore, in order to identify potential DNRA microorganisms in wastewater ecosystems, an anoxic semi-continuous sequencing batch reactor (SBR) maintained at a high C/NO3− ratio was applied to monitor subsequent microbial community adaptation when using activated sludge as the inoculum. In a direct comparison, a low C/NO3− reactor was operated under otherwise the same environmental conditions and using the same inoculum. The objectives of the present study were (i) to investigate the presence and activity of the DNRA pathway under low and high C/NO3− anoxic sludge ecosystems and (ii) to identify the composition of the resulting microbial communities using an Illumina MiSeq 16S rRNA sequencing analysis. In order to trace the occurrence of DNRA, the stable-isotope tracers 15NO3− and 15NO2− were utilized to track the formation of 15NH4+ in low and high C/NO3− SBR sludge samples.
Materials and Methods
Operation of the reactors
The two reactors were maintained under low and high C/NO3− ratios, respectively, and operated in the semi-continuous sequencing batch mode, which provided environments for the growth of microorganisms favored under each condition. During each cycle, the systems were set to the continuous feeding batch mode in order to maintain stable concentrations of substrates in the reactors, while also retaining a large amount of biomass sludge in each SBR. Both SBRs were started with an inoculum (1,393±42 mg mix liquor suspended solids [MLSS] L−1) from a municipal wastewater treatment plant operated under an anaerobic/oxic/anoxic/oxic (AOAO) process exhibiting good denitrification efficiency with a nitrogen removal rate of approximately 60%. SBRs were run in 1-L working volume reactors with six-blade turbine stirrers, and were maintained at 20°C in a control temperature room. Mixing was performed at 150 rpm in order to maintain a homogenous state inside the systems. During days 1–15 of the operation, the reactor cycle was set to 6 h, which comprised 320 min of continuous feeding and mixing, 30 min of sludge settling, and 10 min of effluent withdrawal. Only unsettled sludge was removed with the discharged effluent during the end of the cycle. The volume exchanged per cycle was set at 1/4 of the reactor working volume, whereas the flow rate was set at 47 mL h−1 (equal to a dilution rate of 0.047 h−1), resulting in a hydraulic retention time (HRT) of 24 h. After day 15, the cycle was changed to 12 h (680 min of continuous feeding and mixing) while the exchanged volume remained the same; therefore, the flow rate was adjusted to 22 mL h−1 during this period (equal to a dilution rate of 0.022 h−1) with 48 h of HRT. The flow rates of the influent and effluent were controlled by peristaltic pumps (Masterflex; Cole-Parmer, Chicago, IL, USA), whereas the reactor cycle was set using timers. The solid retention times (SRT) of each reactor during the operation were 3.40±0.45 and 2.52±0.97 d for the low and high C/NO3− systems, respectively.
Glucose was selected as the carbon and energy source for microbial growth, and its concentration was varied in order to achieve COD/NO3−-N ratios of 4/1 and 8/1 for the low and high C/NO3− SBRs, respectively (for details, see Table S1). NO3− was supplied at the same concentration in both systems. The medium fed to the reactors during the 6-h cycle period was composed of the following (L−1): 90 mg of MgSO4·7H2O, 160 mg of MgCl2·6H2O, 42 mg of CaCl2·2H2O, 122 mg of peptone, 20 mg of yeast extract, 50 mg of NH4Cl, 11.33 mg of KH2PO4, 25.67 mg of Na2HPO4·12H2O (15), and 0.3 mL of nutrient solution (40). The concentrations of all nutrients were doubled during the 12-h cycle in order to obtain the same loading as previously. Media were autoclaved and later supplemented with filter-sterilized glucose solution in order to meet the specified COD/NO3−-N ratio for each reactor. Media were then flushed with argon (Ar) gas for 30–60 min (depending on the volume prepared) before being added to the system.
Chemical analysis
Water samples were collected daily from both SBRs in order to monitor changes in inorganic nitrogen and organic carbon in the systems. Samples were initially filtered through a glass microfiber filter with a pore size of 0.7 μm (Grade GF/F; GE Healthcare, Little Chalfont, UK) before the chemical analysis. The concentrations of NO3−, NO2−, and SO42− were analyzed by ion chromatography (IC) equipped with an anion column (861 Advanced Compact IC; Metrohm, Herisau, Switzerland), whereas NH4+ was measured by IC with a cation column (761 Advanced Compact IC; Metrohm). Organic carbon was monitored in the form of dissolved organic carbon (DOC) using a total carbon analyzer (TOC-V; Shimadzu, Kyoto, Japan). Changes in biomass were quantified in terms of MLSS as described by the Standard Methods for the Examination of Water and Wastewater (3). The pH of each withdrawn sample was measured using a pH meter.
Stable isotope incubations
The stable isotope tracers, 15NO3− and 15NO2−, were applied to verify the occurrence and extent of DNRA in the low and high C/NO3− SBR samples. This was performed by monitoring the amount of 15NH4+ produced from the dissimilatory 15NO3− or 15NO2− reduction. Sludge for the incubations was taken from the reactors at the end of the operation (on days 60 and 54 for the low and high C/NO3− systems, respectively). Experiments were conducted in the batch mode and samples were collected in time series. 15NO3− and 15NO2− were used in the form of Na15NO3 and Na15NO2 (NLM-157-PK and NLM- 658-PK, respectively, both with 98%+ 15N atom; Cambridge Isotope Laboratories). Before the incubations, sludge taken from the reactors was centrifuged to remove the original medium, washed once with fresh medium (same composition as that prepared for SBRs, excluding NH4Cl), and then placed into 100-mL serum bottles along with new medium. Serum bottles were closed with butyl rubber stoppers and sealed with aluminum caps before flushing with Ar gas for 10 min. The incubations were then started by injecting either 15NO3− or 15NO2− into the serum bottles to a final concentration of 20 mg N L−1, along with 14NH4Cl to a final concentration of 100 mg N L−1 and glucose as appropriate to make the same COD/NO3−-N ratio as in the reactors. All stock solutions were flushed with Ar gas before use. Samples were placed on a rotary shaker set at 150 rpm and temperature was maintained at 20°C. All incubations were performed in triplicate for the sludge of each reactor. The measurement of 15NH4+ produced from the dissimilatory 15NO3− or 15NO2− reduction was conducted as previously described (19). Since the original medium from the reactor had been washed off the sludge before the incubations, the amounts of 14NO3− and 14NO2− in the samples were considered to be negligible and changes in 15NO3− and 15NO2− were measured using the IC method (861 Advanced Compact IC; Metrohm).
Illumina MiSeq 16S rRNA sequencing analysis of microbial communities
The microbial communities of the low and high C/NO3− SBRs were examined using the Illumina MiSeq 16S rRNA sequencing method. Samples analyzed included sludge collected on days 27, 30, 38, 42, and 48 from both reactors as well as the inoculum used to start the systems. Total microbial DNA was extracted from samples using the FastDNATM SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) as described by the manufacturer’s protocol. Extracted DNA was then used as a template for PCR amplification with the 341F and 805R primer pair (targeting the V3 and V4 regions of the 16S rRNA gene). The PCR reaction mixture (25 μL) was prepared from a TaKaRa Ex TaqTM kit (Takara, Otsu, Japan) with 12 ng of the DNA template, 0.2 μM of each dNTP, 2 mM MgCl2, 0.2 μM of each primer, and 1.25 U of TaKaRa Ex TaqTM. The PCR thermal steps were as follows: 94°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and then a final 72°C for 5 min. Four PCR reactions were conducted per sample to reduce bias during PCR amplification. PCR products were then checked by 1.5% (w/v) agarose gel electrophoresis in order to ensure that the correct-sized products were amplified (approximately 460 bp).
Four replicates of each sample were subsequently combined and purified with a NucleoSpin® Gel and PCR Clean-up kit (Macherey- Nagel, Bethlehem, PA, USA) according to the manufacturer’s protocol. The concentrations of the PCR products were examined with a spectrophotometer (NanoDropTM 2000c Spectrophotometer; Thermo Fisher Scientific, Waltham, MA, USA). The 16S rRNA amplicons obtained were then prepared in a 50-μL final volume reaction containing 5 μL of each Illumina Nextera XT index primer (Illumina, San Diego, CA, USA) and 2X KAPA HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA) to index each sample. The PCR thermal steps used were as follows: 94°C for 3 min, followed by 8–10 cycles of 98°C for 20 s, 55°C for 30 s, and 72°C for 30 s, and then a final 72°C for 5 min. The indexed PCR amplicons were then purified with Agencourt AMPure XP beads (Agencourt Bioscience, La Jolla, CA, USA), pooled, and diluted to a final loading concentration of 4 pM. DNA paired-end sequencing was conducted on the Illumina MiSeq platform with MiSeq Reagent Kits v3 (600 cycles; Illumina) at the Omics Sciences and Bioinformatics Center (Chulalongkorn University, Bangkok, Thailand). Sequences were assembled by merging forward and reverse reads, and the operational taxonomic unit (OTU) picking process was performed using QIIME software (version 1.9.0) with an open reference method. Taxonomic assignment from the OTUs to related taxa was performed using UCLUST against the Greengenes database (version 13_8, with a 97% sequence identity threshold). Sequence reads in this study may be accessed at the NCBI Short Read Archive (SRA) under accession numbers SRX3440460–SRX3440470. OTUs with ≥1% and ≥2% maximum relative abundance were selected as representatives for class-level and genus-level analyses, respectively. The beta-diversity of each sample was evaluated in QIIME, and the results obtained were used for the principal coordinate analysis (PCoA) using the weighted UniFrac distance metric (24). Statistical analyses of the results obtained were performed using a two-sample t-test on SPSS Statistic ver. 22 (IBM, Armonk, NY, USA).
Results and Discussion
Competition between denitrification and DNRA in low and high C/NO3− SBRs
In the operation of the reactors, semi-continuous SBRs enriched with NO3−-reducing communities exhibited the ability to maintain a stable C/NO3− ratio during the feeding period (see Fig. S1) as well as the biomass within the systems. Apart from the C/NO3− ratio, the applied dilution rate is another factor known to affect competition for NO3− in a reactor setting, in which DNRA microorganisms appear to require a low dilution rate for growth (27). This feature may be related to the kinetic aspect of these microorganisms because the requirement for a low dilution rate indicates a long generation time for growth (23), the need for a low NO3− concentration to kinetically compete with denitrifiers, or both. Therefore, the dilution rate used in the present study was selected from a range that was expected to allow DNRA to occur. All other operating parameters, besides the glucose concentration, were the same in the low and high C/NO3− SBRs in order to allow the C/NO3− ratio to be the determining factor for the successful NO3−-reducing pathway in each ecosystem.
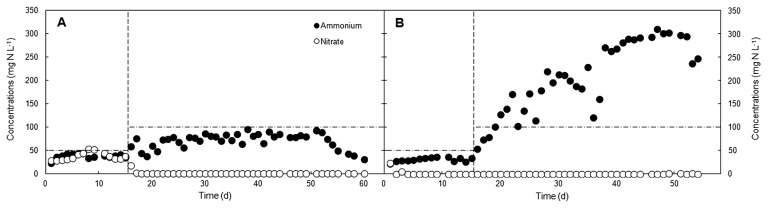
Competition between the two NO3− reduction pathways was initially verified by observing changes in the inorganic nitrogen profiles of the low and high C/NO3− SBRs (Fig. 1). During the initial phase of the low C/NO3− reactor operation, NO3− was detected in the system in the range of 17.7–52.5 mg N L−1 (47.8–82.3% NO3− conversion) and was consumed to less than 1.0 mg N L−1 (>99% NO3− conversion) after day 17, which suggested that NO3− had become limited for heterotrophic growth or that it was used by autotrophic NO3− reducers. NO2− was only detected on day 4 (1.7 mg N L−1) and then remained below the detection limit (see Fig. S2). The level of NH4+ (23.2–95.1 mg N L−1) was always lower than the added NH4+ concentration, and, thus, there was no obvious indication of DNRA activity, or at least not at a level that exceeded the process of NH4+ assimilation. Therefore, the major NO3−- reducing pathway under the COD/NO3−-N ratio of 4/1 was assumed to be denitrification because depleted inorganic nitrogen was most likely converted to nitrogenous gases. The existence of DNRA in this system was subsequently verified with stable-isotope tracers (see below).
Fig. 1.
Changes in NO3− and NH4+ concentrations in (A) low and (B) high C/NO3− SBRs. Vertical lines indicate the point of change in the SBR cycle, horizontal lines indicate the level of NH4+ supplied to the reactors.
In the high C/NO3− SBR, the COD/NO3−-N ratio was started at 8/1 and the system was observed in order to establish whether it has the ability to sustain the growth of DNRA microorganisms. After 4 d of operation, the NO3− level was depleted (<0.2 mg N L−1). This rapid consumption of NO3− was likely to be due to the higher glucose concentration in the high C/NO3− reactor than in the low C/NO3− reactor, which, in turn, required electron acceptors (NO3− and NO2−) to complete the reactions catalyzed by the microorganisms. Therefore, NO3− became a limiting substrate in this system, whereas NO2− was undetectable throughout the experiment, suggesting the rapid conversion of NO2− to other end products. The amount of NH4+ in the high C/NO3− SBR was increased to a markedly higher level than the supplied concentration, possibly accounting for 30.1–64.4% and 68.6–100% of the NO3− conversion during days 20–37 and 38–54, respectively. The active formation of NH4+ most likely indicates the occurrence of DNRA in this high C/NO3− system. Judging from the portion of NO3− converted to NH4+ during days 20–37, DNRA microorganisms and denitrifiers appeared to have a relatively similar share in NO3− reduction. However, after day 37, DNRA microorganisms appeared to be the principal NO3− reducers in the reactor, with a minor fraction of reduced NO3− being released as gases (<14.5% during peak NH4+-forming activity). The diverged nitrogen profiles observed in the two SBRs indicated that the difference in the ratios selected for the low and high C/NO3− reactors induced partitioning in the pathways of microbial NO3− reduction. Since the COD/NO3−-N of 8/1 already supported the growth and activity of DNRA microorganisms, the ratio was not increased from this value. COD/NO3−-N of 7.7 was successfully used to enrich a DNRA community in an acetate-fed continuous culture (36). This range of COD/NO3−-N values appears to be sufficiently high to promote the reduction of NO3− to NH4+. However, other parameters, such as the type of organic carbon and its fermentability, also need to be considered as additional factors that affect the COD/NO3−-N threshold partitioning the pathways of NO3− reduction.
With respect to the cell biomass grown in the two SBRs, the MLSS in the high C/NO3− system was greater (3,008±996 mg L−1) than that observed in the low C/NO3− system (2,740±272 mg L−1). Since the same amount of NO3− was supplied to both reactors, the higher cell mass in the high C/NO3− system may be due to the higher mole of organic carbon required per mole of NO3− for DNRA than for denitrification (32), thereby providing a greater carbon and energy source for microbial growth. It may also have been caused by the growth of other microorganisms apart from NO3− reducers because an organic carbon concentration above the stoichiometric requirement for NO3− reduction is expected to support other anaerobic heterotrophs, such as fermentative bacteria and SO42− reducers. Sulfide (S2−) produced from SO42− reduction may sustain DNRA microorganisms by acting as an electron donor (6). However, the effects of S2− in the high C/NO3− reactor may be small considering the amount of glucose added, with SO42− being supplied in the media at 24.6±0.66 mg S L−1. Therefore, if all the SO42− supplied was reduced to S2−, 1.35% (2.69±0.07 mg N L−1) of the added NO3− would be used by S2−-driven DNRA (see Supplementary for details), and, hence, its effects on this process were expected to be negligible relative to those on the organic carbon-driven reaction.
Potential activity of DNRA in sludge from low and high C/NO3− SBRs
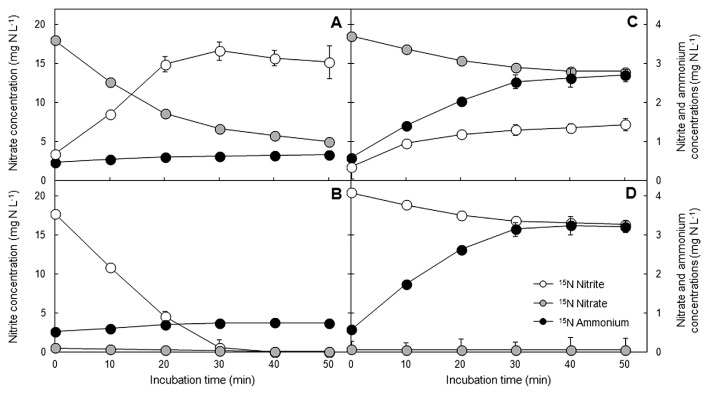
Batch incubations with low C/NO3− sludge revealed that 15NO3− and 15NO2− were mostly reduced to end products that did not remain in the liquid phase (Fig. 2A, B), and, thus, they were assumed to be converted into nitrogenous gases. This was mostly attributed to the activity of denitrifiers, which produce nitric oxide (NO), nitrous oxide (N2O), and dinitrogen (N2) gases from the reduction of NO3− and NO2−. However, a slight increase in 15NH4+ was measured with the 15NO3− and 15NO2− tracers, although at small concentrations only (0.19 and 0.23 mg N L−1, respectively). This weak activity indicated the presence of a small amount of DNRA microorganisms in the low C/NO3− community. 15NH4+ and 15NO3−/15NO2− assimilation in all the incubations was expected to be suppressed due to the presence of a high concentration of 14NH4+ (100 mg N L−1). Therefore, this process was not taken into account for the balance of nitrogen conversion, and all of the 15NH4+ produced was assumed to be left in the media.
Fig. 2.
Changes in 15NO3−, 15NO2−, and 15NH4+ during stable-isotope tracer experiments; (A, C) 15NO3− and (B, D) 15NO2− incubations with (A, B) low and (C, D) high C/NO3− sludge.
Regarding high C/NO3− sludge, a marked increase in the level of 15NH4+ was observed when 15NO3− or 15NO2− was added (Fig. 2C, D). Therefore, DNRA activity was confirmed by the evidence of NH4+ production from the reduction of NO3− and NO2−. The portion of consumed 15NO3−/15NO2− not detected in the liquid phase was assumed to be converted to gaseous products. The addition of 15NO2− instead of 15NO3− did not significantly change the activity of DNRA microorganisms in the low and high C/NO3− sludge incubations. Hence, NO2− does not appear to have had a different effect on the nitrogen conversion pathways from that observed for NO3−, at least for the samples and conditions tested in this experiment.
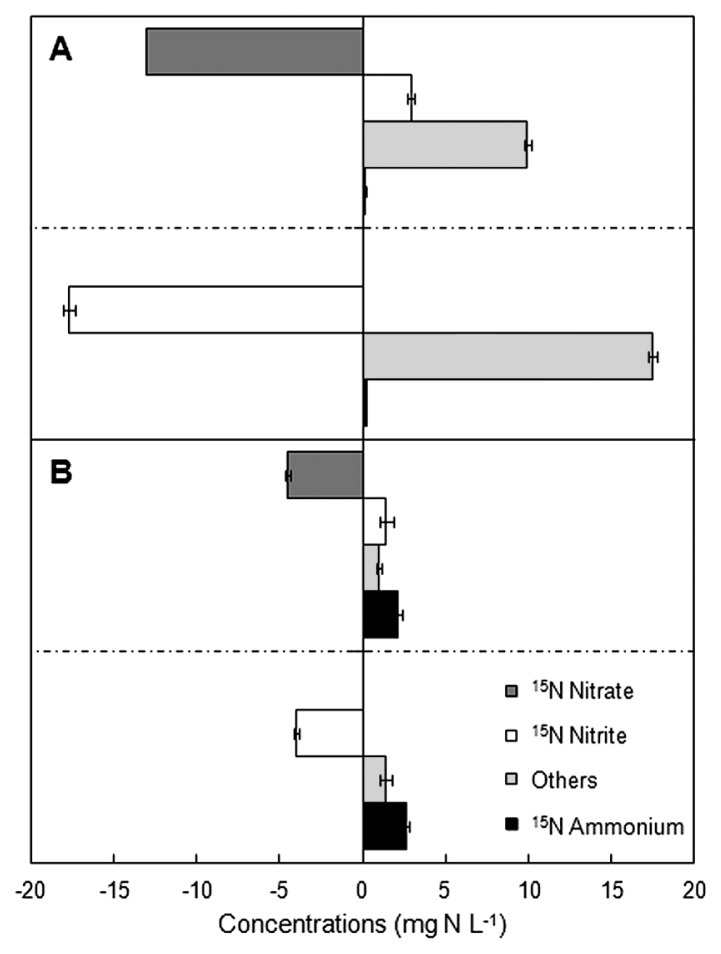
Net changes in 15N-nitrogen for all incubations (calculated based on the initial and final concentrations of 15NO3−, 15NO2−, and 15NH4+) are shown in Fig. 3. The main products of the reduction of 15NO3− and 15NO2− in low C/NO3− sludge were in gaseous forms (Fig. 3A), and, thus, denitrifiers were assumed to be the main contributors to nitrogen conversion in these samples. The 15NH4+ produced from DNRA microorganisms inhabiting the low C/NO3− SBR was 1.4 and 1.3% of the amount of 15NO3− and 15NO2− consumed, respectively, indicating a small role for these microorganisms in the denitrifier-dominated community. In contrast, the main product in high C/NO3− sludge was 15NH4+ in the 15NO3− and 15NO2− incubations (Fig. 3B). The proportions of nitrogenous gases produced in the high C/NO3− incubations (21.9 and 34.5% of consumed 15NO3− and 15NO2−, respectively) were lower than those converted to 15NH4+. However, these gases may not come exclusively from the activity of denitrification. Previous studies revealed that DNRA also released NO and N2O (30, 39, 42); however, the exact production pathways remain unclear. Since high C/NO3− sludge was predominated by DNRA activity, the nitrogenous gases produced may come from denitrification, DNRA, or both.
Fig. 3.
Net changes in 15N-nitrogen in 15NO3− and 15NO2− incubations with (A) low and (B) high C/NO3− sludge. Data are shown with the standard deviation (error bar) derived from triplicate incubations of the sludge of each reactor.
At the end of the incubations, the net 15NH4+ generated in the high C/NO3− sludge was 46.6 and 65.5% of the consumed 15NO3− and 15NO2−, respectively. The extent of 15NH4+ formed here suggested the competitive potential of DNRA microorganisms in activated sludge when under a continuous supply of high C/NO3− loading. Additionally, NO3− and NO2− had the ability to induce DNRA in a similar manner; however, these two nitrogen species were previously reported to exert different stimulating effects on the pathway in Escherichia coli (26, 41). In conclusion, the results from stable isotope tracers confirmed that NH4+ detected in the high C/NO3− SBR was mainly the product of DNRA, and that the addition of glucose at a COD/NO3−-N ratio of 8/1 may be selected for the growth and activity of DNRA microorganisms; however, denitrifiers may still co-exist to a certain level.
Microbial communities of low and high C/NO3− SBRs
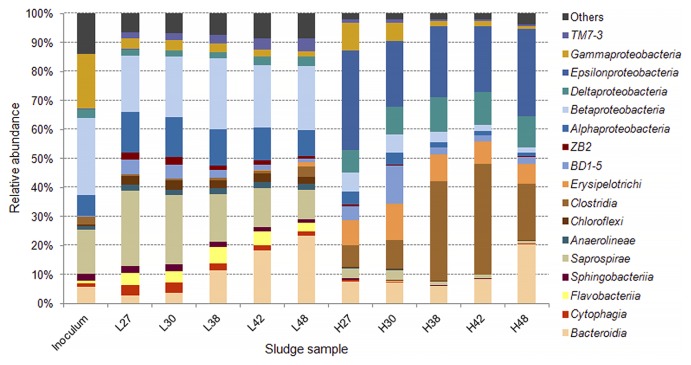
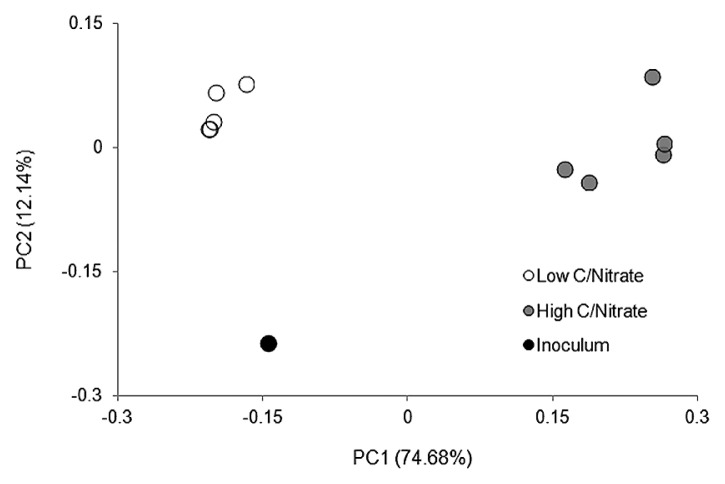
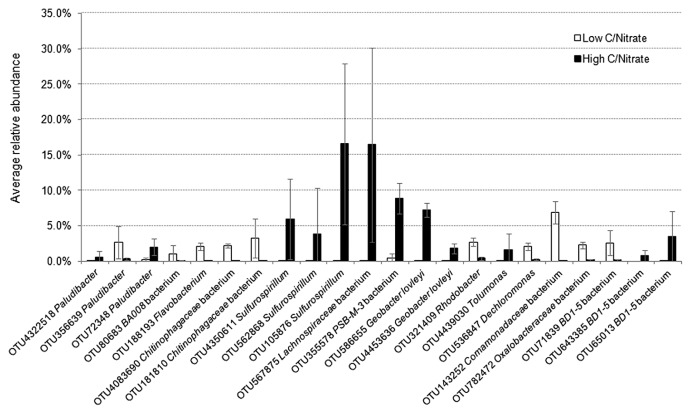
An Illumina MiSeq 16S rRNA analysis revealed that from all the samples collected, 98.4–99.8% of sequences were identified as Bacteria and ≤0.01% as Archaea, while it was not possible to designate 0.2–1.6% of the reads. The OTUs assigned from the sequence reads were classified into 17 main classes (Fig. 4) belonging to seven phyla, including Proteobacteria, Firmicutes, Bacteroidetes, Chloroflexi, and the candidate divisions GN02, OD1, and TM7. Microbial richness, as estimated by the Chao1 index, and microbial diversity, as calculated by the Shannon index, revealed a significant difference (P<0.05) between the low and high C/NO3− microbial populations (see Table S2). Lower richness and biodiversity were observed in high C/NO3− samples, whereas low C/NO3− populations were more diverse with higher community richness. Therefore, a higher C/NO3− ratio appeared to allow certain bacteria to thrive with elevated proportions, while the lower C/NO3− ratio nurtured diverse bacteria that, among the dominating ones, had relatively fair shares of the microbial composition, as indicated by the relative abundance of the major OTUs in the system (see Fig. S3). The relationships between the inoculum and the low and high C/NO3− microbial communities were revealed in the PCoA plot (Fig. 5) using a beta-diversity measure of each sample. The PCoA plot showed distinct clustering within the low and high C/NO3− samples, with the distance between the beta-diversities of the two reactors’ populations conveying the differences in microbial compositions among these two ecosystems. The average relative abundance of the major taxa identified (Fig. 6) indicated that the microbial communities of the low and high C/NO3− SBRs were distinct from each other with rarely shared OTUs among them, which reflects partitioning in the NO3−-reducing activities observed in the two systems.
Fig. 4.
Relative abundance of dominant classes in the inoculum and low C/NO3− (L27, L30, L38, L42, and L48) and high C/NO3− (H27, H30, H38, H42, and H48) sludge samples.
Fig. 5.
PCoA plot of beta-diversity measures of the inoculum and low and high C/NO3− microbial communities.
Fig. 6.
Average relative abundance of top 22 OTUs identified in low and high C/NO3− SBRs (average from five sampling days). Data are shown with the standard deviation (error bar) among the sampling days of each sludge sample.
Among the top OTUs identified in the high C/NO3− reactor, those with the potential of being DNRA microorganisms included those closely related to the genus Sulfurospirillum, the family Lachnospiraceae, the proposed genus PSB-M-3, and the species Geobacter lovleyi. These taxa were present at an average abundance of ≤0.4% (from the five sampling days) in the low C/NO3− system, which emphasized their roles in shaping the NH4+-producing community in the high C/NO3− SBR. The OTU affiliated with Sulfurospirillum (OTU105876) was one of the OTUs detected with the highest average abundance in the high C/NO3− reactor (16.5%). The genus Sulfurospirillum (belonging to the class Epsilonproteobacteria) is known to exhibit versatile metabolism ranging from SO42− reduction, S2− oxidation, and DNRA (18, 25, 31). However, the amounts of NO3− and organic carbon available in the high C/NO3− SBR were expected to promote NO3− reduction to NH4+ in these microorganisms, while the influence of sulfur compounds was expected to be minimal because the level of sulfur-driven NO3− reduction in the reactor was small. Three OTUs related to Sulfurospirillum were found at a high abundance in the high C/NO3− system, but were present at ≤0.02% of the total community in the low C/NO3− reactor, which indicated that the high C/NO3− condition was highly selective for the growth of these populations. The family Lachnospiraceae (in the class Clostridia of the phylum Firmicutes), the other highest average abundant OTU (16.5%) (OTU567875), is not known to contain DNRA microorganisms, but was previously found in anoxic microcosms performing DNRA (23). Apart from the highest OTU, another top OTU in the high C/NO3− SBR was related to PSB-M-3 (OTU355578) in the family Erysipelotrichaceae of the phylum Firmicutes, with an average abundance of 8.9%. The high proportions of the taxa affiliated with Lachnospiraceae and PSB-M-3 suggest their essential role in the high C/NO3− ecosystem; however, further investigations are needed in order to elucidate their actual ecological function in NO3−-reducing communities.
The OTU assigned to the species G. lovleyi (OTU586655), a member of the class Deltaproteobacteria, was detected with an average abundance of 7.2% in the high C/NO3− reactor. G. lovleyi is known to conduct DNRA (33) and taxa related to this species have also been found to dominate DNRA enrichment cultures fed with acetate (36–38). Previous findings and the present results indicate the common occurrence of these DNRA microorganisms when the conditions favor NO3− reduction to NH4+. Beside those with DNRA potential, the remaining major OTUs were found to be related to bacteria with fermentative abilities, including the OTUs assigned to the class BD1-5 (OTU65013), genus Tolumonas (OTU4439030), and genus Paludibacter (OTU72348). There is no direct evidence to show that these taxa perform NO3− reduction (9, 16, 35), and, thus, they most likely acted as contributors supplying simpler fermented products to the rest of the community.
In the low C/NO3− SBR, the OTU detected with the highest average abundance (6.9%) was assigned to the family Comamonadaceae (OTU143252) in the class Betaproteobacteria. Several members of this family have been recognized as denitrifying bacteria, such as Comamonas and Acidovorax (43). Other OTUs affiliated with known denitrifiers include those related to Rhodobacter (OTU321409), Dechloromonas (OTU536847), and Flavobacterium (OTU188193) (5, 8, 11). OTUs related to the families Chitinophagaceae (OTU181810 and OTU4083690) and Oxalobacteraceae (OTU782472) were also identified in the low C/NO3− community, in which some members are known to reduce NO3− or perform complete denitrification (20). The detection of these major OTUs in the low C/NO3− reactor was in accordance with the denitrifying activity observed in the system. Additionally, certain denitrifiers, such as Shewanella loihica and several nirK-containing bacteria, have been found to contain the DNRA pathway in their genomes (17, 44). Therefore, the weak activity of DNRA detected in the low C/NO3− SBR may be attributed to one of these denitrifiers because no other known DNRA microorganism was identified. Between the two NO3−-reducing ecosystems, smaller changes in the microbial community were observed for the low C/NO3− reactor (in which the enriched populations were still denitrifiers), while the high C/NO3− condition was enriched for a markedly different microbial composition from the inoculum (see Fig. S4). This result demonstrated that the selected C/NO3− ratios applied to the two SBRs were already critical for the partition of microbial populations to those with different functional abilities.
Conclusion
In the high C/NO3− SBR, DNRA was observed by the marked increase in the NH4+ concentration. Nitrogen conversion, monitored via 15NO3− and 15NO2− tracers, revealed that DNRA microorganisms were the major contributors to NO3−/NO2− reduction under a high COD/NO3−-N ratio of 8/1, at which NO3− and NO2− induce DNRA in a similar manner. With a low COD/NO3−-N ratio of 4/1, denitrifiers were the major NO3−/NO2− reducers. Additionally, the high C/NO3− conditions enriched for microbial populations markedly differed from those in the low C/NO3− reactor and the inoculum. These populations were comprised of OTUs closely related to known DNRA microorganisms (Sulfurospirillum and G. lovleyi) as well as fermentative bacteria and those that may be capable of both functions. These bacteria were rarely present in the low C/NO3− system, which harbored a community of denitrifiers. Therefore, the C/NO3− ratios applied to each SBR provided an environment that partitioned the pathways of NO3− reduction to either nitrogenous gases or NH4+ as the end product, which was the result of the distinctive microbial composition found in each SBR system.
Supplementary Material
Acknowledgements
This research was supported by the Overseas Research Experience Scholarship for Graduate Students and the 90th Anniversary of Chulalongkorn University Scholarship granted by the Graduate School, Chulalongkorn University, as well as the Chula Engineering’s Promoting Research Grant from the Faculty of Engineering, Chulalongkorn University. This work was also part of the program of Research Network of NANOTEC (RNN), NSTDA, Ministry of Science and Technology, Thailand. Part of the research was conducted at the Department of Urban Engineering, the University of Tokyo, Japan. The other part was performed at the Center of Excellence for Marine Biotechnology, Department of Marine Science, Chulalongkorn University, Thailand.
References
- 1.Akunna J.C., Bizeau C., Moletta R. Nitrate and nitrite reductions with anaerobic sludge using various carbon sources: glucose, glycerol, acetic acid, lactic acid and methanol. Water Res. 1993;27:1303–1312. [Google Scholar]
- 2.Akunna J.C., Bizeau C., Moletta R. Nitrate reduction by anaerobic sludge using glucose at various nitrate concentrations: ammonification, denitrification and methanogenic activities. Environ Technol. 1994;15:41–49. [Google Scholar]
- 3.American Public Health Association (APHA), American Water Works Association (AWWA), and Water Environment Federation (WEF) Standard Methods for the Examination of Water and Wastewater. 20th ed. American Public Health Association; Washington DC: 1998. [Google Scholar]
- 4.Barber W.P., Stuckey D.C. Nitrogen removal in a modified anaerobic baffled reactor (ABR): 1, denitrification. Water Res. 2000;34:2413–2422. [Google Scholar]
- 5.Betlach M.R., Tiedje J.M. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl Environ Microbiol. 1981;42:1074–1084. doi: 10.1128/aem.42.6.1074-1084.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunet R.C., Garcia-Gil L.J. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol Ecol. 1996;21:131–138. [Google Scholar]
- 7.Burgin A.J., Hamilton S.K. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ. 2007;5:89–96. [Google Scholar]
- 8.Byrne M.D., Nicholas D.J.D. A membrane-bound dissimilatory nitrate reductase from Rhodobacter sphaeroides f.sp. denitrificans. Biochim Biophys Acta. 1987;915:120–124. [Google Scholar]
- 9.Caldwell M.E., Allen T.D., Lawson P.A., Tanner R.S. Tolumonas osonensis sp. nov., isolated from anoxic freshwater sediment, and emended description of the genus Tolumonas. Int J Syst Evol Microbiol. 2011;61:2659–2663. doi: 10.1099/ijs.0.023853-0. [DOI] [PubMed] [Google Scholar]
- 10.Chutivisut P., Pungrasmi W., Powtongsook S. Denitrification and dissimilatory nitrate reduction to ammonium (DNRA) activities in freshwater sludge and biofloc from Nile tilapia aquaculture systems. J Water Environ Technol. 2014;12:347–356. [Google Scholar]
- 11.Coates J.D., Chakraborty R., Lack J.G., O’onnor S.M., Cole K.A., Bender K.S., Achenbach L.A. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature. 2001;411:1039–1043. doi: 10.1038/35082545. [DOI] [PubMed] [Google Scholar]
- 12.Cole J.A., Brown C.M. Nitrite reduction to ammonia by fermentative bacteria: a short circuit in the biological nitrogen cycle. FEMS Microbiol Lett. 1980;7:65–72. [Google Scholar]
- 13.Cytryn E., Gelfand I., Barak Y., van Rijn J., Minz D. Diversity of microbial communities correlated to physiochemical parameters in a digestion basin of a zero-discharge mariculture system. Environ Microbiol. 2003;5:55–63. doi: 10.1046/j.1462-2920.2003.00392.x. [DOI] [PubMed] [Google Scholar]
- 14.Du Preez L.A., Maree J.P. Pilot-scale biological sulphate and nitrate removal utilizing producer gas as energy source. Water Sci Technol. 1994;30:275–285. [Google Scholar]
- 15.Ginige M.P., Keller J., Blackall L.L. Investigation of an acetate-fed denitrifying microbial community by stable isotope probing, full-cycle rRNA analysis, and fluorescent in situ hybridization-microautoradiography. Appl Environ Microbiol. 2005;71:8683–8691. doi: 10.1128/AEM.71.12.8683-8691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanke A., Hamann E., Sharma R., et al. Recoding of the stop codon UGA to glycine by a BD1-5/SN-2 bacterium and niche partitioning between Alpha- and Gammaproteobacteria in a tidal sediment microbial community naturally selected in a laboratory chemostat. Front Microbiol. 2014;5:231. doi: 10.3389/fmicb.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helen D., Kim H., Tytgat B., Anne W. Highly diverse nirK genes comprise two major clades that harbour ammonium-producing denitrifiers. BMC Genomics. 2016;17:155. doi: 10.1186/s12864-016-2465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubert C., Voordouw G. Oil field souring control by nitrate-reducing Sulfurospirillum spp. that outcompete sulfate-reducing bacteria for organic electron donors. Appl Environ Microbiol. 2007;73:2644–2652. doi: 10.1128/AEM.02332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isobe K., Suwa Y., Ikutani J., et al. Analytical techniques for quantifying 15N/14N of nitrate, nitrite, total dissolved nitrogen and ammonium in environmental samples using a gas chromatograph equipped with a quadrupole mass spectrometer. Microbes Environ. 2011;26:46–53. doi: 10.1264/jsme2.me10159. [DOI] [PubMed] [Google Scholar]
- 20.Kämpfer P., Lodders N., Falsen E. Hydrotalea flavagen. nov., sp. nov., a new member of the phylum Bacteroidetes and allocation of the genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferruginibacter, Filimonas and Hydrotalea to the family Chitinophagaceae fam. nov. Int J Syst Evol Microbiol. 2011;61:518–523. doi: 10.1099/ijs.0.023002-0. [DOI] [PubMed] [Google Scholar]
- 21.Kaspar H.F., Tiedje J.M., Firestone R.B. Denitrification and dissimilatory nitrate reduction to ammonium in digested sludge. Can J Microbiol. 1981;27:878–885. doi: 10.1139/m81-139. [DOI] [PubMed] [Google Scholar]
- 22.Kraft B., Strous M., Tegetmeyer H.E. Microbial nitrate respiration—genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–117. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Kraft B., Tegetmeyer H.E., Sharma R., Klotz M.G., Ferdelman T.G., Hettich R.L., Geelhoed J.S., Strous M. The environmental controls that govern the end product of bacterial nitrate respiration. Science. 2014;345:676–679. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- 24.Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luijten M.L.G.C., de Weert J., Smidt H., Boschker H.T.S., de Vos W.M., Schraa G., Stams A.J.M. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int J Syst Evol Microbiol. 2003;53:787–793. doi: 10.1099/ijs.0.02417-0. [DOI] [PubMed] [Google Scholar]
- 26.Page L., Griffiths L., Cole J.A. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch Microbiol. 1990;154:349–354. doi: 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- 27.Rehr B., Klemme J.H. Competition for nitrate between denitrifying Pseudomonas stutzeri and nitrate ammonifying enterobacteria. FEMS Microbiol Lett. 1989;62:51–57. [Google Scholar]
- 28.Robertson E.K., Roberts K.L., Burdorf L.D.W., Cook P., Thamdrup B. Dissimilatory nitrate reduction to ammonium coupled to Fe(II) oxidation in sediments of a periodically hypoxic estuary. Limnol Oceanogr. 2016;61:365–381. [Google Scholar]
- 29.Silver W.L., Thompson A.W., Reich A., Ewel J.J., Firestone M.K. Nitrogen cycling in tropical plantation forests: potential controls on nitrogen retention. Ecol Appl. 2005;15:1604–1614. [Google Scholar]
- 30.Smith M.S. Nitrous oxide production by Escherichia coli is correlated with nitrate reductase activity. Appl Environ Microbiol. 1983;45:1545–1547. doi: 10.1128/aem.45.5.1545-1547.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolz J.F., Ellis D.J., Blum J.S., Ahmann D., Lovley D.R., Oremland R.S. Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the ɛ-Proteobacteria. Int J Syst Bacteriol. 1999;49:1177–1180. doi: 10.1099/00207713-49-3-1177. [DOI] [PubMed] [Google Scholar]
- 32.Strohm T.O., Griffin B., Zumft W.G., Schink B. Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol. 2007;73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung Y., Fletcher K.E., Ritalahti K.M., Apkarian R.P., Ramos-Hernández N., Sanford R.A., Mesbah N.M., Löffler F.E. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl Environ Microbiol. 2006;72:2775–2782. doi: 10.1128/AEM.72.4.2775-2782.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiedje J.M., Sexstone A.J., Myrold D.D., Robinson J.A. Denitrification: ecological niches, competition and survival. Antonie van Leeuwenhoek. 1983;48:569–583. doi: 10.1007/BF00399542. [DOI] [PubMed] [Google Scholar]
- 35.Ueki A., Akasaka H., Suzuki D., Ueki K. Paludibacter propionicigenes gen. nov., sp. nov., a novel strictly anaerobic, Gramnegative, propionate-producing bacterium isolated from plant residue in irrigated rice-field soil in Japan. Int J Syst Evol Microbiol. 2006;56:39–44. doi: 10.1099/ijs.0.63896-0. [DOI] [PubMed] [Google Scholar]
- 36.van den Berg E.M., van Dongen U., Abbas B., van Loosdrecht M.C.M. Enrichment of DNRA bacteria in a continuous culture. ISME J. 2015;9:2153–2161. doi: 10.1038/ismej.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Berg E.M., Boleij M., Kuenen J.G., Kleerebezem R., van Loosdrecht M.C.M. DNRA and denitrification coexist over a broad range of acetate/N-NO3− ratios, in a chemostat enrichment culture. Front Microbiol. 2016;1842;7 doi: 10.3389/fmicb.2016.01842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berg E.M., Rombouts J.L., Kuenen J.G., Kleerebezem R., van Loosdrecht M.C.M. Role of nitrite in the competition between denitrification and DNRA in a chemostat enrichment culture. AMB Express. 2017;7:91. doi: 10.1186/s13568-017-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vermeiren J., Van de Wiele T., Verstraete W., Boeckx P., Boon N. Nitric oxide production by the human intestinal microbiota by dissimilatory nitrate reduction to ammonium. J Biomed Biotechnol. 2009 doi: 10.1155/2009/284718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vishniac W., Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H., Gunsalus R.P. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J Bacteriol. 2000;182:5813–5822. doi: 10.1128/jb.182.20.5813-5822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welsh D., Castadelli G., Bartoli M., Poli D., Careri M., de Wit R., Viaroli P. Denitrification in an intertidal seagrass meadow, a comparison of 15N-isotope and acetylene-block techniques: dissimilatory nitrate reduction to ammonia as a source of N2O? Mar Biol (Heidelberg, Ger) 2001;139:1029–1036. [Google Scholar]
- 43.Willems A. The family Comamonadaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer; Berlin, Heidelberg: 2014. pp. 777–851. [Google Scholar]
- 44.Yoon S., Cruz-Garcia C., Sanford R., Ritalahti K.M., Löffler F.E. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J. 2015;9:1093–1104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.